Abstract
Background
The benefits of breastfeeding infants are well characterized, including those on the immune system. However, determining the mechanism by which human breast milk (HBM) elicits effects on immune response requires investigation in an appropriate animal model.
Objective
The primary aim of this study was to develop a novel porcine model and to determine the differential effects of feeding HBM and a commercial milk formula (MF) on immune response and gastrointestinal microbial colonization in a controlled environment.
Methods
Male piglets were fed HBM (n = 26) or MF (n = 26) from day 2 through day 21. Piglets were vaccinated (n = 9/diet group) with cholera toxin and cholera toxin subunit B (CTB) and tetanus toxoid at 21 d or were fed placebo (n = 6/diet group) and then weaned to a standard solid diet at the age of 21 d. Humoral and cell-mediated immune responses were assessed from blood on days 35 and 48. Immune response was further examined from tissues, including mesenteric lymph nodes (MLNs), Peyer's patches (PPs), and spleen. The colonization of gut microbiota was characterized from feces on days 16 and 49.
Results
Serum antibody titers in piglets fed HBM were 4-fold higher (P < 0.05) to CTB and 3-fold higher (P < 0.05) to tetanus toxoid compared with piglets fed MF on day 48. Compared with MF, the numbers of immunoglobulin A antibody–producing cells to CTB were 13-fold higher (P < 0.05) in MLNs and 11-fold higher (P < 0.05) in PPs in the HBM diet group on day 51. In addition, significantly increased T cell proliferation was observed in the HBM group relative to the MF group. Furthermore, microbial diversity in the HBM group was lower (P < 0.05) than in the MF group.
Conclusions
This porcine model appears to be valid for studying the effects of early postnatal diet on immune responses and the gastrointestinal microbiome. Our results lay the groundwork for future studies defining the role of infant diet on microbiota and immune function.
Keywords: gut-associated lymphoid tissue, humoral immunity, cell-mediated immunity, intestine, microbiota
Introduction
Many of the studies reporting on beneficial outcomes (reduced allergies, upper respiratory tract infections, and ear infections) of feeding with human breast milk (HBM) are epidemiologic in nature (1–5). Mechanisms that drive the association between breastfeeding and immune function are not fully elucidated. Studies have shown that neonatal immune system development is influenced by postnatal diet. Children vaccinated against Haemophilus influenzae type b and pneumococcal serotypes 6B and 14 and who experienced a longer duration of exclusive breastfeeding showed enhanced antibody response to vaccination at 13 mo of age (6). Interestingly, children aged 18 mo showed higher serum titers of IgG, IgA, and IgM if fed HBM exclusively for >13 wk compared with <13 wk (7). Hahn-Zoric et al. (8) also showed higher antibody responses to tetanus, diphtheria, live poliovirus, and H. influenzae type b in breastfed infants than in those consuming formula. However, other studies of vaccine response to tetanus toxoid (TT) and H. influenzae type b vaccines provide mixed evidence with regard to feeding with HBM (9, 10).
Gut microbiota composition has been shown to be related to an array of intestinal functions, including digestion, immune response, metabolite production, and disease susceptibility (11–19). For example, dietary lactose tolerance may be mediated by the presence of bacteria with greater fermentative capacity (20). In addition, postnatal diet shifts populations of intestinal microbiota that are associated with altered tryptophan metabolism (21), which may have consequences for allergic manifestation in infants (21–23). Previously, changes in the gut microbiota were observed between breastfed and formula-fed infants. However, these studies were limited in terms of understanding the immune response in relation to changes in the microbiota (21, 24, 25). Thus, it is prudent to investigate the acute and longitudinal effects of postnatal diet on immune system outcomes potentially mediated by the intestinal microbiome.
Due to the inherent limitations in obtaining tissues from human infants, an animal model can be used to gain a mechanistic understanding of the health outcomes related to postnatal diet. However, studies utilizing an animal model are often confounded by suckling compared with bottle feeding and by animal housing (e.g., farm, vivarium) (26, 27). These environmental and technical factors limit interpretations that can be derived from studying the effects of diet on immunity. Furthermore, previous studies have been designed to rear piglets with sow milk to emulate the condition of breastfeeding, even though the compositions of sow milk and HBM vastly differ (28). However, allowing piglets to suckle may not accurately recapitulate the effects of infant breastfeeding due to limitations such as housing environment, exposure to mother's pen, and inability to determine calorie intake. Therefore, we hypothesized that an HBM-fed piglet model that allows the study of the immune system under controlled dietary and environment conditions will eliminate the environmental limitations of a sow-fed model. Developing an animal model of feeding with HBM is fundamental to bridging the gap in understanding the effects of postnatal diet on immune system development, to further translational research initiatives focused on optimizing infant health. Because pigs are similar to humans in terms of gastrointestinal tract development (29–31), the neonatal porcine model is considered to be a good model for investigating the relation between gastrointestinal tract development and postnatal diet.
A primary aim of this study was to develop a novel porcine model to understand the relation between diet, HBM feeding, and immune response and the resulting coincident microbial colonization in a controlled environment. The present study focuses solely on the differences between HBM and infant formula diets in relation to both humoral and cell-mediated immunity outcomes under controlled environmental and feeding conditions. In addition, we explored diet-induced differences in fecal microbiota composition and its relation to postnatal dietary effects on gut health and the resulting immune system development.
Methods
Animal experiments
Animal maintenance and experimental treatments were conducted in accordance with the ethical guidelines for animal research established and approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. Piglet experimental conditions were performed as previously described (32).
Study design
Two-day-old White Dutch Landrace Duroc male piglets (Metz Farm, Russellville, Arkansas) were transferred to individual housing at the vivarium in Arkansas Children's Nutrition Center. Piglets were randomly assigned to consume HBM (n = 26) or an isocaloric dairy milk–based formula (MF; n = 26) (Figure 1). HBM was obtained from the Mothers’ Milk Bank of North Texas, and Similac Advance powder was obtained from Ross Products (Abbot Laboratories). Both HBM and MF diets were modified (Supplemental Table 1) (31) to meet the energy and nutrient recommendations of the NRC for growing pigs (33). In addition, diet composition and nutritional contents were analyzed (Eurofins, Des Moines, Iowa). With the transition to the Arkansas Children's Nutrition Center, piglets were trained to drink from rubber nipples and were fed 1.047 MJ ⋅ kg−1 ⋅ d−1 of either HBM or MF. Piglets were fed with warmed diet every 2 h in the first week of the study, every 4 h in the second week of the study, and every 6 h in the third week of the study through day 21. At day 14, solid “starter pig food” was slowly introduced until day 21 and all piglets had transitioned completely to an ad libitum solid diet (Teklad diet, TD 140608; Harlan) (Supplemental Table 2). Piglet weights and diet consumption were recorded daily for the duration of the study. Fecal samples were collected at day 16 and again at day 49 to provide microbial composition and relative abundance values. Eleven randomly selected piglets from each diet group were killed at 21 d. The remaining piglets from each diet group (HBM, n = 15; MF, n = 15) were further randomly assigned to the nonimmunized control group (n = 6) or an immunization group (n = 9). Initial immunization was conducted at day 21 and a booster immunization was given at day 35. The immunization regimen included oral administration of 100 µg cholera toxin (CT; C8052; Millipore Sigma) and 100 µg CT subunit B (CTB; C9903; Millipore Sigma) by gavage in 2 mL saline with 0.2 M sodium bicarbonate. Eight hours before immunization, piglets were feed deprived and received omeprazole (20 mg, orally; Arkansas Children's Hospital pharmacy) to promote optimal absorption and prevent degradation of CT. Piglets continued to be feed deprived for 1 h postimmunization. The DAPTACEL [diphtheria, tetanus, pertussis (DTaP)] vaccine (0.5 mL; Arkansas Children's Hospital pharmacy) was administered intramuscularly in the shank on the same day. Piglets in the nonimmunized control group also received omeprazole and were feed deprived before and 1 h postgavage with PBS (detailed methods outlined in the online supplemental material).
FIGURE 1.
