Abstract
Introduction
Emerging evidence has indicated that long non-coding RNAs (lncRNAs) play vital roles in multiple myeloma (MM) development and progression. However, the underlying mechanism of PVT1 in MM remains unclear.
Material and methods
QRT-PCR was used to detect the expression of PVT1 and miR-203a in MM samples and cell lines. The effects of PVT1 on MM cell proliferation and apoptosis were determined by CCK8 assay and flow cytometer assay, respectively. Bioinformatics methods were used to identify the downstream target miRNAs of PVT1.
Results
We found that the expression of PVT1 was upregulated in MM samples and cell lines (p < 0.05), while the expression of miR-203a was downregulated in MM samples and cell lines (p < 0.05). There was a negative correlation between PVT1 expression and miR-203a expression in MM samples (p < 0.05). In in vitro function assays, we found that PVT1 inhibition suppressed MM cell proliferation and induced MM cell apoptosis (p < 0.05). The bioinformatics approach predicted that PVT1 sponge miR-203a would modulate MM cells. Rescue experiments confirmed the recovering roles of miR-203a for PVT1 on MM progression.
Conclusions
In the present study, we found that lncRNA PVT1 could promote MM cell proliferation and induce cell apoptosis by inhibiting miR-203a expression. Therefore, PVT1 may represent a potential therapeutic target for the treatment of MM patients.
Keywords: multiple myeloma, miR-203a, PVT1, proliferation, apoptosis
Introduction
Multiple myeloma (MM) is a fatal hematological cancer and is also a genetically complex malignancy with a significant heritable basis [1]. The disease is characterized by the clonal proliferation of malignant plasma cells in the bone marrow microenvironment, osteolytic bone destruction and pathological fractures [2]. During the past decades, the incidence of MM has continued to rise in the world [3]. Despite recent progress in the understanding of MM pathobiology and the availability of innovative drugs which have improved the clinical outlook, MM remains an incurable disease [4], and the complex biological and molecular mechanisms underlying MM have not yet been fully elucidated.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules, 19–25 nucleotides in length, that regulate gene expression post-transcriptionally by targeting mRNAs [5]. Increasing studies have shown that miRNAs are aberrantly expressed and act as oncogenes or tumor suppressors in tumor progression [6]. Recent studies showed that miRNAs were dysregulated in MM progression. For example, Hua et al. reported that miR-32 expression was increased in MM tissues and promoted tumor progression by targeting FBXW7 [7]. Wu et al. found that miR-34a overexpression inhibited MM cancer stem cell growth in mice by suppressing TGIF [8]. Qin et al. observed that miR-137 inhibition induced drug resistance and chromosomal instability by targeting AURKA in MM [9]. However, it is still unclear how miRNAs regulate MM progression.
Long non-coding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides with limited protein coding ability and function as new regulators that modulate gene expression transcriptionally or post-transcriptionally [10]. Recent studies showed that lncRNAs play important roles in MM progression. For example, Shen et al. observed that upregulated lncRNA PCAT1 was closely related to clinical diagnosis of MM as a predictive biomarker in serum [11]. Cho et al. reported that lncRNA MALAT1 was overexpressed in MM samples and might serve as a biomarker to predict disease progression [12].
Plasmacytoma variant translocation 1 (PVT1) is encoded by the PVT1 gene and is located downstream of the MYC gene [13]. Recently, numerous studies have shown that PVT1 was overexpressed in human cancers. For example, Huang et al. found that increased expression of lncRNA PVT1 was associated with poor prognosis of pancreatic cancer patients [14]. Song et al. found that PVT1 promoted glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma [15]. Zhang et al. reported that PVT1 facilitated cervical cancer progression by inhibiting miR-128-3p expression [16]. However, the expression and underlying molecular mechanisms of PVT1 on MM tumorigenesis remain unclear.
The present study showed that PVT1 was increased in MM samples and cell lines, while miR-203a was downregulated. PVT1 was identified as a molecular sponge of miR-203a. PVT1 inhibition suppressed MM cell proliferation and induced cells apoptosis, while this effect was reversed by miR-203a inhibitors. Taken together, these data suggest that PVT1 acted as an oncogene and served as a potential therapeutic target for the treatment of MM patients.
Material and methods
Patients and sample collection
Thirty-three MM patients and 14 healthy control samples were collected between April 2015 and August 2016 at the Department of Hematology, The First Affiliated Hospital of Xinxiang Medical University. The study protocol was approved by the Ethics Committee of Xinxiang Medical University. Informed written consent was taken from all the patients and healthy controls.
