Abstract
Mobile genetic elements play an important role in the dissemination of antibiotic resistant bacteria among human and environmental sources. Therefore, the aim of this study was to determine the occurrence and patterns of integrons and insertion sequences of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from different sources in Navarra, northern Spain. A total of 150 isolates coming from food products, farms and feeds, aquatic environments, and humans (healthy people and hospital inpatients), were analyzed. PCRs were applied for the study of class 1, 2, and 3 integrons (intI1, intI2, and intI3), as well as for the determination of insertion sequences (IS26, ISEcp1, ISCR1, and IS903). Results show the wide presence and dissemination of intI1 (92%), while intI3 was not detected. It is remarkable, the prevalence of intI2 among food isolates, as well as the co-existence of class 1 and class 2 (8% of isolates). The majority of isolates have two or three IS elements, with the most common being IS26 (99.4%). The genetic pattern IS26–ISEcp1 (related with the pathogen clone ST131) was present in the 22% of isolates (including human isolates). In addition, the combination ISEcp1–IS26–IS903–ISCR1 was detected in 11 isolates being, to our knowledge, the first study that describes this genetic complex. Due to the wide variability observed, no relationship was determined among these mobile genetic elements and β-lactam resistance. More investigations regarding the genetic composition of these elements are needed to understand the role of multiple types of integrons and insertion sequences on the dissemination of antimicrobial resistance genes among different environments.
Keywords: ESBL-producing E. coli, β-lactamase genes, antimicrobial resistance, integrons, insertion sequences
1. Introduction
Antimicrobial resistance (AMR) has become a public health problem, reaching alarming levels in many parts of the world [1,2]. In recent years, resistances in the Enterobacteriaceae family have increased significantly because of the extensive use of antibiotics in human treatment, veterinary, and agriculture, leading to the selection and global spread of resistant clones [3,4]. In particular, the dissemination of extended-spectrum β-lactamases (ESBLs) have increased dramatically in the recent years, becoming a serious global threat [5,6].
Several genetic mechanisms have been involved in the acquisition and dispersion of antimicrobial resistances. The commonly called “mobilome” [7,8] is composed of a variety of mobile genetic elements (MGEs), including plasmids, transposons (Tn), insertion sequences (IS), integrons (intI), and introns. Conjugation, transformation, and transduction are the main mechanisms for the horizontal transfer of MGEs [9,10].
Integrons are DNA elements capable of capturing gene cassettes (including antimicrobial resistance genes) and disseminating them using an MGE [11]. Integrons are usually composed of two conserved segments (termed 5′-conserved region (5′-CS) and 3′-conserved region (3′-CS)) separated by a variable region which contains the gene cassettes. The 5′-CS end includes (i) the int gene coding for an integrase, that belongs to a distinct family of the tyrosine-recombinase; (ii) a primary recombination site (attI); and (iii) a promoter (Pc), which ensures the transcription of the cassette genes. On the other hand, the 3′-CS region is formed by (i) a truncated gene of resistance to quaternary ammonium compounds (qacEΔ1); (ii) a sulfonamide resistance gene (sul1); and (iii) an unknown function sequence (orf5) [12]. Class 1 (intI1) and class 2 (intI2) integrons are the most commonly involved in antibiotic resistances [13,14,15,16,17], while limited work has shown the presence of class 3 (intI3) in Enterobacteriaceae. The gene intI3 was reported for the first time in a carbapenem-resistant Serratia marcescens strain [18] and has been also detected in Klebsiella pneumoniae isolates [19] and other Enterobacteriaceae [20]. In addition, bla ESBL genes have been associated with insertion sequences. These IS are the smallest transposable elements (<2.5 kb), and are classified into families according to different characteristics, with transposases (enzymes that catalyze the IS movement) being the main classification system used [21,22]. It has been well documented that IS26, ISEcp1, ISCR1, and IS903, in association with class 1 integrons, are the most involved elements in the antimicrobial resistance to β-lactamics [23,24,25,26,27].
Therefore, the investigation of these elements might be critical, in order to predict the potential spread of ESBL-producing strains. In this context, the aim of this study was to evaluate the presence of different types of integrons (intI1, intI2, and intI3) and insertion sequences (ISEcp1, IS26, ISCR1, and IS903) in a collection of 150 ESBL-producing E. coli isolated from different sources in Navarra, Spain.