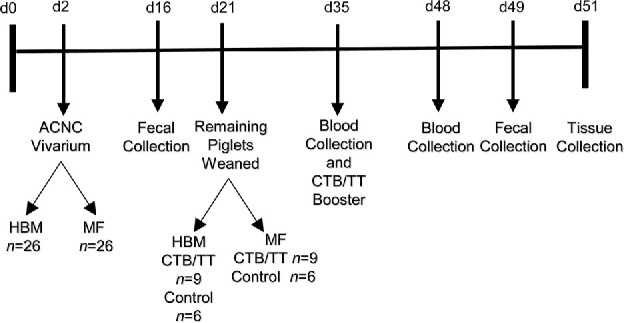
Two-day-old White Dutch Landrace Duroc male piglets were fed HBM (n = 26) or MF (n = 26) at the vivarium. Eleven piglets in each diet group were killed at 21 d, and tissues were obtained. The remaining piglets were weaned to an ad libitum diet at 21 d. In each diet group, piglets were immunized (n = 9) against CT+CTB and DTaP on day 21 and given a booster dose on day 35. Blood was drawn at 35 and 48 d. Tissues were obtained after killing at 51 d. ACNC, Arkansas Children's Nutrition Center; CT, cholera toxin; CTB, cholera toxin subunit B; d, day; DTaP, diphtheria, tetanus, and pertussis vaccine; HBM, human breast milk; MF, milk formula; TT, tetanus toxoid.
Sample collection and processing
Blood samples were collected on day 35, before the immunization booster vaccine, and again at day 48. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood samples collected (Fico/Lite; density, 1.077 g/L; Atlanta Biologicals) at both time points. As previously mentioned, a subset of piglets were killed at 21 d, or subsequently at 51 d for the remaining cohort, by exsanguination after anesthetization with isoflurane. Organs and tissues, including spleen, colon, small intestine, cecum, and prostate, were collected and weighed. In addition, individual segments of intestine were measured for length and weight after rinsing the contents out with 1× PBS. Peyer's patches (PPs), mesenteric lymph nodes (MLNs), spleen, and cervical lymph nodes (CLNs) were harvested using aseptic technique (detailed methods outlined in the online supplement material). Cells were washed and counted by Trypan blue exclusion before preparation for cell proliferation or ELISpot assays.
ELISA
CTB- and TT-specific antibody titers were assessed as well as total (nonspecific) serum IgA and IgG titers (detailed methods outlined in online supplemental material) by ELISA. Absorbance was measured on a Bio-Rad Benchmark Plus spectrophotometer at 450 nm.
ELISpot
To understand the humoral immune response elicited, secretions of antibodies for CTB-IgA and TT-IgG from MLNs, PPs, spleen, and CLNs at 51 d and PBMCs at 35 and 48 d were also assessed using an in-house-developed ELISpot assay (Supplemental Methods). Distinct antibody secretion spots were counted using a Nikon 7645 microscope or a Nikon NI-150 Fiber Optic Illuminator.
Lymphocyte cell proliferation
Cells were maintained in complete media (Roswell Park Memorial Institute media containing 1 mM sodium pyruvate, 1 mM nonessential amino acids, 1 mM HEPES, 0.5 mM 2-mercaptoethanol, 2mM l-glutamine, 10% FBS, and 50 µg gentamycin/mL). To understand the effects of diet on cell-mediated immune response, cells (2 × 105, duplicate/treatment) from MLNs, PPs, spleen, or PBMCs were stimulated by adding one of the following: 5 µg concanavalin A (ConA)/mL (C5275; Millipore Sigma), 50 ng phorbol 12-myristate-13-acetate (PMA)/mL (P8139, Millipore Sigma), 750 ng ionomycin (PMA+ionomycin)/mL (I-24222; Thermo Fisher Scientific), 1 µg LPS/mL (SC3535; Santa Cruz Biotechnology), 5 µg CTB/mL, or 3 μg TT/mL. CLN cells were assayed by using the aforementioned method and served as controls. Cells were incubated for 72 h (37°C, 5% CO2). Alamar blue (20 µL; DAL1100; Thermo Fisher Scientific) was added to the wells, and after 24 h the absorbance was read on a Bio-Rad Benchmark Plus spectrophotometer at 570 and 600 nm. The percentage difference in reduction of Alamar blue by antigen-treated compared with control untreated cells was calculated in order to determine the level of proliferation as described previously (34). The formula used to calculate the percentage difference is shown in the Supplemental Methods.
Microbiota analysis
Feces were homogenized, assessed for 16S ribosomal RNA (rRNA) gene amplicon sequencing (35), and analyzed by using QIIME 1.9.1 as described previously (Supplemental Methods) (36). Dietary group differences of α- and β-diversity and differential abundance were assessed in R using DAME (Dynamic Assessment of Microbial Ecology) (37).
Histomorphometric analyses
PP width and height and germinal center formation were assessed as previously described (21).
Statistical analysis
To model body weight and calorie intake over time, a mixed-effects regression model with a repeated-measures test was used. Restricted cubic splines for age were computed and interacted with diet category to better model the association between energy intake and age. All of the models were fitted via maximum likelihood estimation. From each fitted model, marginal effects and δ-method SEs were computed at each age. Computed marginal effects at each day were compared across diet categories with the use of Bonferroni-adjusted Wald tests. Data from day 22 onward were initially analyzed for diet, immunization, and interaction. For body and organ weights and histology, proliferation, and microbiome data, no immunization or interactions were observed; thus, data were analyzed for dietary group differences. Means of organ weights and lengths, antibody titers, and proliferation data were compared between HBM and MF diet groups by Mann-Whitney U test. Antibody titers are shown in log10. Proliferation data from immunized and nonimmunized piglets were subsequently pooled when neither interaction nor immunization was determined to be significant by 2-factor ANOVA. Data from the immunized piglets in the ELISpot assay were normalized to observations in the nonimmunized (control) piglets at 51 d, and negative binomial regression was used to assess differences between diet groups for the number of antibody-secreting cells (ASCs)/106 cells. Mann-Whitney U test and Permutational multivariate analysis of variance were used to assess microbiota differences between diet groups for α- and β-diversity, respectively. Pairwise comparisons of diet groups were assessed by using negative binomial regression as used by the DESeq2 Bioconductor package (38). Significance was defined at α < 0.05.
Results
Energy intake and body weight
Energy consumption is reported for piglets from days 3 through 48 (Supplemental Figure 1). There was no immunization or interaction effect observed; thus, data were pooled across immunization groups for subsequent energy and body-weight analyses. From days 3 through 48, there were no significant changes in energy intake between diet groups. However, there were acute exceptions observed on days 35, 37, and 38. Energy intake was lower in the HBM diet group than in the MF group at days 35 and 37, and energy intake was higher on day 38 in the HBM group than in the MF group.
Longitudinally, there were no significant diet effects on body-weight gain (Supplemental Figure 1), which is an expected outcome because diets were isocaloric (Supplemental Table 1). There were no significant group differences between organ weight or gastrointestinal length measures at day 21 (Supplemental Figure 2). On day 51, weights of the prostate and small intestine were lower in the HBM diet group than in the MF group (Supplemental Figure 2).
Humoral immune response
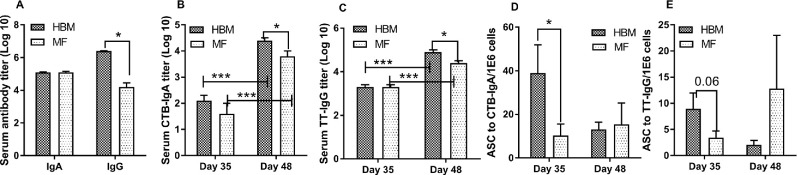
We assessed total antibody responses from serum on day 48 and found no differences in total IgA serum titer between diet groups (Figure 2A). However, on day 48, serum titer levels of total IgG in the HBM group were significantly higher than in the MF group (Figure 2A). We further analyzed serum samples for CTB- and TT-specific antibody response on days 35 and 48. In both HBM-fed and MF-fed groups, immunization elicited CTB-specific IgA and TT-specific IgG antibody titers of log2.2 or greater and titers from nonimmunized piglets (controls) were log1 or less, indicating that the immunization regimen elicited the anticipated antibody response as designed (data not shown). In both HBM- and MF-fed piglets, we observed significant increases in serum antibody titers at day 48 compared with day 35 in response to CTB and TT (Figure 2B, C). When diet groups were compared at each day, no group differences were observed on day 35 in CTB or TT antibody responses. However, on day 48, titers of CTB-specific IgA and TT-specific IgG were significantly higher in the HBM group compared with the MF group (Figure 2B, C). PBMCs isolated from blood samples on days 35 and 48 were stimulated ex vivo with CTB and TT and numbers of ASCs were measured. At day 35, there was a significantly higher number of ASCs for CTB in HBM-fed relative to MF-fed piglets (Figure 2D). For TT, there was a trend toward higher ASCs in the HBM group relative to the MF group, but this did not achieve significance (P = 0.06) (Figure 2E). On day 48, there were no significant differences between diet groups to CTB or TT (Figure 2D, E).