Cell culture and transfection
Five MM cell lines (NCI-H929, U266, SKMM1 KMS-12-BM, and INA-6) and normal plasma cells (nPCs) were purchased from the American Type Culture Collection. These cell lines were maintained in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Gibco) and 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C in a 5% humidified CO2 atmosphere.
The miR-203a mimics, miR-203a inhibitors and PVT1 siRNAs (si-PVT1) were purchased from Ribobio (Guangzhou, China). miR-203a mimics, miR-203a inhibitors and PVT1 siRNA were transfected into cells by using Lipofectamine 2000 Reagent (Thermo Fisher Scientific) in a 6-well cell culture plate according to the manufacturer’s protocol.
Reverse transcription and quantitative real-time PCR (qRT-PCR) analysis
All RNAs were extracted from bone marrow mononuclear cells by TRIzol reagent (Invitrogen) referring to the manufacturer’s instructions and then stored at –80°C. The isolated RNA was reverse transcribed to obtain complementary DNA by PrimeScript RT reagent Kit (Invitrogen). The total obtained complementary DNA was operated to real-time PCR using a standard SYBR Green PCR kit (Applied Biosystems). All processes of PCR were performed in an ABI Prism 7500 system (Applied Biosystems). To determine the relative RNA expression, the 2–ΔΔCT method was used. The expression levels of miR-203a and PVT1 were respectively normalized by U6 and GAPDH served as a control. The primers for genes were as follows: PVT1 forward 5′-TGAGAACTGTCCTTACGTGACC-3′, reverse 5′-AGAGCACCAAGACTGGCTCT-3′; miR-203a forward 5′-CACCATAAAGACAGGAACCTG-3′, reverse 5′-GGAGGTGCCATCAATACCTGC-3′.
Cell proliferation assay
Cell proliferation was explored using a cell proliferation kit, Cell Counting Kit-8 (CCK8; Dojindo), according to the manufacturer’s instructions. Cells were seeded into 96-well tissue culture plates at a density of 2 × 103 cells/well the day before transfection. Cell growth was analyzed at a wavelength of 450 nm at 24, 48, and 72 h after transfection using Envision (PerkinElmer).
Flow cytometry analysis
Cells were cultivated in 6-well plates before transfection. Transfected cells were harvested and double stained with FITC-Annexin V and PI using the FITC Annexin V Apoptosis Detection Kit (TransGen) according to the manufacturer’s protocol. Flow cytometry was used to detect cell apoptosis. In the images, cells were classified as dead cells, early apoptosis cells, living cells and late apoptosis cells.
Dual luciferase reporter assay
Transient transfection of the lncRNA-PVT1 reporter plasmid and the internal control Renilla luciferase plasmid was carried out with the appropriate plasmids using Lipofectamine 3000 (Invitrogen). The relative luciferase activity was normalized to Renilla luciferase activity 48 h after transfection, and luciferase activity was measured using a dual-luciferase reporter gene assay system (Promega) according to the manufacturer’s protocol.
Statistical analysis
Data are presented as mean ± SD of three independent experiments and processed using SPSS 17.0 statistical software (SPSS). All data are evaluated as the mean ± standard deviation (SD). For comparisons, one-way analyses of variance, χ2 tests, and two-tailed Student’s t-tests were performed, as appropriate. P < 0.05 was considered statistically significant.