2. Materials and Methods
2.1. Sample Collection
A total of 150 ESBL-producing Escherichia coli were selected from a wide collection of ESBL-producing Enterobacteriaceae, isolated in Navarra from different environments: food products (n = 48), farms and feeds (n = 20), rivers and wastewater treatment plants (WWTPs) (n = 33) and human origins, including healthy volunteers (n = 13) and hospital inpatients (n = 36). Clinical isolates from hospital inpatients were provided by Clínica Universidad de Navarra, and were collected from January 2009 to December 2012 [5]. Food and environmental samples were collected from different locations in Navarra in two sampling periods (2010–2013 [5,28,29]; 2015–2016 [30]) and, finally, isolates from healthy people were collected from September 2015 to September 2016 (data not published). All samples were already identified, and phenotypically and genotypically characterized, in order to know the antimicrobial susceptibility pattern, the types of β-lactamase genes, and the phylogenetic group [28,29,30]. Isolates were selected according to the following criteria: they must show multidrug-resistant phenotype (to at least three different classes of antimicrobials) and must carry at least one ESBL gene. The main characteristics of the selected isolates, regarding type of ESBL, is shown in Table 1. Resistance profiles and complete information of each isolate is presented in the Supplementary Material, Tables S1–S6).
Table 1.
Genotypic characteristics of extended-spectrum β-lactamase (ESBL)-producing E. coli according to their origin [5,28,29,30].
| Sample Origin | Percentages of Detected bla Genes | |||||
|---|---|---|---|---|---|---|
| bla CTX-M-14 | bla CTX-M-15 | bla CTX-M-1 | bla TEM-42 | bla TEML-171 | bla SHV-12 | |
| Hospital inpatients | 41.7 | 61.1 | 8.3 | 11.1 | NA | 5.5 |
| Healthy people | 46.2 | 30.8 | 15.4 | 0 | 46.2 | 0 |
| WWTP and rivers | 33.3 | 30.3 | 18.2 | 6 | NA | 6 |
| Food | 32.7 | 4.1 | 18.3 | 12.3 | 31.8 | 35.6 |
| Farms and feeds | 31.6 | 5.26 | 47.4 | 26.3 | 5 | 21 |
NA: Not analyzed.
2.2. DNA Extraction and Detection of Integrons
DNA extraction was performed with DNeasy® Blood & Tissue kit (Qiagen, Barcelona, Spain), using a pre-treatment protocol for Gram-negative bacteria, and following the manufacturer’s instruction. The quantity and quality of the DNA was analyzed using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Detection of class 1, class 2, and class 3 integrons in ESBL-producing E. coli was performed according to PCR, as described by Mazel et al. [31], and using only the primers shown in Table 2.
Table 2.
Primers used for the detection of integrons.
| Primer | Sequence (5′-3′) | Amplicon Size (pb) | T (°C ) 3 | GenBank Accession No | Reference |
|---|---|---|---|---|---|
| intI1-Fw 1 | GGTCAAGGATCTGGATTTCG | 483 | 62 | U49101 | [31] |
| intI1-Rv 2 | ACATGCGTGTAAATCATCGTC | 483 | 62 | U49101 | [31] |
| intI2-Fw 1 | CACGGATATGCGACAAAAAGGT | 789 | 62 | L10818 | [31] |
| intI2-Rv 2 | TAGCAAACGAGTGACGAAATG | 789 | 62 | L10818 | [31] |
| intI3-Fw 1 | AGTGGGTGGCGAATGAGTG | 600 | 60 | D50438 | [31] |
| intI3-Rv 2 | TGTTCTTGTATCGGCAGGTG | 600 | 60 | D50438 | [31] |
1 Fw: forward; 2 Rv: reverse; 3 T (°C): annealing temperature.
DNA amplification was performed in a DNA thermal cycler GeneAmp® PCR system 2700 (Applied Biosystems Division, Foster City, CA, USA) in a final volume of 25 µL containing 2 µL of DNA extract mixed with 2.5 µL of 10× buffer (Bioline, London, UK), 5 µL of dNTPs (Bioline, London, UK), 1.5 µL of MgCl2 50 mM (Bioline, London, UK), 2 µL of each primer Sigma-Aldrich, Steinheim, Germany), and 1.5 U of Inmolase™ DNA polymerase (Bioline, London, UK). The conditions of the amplification were as follows: initial denaturation at 94 °C for 10 min, followed by 30 cycles of DNA denaturation at 94 °C for 45 s, primer annealing at 62 °C (intI1 and intI2) or 60 °C (intI3) for 35 s, primer extension at 72 °C for 2 min, and a final elongation at 72 °C for 7 min. Positive and negative controls [17] were included in all PCR assays, and 1 kb ladder (Invitrogen) was used as a molecular size standard. After amplification, PCR products were separated by electrophoresis on 1% agarose gel in 1× TBE buffer, stained with ethidium bromide and visualized by UV transillumination. E. coli C828, K. pneumoniae C933 (provided both by Centro de Investigación Biomédica de la Rioja) and E. coli isolated from hospital inpatients, confirmed as carrying intI2 by DNA sequencing, were used as positive controls for intI1, intI3, and intI2, respectively.
