Abstract
Introduction
Imatinib mesylate is the drug of choice for patients with chronic myeloid leukemia (CML). Imatinib pharmacokinetics is affected by a number of transport proteins and enzymes.
Material and methods
In the present study we evaluated the association of eight polymorphisms in the seven genes CYP3A5*3 (rs776746), CYP3A4*1 (rs2740574), CYP2C9*3 (rs1057910), SLC22A1 (rs683369), ABCB1 (rs1045642, rs1128503), ABCG2 (rs2231142) and ABCC2 (rs717620) with imatinib plasma level and achieving an optimal clinical response in 112 CML patients (53 men and 59 women).
Results
No association was found between the examined polymorphisms in rs776746, rs2740574, rs1057910, rs683369, rs1045642, rs1128503, rs2231142, rs717620 and the achieved imatinib plasma level. The influence of rs776746 (CYP3A5*3) on the achievement of a complete cytogenetic response (CCyR) at 6 months was borderline non-significant (p = 0.06). Furthermore, no association was demonstrated between rs776746 polymorphisms and the achievement of a major molecular response (MMR) at 12 or 18 months. Polymorphisms rs776746, rs2740574, rs1057910, rs683369, rs1045642, rs1128503, rs2231142, rs717620 showed no impact on the optimal therapeutic response.
Conclusions
Despite the results of some other studies, no other polymorphism we analyzed was associated with imatinib plasma level or clinical response. The treatment outcomes cannot be predicted using the candidate gene approach and treatment decisions cannot be made according to the polymorphisms investigated in this study.
Keywords: clinical response, imatinib, chronic myeloid leukemia, genetic polymorphisms
Introduction
The discovery of imatinib mesylate substantially changed the results of the treatment and survival of patients with chronic myeloid leukemia (CML) [1, 2]. The final clinical response in CML patients is affected by many factors which may be divided into three basic groups. The first group involves age-related factors (the patient’s age, overall health, associated diseases and cooperation). The second group includes disease-related factors (CML phase, prognostic risk score, cytogenetic finding, transcript type, presence of tyrosine kinase mutations, and pharmacokinetics of imatinib). The last group involves the treatment of CML and comprises the choice of an appropriate tyrosine kinase inhibitor (TKI) and dealing with adverse effects. Despite the remarkable progress in the treatment of CML, approximately 20–30% of patients still remain resistant to imatinib therapy. The known causes of imatinib resistance include BCR-ABL inhibition, development of kinase domain mutations, BCR-ABL1 gene amplification and overexpression or clonal evolution [3–7].
However, many cases of resistance development are unclear.
Recently, increased attention has been paid to the pharmacokinetics of imatinib mesylate because it could clarify some cases of therapeutic failures. Imatinib mesylate is predominantly metabolized in the liver to an active metabolite via cytochrome CYP3A4 and CYP3A5. Other cytochromes are of very little importance in the metabolism of imatinib (CYP1A2, CYP2D6, CYP2C9, CYP2C19) [8–10]. Various ethnic groups show significantly different representation of CYP3A5 and CYP3A4 allele variants, which explains the variable pharmacokinetics of TKI in patients. The TKI transport mechanisms can be divided into a group of proteins which ensure transport into the cell and those which ensure transport out of the cell. Human organic cation transporter 1 (hOCT1), a membrane transporter of imatinib into the cell that is encoded by the SLC22A gene, plays an important role in BCR-ABL kinase inhibition. Conversely, the proteins ABCB1 (MDR-1) and ABCG2 (BCRP) rank among important transporters which remove imatinib from the cell [8–10].
Our study investigated whether the diverse pharmacokinetics of imatinib mesylate caused by polymorphisms in the CYP3A5*3, CYP3A4*1, and CYP2C9*3 enzymes or by polymorphisms in the transporter proteins SLC22A1, ABCB1, ABCG2 and ABCC2 affects plasma levels of imatinib and achievement of an optimal clinical response. It is likely that pharmacogenetics, genetic variability in the metabolism of therapeutic agents, plays a role in the prediction of survival in other cancers too [11–13].