FIGURE 2.
Total IgA and IgG serum titers (n = 15/diet group) for day 48 (A), CTB-specific serum IgA (n = 9/diet group) (B), TT-specific serum IgG (n = 9/diet group) (C), CTB-specific ASCs from PBMCs (n = 9/diet group) (D), and TT-specific ASCs from PBMCs on days 35 and 48 (n = 9/diet group) (E). Data were evaluated by using Mann-Whitney U test to compare diet groups and time points. *,***Different between bracketed means: *P < 0.05, ***P < 0.001. ASC, antibody-secreting cell; CTB, cholera toxin subunit B; HBM, human breast milk; MF, milk formula; PBMC, peripheral blood mononuclear cell; TT, tetanus toxoid.
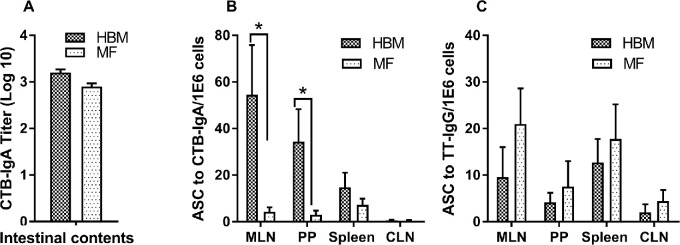
To understand intestinal antibody response, intestinal contents collected at the termination of the experiment on day 51 were used to measure CTB-specific IgA response. No significant differences in the production of CTB-specific IgA between diet groups were observed (Figure 3A). Furthermore, we determined the local immune response from the gut (e.g., MLNs, PPs) and systemic response from spleen cells on day 51 to CTB and TT ex vivo. Antibody secretions of IgA to CTB and IgG to TT were measured by ELISpot assay. Data from control, nonimmunized piglets were below the detection limit for ASCs. The number of ASCs for CTB was significantly higher in the HBM group than in the MF group from MLNs and PPs in immunized piglets (Figure 3B). No significant differences in ASCs were observed between diet groups with spleen cells to CTB. In addition, the number of ASCs to TT did not show any difference between the diet groups (Figure 3C). CLN tissues served as a negative control and, as expected, did not secrete anti-CTB antibodies. A very low number of ASCs were detected to TT (Figure 3C).
FIGURE 3.
IgA titers in the small intestine [duodenum (HBM: n = 2; MF: n = 2), jejunum (HBM: n = 6; MF: n = 6), and ileum (HBM: n = 6; MF: n = 6)] of immunized piglets fed HBM or MF on day 51 (A); CTB-specific ASCs from MLNs, PPs, spleen, and CLNs (n = 9/diet group) (B); and TT-specific ASCs from MLNs, PPs, spleen, and CLNs on day 51 (n = 9/diet group) (C). Values are means ± SEMs. Data were evaluated by Mann-Whitney U test. *Different between groups, P < 0.05. ASC, antibody-secreting cell; CLN, cervical lymph node; CTB, cholera toxin subunit B; HBM, human breast milk; MF, milk formula; MLN, mesenteric lymph node; PP, Peyer's patch; TT, tetanus toxoid.
Cell-mediated immunity
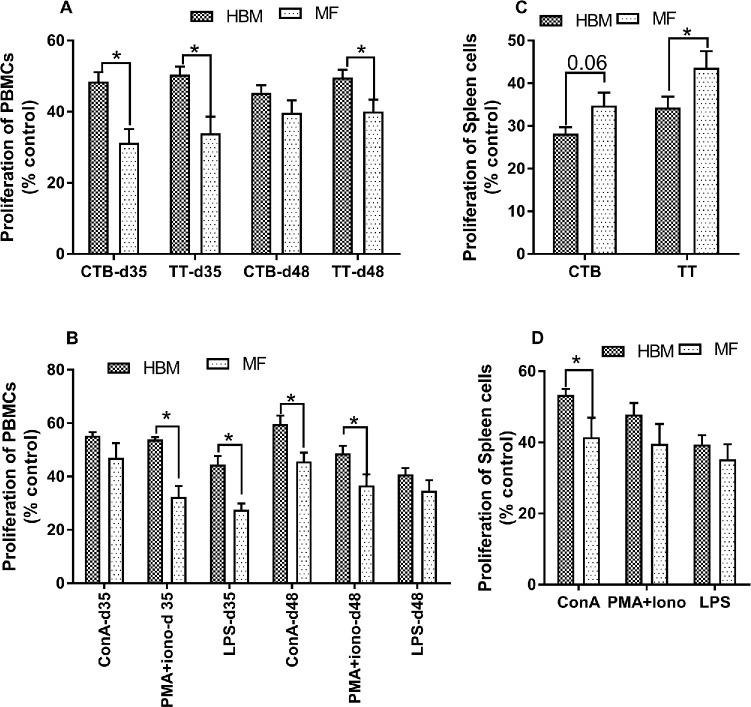
Cell-mediated immunity was examined from PBMCs, MLNs, PPs, and spleen by measuring cell proliferation after exposure to CTB, TT, and nonspecific mitogens (ConA, LPS, and PMA+ionomycin). Cells without any antigens or mitogens served as controls, and data were normalized to cells alone. No immunization effect was observed in proliferation; therefore, data were pooled from immunized and control, nonimmunized groups. PBMCs from HBM-fed piglets showed significantly increased proliferation relative to MF-fed piglets in response to CTB challenge on day 35 but not on day 48 (Figure 4A). In addition, PBMCs stimulated with TT from the HBM group showed increased proliferation on days 35 and 48 relative to the MF group (Figure 4A). PBMCs from the HBM-fed group showed significantly increased proliferation to nonspecific mitogens relative to the MF group on both days 35 and 48, except to ConA on day 35 and LPS on day 48 (Figure 4B). In addition, spleen cells from the HBM-fed group showed significantly decreased proliferation to TT on day 51 relative to the MF group (Figure 4C). Interestingly, spleen cells from the HBM group showed increased proliferation to ConA on day 21 (Figure 4D) relative to the MF group. No difference in proliferation was noted from MLNs and PPs; with the exception of day 21 in which PMA showed increased proliferation of T cells in the MF group relative to the HBM group in MLNs. ConA increased proliferation on day 51 in the MF group relative to the HBM group from PPs (Supplemental Figure 3).
FIGURE 4.
Proliferative response elicited from PBMCs and spleen cells to CTB and TT in piglets (n = 15/diet group) fed HBM or MF (A), mitogen-specific proliferation from PBMCs on days 35 and 48 (B), CTB- and TT-specific proliferation from spleen cells on day 51 (C), and mitogen-specific proliferation of spleen cells on day 21 (D). Values are means ± SEMs. Data were evaluated by Mann-Whitney U test. *Different between groups, P < 0.05. ConA, concanavalin A; CTB, cholera toxin subunit B; HBM, human breast milk; MF, milk formula; PBMC, peripheral blood mononuclear cell; PMA+Iono, phorbol 12-myristate-13-acetate + ionomycin; TT, tetanus toxoid; % control, percentage difference in reduction of Alamar blue by antigen-treated compared with control untreated cells calculated in order to determine the level of proliferation.
Histomorphometric analyses
To understand if the neonatal diet has an impact on PP morphology and germinal center development, histomorphometric analyses were carried out. We did not observe any difference in PP width or height or in germinal center formation on days 21 and 51, regardless of postnatal diet or immunization status (Supplemental Figure 4).