2.3. Detection of Insertion Sequences
DNA extracts were examined for the detection of different insertion sequences associated with ESBL genes, performing PCRs assays using the specific primers and conditions showed in Table 3 [27,32,33].
Table 3.
Primers and conditions used for the amplification of insertion sequences.
| Primer 1 | Sequence (5′-3′) | Amplicon Size (pb) | T (°C) 3 | GenBank Accession No. | Reference |
|---|---|---|---|---|---|
| ISEcp1-Fw 1 | ATCTAACATCAAATGCAGG | 1381 | 60 | AJ972954 | [27] |
| ISEcp1-Rv 2 | AGACTGCTTCTCACACAT | 1381 | 60 | AJ972954 | [27] |
| IS26-Fw 1 | TCACTCCACGATTTACCGCT | 557 | 61 | AF205943 | [27] |
| IS26-Rv 2 | CTTACCAGGCGCATTTCGCC | 557 | 61 | AF205943 | [27] |
| ISCR1-Fw 1 | TCGCTGCGAGGATTGTCATC | 1100 | 60 | AF174129 | [32] |
| ISCR1-Rv 2 | CTCGCTTGAGGCGTTGCAT | 1100 | 60 | AF174129 | [32] |
| IS903-Fw 1 | CATATGAAATCATCTGCGC | 473 | 56 | EU056266 | [33] |
| IS903-Rv 2 | CCGTAGCGGGTTGTGTTTTC | 473 | 56 | EU056266 | [33] |
1 Fw: forward; 2 Rv: reverse; 3 T (°C): annealing temperature.
The PCRs were performed in a final volume of 25 µL containing 2 µL of DNA extract mixed with 2.5 µL of 10× buffer (Bioline, London, UK), 5 µL of dNTPs (Bioline, London, UK), 1.5 µL of MgCl2 50 mM (Bioline, London, UK), 2 µL of each primer (Sigma-Aldrich, Madrid, Spain), and 1.5 U of Inmolase™ DNA polymerase (Bioline, London, UK), in a DNA thermal cycler GeneAmp® PCR system 2700 (Applied Biosystems Division, Foster City, CA, USA). Amplification conditions were modified in order to improve the specificity using an initial denaturation at 94 °C for 12 min, followed by 35 cycles of DNA denaturation at 94 °C for 1 min, and primer annealing temperature depending on the IS (Table 3), primer extension at 72 °C for 2 min, and a final elongation at 72 °C for 10 min. PCR products were separated by electrophoresis on 1% agarose gels and were visualized under UV light after staining with ethidium bromide.
2.4. Sequence Analysis
Amplicons obtained in the different PCRs were sequenced to confirm the presence of integrons and insertion sequences. Bidirectional DNA sequence analysis was performed by the Macrogen EZ-Seq purification service (Macrogen Europe, Amsterdam, The Netherlands). Searches for DNA and protein homologies were carried out using the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the BLAST program and the alignment of DNA and amino acids sequences were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).
2.5. Statistical Analysis
The results were subjected to statistical processing with the SPSS 15 software (SPSS Inc., Chicago, IL, USA), applying the chi-square test with a level of significance of p < 0.05.
3. Results and Discussion
3.1. Distribution of Integrons in ESBL-Producing E. coli
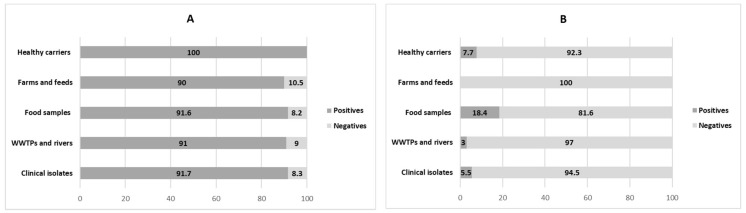
The occurrence and types of integrons, according to the isolate origin, is presented in Figure 1. As expected, class 1 showed the highest dissemination, being present in 92% of the isolates (n = 138) and in all environments, without significant differences among them (p < 0.05). Class 1 integrons have been reported as the most ubiquitous type among enteric bacteria [34,35,36]. In a similar way, Solberg et al. [37] reported the presence of class 1 integron in 70% of E. coli causing community-acquired infections. According to Roe et al. [38], the occurrence of class 1 integrons in healthy people suggests a possible acquisition of resistance genes circulating in different environments by a constant horizontal exchange of these genes. By contrast, class 2 integron was found in only 13 strains (8.5%), in accordance with the study of Ozgumus et al. [39], who found this class of integron in pathogenic, environmental, and commensal E. coli with a lower frequency than class 1. Finally, intI3 was not detected, similarly to the report by Vinué et al. [40]. In fact, limited studies describe the presence of class 3 integron in E. coli [14,41] and, to date, there are no published data reporting the presence of this integron in ESBL-producing E. coli isolated from Spain.