Material and methods
Cohort of patients
In this study we performed a retrospective evaluation of 112 patients with CML – chronic phase (97%) and accelerated phase (3%) – receiving treatment in two hospitals in the Czech Republic. The cohort included 53 Caucasian men (47%) and 59 Caucasian women (53%) with a median age at diagnosis of 56 years (range: 19–84). The CML was diagnosed in these hospitals between 5/1997 and 12/2012 and the median of patient monitoring was 104 months (M) (range: 29–236). The information of current medical condition and survival of the patients was completed in 6/2016. The patients gave informed consent to the analysis of genetic polymorphisms. The characteristics of the patients under evaluation are described in Table I. Patients who had received imatinib 400 mg/day for at least 18 months were eligible for inclusion in our study. Patients whose imatinib dose was reduced (< 400 mg/day for > 2 weeks) were not included in the evaluation. Furthermore, we did not assess patients with imatinib resistance caused by a mutation in the kinase domain.
Table I.
Patients’ and disease characteristics of study cohort (n = 112)
| Parameter | Value |
|---|---|
| Age at diagnosis, median (range) [years] | 56 (range: 19–84) |
| Disease stage, n (%): | |
| Chronic phase | 109 (97) |
| Accelerated phase | 3 (3) |
| Blastic phase | 0 |
| Gender, n (%): | |
| Male | 53 (47) |
| Female | 59 (53) |
| Sokal risk group, n (%): | |
| Low | 34 (30) |
| Intermediate | 60 (54) |
| High | 11 (10) |
| Not applicable | 7 (6) |
| Hasford risk group, n (%): | |
| Low | 43 (39) |
| Intermediate | 44 (40) |
| High | 18 (15) |
| Not applicable | 7 (6) |
| Cytogenetics, n (%): | |
| t (9;22) only | 109 (97) |
| Additional abnormalities | 3 (3) |
| Preview treatment to imatinib, n (%): | |
| Interferon | 11 (10) |
| Hydroxyurea > 1 months | 3 (3) |
| Response to imatinib, n (%): | |
| CHR at 3 months | 108 (96) |
| CCyR at 6 months | 63 (56) |
| MMR at 12 months | 61 (54) |
| Current status, n (%): | |
| Alive | 107 (95.5) |
| Dead | 5 (4.5) |
Both hospitals evaluated optimal clinical responses in accordance with the recommendations of the European LeukemiaNet (ELN) from 2013 [6, 7]. At 3 months of treatment, a complete hematologic response (CHR) was assessed. CHR was defined as the number of thrombocytes < 450 × 109/l, the number of leukocytes < 10 × 109/l, the absence of young forms of leukocytes and the number of basophils < 5% in a complete blood count and a peripheral smear. Simultaneously, the spleen could not be palpable during the physical examination of the patients. A cytogenetic response was assessed according to the number of the mitoses detected by standard cytogenetic methods in Ph+ cells in the bone marrow. Complete cytogenetic response (CCyR) is defined as the absence of Ph+ mitoses in the bone marrow. Molecular response and its depth are assessed according to the BCR-ABL1 transcript level, which was determined by means of a quantitative polymerase chain reaction method using a reverse transcriptase (RT-PCR). The result was expressed as a ratio of BCR-ABL1 to ABL (or to another control gene) × 100% and was converted to the International Scale (IS). Major molecular response (MMR) is defined as BCR-ABL1/control gene ≤ 0.1%.
Imatinib plasma level determination
The imatinib plasma level was determined by high-performance liquid chromatography and the analysis was validated for a detection limit of 10 ng/ml. Peripheral blood of the patients was taken 24 ±2 h prior to another imatinib dose. The imatinib plasma level was determined at least 12 months after imatinib therapy commencement. Because our study is retrospective, the taking of the imatinib plasma levels in all patients was not performed at exactly the same time from the beginning of the imatinib therapy.
Genotyping
DNA was obtained from peripheral blood and the sample was then frozen at –80°C until the completion of the analysis. Single-nucleotide polymorphisms (SNP) were detected by means of real-time allele-discriminating PCR using TaqMan dual labeled hydrolysis probes. Commercial genotyping assays and a commercial master mix were used for detection. The PCR reaction comprises three steps: activation of DNA polymerase, denaturation and primer annealing. The obtained amplification curves of the genotypes of the selected polymorphisms in genes were evaluated by means of the Rotor Gene 6000 Series software, which is part of the Rotor Gene 6000 thermal cycler manufactured by Qiagen. The analysis of the data was performed in the controlling and analytical software of the cycler under an ‘allelic discrimination’ mode. The limit distinguishing the positive and negative signal of the individual alleles was set by comparison with a known genotype.