Microbial diversity and abundance
α-Diversity
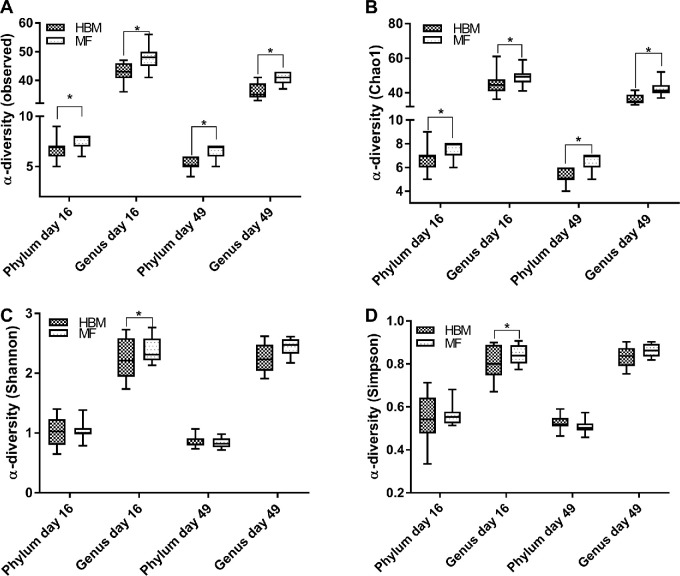
Previous studies have shown microbiota changes in breastfed compared with formula-fed infants (29–32), and the microbiome is believed to be a critical regulator of host immune system development (14, 39–42). Thus, differences in microbiota between diet groups were evaluated. No immunization effect on α-diversity was observed; thus, we pooled the control and immunized groups into their respective diets. We observed significantly less diversity in the HBM group than in the MF group at days 16 and 49 in both the observed and Chao1 indexes (Figure 5A, B) at the phylum level. However, these differences were not exhibited in the Shannon or Simpson indexes for the same time points for phylum level (Figure 5C, D). Genus-level α-diversity was also affected by diet group. At day 16, the HBM group showed less α-diversity than the MF group across all 4 indexes (Figure 5). The trend of significantly less α-diversity seen in the HBM group compared with the MF group at the genus level continued at day 49 in both the observed and Chao1 indexes (Figure 5).
FIGURE 5.
α-Diversity boxplots for piglets at the phylum and genus levels for observed (A), Chao1 (B), Shannon (C), and Simpson (D) indexes at 16 d in HBM (n = 14) and MF (n = 15) diet groups and at 49 d in the HBM (n = 11) and MF (n = 12) diet groups. Upper and lower hinges (boxes) are minimum and maximum values within the IQR and the solid lines are median values. Data were analyzed by Mann-Whitney U test. *Different between groups, P < 0.05. HBM, human breast milk; MF, milk formula.
β-Diversity
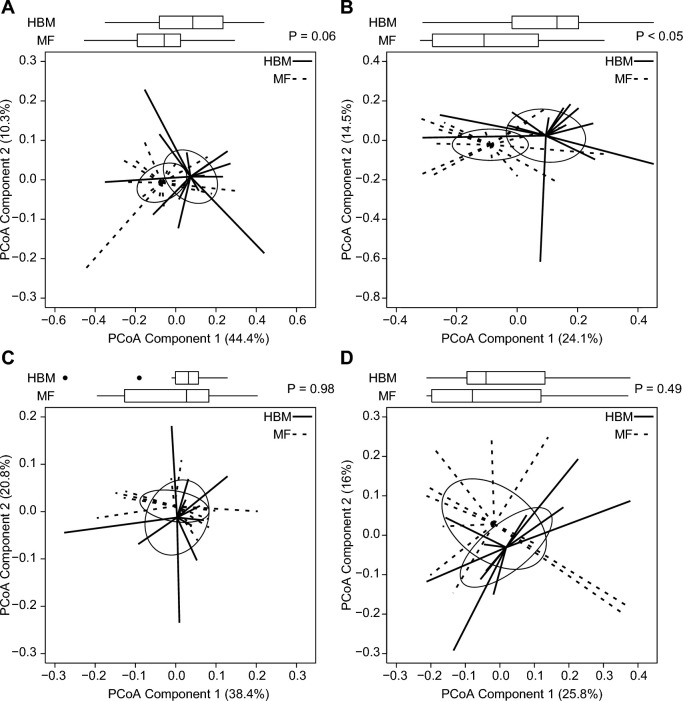
To determine the differences between piglets relative to dietary group assignment, β-diversity was assessed by using the Bray-Curtis dissimilarity (Figure 6). No effect was observed between groups at day 16 or day 49 at the phylum level using principal coordinates analysis. However, genus-level diversity between diet groups was significant at day 16 (Figure 6B) but not at day 49.
FIGURE 6.
Box and spider plots represent Bray-Curtis estimates of β-diversity for piglets for phylum (A) and genus (B) in HBM (n = 14) and MF (n = 15) diet groups at 16 d and for phylum (C) and genus (D) in HBM (n = 11) and MF (n = 12) diet groups at 49 d. Box plot outliers are shown as black dots outside the whiskers. Solid and dashed lines extend from diet group medoids to the PCoA score for an individual piglet. Ellipses represent the SE of the point scores along the 2 components. P values represent group differences among PCoA scores along component 1 (Mann-Whitney U test). HBM, human breast milk; MF, milk formula; PCoA, principal coordinates analysis.
Relative abundance
In addition to microbial diversity, the relative abundance of taxa between diet groups was quantified by using 16s rRNA sequencing. Fecal samples collected at day 16 in both the HBM and MF groups detected 9 phylogenic groups of microbiota (Table 1). Bacteriodetes, Firmicutes, and Proteobacteria were the most abundant phyla in both diet groups at this time point. Of the 9 phyla shown at day 16, Verrucomicrobia was the only phylum to reach a significantly higher abundance in the HBM group compared with the MF group. To determine relative abundance after postneonatal diet, 16s rRNA sequencing was repeated using fecal samples taken on day 49. Interestingly, in both diet groups at day 49, only 7 different microbial phyla were detected as opposed to the 9 phyla seen on day 16 (Table 2). At day 49, relative abundances of Proteobacteria and Cyanobacteria were less in the HBM group than in the MF group. Although Verrucomicrobia was the only phylum to have a significantly different abundance at day 16, it was not detected in either diet group at day 49. Furthermore, Actinobacteria abundance decreased from day 16 to day 49 in both diet groups.
TABLE 1.
Relative abundance of phyla in piglets fed HBM or MF at days 16 and 491
| Day 16 | Day 49 | |||||||
|---|---|---|---|---|---|---|---|---|
| Phylum | HBM (n = 14) | MF (n = 15) | Fold-change (log2)2 | P (adjusted) | HBM (n = 11) | MF (n = 12) | Fold-change (log2)2 | P (adjusted) |
| Verrucomicrobia | 3.11 ± 1.3 | 0.09 ± 0.0 | 5.30 | <0.01 | — | — | — | — |
| Lentisphaerae | 0.06 ± 0.0 | 0.02 ± 0.0 | 0.70 | 0.83 | 0.20 ± 0.1 | 0.12 ± 0.0 | −0.58 | 0.88 |
| Actinobacteria | 2.63 ± 1.1 | 3.53 ± 0.6 | −1.30 | 0.14 | 0.01 ± 0.0 | 0.02 ± 0.0 | 0.16 | 0.88 |
| Proteobacteria | 5.05 ± 1.5 | 4.64 ± 1.1 | 0.30 | 0.83 | 1.23 ± 0.4 | 2.40 ± 0.4 | −0.77 | <0.05 |
| Firmicutes | 36.53 ± 5.0 | 50.53 ± 5.1 | −0.10 | 0.83 | 34.61 ± 3.3 | 34.07 ± 3.1 | 0.29 | 0.12 |
| Bacteroidetes | 47.26 ± 3.9 | 38.82 ± 4.7 | 0.10 | 0.83 | 63.21 ± 3.6 | 62.33 ± 3.4 | 0.17 | 0.74 |
| Tenericutes | 2.56 ± 0.8 | 1.25 ± 0.5 | 1.20 | 0.62 | 0.73 ± 0.2 | 0.54 ± 0.2 | 0.92 | 0.32 |
| Fusobacteria | 2.77 ± 0.0 | 0.04 ± 0.0 | −0.40 | 0.83 | — | — | — | — |
| Synergistetes | 0.01 ± 0.0 | 0.04 ± 0.0 | −3.10 | 0.28 | — | — | — | — |
| Cyanobacteria | — | — | — | — | <0.01 | 0.53 ± 0.2 | −8.05 | <0.01 |
1Values in diet columns are means ± SEMs of relative abundances on days 16 and 49. The SEM is not indicated where relative abundance was <0.01% and the em dash (—) indicates <0.005% relative abundance. P values (adjusted) from Wald test indicate differences between diet groups within day. HBM, human breast milk; MF, milk formula.