Figure 1.
Prevalence (percentages) and distribution of (A)class 1 (intI1) and (B) class 2 (intI2) integrons in ESBL-producing E. coli according to their origin. ESBL: Extended spectrum beta-lactamases.
It should be noted that intI2 was mainly detected in food isolates (18.4%), but not in farming environments (p = 0.044). This situation seems a little bit contradictory, but it could be due to the low number of isolates coming from farms and feed (n = 20), compared with food (n = 48). Probably, if we extended the study by increasing n, we would find positive results for this type of integron, as shown in the literature. In any case, our results are comparable to those obtained by Goldstein et al. [42], who demonstrated the presence of class 1 and class 2 integrons in food, livestock, and water contaminated with farm animal feces. In a similar way, intI2 has been detected in poultry products [38].
In addition, it is remarkable that intI1 and intI2 coexist in 8% of the isolates (92.1% of those carrying intI2). Rizk et al. [20] reported the co-existence of more than one type of integron in 36.9% of isolates, and a prevalence of 38% was reported by Kargar et al. [41] in a study performed in 69 multidrug-resistant (MDR) E. coli. By contrast, Kor et al. [43] found only one isolate carrying both integrons among clinical isolates, and Odetoyin et al. [16] reported a prevalence of 2.4% in fecal E. coli isolated from mother–child pairs in Nigeria. The simultaneous existence of multiple integrons represents a great threat for the dissemination of antimicrobial resistance genes among Enterobacteriaceae [31].
3.2. Analysis of Insertion Sequences
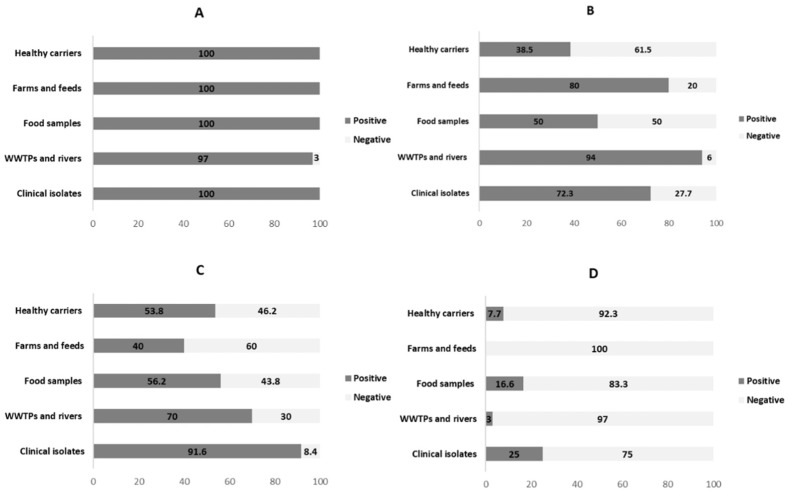
The prevalent type was IS26 (99.4%), followed by ISEcp1 (68%) and IS903 (65.3%), while ISCR1 was detected only in 19 isolates (12.6%). The wide presence of IS26 in almost all multidrug-resistant isolates is a hint that IS26 is not associated with multidrug resistance, but only with ESBL-producing isolates.
The four insertion sequences were present in all environments (Figure 2), except ISCR1, that was not detected in farm and feeds. This latest result contrasts with the reported by Ali et al. [44], that showed the connection between ISCR1 and intI1 in strains isolated from diverse dairy farms in China. However, the wide dissemination of IS among different niches has been reported by other authors. For instance, Cullik et al. [25] showed the association between blaCTX-M with the common elements ISEcp1, IS26, and IS903, in ESBL-producing E. coli isolated in a German Hospital. The ISEcp1 type has been detected in clinical isolates from Korea, and in isolates from healthy or diseased food-producing animals, including swine and avian [45,46].
Figure 2.
Prevalence (percentages) and distribution of insertion sequences in ESBL-producing E. coli according to their origin. (A) IS26; (B) ISEcp1; (C) IS903; (D) ISCR1.