In the present study we evaluated eight polymorphisms in seven genes: rs776746 CYP3A5*3 (6986A>G), rs2740574 CYP3A4*1 (–392A>G), rs1057910 CYP2C9*3 (1075A>C), rs683369 SLC22A1 (480C>G), rs1045642 ABCB1 (3435C>T), rs1128503 ABCB1 (1236 C>T), rs2231142 ABCG2 (421C>A), and rs717620 ABCC2 (–24C>T).
Statistical analysis
A χ2 test was used to evaluate the statistically significant dependences of qualitative variables. Quantitative variables were compared using the Mann-Whitney test and some analyses were completed by means of ANOVA tests. The significance level applied (p-value) was 0.05. Power of all significant and borderline non-significant results was verified and it was higher than 0.8 in all cases.
Results
Clinical response in the cohort
The following responses were achieved in the evaluated cohort: CHR was achieved in 108 (96%) patients at 3 months, CCyR was achieved in 63 (56%) patients at 6 months, CCyR was achieved in 101 (90%) patients at 12 months, MMR was achieved in 61 (54%) patients at 12 months, and MMR was achieved in 79 (70%) patients at 18 months of imatinib therapy. Hematologic toxicities of all grades occurred in 35.5% of subjects during treatment with imatinib (anemia – all degrees, 3.5%; neutropenia – all degrees, 18%; thrombocytopenia, 14%). Non-hematologic toxicities observed during therapy most often included peripheral edemas (24%), bone and muscle pain (20.5%), nausea or vomiting (19%), diarrhea (11%), rash (7%), and hypophosphatemia (7%). Five (5%) patients died. Two of these deaths were associated with CML (blast crisis), 1 patient suffered from progressing CML and concurrent heart failure, and 2 female patients died of breast carcinoma.
Impact of polymorphisms on final imatinib plasma level
It was our aim to prove whether the polymorphisms in the genes affecting the metabolism of imatinib mesylate affect the final imatinib plasma level. The median imatinib plasma levels identified for the individual polymorphism genotypes are presented in Table II. No association between the polymorphism genotypes in rs776746 (CYP3A5*3), rs2740574 (CYP3A4*1), rs1057910 (CYP2C9*3), rs683369 (SLC22A1), rs1045642 and rs1128503 (ABCB1), rs2231142 (ABCG2), rs717620 (ABCC2) and the achieved imatinib plasma level was established. Also, our study did not prove any association between the CC genotypes as compared with the other genotypes of CYP3A5, any association between the GG and CC genotypes as compared with the other genotypes of SLC22A1, any association between the GG or AA genotype as compared with the other genotypes of ABCB1 (3435C>T) and ABCB1 (1236C>T), any association between the GG or TT genotype as compared with the other genotypes of ABCG2, or any association between the TT or CC genotype as compared with the other genotypes of ABCG2.
Table II.