2Fold-change [log2(normalized means)] reference group is MF.
TABLE 2.
Relative abundance of genera in piglets fed HBM or MF at day 161
| Genus | HBM (n = 14) | MF (n = 15) | Fold-change (log2)2 | P (adjusted) |
|---|---|---|---|---|
| Bacteroides | 29.34 ± 4.3 | 22.03 ± 3.8 | 1.40 | 0.02 |
| Unassigned genus (in Ruminococcaceae family) | 11.31 ± 2.2 | 13.36 ± 1.4 | 0.40 | 0.55 |
| Parabacteroides | 6.77 ± 2.7 | 6.94 ± 1.4 | 0.30 | 0.85 |
| Lactobacillus | 1.89 ± 1.2 | 8.35 ± 2.0 | −1.40 | 0.13 |
| Prevotella | 4.80 ± 2.2 | 3.00 ± 1.3 | 1.40 | 0.39 |
| - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - | ||||
| Sharpea | 0.23 ± 0.2 | 0.01 ± 0.0 | 22.50 | <0.01 |
| Akkermansia | 3.11 ± 1.3 | 0.084 ± 0.0 | 6.00 | <0.01 |
| Sutterella | 1.18 ± 0.6 | 0.12 ± 0.1 | 4.60 | <0.01 |
| Dorea | 2.30 ± 1.0 | 0.51 ± 0.1 | 2.60 | <0.01 |
| Megasphaera | 0.10 ± 0.1 | 0.04 ± 0.0 | 5.30 | <0.01 |
| Unassigned genus (in Enterobacteriaceae family) | 0.82 ± 0.6 | 3.18 ± 1.1 | −2.70 | 0.02 |
| Pasteurella | 0.30 ± 0.1 | 0.17 ± 0.1 | 2.40 | 0.04 |
1Values in diet columns are means ± SEMs of relative abundance on day 16. The dashed line separates the 5 most abundant taxa from other taxa that were significantly different (P < 0.05) between diet groups. P values (adjusted) from Wald test indicate differences between diet groups within day. HBM, human breast milk; MF, milk formula.
2Fold-change [log2(normalized means)] reference group is MF.
We further investigated genera differences between the diet groups. At day 16, the most prevalent genera observed in both diet groups were from Bacteroides, an unassigned genus in the Ruminococcaceae family, Parabacteroides, Lactobacillus, and Prevotella. In addition, significantly higher abundances of Sharpea, Akkermansia, Sutterella, Dorea, Megaspaera, and Pasturella were observed in the HBM diet group compared with the MF diet group (Table 2). Also of distinction, the relative abundance of an unassigned genus in the Enterobacteriaceae family was lower in the HBM group than in the MF group at day 16.
At day 49, the 5 most abundant genera detected were Prevotella (Prevotellaceae family), Lactobacillus, Bacteroides, Prevotella (Paraprevotellaceae family), and an unassigned genus in the Ruminococcaceae family (Table 3). The 5 genera exhibiting significantly lower relative abundance at day 49 in the HBM group in comparison to the MF group were YRC22, Phascolarctobacterium, p-75-a5, CF231, and an unassigned genus in the Erysipelotrichaceae family.
TABLE 3.
Relative abundance of genera in piglets fed HBM or MF at day 491
| Genus | HBM (n = 11) | MF (n = 12) | Fold-change (log2)2 | P (adjusted) |
|---|---|---|---|---|
| Prevotella (in Prevotellaceae family) | 26.66 ± 3.5 | 22.65 ± 2.6 | 0.11 | 0.87 |
| Lactobacillus | 9.42 ± 3.2 | 6.60 ± 1.8 | 1.21 | 0.30 |
| Bacteroides | 11.52 ± 2.6 | 9.96 ± 2.8 | 0.74 | 0.30 |
| Prevotella (in Paraprevotellaceae family) | 4.58 ± 1.1 | 2.56 ± 0.8 | 1.54 | 0.21 |
| Unassigned genus (in Ruminococcaceae family) | 3.49 ± 1.3 | 3.23 ± 1.5 | 0.17 | 0.79 |
| - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - | ||||
| YRC22 | <0.01 | 4.14 ± 2.0 | −8.18 | <0.01 |
| Phascolarctobacterium | 0.30 ± 0.1 | 1.47 ± 0.3 | −1.90 | <0.01 |
| p-75-a5 | 0.01 ± 0.0 | 0.21 ± 0.1 | −4.40 | <0.01 |
| CF231 | <0.01 | 3.24 ± 1.7 | −9.14 | <0.01 |
| Eubacterium (in Erysipelotrichaceae family) | 0.03 ± 0.0 | 0.03 ± 0.0 | −2.98 | 0.01 |
1Values in diet columns are means ± SEMs of relative abundance on day 49. The SEM is not indicated where relative abundance was <0.01%. The dashed line separates the 5 most abundant taxa from other taxa that were significantly different (P < 0.05) between diet groups. P values (adjusted) from Wald test indicate differences between diet groups within day. HBM, human breast milk; MF, milk formula.
2Fold-change [log2(normalized means)] reference group is MF.
Discussion
Porcine models for studying the gut and immune system offer several advantages to other animal models, but when the gastrointestinal microbiome is an important variable, the use of the sow-fed piglets offers some challenges. These challenges relate to housing conditions that allow fecal contamination to influence piglet gastrointestinal microbiota composition and relative abundance. A major challenge associated with allowing piglets to suckle from the sow is the intake of microbiota from the maternal environment and outcomes associated with skin-to-skin contact (27, 43). A second challenge is the difficulty in monitoring energy intake in sow feeding compared with bottle feeding. The primary aim of the present study was to use a novel porcine model to determine how the 2 widely used infant diets, HBM and commercially marketed formula (MF), affect development of immune responses. We believe this is the first use of HBM in a pig model and the first head-to-head comparison of HBM and commercial infant formula effects on the immune system in a pig model. The present study controlled for feeding conditions and environment, allowing for differences in immune outcomes and microbial colonization to be attributed specifically to postnatal diet. Therefore, the porcine model of HBM feeding used in the current study is novel because it controls for these challenges and may be particularly relevant to studying the effects of diet on immune response and relative to colonization of microbiota. The results indicate that humoral and cell-mediated immune responses are significantly greater in HBM-fed than in MF-fed piglets, suggesting that breastfeeding may enhance experimentally induced immune responses.
A previous study showed enhanced immune function related to innate immunity in sow-fed piglets relative to formula-fed piglets (44). In addition, the immune response exhibited in the intestinal mucosa differs in piglets depending on whether a formula or sow-milk postnatal diet is consumed. For example, Jensen et al. (45) showed that formula-fed piglets had significantly increased mucosal weights and crypt depth. In another study, formula-fed piglets exhibited reduced lactose-digestive capacity relative to sow milk–fed piglets (46). Previously, we reported increased PP size in sow-fed relative to formula-fed piglets (32). Our results suggest that 4 wk postweaning, the HBM-diet group had a higher IgG response than did the MF group. Furthermore, serum titers of IgA and IgG specific to CTB and TT, as well as total IgG, were significantly lower in formula-fed piglets relative to HBM-fed piglets. This observation agrees with the enhanced immune cell stimulation and proliferation elicited by CTB antigen seen in PBMCs 2 wk after administration of vaccine in the HBM diet group. Although antibody secretion by MLNs and PPs was also increased by HBM feeding, this observation was not reflected in the titer of IgA specific to CTB in the small intestine contents. This latter effect may not have been observed due to the stimulus used to evoke an immune response or because intestinal antibody titers had reached a plateau and were thus equal in both diet groups 4 wk after initial immunization. Alternatively, feed deprivation of piglets before collecting luminal contents may have eliminated diet-induced differences in antibody levels in the lumen. Taken together, these data support the notion that the early effect of postnatal diet influences humoral immune responses in the gut and systemically with regard to vaccine response and highlight that feeding breast milk early in life may enhance the immune response to exogenous challenges. The processes that underlie the differences in immune function when comparing breastfed with formula-fed conditions might involve host IgA, toll-like receptor (TLR) signaling, and shifts in the population and activities of the gut microbiota.