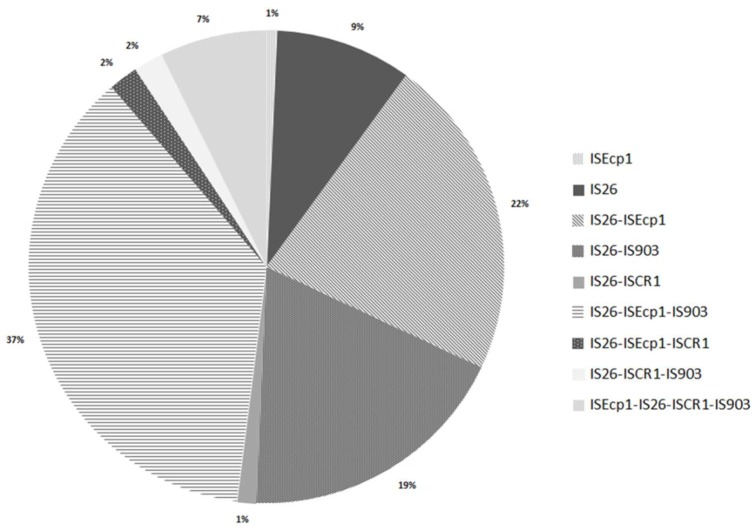
In addition, the frequent co-existence of several insertion sequences in the same strain has been detected, in agreement with other studies [25,26,27,47]. Genetic patterns are presented in Figure 3, showing that the majority of isolates carried two or three IS (42% and 40.7%, respectively), whereas 10% of them carried only one. The three prevalent genetic patterns were IS26–ISEcp1–IS903 (n = 55), IS26–ISEcp1 (n = 33), and IS26–IS903 (n = 28) (37%, 22%, and 19%, respectively). The combination IS26–ISEcp1 has been related with the pathogen clone ST131 [25,48], and it was present in 4 isolates coming from healthy people (n = 2) and hospital inpatients (n = 2), that supposes a possible risk situation for the healthy population. Finally, it is remarkable that 7.3% of isolates contain the four IS (ISEcp1–IS26–IS903–ISCR1), a situation that, to our best knowledge, is being described in the literature for the first time. These isolates come mainly from hospital inpatients (n = 9), but we also found the genetic patterns in isolates from a river (n = 1) and from a chicken hamburger (n = 1). In summary, these results show the complexity of mobile genetic elements, and suggest the facility to acquire different mechanisms to disseminate resistance genes through all environments.
Figure 3.
Genetic patterns and prevalences among the studied ESBL-producing E. coli.
3.3. The Important Role of Horizontal Genetic Elements in the Dissemination of ESBLs
Correlation between the presence of genetic elements and ESBL has been reported by several authors [25,49,50], and our results support this fact (Table 4). IS26 have been observed flanking the open reading frame (orf) regions of β-lactamase genes [51], and prevalences higher than 94% in all ESBL types were observed in this study. Similar results were detected in a study carried out in Kenya with 27 E. coli strains obtained from hospitalized patients, in which over 40% of isolates carrying blaTEM-52, blaSHV-5, or blaCTX-M-14, were linked to the IS26 [50]. Otherwise, Billard-Pomares et al. [52] reported the characterization of a P1 bacteriophage from an ESBL-E. coli strain which had acquired two foreign DNA fragments, one of them being a fragment mobilized by two IS26 elements containing a blaSHV-2 gene. Finally, Doi et al. [53] reported the relation between OXA (Beta lactamase product of blaOXA genes) and IS26 downstream of a class 1 integron in a K. pneumoniae strain. In summary, as Cullik et al. affirm [25], IS26 have an important role in the spread of resistance genes.
Table 4.
Prevalences of insertion sequences and integrons among the different types of ESBL-E. coli producers.