Correlation between candidate genotypes and imatinib plasma levels
| Genotypes | N | % | Median imatinib plasma levels [ng/ml] | Range of imatinib plasma levels [ng/ml] | P-value |
|---|---|---|---|---|---|
| CYP 3A5*3 (rs776746): | |||||
| C – C | 98 | 87 | 841 | 173–2173 | 0.72 |
| T – C | 13 | 12 | 621 | 423–1472 | |
| T – T | 1 | 1 | 975* | 0.39 | |
| CC vs. TC + TT | |||||
| CYP3A4*1 (rs2740574): | 0.99 | ||||
| T – T | 104 | 93 | 881 | 173–2173 | |
| T – C | 8 | 7 | 605 | 423–1094 | |
| CYP2C9*3 (rs1057910): | |||||
| A – A | 95 | 85 | 823 | 210–2173 | 0.99 |
| A – C | 17 | 15 | 1045 | 223–1502 | |
| SLC22A1 (rs683369): | |||||
| C – C | 78 | 70 | 838 | 173–2173 | 0.20 |
| C – G | 27 | 24 | 727 | 374–1724 | |
| G – G | 7 | 6 | 925 | 627–1163 | |
| GG vs. CG + CC | 0.17 | ||||
| CC vs. CG + GG | 0.50 | ||||
| ABCB1 (rs1045642): | |||||
| A – G | 58 | 51 | 881 | 223–2173 | 0. 84 |
| A – A | 32 | 29 | 793 | 319–1724 | |
| G – G | 22 | 20 | 825 | 173–1477 | 0.76 |
| GG vs. AA + AG | 0.79 | ||||
| AA vs. AG + GG | |||||
| ABCB1 (rs1128503): | |||||
| A – A | 18 | 16 | 1039 | 173–1304 | 0.80 |
| A – G | 62 | 55 | 838 | 210–2173 | |
| G – G | 32 | 29 | 727 | 392–1477 | |
| GG vs. AA + AG | 0.65 | ||||
| AA vs. AG + GG | 0.74 | ||||
| ABCG2 (rs2231142): | |||||
| G – G | 87 | 80 | 804 | 439–2173 | 0.47 |
| T – G | 23 | 19 | 910 | 173–1710 | |
| T – T | 2 | 1 | 647* | ||
| GG vs. TG + TT | 0.27 | ||||
| TT vs. GT + GG | 0.79 | ||||
| ABCC2 (rs717620): | |||||
| C – C | 70 | 63 | 889 | 210–2173 | 0.70 |
| C – T | 37 | 33 | 910 | 319–1519 | |
| T – T | 5 | 4 | 763 | 661–1484 | |
| TT vs. CC + CT | 0.85 | ||||
| CC vs. TT + CT | 0.46 | ||||
Not evaluated, minor frequency.
With regard to the number of individual polymorphism genotypes it was possible to evaluate three genes for the quantitative measure of the expression of the polymorphism of rs683369 SLC22A1, rs1045642 and rs1128503 in the ABCB1 gene using an ANOVA test. Even this evaluation did not confirm a statistically significant association of these polymorphisms with the achieved imatinib plasma level.
Impact of polymorphisms on achieving an optimal clinical response
Our study showed that none of the polymorphisms under examination was associated with the achievement of a CHR at 3 months. At 6th month of imatinib treatment the influence of rs776746 (CYP3A5*3) on the achievement of CCyR was borderline non-significant (p = 0.06). There was a significant difference between CC genotype and other genotypes (p = 0.05). We did not evaluate the association of rs776746 and the achievement of CCyR at 12 months due to low incidence of heterozygotes and homozygotes in nonresponder patients. Furthermore, no association was demonstrated between the other polymorphisms and the achievement of CCyR at 6 and 12 months (Table III).
Table III.
Correlation between candidate genotypes and clinical response (CCyR, MMR)
| Referent genotype | N | CCyR at 6M Yes | CCyR at 6M No | P-value | CCyR at 12M Yes | CCyR at 12M No | P-value |
|---|---|---|---|---|---|---|---|
| CYP3A5*3 (rs776746): | |||||||
| C – C | 98 | 59 | 39 | 0.06 | 89 | 9 | ± |
| T – C | 13 | 4 | 9 | 12 | 1 | ||
| T – T | 1 | 0 | 1 | 0.05 | 0 | 1 | 0.90 |
| CC vs. TC + TT | |||||||
| Referent genotype | N | MMR at 12M Yes | MMR at 12M No | P-value | MMR at 18M Yes | MMR at 18M No | P-value |
| CYP3A5*3 (rs776746): | |||||||
| C – C | 98 | 54 | 44 | 0.54 | 70 | 28 | 0.31 |
| T – C | 13 | 7 | 6 | 9 | 4 | ||
| T – T | 1 | 0 | 1 | 0 | 1 | ||
| CC vs. TC + TT | 0.94 | 0.87 | |||||
| ABCG2 (rs2231142): | |||||||
| G – G | 87 | 52 | 35 | 0.06 | 64 | 23 | 0.23 |
| T – G | 23 | 9 | 14 | 14 | 9 | ||
| T – T | 2 | 0 | 2 | 1 | 1 | ||
| GG vs. TG + TT | 0.06 | 0.15 | |||||
| TT vs. GT + GG | 0.39 | 1 | |||||
CCyR – complete hematologic response, MMR – major molecular response ± not evaluated (minor frequency nonresponders).