Results
Relative expression of PVT1 and miR-203a in MM
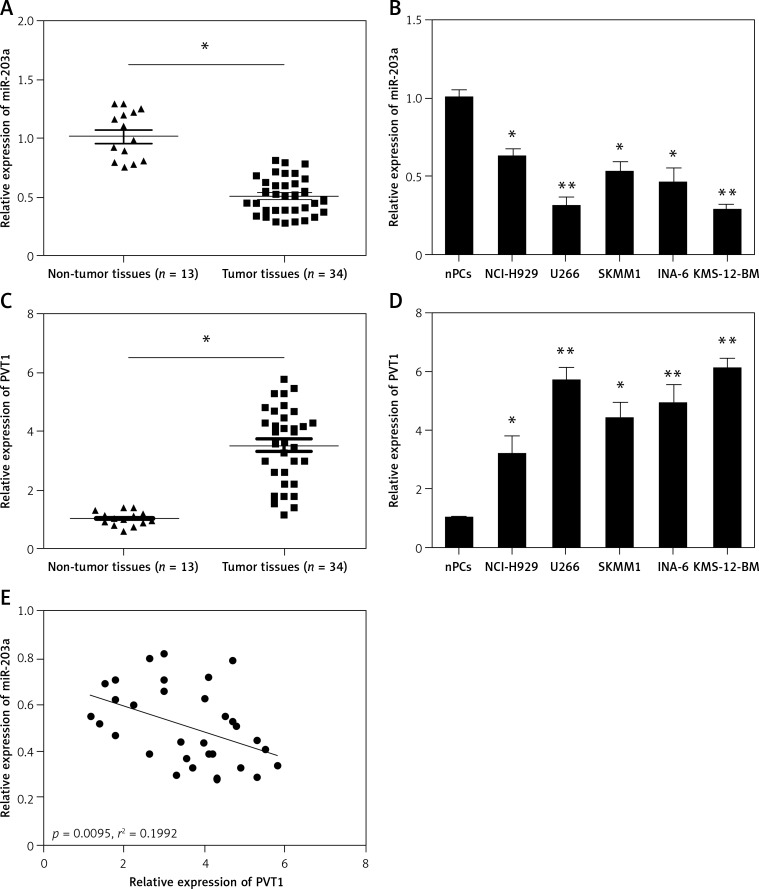
QRT-PCR was used to determine the expression of PVT1 and miR-203a in MM samples and cell lines. Results showed that the expression of PVT1 was upregulated in MM samples lines compared to normal healthy controls (Figure 1 A; p < 0.05). Additionally, we determined PVT1 expression in MM cell lines; the results showed that PVT1 expression in MM cell lines (NCI-H929, U266, SKMM1 KMS-12-BM, and INA-6) was significantly increased compared to normal healthy bone marrow-derived plasma cells (nPCs) (Figure 1 B; p < 0.05). We also determined the expression of miR-203a in MM samples and cell lines. Our data showed that miR-203a expression was significantly decreased in MM samples and cell lines (Figures 1 C and D; p < 0.05). Furthermore, we explored the correlation between miR-203a expression and PVT1 expression in MM samples. As shown in Figure 1 E, there is a negative correlation between miR-203a expression and PVT1 expression in MM samples (r2 = 0.1992, p = 0.0095).
Figure 1.
Relative expression of PVT1 and miR-203a in MM. A – Relative expression of PVT1 was determined by qRT-PCR in MM samples and healthy controls. B – Relative expression of PVT1 was tested by qRT-PCR in MM cell lines and normal plasma cells (nPCs). C – Relative expression of miR-203a was determined by qRT-PCR in MM samples and healthy controls. D – Relative expression of miR-203a was assessed by qRT-PCR in MM cell lines and nPCs. E – The correlation between PVT1 and miR-203a expression level was measured in MM samples using Pearson correlation analysis
*P < 0.05, **p < 0.01.
Knockdown of PVT1 suppressed MM cell proliferation
To explore the biological role of PVT1 in MM, we transfected si-PVT1 or si-NC into MM cells and the knockdown efficiency was confirmed by qRT-PCR (Figure 2 A; p < 0.05). CCK8 assays showed that PVT1 inhibition significantly decreased the proliferation of MM cells (Figure 2 B; p < 0.05). Furthermore, we analyzed the effects of PVT1 on MM cell apoptosis. Flow cytometry assays revealed that the apoptosis of MM cells was greatly enhanced after PVT1 inhibition (Figure 2 C; p < 0.05). These results indicated that PVT1 might promote cell proliferation by reducing MM cell apoptosis.
Figure 2.
PVT1 inhibition suppressed MM cell proliferation. A – PVT1 expression was determined by qRT-PCR in MM cells transfected with si-PVT1 or si-NC. B – The down-regulation of PVT1 inhibited MM cell proliferation as measured using the CCK8 assays. C – The down-regulation of PVT1 induced MM cell apoptosis as measured using the flow cytometry analysis
*P < 0.05, **p < 0.01.