| bla Genes | IS26 | IS903 | ISEcp1 | ISCR1 | intI1 | intI2 |
|---|---|---|---|---|---|---|
| bla CTX-M | 99.2 | 90.3 | 79 | 11.3 | 94 | 6.5 |
| bla TEM | 100 | 89.9 | 88.5 | 16 | 94 | 7.3 |
| bla OXA-1 | 94.5 | 83.3 | 50 | 5.5 | 100 | 0 |
| bla SHV | 100 | 56.5 | 26 | 0 | 95.7 | 21.8 |
Similarly, ISEcp1-like insertion sequences have been observed upstream of orfs encoding members belonging to the CTX-M-1, CTX-M-2, and CTX-M-9 clusters. Kim et al. [45] found the association of ISEcp1 and CTX-M in clinical isolates, especially in strains containing CTX-M-14 (in agreement with the 37% observed in our study). A similar association was found in China by Sun et al. [54] in healthy and sick pets. In addition, Tamang et al. [55] reported that 97.6% blaCTX-M genes (isolated from cattle, farm workers, and the farm environment) possessed the insertion sequence ISEcp1 upstream of blaCTX. On the other hand, our results show that 9 out of 102 isolates carrying ISEcp1 (isolated from WWTP, river, farm soil and feed) were disrupted by IS26. Similar findings have been reported in a German University Hospital [25], where cases of ISEcp1 disrupted by an intact IS26 were detected. In the same way, Wang et al. [48] detected a truncated copy of ISEcp1 gene with an IS26 gene being located upstream in 3 out of 9 ESBL-producing E. coli isolated from fecal samples of food producing animals and healthy humans. Finally, despite the lower prevalence of ISCR1 observed in the present study (12.6%), the aforementioned IS is another important element in the genetic platforms associated with the dissemination of CTX-M genes [22,56,57]. In general, ISCR1 has been associated with CTX-M-2 and CTX-M-9 subtypes [57,58,59], but the majority of our strains carrying this IS were CTX-M-14 and CTX-M-15 producers. That could explain the low number of strains carrying ISCR1. Moreover, ISCR1 mediates the formation of a complex with class 1 integrons [23,57]. From the total of isolates carrying ISCR1, 94.7% contain intI1, and even one of them contained both integrons (intI1 and intI2). However, we have not found a specific association between the isolates containing intI1 and the different ESBLs, due to its wide presence (92% of isolates). On the other hand, CTX-M-14 was present in the 46% of the isolates containing intI2 (the same as SHV-12), whereas TEM and CTX-M-1 were detected in 38.5% and 15.4%, respectively, of intI2 carriers.
Moreover, it must be pointed out that ISEcp1-IS903 is known as one of the major genetic platforms [22,27,54]. Our results showed that IS903 and ISEcp1 were present in 55 isolates in co-existence with IS26 (IS26-ISEcp1-IS903). Similarly, a recent report detected this genetic platform [46] in CTX-M-14-producing E. coli isolated from animals [48]. Furthermore, all the analyzed strains show multidrug-resistant (MDR) phenotype, which means that they are resistant to at least three different classes of antimicrobials [28,29]. Similar results were reported by Woodford et al. [60] that found the plasmid pEK499 harboring 10 genes that confer resistance to eight antibiotic classes and also carrying IS (IS26 and ISEcp1).
Finally, Table 5 summarizes the relationship between the number of IS present in the same isolate, and the number of ESBL types produced by each microorganism. It can be seen that as the number of ESBL enclosed in the same genetic environment increases, the number of insertion sequences present also increases.
Table 5.
Relationship between number of IS in each isolate and the number of expressed ESBLs.
| Number of IS in Each Isolate | N Isolates | N Isolates (%) Producing | |||
|---|---|---|---|---|---|
| 1 ESBL | 2 ESBL | 3 ESBL | 4 ESBL | ||
| 1 | 15 | 46.6 | 40 | 0 | 0 |
| 2 | 63 | 46 | 46 | 8 | 0 |
| 3 | 61 | 49 | 41 | 8.2 | 1.6 |
| 4 | 11 | 0 | 81.8 | 18.2 | 0 |
To sum up, the MDR ESBL-producing E. coli analyzed in the present study carried at least one genetic element (integron and IS). Since the strains were isolated from different sources (clinical isolates, healthy carriers, farms and feeds, food samples, WWTPs and rivers), these data revealed the potential risk for the dissemination of antimicrobial resistances among environmental and human bacteria.
4. Conclusions
In conclusion, this study highlights the high prevalence of different horizontal genetic elements among ESBL-producing E. coli isolates from food, environmental, and human samples. The analysis of integrons, showed that intI1 was present in the majority of strains and in all sources, while the prevalence of intI2 was lower but remarkable in the food isolates. Concerning insertion sequences, the multiple associations, like IS26-ISEcp1, are relevant. Thus, the co-existence of diverse types of integrons and insertion sequences suggest possible risk for the dissemination of resistance genes among different environments and, therefore, additional investigations regarding the genetic composition of these integrons and insertion sequences are encouraged, to understand the role of these mobile elements in the spread of multidrug-resistant bacteria.
Acknowledgments
We are particularly grateful to Yolanda Saénz from the “Centro de Investigación Biomédica de la Rioja” for providing the positives controls for the PCRs.
Supplementary Materials
Table S1: Antimicrobial profiles of ESBL-producing E. coli according to their origin; Table S2: Phenotypic and genotypic characteristics of strains isolated from hospital inpatients (n = 36); Table S3: Phenotypic and genotypic characteristics of aquatic strains included in the study (n = 33); Table S4: Phenotypic and genotypic characteristics of strains isolated from food (n = 48); Table S5: Phenotypic and genotypic characteristics of strains from farm origin included in the study (n = 20); Table S6: Phenotypic and genotypic characteristics of strains isolated from healthy people (n = 13).