Humoral immune response can be initiated by binding of pattern recognition receptors to pathogen-associated molecular patterns. In this respect, TLR signaling pathways are critical in the induction of the adaptive immune response (47). For example, TLR5 knockout in mice results in decreased antibody titers to trivalent inactivated influenza vaccine and restoration of TLR5-mediated sensing of microbial flagellin-induced IgG responses similar to wild-type mice (48). In addition, stimulation of TLR4 in monocytes increases IgG titer (49), and the production of IgG is reduced by TLR4 knockout (50). Moreover, there is cross-talk between TLR4 and nucleotide-binding oligomerization domain–containing protein 2 (NOD2) (51), which enhances the magnitude of both humoral and cell-mediated immune responses. Interestingly, pattern recognition receptors interact with the gut microbiota and then confer downstream signaling to inhibit or promote the humoral immune response (52–54). In fact, administration of vancomycin decreases intestinal Bacteroidetes and decreases serum IgA and IgG titers, and specific genera within the Bacteroidetes phylum are known TLR agonists (55). Thus, microbiota composition was measured and an elevation in the abundance of Bacteroides was noted in the HBM group. Several studies have shown differences in the gut microbiota when comparing breastfeeding with formula feeding (25, 56, 57), suggesting that the gut microbiota may play a role in immune response (13, 39, 40, 58–60). Results noted here indicate that HBM enhances the humoral immune response when compared with formula feeding, which may be due to the postnatal diet, intestinal microbiota composition, or both (39, 42, 50, 61–64). In summary, these studies suggest that immune cell signaling and subsequent response may be modulated on the basis of the presence of specific microbiota and cross-talk with the host.
Interestingly, previous literature has shown that fecal transplant significantly improves clinical manifestation of infantile allergic colitis and was also concurrent with an increase in Bacteroides in feces (65). From a mechanistic standpoint, Bacteroides have been shown to induce development of T-regulatory cells by promoting the expression of IL-10 (66) which mitigates proinflammatory signaling via production of polysaccharide A (67). Exposure of naïve T cells to commensal gut microbiota has been suggested to divert from the generation of colitogenic effector cells (68). The literature supports our results, because T cell stimulation (69) was enhanced in PBMCs and spleen by the HBM diet relative to formula. Therefore, intestinal colonization by Bacteroides may affect cell-mediated immunity systemically and constitutes a potential mechanism by which postnatal diet exhibits effects on the immune system. Although we did not observe major differences in T cell proliferation in the gut tissues, it is possible that cells from gut regions reached the threshold for stimulants to observe differences between diet groups. Future studies should evaluate the specific role of the microbiota in immune response.
In addition to Bacteroides, distinct dietary differences in the relative abundances of several other genera and phyla were noted in feces in the postnatal and weaning periods. The relative abundance of the Verrucomicrobia phylum and Akkermansia genus were higher with HBM diet. Akkermansia has been linked to altered physiologic processes. Specifically, it has been associated with regulation and maintenance of intestinal barrier integrity (70); the implication is that HBM may regulate gut permeability via promoting growth of specific microbiota such as Akkermansia and others. In addition, the gut microbiota has been shown to utilize IgA for colonization (61). Cumulatively, this study and the body of literature suggest that there is cross-talk between the immune system and microbiota composition during postnatal feeding. This, in turn, could possibly alter processes that affect chronic disease risk later in life. Future studies should aim to determine the cross-talk between host and microbiota and its impact on immune function.
Strengths and limitations
There is a demonstrated need for reliable models in which to study how early diet mediates immune response. Piglets are considered the model most similar to human infants to understand immune system function (30, 45, 71–73). However, in previous porcine models, there have been technical limitations that have contributed to the limited generalizability of results. The diets used in this study were matched for energy density and macronutrient composition. It is possible that the effects of postnatal diet observed in this study were due to the presence or absence of immunomodulatory components such as prebiotics (60), oligosaccharides (74, 75), immunoglobulins, and lactoferrin (76), but it is beyond the scope of the current study to infer correlation between any specific component in the diet and the HBM effects observed. This study shows the feasibility of using a porcine model of feeding HBM under controlled conditions, with the limitations that piglets were fed 48 h of sow milk, only male piglets were analyzed, and the donor human milk was pasteurized. The findings in the present study confirm that the gut microbiota are more diverse in infants who consume formula compared with HBM (12, 25, 56, 57, 77), indicating a strength of this animal model. However, there is a distinct difference between colonization of piglet and infant intestine with respect to the relative abundance of certain taxa. The infant gut microbiota is highly abundant in Actinobacteria, of which Bifidobacterium is among the most abundant genus (78, 79). However, detection of this phylum and genus was very low in the piglet intestine, even when fed HBM. There is speculation that the intestines of some infants do not permit growth of Bifidobacterium (80). Thus, there is an opportunity to utilize the piglet HBM model to discern the necessary habitable conditions for intestinal colonization of Actinobacteria in future research.
Conclusions
The postnatal diet influences gut and systemic immune responses. Specifically, humoral and cell-mediated immune responses are higher in breast milk–fed than in commercially marketed formula–fed piglets. The influence of postnatal diet on intestinal microbiota composition constitutes a plausible and likely mechanism by which HBM confers beneficial effects on the development of the local gut and systemic immune response. Although there are differences in perinatal development of the intestinal tract between piglets and infants, the porcine HBM model appears to offer advantages over the sow-fed model to eliminate the limitations associated with housing environment and diet intake. Future studies in sow-fed and HBM-fed piglets under same environmental conditions would further validate the model. Future studies should aim to identify specific gut bacterial signals or diet components that modulate immune function.
Supplementary Material
Acknowledgments
We thank Matt Fergusson, Bobby Fay, and Trae Pittman for vivarium help with the piglets. The authors’ responsibilities were as follows—LY and TMB: designed the research; LY, JJM, and KSM: wrote the manuscript; LY, AKB, and MKS: conducted technical aspects of the research; MAC: conducted statistical analyses; SVC, BDP, and KS: helped with microbiome data acquisition; TL: conducted histomorphometric analyses; LY: had primary responsibility for the final content and for editing the manuscript; and all authors: read and approved the final manuscript.
Notes
Supported by the USDA–Agricultural Research Service Project 6026-51000-010-05S and the NIH (P20GM121293).
Author disclosures: JJM, TMB, AKB, KSM, MAC, TL, MKS, SVC, BDP, KS, and LY, no conflicts of interest.
Supplemental Tables 1 and 2, Supplemental Methods, and Supplemental Figures 1–4 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: ASC, antibody-secreting cell; CLN, cervical lymph node; ConA, concanavalin A; CT, cholera toxin; CTB, cholera toxin subunit B; HBM, human breast milk; MF, milk formula; MLN, mesenteric lymph node; PBMC, peripheral blood mononuclear cell; PMA, phorbol 12-myristate-13-acetate; PP, Peyer's patch; rRNA, ribosomal RNA; TLR, Toll-like receptor; TT, tetanus toxoid.