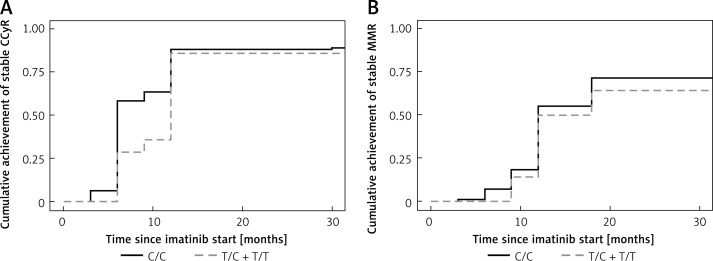
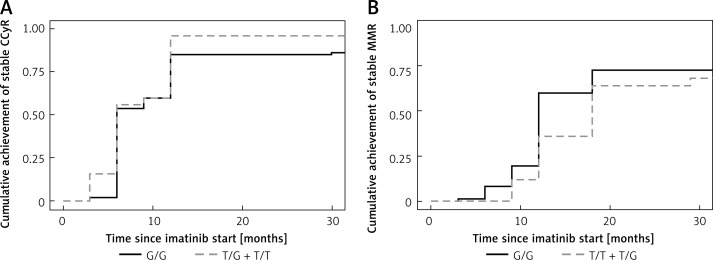
The assessment of MMR at 12 months has shown a possible influence of the polymorphism rs2231142 in the gene ABCG2 but without statistical significance (p = 0.06). The same implication resulted from the evaluation of the association between the GG genotype and the other (TG and TT) genotypes (p = 0.06). The other polymorphisms did not show any influence on the achievement of MMR at 12 months. Moreover, none of the polymorphisms under investigation showed any association with the achievement of MMR at 18 months (Table III). We did not prove any differences between homozygotes and heterozygotes, when we evaluated the cumulative achievement of a stable response (CCyR) or MMR in rs776746 (CYP3A5*3) and rs2231142 (ABCG2) (Figures 1, 2).
Figure 1.
A – Cumulative incidence of achievement of stable CCyR in CYP3A5*3 (rs776746) (p = 0.85). B – Cumulative incidence of achievement of stable MMR in CYP3A5*3 (rs776746) (p = 0.38)
Figure 2.
A – Cumulative incidence of achievement of stable CCyR in ABCG2 (rs2231142) (p = 0.26). B – Cumulative incidence of achievement of stable MMR in ABCG2 (rs2231142) (p = 0.33)
Discussion
The first study to investigate the significance of genetic polymorphisms for the imatinib plasma level was published by Takahashi et al. in 2010 [14]. In this study, the polymorphisms of CYP3A5*3 (6986A>G), SLC22A1 (156T>C, 480G>C, 1022C>T and 1222A>G), ABCB1 (1236T>C, 2677G>T, 3435T>C), ABCC2 (–24C>T) and ABCG2 (421C>A) were analyzed in 67 patients. Only one correlation between the imatinib plasma level and polymorphism was confirmed, namely with ABCG2 polymorphism. The detected imatinib plasma level was statistically higher than the level of non-CC alleles in ABCG2 polymorphism (p = 0.015). Another study dealing with the evaluation of certain polymorphisms and their impact on the imatinib plasma level was presented by Seong et al. in 2012 and assessed 10 genotypes [15]. However, none of the examined polymorphisms showed a correlation between a certain genotype and an imatinib plasma level and this observation corresponds to our findings.
Several studies have reported that certain variants of polymorphisms may predict a clinical response. The first of these studies was published by Kim et al. in 2009 and described the investigation of 16 SNPs in the five candidate genes CYP3A5*3, SLC22A1, ABCB1, ABCC2, and AGP in 229 patients [16]. The analysis established that the GG genotype of ABCG2 (rs2231142) had a negative impact on achieving CCyR (p = 0.03), whereas the AC or CC genotype had a positive influence on achieving MMR. Seong’s study suggests that the individual ABCG2 genotypes did not affect the achievement of a cytogenetic response, while molecular responses achieved by individual genotypes (p = 0.02) showed significant differences, with the AC or CC genotype once again accounting for a lower number of patients with MMR [15]. In 2013 Au et al. published an analysis of 215 patients which proved the significance of the individual ABCG2 genotype variants on the achievement of MMR (p = 0.004) [17]. This observation is similar to results of this study showing borderline non-significant tendency of the ABCG2 polymorphism to influence the achievement of MMR at 12 months (p = 0.06), even though our findings are not as conclusive as those presented in the above-mentioned studies.