PVT1 negatively regulated miR-203a expression
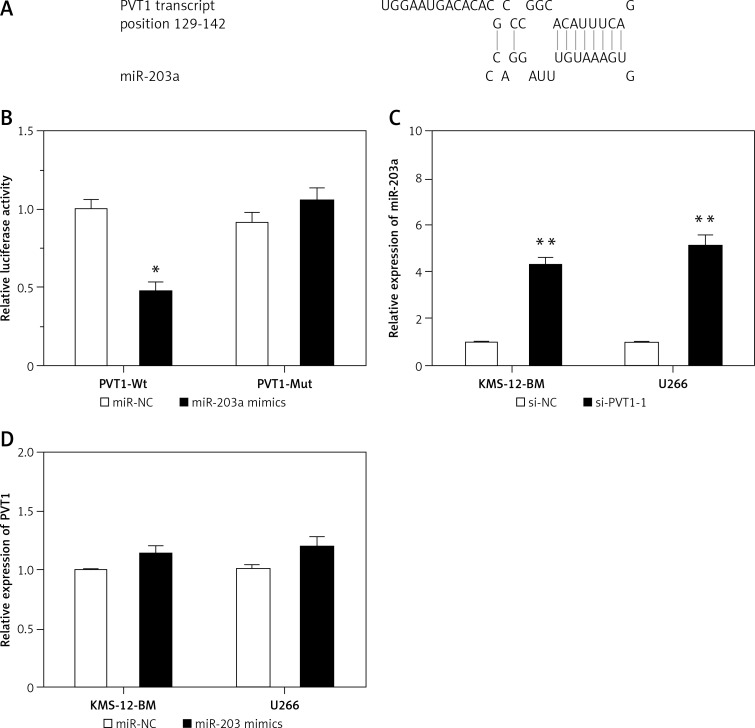
Recent studies showed that PVT1 might function as a competing endogenous RNA (ceRNA) in tumor progression. To search the target miRNAs of PVT1, DIANA-LncBase and miRcode software were used. We found that miR-203a had a binding site with PVT1 (Figure 3 A). The luciferase reporter assay showed that co-transfection of miR-203a mimics and PVT1-Wt significantly reduced the luciferase activity, but co-transfection of miR-203a mimics and PVT1-Mut did not affect the luciferase activity (Figure 3 B; p < 0.05). In addition, our data showed that si-PVT1 markedly increased the expression of miR-203a in MM cells compared to si-NC cells (Figure 3 C; p < 0.05). However, ectopic overexpression of miR-203a did not affect PVT1 expression in MM cells (Figure 3 D; p > 0.05).
Figure 3.
PVT1 negatively regulated miR-203a expression. A – Predicted miR-203a binding sites within the 3′-UTR of PVT1 mRNA. B – Dual-luciferase reporter assay revealed that miR-203a mimics decreased luciferase activity of PVT1-Wt, but not of PVT1-Mut. C – Relative expression of miR-203a in si-PVT1 transfected MM cells. D – Relative expression of PVT1 in miR-203a mimics transfected MM cells
*P < 0.05, **p < 0.01.
MiR-203a reversed the promoting effect of PVT1 on growth of MM cells
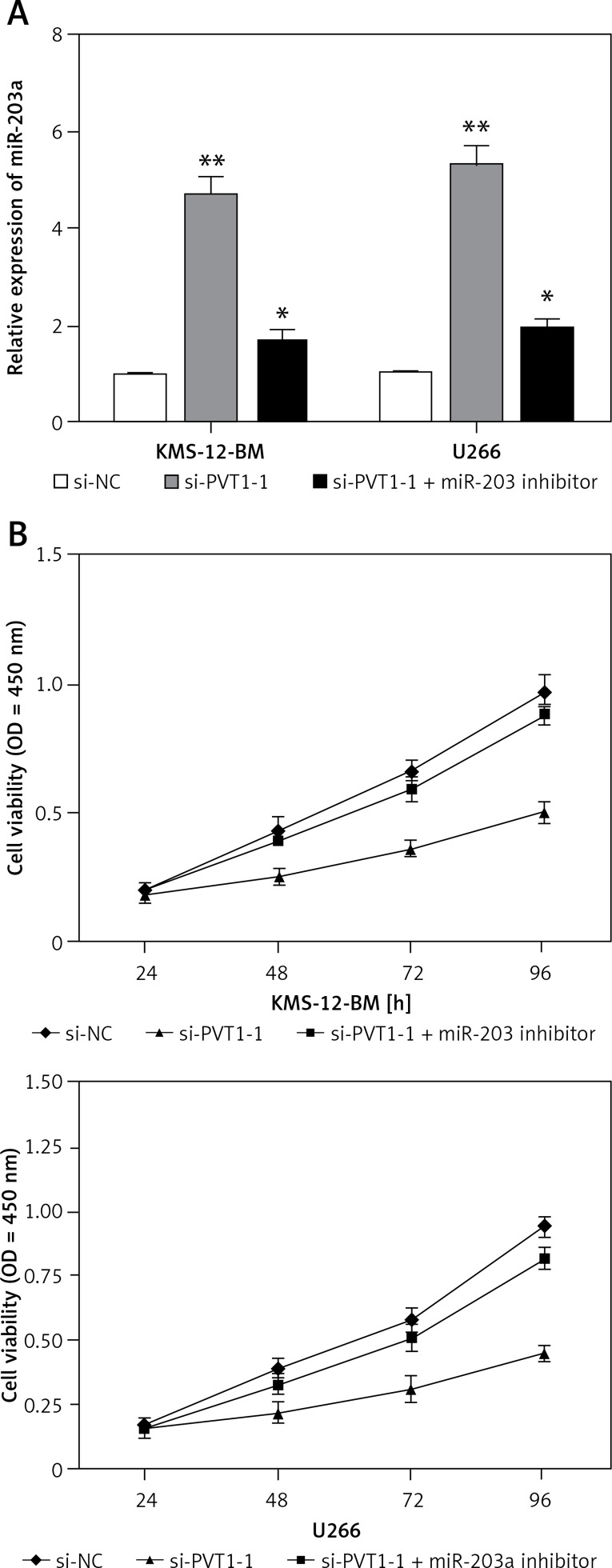
To explore whether PVT1 exerted biological functions through miR-203a, we used rescue experiments via suppressing miR-203a expression in PVT1 inhibited MM cells, and the expression of miR-203a was determined by qRT-PCR (Figure 4 A; p < 0.05). CCK8 assays showed that MM cell proliferation was significantly decreased in PVT1 knockdown MM cells, whereas miR-203a inhibitors partially reversed the reduction of proliferation (Figure 4 B; p < 0.05). These results showed that PVT1 promotes tumor cell growth in part via competitively binding miR-203a.
Figure 4.
MiR-203a reversed the promoting effect of PVT1 on the growth of MM cells. A – Expression of miR-203a detected by qRT-PCR in MM cells transfected with si-PVT1 or si-PVT1+miR-203a inhibitor. B – CCK8 assays were used to determine proliferation of MM cells transfected with si-PVT1 or si-PVT1 + miR-203a inhibitor
*P < 0.05, **p < 0.01.
Discussion