References
- 1. Beaudry M, Dufour R, Marcoux S. Relation between infant feeding and infections during the first six months of life. J Pediatr 1995;126(2):191–7. [DOI] [PubMed] [Google Scholar]
- 2. Beaudry M, Dufour R, Marcoux S. [Breast feeding and protection against infection in industrialized countries.] Arch Pediatr 1996;3(Suppl 1):126s–7s (in French). [DOI] [PubMed] [Google Scholar]
- 3. Hanson LA, Korotkova M. The role of breastfeeding in prevention of neonatal infection. Semin Neonatol 2002;7(4):275–81. [DOI] [PubMed] [Google Scholar]
- 4. Hanson LA, Korotkova M, Telemo E. Breast-feeding, infant formulas, and the immune system. Ann Allergy Asthma Immunol 2003;90(6 Suppl 3):59–63. [DOI] [PubMed] [Google Scholar]
- 5. Hanson LA, Hofvander Y, Lindquist B, Zetterstrom R. Breast-feeding as a protection against gastroenteritis and other infections. Acta Paediatr Scand 1985;74(5):641–2. [DOI] [PubMed] [Google Scholar]
- 6. Silfverdal SA, Ekholm L, Bodin L. Breastfeeding enhances the antibody response to Hib and Pneumococcal serotype 6B and 14 after vaccination with conjugate vaccines. Vaccine 2007;25(8):1497–502. [DOI] [PubMed] [Google Scholar]
- 7. Silfverdal SA, Bodin L, Ulanova M, Hahn-Zoric M, Hanson LA, Olcen P. Long term enhancement of the IgG2 antibody response to Haemophilus influenzae type b by breast-feeding. Pediatr Infect Dis J 2002;21(9):816–21. [DOI] [PubMed] [Google Scholar]
- 8. Hahn-Zoric M, Fulconis F, Minoli I, Moro G, Carlsson B, Bottiger M, Raiha N, Hanson LA. Antibody responses to parenteral and oral vaccines are impaired by conventional and low protein formulas as compared to breast-feeding. Acta Paediatr Scand 1990;79(12):1137–42. [DOI] [PubMed] [Google Scholar]
- 9. Stephens S, Kennedy CR, Lakhani PK, Brenner MK. In-vivo immune responses of breast- and bottle-fed infants to tetanus toxoid antigen and to normal gut flora. Acta Paediatr Scand 1984;73(4):426–32. [DOI] [PubMed] [Google Scholar]
- 10. Watemberg N, Dagan R, Arbelli Y, Belmaker I, Morag A, Hessel L, Fritzell B, Bajard A, Peyron L. Safety and immunogenicity of Haemophilus type b-tetanus protein conjugate vaccine, mixed in the same syringe with diphtheria-tetanus-pertussis vaccine in young infants. Pediatr Infect Dis J 1991;10(10):758–63. [DOI] [PubMed] [Google Scholar]
- 11. Catassi C, Bonucci A, Coppa GV, Carlucci A, Giorgi PL. Intestinal permeability changes during the first month: effect of natural versus artificial feeding. J Pediatr Gastroenterol Nutr 1995;21(4):383–6. [DOI] [PubMed] [Google Scholar]
- 12. Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 2003;91(441):48–55. [DOI] [PubMed] [Google Scholar]
- 13. Andersson Y, Hammarstrom ML, Lonnerdal B, Graverholt G, Falt H, Hernell O. Formula feeding skews immune cell composition toward adaptive immunity compared to breastfeeding. J Immunol 2009;183(7):4322–8. [DOI] [PubMed] [Google Scholar]
- 14. Le Huerou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev 2010;23(1):23–36. [DOI] [PubMed] [Google Scholar]
- 15. Dube K, Schwartz J, Mueller MJ, Kalhoff H, Kersting M. Iron intake and iron status in breastfed infants during the first year of life. Clin Nutr 2010;29(6):773–8. [DOI] [PubMed] [Google Scholar]
- 16. Poroyko V, Morowitz M, Bell T, Ulanov A, Wang M, Donovan S, Bao N, Gu S, Hong L, Alverdy JC et al. Diet creates metabolic niches in the “immature gut” that shape microbial communities. Nutr Hosp 2011;26(6):1283–95. [DOI] [PubMed] [Google Scholar]
- 17. Thompson AL. Developmental origins of obesity: early feeding environments, infant growth, and the intestinal microbiome. Am J Hum Biol 2012;24(3):350–60. [DOI] [PubMed] [Google Scholar]
- 18. Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv Nutr 2012;3(Suppl):450S–5S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bouzerzour K, Morgan F, Cuinet I, Bonhomme C, Jardin J, Le Huerou-Luron I, Dupont D. In vivo digestion of infant formula in piglets: protein digestion kinetics and release of bioactive peptides. Br J Nutr 2012;108(12):2105–14. [DOI] [PubMed] [Google Scholar]
- 20. Zhong Y, Priebe MG, Vonk RJ, Huang CY, Antoine JM, He T, Harmsen HJ, Welling GW. The role of colonic microbiota in lactose intolerance. Dig Dis Sci 2004;49(1):78–83. [DOI] [PubMed] [Google Scholar]
- 21. Saraf MK, Piccolo BD, Bowlin AK, Mercer KE, LeRoith T, Chintapalli SV, Shankar K, Badger TM, Yeruva L. Formula diet driven microbiota shifts tryptophan metabolism from serotonin to tryptamine in neonatal porcine colon. Microbiome 2017;5(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van der Leek AP, Yanishevsky Y, Kozyrskyj AL. The microbiome. Front Immunol 2017;8:1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feldberg W, Smith AN. Release of histamine by tryptamine and 5-hydroxytryptamine. J Physiol 1953;122(Suppl):62. [PubMed] [Google Scholar]
- 24. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 2011;108(Suppl 1):4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Z, Roy NC, Guo Y, Jia H, Ryan L, Samuelsson L, Thomas A, Plowman J, Clerens S, Day L et al. Human breast milk and infant formulas differentially modify the intestinal microbiota in human infants and host physiology in rats. J Nutr 2016;146(2):191–9. [DOI] [PubMed] [Google Scholar]
- 26. Kubasova T, Davidova-Gerzova L, Merlot E, Medvecky M, Polansky O, Gardan-Salmon D, Quesnel H, Rychlik I. Housing systems influence gut microbiota composition of sows but not of their piglets. PLoS One 2017;12(1):e0170051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson CL, Wang B, Holmes AJ. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. ISME J 2008;2(7):739–48. [DOI] [PubMed] [Google Scholar]
- 28. Salcedo J, Frese SA, Mills DA, Barile D. Characterization of porcine milk oligosaccharides during early lactation and their relation to the fecal microbiome. J Dairy Sci 2016;99(10):7733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moughan PJ, Birtles MJ, Cranwell PD, Smith WC, Pedraza M. The piglet as a model animal for studying aspects of digestion and absorption in milk-fed human infants. World Rev Nutr Diet 1992;67:40–113. [DOI] [PubMed] [Google Scholar]
- 30. Darragh AJ, Moughan PJ. The three-week-old piglet as a model animal for studying protein digestion in human infants. J Pediatr Gastroenterol Nutr 1995;21(4):387–93. [DOI] [PubMed] [Google Scholar]
- 31. Helm RM, Golden C, McMahon M, Thampi P, Badger TM, Nagarajan S. Diet regulates the development of gut-associated lymphoid tissue in neonatal piglets. Neonatology 2007;91(4):248–55. [DOI] [PubMed] [Google Scholar]
- 32. Yeruva L, Spencer NE, Saraf MK, Hennings L, Bowlin AK, Cleves MA, Mercer K, Chintapalli SV, Shankar K, Rank RG et al. Formula diet alters small intestine morphology, microbial abundance and reduces VE-cadherin and IL-10 expression in neonatal porcine model. BMC Gastroenterol 2016;16:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Research Council. Nutrient requirements of swine. 11th revised ed Washington (DC): National Academies Press; 2012. [Google Scholar]
- 34. Ahmed SA, Gogal RM Jr., Walsh JE. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay. J Immunol Methods 1994;170(2):211–24. [DOI] [PubMed] [Google Scholar]
- 35. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Piccolo BD, Wankhade UD, Chintapalli SV, Bhattacharyya S, Luo C, Shankar K. Dynamic Assessment of Microbial Ecology (DAME): a Web app for interactive analysis and visualization of microbial sequencing data. Bioinformatics 2018;34(6):1050–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ardeshir A, Narayan NR, Mendez-Lagares G, Lu D, Rauch M, Huang Y, Van Rompay KK, Lynch SV, Hartigan-O'Connor DJ. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med 2014;6(252):252ra120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Atarashi K, Umesaki Y, Honda K. Microbiotal influence on T cell subset development. Semin Immunol 2011;23(2):146–53. [DOI] [PubMed] [Google Scholar]