In their studies Kim et al., Takahashi et al. and Seong et al. also examined the polymorphisms in the gene ABCB1 without proving the impact of this polymorphism on a clinical response. This finding is in agreement with our observations. On the other hand, other studies by Au et al. and Salimizand et al. confirmed an association between the CC genotype of the ABCB1 3435C allele and a worse response to imatinib [17, 18]. In 2015 Wang et al. published a meta-analysis of 10 studies which involved 987 patients with CML and evaluated the significance of the polymorphisms rs1045642, rs1128503 and rs2032582 in ABCB1 (MDR1). The meta-analysis did not demonstrate an association between achieving optimal response and the genotypes of a certain polymorphism [19].
The available published results on the significance of the polymorphism of the hepatic enzymes CYP3A5 and CYP3A4 are not unambiguous. Kim’s study suggests that the AA genotype in CYP3A5 (rs776746) is associated with a lower probability of achieving MMR (p = 0.01). This observation corresponds with our findings which demonstrated the implied significance of CYP3A5*3 polymorphism for achieving CCyR at 6 months of treatment (p = 0.06). The recent, less extensive analysis by Bedewy et al. were published in 2013. It evaluated 78 CML patients and confirmed the impact of polymorphisms in the CYP3A5 gene on achieving an optimal response [20]. On the other hand, Seong’s analysis examined six different polymorphisms in the CYP3A4*18, CYP3A5*3, CYP2C9*3, CYP2C19*2, CYP2C19*3 and CYP2D6*10B genes and did not establish any impact of those polymorphisms.
Angelini’s study from 2012 included 189 patients and evaluated 20 polymorphisms. Of hepatic enzymes, only the polymorphism in the gene CYP3A4 was assessed. It was not confirmed to affect the achievement of an optimal response [21]. The evaluation of the whole cohort presented in the study implied the impact of the polymorphism in ABCB1 (MDR1; rs60023214) on achieved MMR (p = 0.06). The statistically significant impact on the achievement of MMR (p = 0.005) was confirmed, when this analysis was performed only on a cohort of Caucasians who had different patterns of allele frequencies. However, our analysis did not prove the association of the rs1045642 and rs1128503 polymorphisms in ABCB1 with the achievement of an optimal response.
In conclusion, our results support the theory that there is an association between CYP3A5 genotype (allele *3) and clinical response to imatinib mesylate. Surprisingly, no association between imatinib plasma level and genotype was found. Despite the results of some other studies, no other polymorphism we analyzed was associated with clinical response or imatinib plasma level and further analyses in a larger patient cohort seem fully warranted. Based on our results and inconsistency among different studies, we believe that it is not possible to predict therapeutic outcome and make treatment decisions according to polymorphisms involved in this study, and other risk factors (i.e., Sokal or Euro score) are more important.