Multiple myeloma (MM) is a neoplasm of terminally differentiated B cells (plasma cells), in which chromosome translocations frequently result in oncogene formation under the control of immunoglobulin enhancers [3]. MicroRNAs (miRNAs) are a class of small noncoding RNAs that control gene expression by targeting mRNAs and triggering either translation repression or RNA degradation [6]. MiRNAs may contribute to cancer development and progression, and are expressed differentially in normal tissues and cancers [17].
In the present study, qRT-PCR showed that PVT1 expression was increased in MM samples and cell lines. PVT1 suppression inhibited MM cell proliferation and induced cell apoptosis. These findings indicated that PVT1 acted as an oncogene in MM processes. Altogether, these findings prompt us to investigate the role of PVT1 in MM progression. To date, a variety of oncogenic pathways have been identified as directly regulated by PVT1. For example, Chen et al. found that PVT1 facilitated invasion through upregulation of MMP9 in lung cancer cell [18]. Zhou et al. reported that PVT1 modulated thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR) [19]. Zhang et al. reported that PVT1 facilitated cervical cancer progression through negative modulation of miR-128-3p [20]. Those studies indicated that PVT1 plays important roles in tumor progression.
LncRNAs have recently been reported to act as “miRNA sponges”, which modulate the expression of target genes [21]. For example, Wang et al. reported that lncRNA XIST exerted oncogenic functions in human glioma by targeting miR-137 [22]. Zhu et al. reported that lncRNA TUG1 promoted cervical cancer progression by regulating the miR-138-5p-SIRT1 axis [23]. In the present study, we explored whether PVT1 has similar effects on miR-203a in MM. QRT-PCR showed that miR-203a expression was significantly downregulated in MM samples and cell lines. PVT1 expression was negatively correlated with miR-203a expression in MM samples. The bioinformatics approach showed that miR-203a had a binding site with PVT1. The luciferase reporter assay indicated that co-transfection of miR-203a mimics and PVT1-Wt significantly reduced the luciferase activity. In addition, we found that si-PVT1 markedly increased the expression of miR-203a, but overexpression of miR-203a did not affect PVT1 expression in MM cells. Moreover, rescue experiments confirmed that miR-203a reversed the promoting effect of PVT1 on the growth of MM cells. These data suggested that PVT1 promotes tumor cell growth in part via competitively binding miR-203a.