- 41. Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity 2009;31(3):368–76. [DOI] [PubMed] [Google Scholar]
- 42. Feng T, Elson CO. Adaptive immunity in the host-microbiota dialog. Mucosal Immunol 2011;4(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen X, Xu J, Ren E, Su Y, Zhu W. Co-occurrence of early gut colonization in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe 2018;49:30–40. [DOI] [PubMed] [Google Scholar]
- 44. Liu KY, Comstock SS, Shunk JM, Monaco MH, Donovan SM. Natural killer cell populations and cytotoxic activity in pigs fed mother's milk, formula, or formula supplemented with bovine lactoferrin. Pediatr Res 2013;74(4):402–7. [DOI] [PubMed] [Google Scholar]
- 45. Jensen AR, Elnif J, Burrin DG, Sangild PT. Development of intestinal immunoglobulin absorption and enzyme activities in neonatal pigs is diet dependent. J Nutr 2001;131(12):3259–65. [DOI] [PubMed] [Google Scholar]
- 46. Thymann T, Burrin DG, Tappenden KA, Bjornvad CR, Jensen SK, Sangild PT. Formula-feeding reduces lactose digestive capacity in neonatal pigs. Br J Nutr 2006;95(6):1075–81. [DOI] [PubMed] [Google Scholar]
- 47. Kawai T, Akira S. TLR signaling. Cell Death Differ 2006;13(5):816–25. [DOI] [PubMed] [Google Scholar]
- 48. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014;41(3):478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tukhvatulin AI, Dzharullaeva AS, Tukhvatulina NM, Shcheblyakov DV, Shmarov MM, Dolzhikova IV, Stanhope-Baker P, Naroditsky BS, Gudkov AV, Logunov DY et al. Powerful complex immunoadjuvant based on synergistic effect of combined TLR4 and NOD2 activation significantly enhances magnitude of humoral and cellular adaptive immune responses. PLoS One 2016;11(5):e0155650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Nunez G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016;44(3):647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim H, Zhao Q, Zheng H, Li X, Zhang T, Ma X. A novel crosstalk between TLR4- and NOD2-mediated signaling in the regulation of intestinal inflammation. Sci Rep 2015;5:12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. de Bruyn M, Vermeire S. NOD2 and bacterial recognition as therapeutic targets for Crohn's disease. Expert Opin Ther Targets 2017;21(12):1123–39. [DOI] [PubMed] [Google Scholar]
- 53. Li YY, Pearson JA, Chao C, Peng J, Zhang X, Zhou Z, Liu Y, Wong FS, Wen L. Nucleotide-binding oligomerization domain-containing protein 2 (Nod2) modulates T1DM susceptibility by gut microbiota. J Autoimmun 2017;82:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu D, Zhang Y, Liu Y, Hou L, Li S, Tian H, Zhao T. Berberine modulates gut microbiota and reduces insulin resistance via the TLR4 signaling pathway. Exp Clin Endocrinol Diabetes 2018; doi: 10.1055/s-0043-125066. [DOI] [PubMed] [Google Scholar]
- 55. van Opstal E, Kolling GL, Moore JH II, Coquery CM, Wade NS, Loo WM, Bolick DT, Shin JH, Erickson LD, Warren CA. Vancomycin treatment alters humoral immunity and intestinal microbiota in an aged mouse model of Clostridium difficile infection. J Infect Dis 2016;214(1):130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee SA, Lim JY, Kim BS, Cho SJ, Kim NY, Kim OB, Kim Y. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr Res Pract 2015;9(3):242–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fan W, Huo G, Li X, Yang L, Duan C. Impact of diet in shaping gut microbiota revealed by a comparative study in infants during the six months of life. J Microbiol Biotechnol 2014;24(2):133–43. [DOI] [PubMed] [Google Scholar]
- 58. Arslanoglu S, Moro GE, Schmitt J, Tandoi L, Rizzardi S, Boehm G. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J Nutr 2008;138(6):1091–5. [DOI] [PubMed] [Google Scholar]
- 59. Battersby AJ, Gibbons DL. The gut mucosal immune system in the neonatal period. Pediatr Allergy Immunol 2013;24(5):414–21. [DOI] [PubMed] [Google Scholar]
- 60. Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes 2017;8(2):143–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 2018;360(6390):795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Parigi SM, Eldh M, Larssen P, Gabrielsson S, Villablanca EJ. Breast milk and solid food shaping intestinal immunity. Front Immunol 2015;6:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009;9(5):313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16(6):341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu SX, Li YH, Dai WK, Li XS, Qiu CZ, Ruan ML, Zou B, Dong C, Liu YH, He JY et al. Fecal microbiota transplantation induces remission of infantile allergic colitis through gut microbiota re-establishment. World J Gastroenterol 2017;23(48):8570–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011;332(6032):974–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Burgess JN, Pant AB, Kasper LH, Colpitts Brass S. CD4(+) T cells from multiple sclerosis patients respond to a commensal-derived antigen. Ann Clin Transl Neurol 2017;4(11):825–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011;478(7368):250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Boilard E, Surette ME. Anti-CD3 and concanavalin A-induced human T cell proliferation is associated with an increased rate of arachidonate-phospholipid remodeling: lack of involvement of group IV and group VI phospholipase A2 in remodeling and increased susceptibility of proliferating T cells to CoA-independent transacyclase inhibitor-induced apoptosis. J Biol Chem 2001;276(20):17568–75. [DOI] [PubMed] [Google Scholar]
- 70. Chelakkot C, Choi Y, Kim DK, Park HT, Ghim J, Kwon Y, Jeon J, Kim MS, Jee YK, Gho YS et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med 2018;50(2):e450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dawson HD, Smith AD, Chen C, Urban JF Jr.. An in-depth comparison of the porcine, murine and human inflammasomes: lessons from the porcine genome and transcriptome. Vet Microbiol 2017;202:2–15. [DOI] [PubMed] [Google Scholar]
- 72. Dawson HD, Loveland JE, Pascal G, Gilbert JG, Uenishi H, Mann KM, Sang Y, Zhang J, Carvalho-Silva D, Hunt T et al. Structural and functional annotation of the porcine immunome. BMC Genomics 2013;14:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Odle J, Lin X, Jacobi SK, Kim SW, Stahl CH. The suckling piglet as an agrimedical model for the study of pediatric nutrition and metabolism. Annu Rev Anim Biosci 2014;2:419–44. [DOI] [PubMed] [Google Scholar]
- 74. Comstock SS, Li M, Wang M, Monaco MH, Kuhlenschmidt TB, Kuhlenschmidt MS, Donovan SM. Dietary human milk oligosaccharides but not prebiotic oligosaccharides increase circulating natural killer cell and mesenteric lymph node memory T cell populations in noninfected and rotavirus-infected neonatal piglets. J Nutr 2017;147(6):1041–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Comstock SS, Wang M, Hester SN, Li M, Donovan SM. Select human milk oligosaccharides directly modulate peripheral blood mononuclear cells isolated from 10-d-old pigs. Br J Nutr 2014;111(5):819–28. [DOI] [PubMed] [Google Scholar]
- 76. Comstock SS, Reznikov EA, Contractor N, Donovan SM. Dietary bovine lactoferrin alters mucosal and systemic immune cell responses in neonatal piglets. J Nutr 2014;144(4):525–32. [DOI] [PubMed] [Google Scholar]
- 77. Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 2011;17(6):478–82. [DOI] [PubMed] [Google Scholar]
- 78. Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 2012;7(5):e36957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lewis ZT, Mills DA. Differential establishment of Bifidobacteria in the breastfed infant gut. Nestle Nutr Inst Workshop Ser 2017;88:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tannock GW, Lee PS, Wong KH, Lawley B. Why don't all infants have Bifidobacteria in their stool? Front Microbiol 2016;7:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.