Acknowledgments
This work was supported by the program PROGRES Q40/08.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.de Lavallade H, Apperley J, Khorashad J, et al. Imatinib for newly diagnosed patients with chronic myeloid leukemia: incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol. 2008;26:3358–63. doi: 10.1200/JCO.2007.15.8154. [DOI] [PubMed] [Google Scholar]
- 2.Hughes T, Hochhaus A, Branford S, et al. IRIS investigators Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic myeloid leukemia: an analysis from the International Randomized Study of Interferon and STI571 (IRIS) Blood. 2010;116:3758–65. doi: 10.1182/blood-2010-03-273979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marin D, Bazeos A, Mahon F, et al. Adherence is the critical factor for achieving molecular responses in patients with chronic myeloid leukemia who achieve complete cytogenetic responses on imatinib. J Clin Oncol. 2010;28:2381–8. doi: 10.1200/JCO.2009.26.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apperley J. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1018–29. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 5.Apperley J. Part II: management of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8:1116–28. doi: 10.1016/S1470-2045(07)70379-0. [DOI] [PubMed] [Google Scholar]
- 6.Baccarani M, Cortes J, Pane F, et al. European Leukemia Net. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009;27:6041–51. doi: 10.1200/JCO.2009.25.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baccarani M, Deininger M, Rosti G, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122:872–84. doi: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng B, Lloyd P, Schran H. Clinical pharmacokinetics of imatinib. Clin Pharmacokinet. 2005;44:879–94. doi: 10.2165/00003088-200544090-00001. [DOI] [PubMed] [Google Scholar]
- 9.Cortes JE, Egorin MJ, Guilhot F, et al. Pharmacokinetic/pharmacodynamic correlation and blood-level testing in imatinib therapy for chronic myeloid leukemia. Leukemia. 2009;23:1537–44. doi: 10.1038/leu.2009.88. [DOI] [PubMed] [Google Scholar]
- 10.Bruhn O, Cascorbi I. Polymorphisms of the drug transporters ABCB1, ABCG2, ABCC2 and ABCC3 and their impact on drug bioavailability and clinical relevance. Expert Opinion. 2014;10:1337–54. doi: 10.1517/17425255.2014.952630. [DOI] [PubMed] [Google Scholar]
- 11.Zhai X, Wang H, Zhu X, et al. Gene polymorphisms of ABC transporters are associated with clinical outcomes in children with acute lymphoblastic leukemia. Arch Med Sci. 2012;8:659–671. doi: 10.5114/aoms.2012.30290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosseini M, Houshmand M, Ebrahimi A. MTHFR polymorphisms and breast cancer risk. Arch Med Sci. 2011;7:134–7. doi: 10.5114/aoms.2011.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mrozikiewicz-Rakowska B, Malinowski M, Nehring P, et al. The MDR1/ABCB1 gene rs1045642 polymorphism in colorectal cancer. Arch Med Sci. doi: 10.5114/aoms.2017.70329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi N, Miura M, Scott SA, et al. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Gene. 2010;55:731–7. doi: 10.1038/jhg.2010.98. [DOI] [PubMed] [Google Scholar]
- 15.Seong S, Lim M, Sohn S, et al. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol. 2013;24:756–60. doi: 10.1093/annonc/mds532. [DOI] [PubMed] [Google Scholar]
- 16.Kim D, Sriharsha L, Xu W, et al. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2009;15:4750–8. doi: 10.1158/1078-0432.CCR-09-0145. [DOI] [PubMed] [Google Scholar]
- 17.Au A, Aziz Baba A, Goh A, et al. Association of genotypes and haplotypes of multi-drug transporter genes ABCB1 and ABCG2 with clinical response to imatinib mesylate in chronic myeloid leukemia patients. Biomed Pharmacother. 2014;68:343–9. doi: 10.1016/j.biopha.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Salimizand H, Amini S, Abdi M, et al. Concurrent effects of ABCB1 C3435T, ABCG2 C421A, and XRCC1 Arg194Trp genetic polymorphisms with risk of cancer, clinical output, and response to treatment with imatinib mesylate in patients with chronic myeloid leukemia. Tumour Biol. 2016;1:791–8. doi: 10.1007/s13277-015-3874-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Liu H, Li F, et al. Multidrug resistance gene (MDR1) polymorphisms may not be directly associated with response to imatinib in chronic myeloid leukemia. Genet Mol Res. 2015;14:14967–78. doi: 10.4238/2015.November.24.4. [DOI] [PubMed] [Google Scholar]
- 20.Bedewy A, El-Maghraby S. Do SLCO1B3 (T334G) and CYP3A5*3 polymorphisms affect response in Egyptian chronic myeloid leukemia patients receiving imatinib therapy? Hematology. 2013;18:211–6. doi: 10.1179/1607845412Y.0000000067. [DOI] [PubMed] [Google Scholar]
- 21.Angelini S, Soverini S, Ravegnini G, et al. Association between imatinib transporters and metabolizing enzymes genotype and response in newly diagnosed chronic myeloid leukemia patients receiving imatinib therapy. Haematologica. 2013;98:193–200. doi: 10.3324/haematol.2012.066480. [DOI] [PMC free article] [PubMed] [Google Scholar]