In conclusion, this study demonstrated that PVT1 promoted cell proliferation by acting as a ceRNA to sponge miR-203a expression in MM. Therefore, our finding showed that a new axis of PVT1/miR-203a is involved in carcinogenesis of MM, which may provide a new perspective for MM therapy.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Kumar SK, Rajkumar SV, Dispenzieri A, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111:2516–20. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker N. Epidemiology of multiple myeloma. Recent Results Cancer Res. 2011;183:25–35. doi: 10.1007/978-3-540-85772-3_2. [DOI] [PubMed] [Google Scholar]
- 3.Fu J. Cx43 expressed on bone marrow stromal cells plays an essential role in multiple myeloma cell survival and drug resistance. Arch Med Sci. 2017;13:236–45. doi: 10.5114/aoms.2017.64722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner H, Gondos A, Pulte D. Recent major improvement in long-term survival of younger patients with multiple myeloma. Blood. 2008;111:2521–6. doi: 10.1182/blood-2007-08-104984. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nature Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Hua J, Ding T, Yang L. Dysfunction of microRNA-32 regulates ubiquitin ligase FBXW7 in multiple myeloma disease. Oncot Targets Ther. 2016;9:6573–9. doi: 10.2147/OTT.S105945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S, He X, Li M, et al. MiRNA-34a overexpression inhibits multiple myeloma cancer stem cell growth in mice by suppressing TGIF2. Am J Transl Res. 2016;8:5433–43. [PMC free article] [PubMed] [Google Scholar]
- 9.Qin Y, Zhang S, Deng S, et al. Epigenetic silencing of miR-137 induces drug resistance and chromosomal instability by targeting AURKA in multiple myeloma. Leukemia. 2017;31:1123–35. doi: 10.1038/leu.2016.325. [DOI] [PubMed] [Google Scholar]
- 10.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 11.Shen X, Zhang Y, Wu X, et al. Upregulated lncRNA-PCAT1 is closely related to clinical diagnosis of multiple myeloma as a predictive biomarker in serum. Cancer Biomark. 2017;18:257–63. doi: 10.3233/CBM-160158. [DOI] [PubMed] [Google Scholar]
- 12.Cho SF, Chang YC, Chang CS, et al. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014;14:809. doi: 10.1186/1471-2407-14-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez ML, DiStefano JK. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One. 2011;6:e18671. doi: 10.1371/journal.pone.0018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C, Yu W, Wang Q, et al. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106:143–9. [PubMed] [Google Scholar]
- 15.Song J, Wu X, Liu F, et al. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun. 2017;490:217–24. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 16.Zhang D, Sun G, Zhang H, Wang X, Tian J, Li Y. Long non-coding RNA PVT1 facilitates cervical cancer progression through negative modulation of miR-128-3p. Int J Clin Exp Pathol. 2017;10:4522–9. [Google Scholar]
- 17.Wan C, Shen Y, Yang T, et al. Diagnostic value of microRNA for pancreatic cancer: a meta-analysis. Arch Med Sci. 2012;8:749–55. doi: 10.5114/aoms.2012.31609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Zhu H, Yin L, et al. lncRNA-PVT1 facilitates invasion through upregulation of MMP9 in nonsmall cell lung cancer cell. DNA Cell Biol. 2017;36:787–93. doi: 10.1089/dna.2017.3725. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Q, Chen J, Feng J, Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR) Tumor Biol. 2016;37:3105–13. doi: 10.1007/s13277-015-4149-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhang D, Sun G, Zhang H, Wang X, Tian J, Li Y. Long non-coding RNA PVT1 facilitates cervical cancer progression through negative modulation of miR-128-3p. Int J Clin Exp Pathol. 2017;10:4522–9. [Google Scholar]
- 21.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–50. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Yuan J, Li L, Yang Y, Xu X, Wang Y. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am J Transl Res. 2017;9:1845–55. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Shi H, Liu H, Wang X, Li F. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017;8:65253–64. doi: 10.18632/oncotarget.18224. [DOI] [PMC free article] [PubMed] [Google Scholar]