Author Contributions
L.P.-E. performed the experiments, analyzed the data and wrote the paper. M.B. performed some PCRs. D.G. and A.I.V. conceived, designed the experiments, supervised data analysis and reviewed the manuscript.
Funding
This research was funded by a grant of the “Asociación de Amigos de la Universidad de Navarra”, obtained in September 2016.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.WHO . Antimicrobial Resistance. Global Report on Surveillance. WHO; Geneva, Switzerland: 2014. pp. 383–394. [Google Scholar]
- 2.Roca I., Akova M., Baquero F., Carlet J., Cavaleri M., Coenen S., Cohen J., Findlay D., Gyssens I., Heure O.E., et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X.J., Lyu Y., Li Y., Xue F., Liu J. Trends in antimicrobial resistance against Enterobacteriaceae strains isolated from blood: A 10-year epidemiological study in mainland China (2004–2014) Chin. Med. J. 2017;130:2050–2055. doi: 10.4103/0366-6999.213407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaikh S., Fatima J., Shakil S., Rizvi S.M.D., Kamal M.A. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J. Biol. Sci. 2015;22:90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ojer-Usoz E., González D., Vitas A.I. Clonal diversity of ESBL-producing Escherichia coli isolated from environmental, human and food samples. Int. J. Environ. Res. Public Health. 2017;14:676. doi: 10.3390/ijerph14070676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sikkema R., Koopmans M. One Health training and research activities in Western Europe. Infect. Ecol. Epidemiol. 2016;6:33703. doi: 10.3402/iee.v6.33703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Tettelin H., Riley D., Cattuto C., Medini D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Norman A., Hansen L.H., Sørensen S.J. Conjugative plasmids: Vessels of the communal gene pool. Philos. Trans. R. Soc. B Biol. Sci. 2009;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodford N., Turton J.F., Livermore D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011;35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 11.Stokes H.W., Hall R.M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: Integrons. Mol. Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 12.Galani I., Souli M., Koratzanis E., Chryssouli Z., Giamarellou H. Molecular characterization of an Escherichia coli clinical isolate that produces both metallo-β-lactamase VIM-2 and extended-spectrum β-lactamase GES-7: Identification of the In8 integron carrying the blaVIM-2 gene. J. Antimicrob. Chemother. 2006;58:432–433. doi: 10.1093/jac/dkl240. [DOI] [PubMed] [Google Scholar]
- 13.Fluit A.C., Schmitz F.J. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 14.Kaushik M., Kumar S., Kapoor R.K., Virdi J.S., Gulati P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents. 2018;51:167–176. doi: 10.1016/j.ijantimicag.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Machado E., Canto R., Baquero F., Gala J., Coque T.M. Integron Content of Extended-Spectrum β-Lactamase-Producing Escherichia coli Strains over 12 Years in a Single Hospital in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005;49:1823–1829. doi: 10.1128/AAC.49.5.1823-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odetoyin B.W., Labar A.S., Lamikanra A., Aboderin A.O., Okeke I.N. Classes 1 and 2 integrons in faecal Escherichia coli strains isolated from mother-child pairs in Nigeria. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0183383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saenz Y., Brinas L., Dominguez E., Ruiz J., Zarazaga M., Vila J., Torres C. Mechanisms of Resistance in Multiple-Antibiotic-Resistant Escherichia coli Strains of Human, Animal, and Food Origins. Antimicrob. Agents Chemother. 2004;48:3996–4001. doi: 10.1128/AAC.48.10.3996-4001.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arakawa Y., Murakami M., Suzuki K., Ito H., Wacharotayankun R., Kato N., Ohta M. A Novel Integron-Like Element Carrying the Metallo-β-Lactamase Gene bla IMP. Microbiology. 1995;39:1612–1615. doi: 10.1128/AAC.39.7.1612.Updated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Correia M., Boavida F., Grosso F., Salgado M.J., Lito L.M., Cristino J.M., Mendo S., Duarte A. Molecular characterization of a new class 3 integron in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2003;47:2838–2843. doi: 10.1128/AAC.47.9.2838-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizk D.E., El-Mahdy A.M. Emergence of class 1 to 3 integrons among members of Enterobacteriaceae in Egypt. Microb. Pathog. 2017;112:50–56. doi: 10.1016/j.micpath.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Mahillon J., Chandler M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998;62 doi: 10.1016/B978-0-12-374984-0.00799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao W.H., Hu Z.Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013;39:79–101. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arduino S.M., Roy P.H., Jacoby G.A., Betina E., Pineiro S.A., Centron D., Arduino S.M., Roy P.H., Jacoby G.A., Orman B.E., et al. blaCTX-M-2 Is Located in an Unusual Class 1 Integron (In35) Which Includes Orf513 blaCTX-M-2 Is Located in an Unusual Class 1 Integron (In35) Which Includes Orf513. Antimicrob. Agents Chemother. 2002;46:2303–2306. doi: 10.1128/AAC.46.7.2303-2306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng C., Sun J., Zheng F., Lu W., Yang Q., Rui Y. New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol. 2016;16 doi: 10.1186/s12866-016-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullik A., Pfeifer Y., Prager R., Von Baum H., Witte W. A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J. Med. Microbiol. 2010;59:580–587. doi: 10.1099/jmm.0.016188-0. [DOI] [PubMed] [Google Scholar]
- 26.Diestra K., Juan C., Curiao T., Moya B., Miro E., Oteo J., Coque T.M., Perez-Vazquez M., Campos J., Canton R., et al. Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 2008;63:60–66. doi: 10.1093/jac/dkn453. [DOI] [PubMed] [Google Scholar]
- 27.Eckert C., Gautier V., Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 2006;57:14–23. doi: 10.1093/jac/dki398. [DOI] [PubMed] [Google Scholar]
- 28.Ojer-Usoz E., González D., Vitas A.I., Leiva J., García-Jalón I., Febles-Casquero A., de la Escolano M.S. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013;93:316–321. doi: 10.1016/j.meatsci.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Ojer-Usoz E., González D., García-Jalón I., Vitas A.I. High dissemination of extended-spectrum β-lactamase-producing Enterobacteriaceae ineffluents from wastewater treatment plants. Water Res. 2014;56:37–47. doi: 10.1016/j.watres.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 30.Vitas A.I., Naik D., Pérez-Etayo L., González D. Increased exposure to extended-spectrum β-lactamase-producing multidrug-resistant Enterobacteriaceae through the consumption of chicken and sushi products. Int. J. Food Microbiol. 2018;269:80–86. doi: 10.1016/j.ijfoodmicro.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 31.Mazel D., Dychinco B., Webb V., Davies J. Antibiotic resistance in the ECOR collection: Integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 2000;44:1568–1574. doi: 10.1128/AAC.44.6.1568-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novais Â., Cantón R., Valverde A., Machado E., Galán J.C., Peixe L., Carattoli A., Baquero F., Coque T.M. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-α, and IncFI groups. Antimicrob. Agents Chemother. 2006;50:2741–2750. doi: 10.1128/AAC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel L., Decousser J., Nordmann P. Insertion Sequence ISEcp1B Is Involved in Expression and Mobilization of a blaCTX-M β-Lactamase Gene. Antimicrob. Agents Chemother. 2003;47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng Y., Bao X., Ji L., Chen L., Liu J., Miao J., Chen D., Bian H., Li Y., Yu G. Resistance integrons: Class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015;14 doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall R.M., Collis C.M. Mobile gene cassettes and integrons: Capture and spread of genes by site-specific recombination. Mol. Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 36.Machado E., Coque T.M., Cantón R., Sousa J.C., Peixe L. Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 2008;62:296–302. doi: 10.1093/jac/dkn179. [DOI] [PubMed] [Google Scholar]
- 37.Solberg O., Ajiboye R., Riley L. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J. Clin. Microbiol. 2006;44:1347–1351. doi: 10.1128/JCM.44.4.1347-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roe M.T., Vega E., Pillai S.D. Antimicrobial Resistance Markers of Class 1 and Class 2 Integron-bearing Escherichia coli from Irrigation Water and Sediments. Emerg. Infect. Dis. 2003;9:5–9. doi: 10.3201/eid0907.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozgumus O.B., Sandalli C., Sevim A., Celik-Sevim E., Sivri N. Class 1 and class 2 integrons and plasmid-mediated antibiotic resistance in coliforms isolated from ten rivers in northern Turkey. J. Microbiol. 2009;47:19–27. doi: 10.1007/s12275-008-0206-z. [DOI] [PubMed] [Google Scholar]
- 40.Vinué L., Sáenz Y., Somalo S., Escudero E., Moreno M.Á., Ruiz-Larrea F., Torres C. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 2008;62:934–937. doi: 10.1093/jac/dkn331. [DOI] [PubMed] [Google Scholar]
- 41.Kargar M., Mohammadalipour Z., Doosti A., Lorzadeh S., Japoni-Nejad A. High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in southwest of Iran. Osong Public Health Res. Perspect. 2014;5:193–198. doi: 10.1016/j.phrp.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldstein C., Lee M.D., Sanchez S., Phillips B., Register B., Grady M., Liebert C., Summers A.O., White D.G., Maurer J.J., et al. Incidence of Class 1 and 2 Integrases in Clinical and Commensal Bacteria from Livestock, Companion Animals, and Exotics. Antimicrob. Agents Chemother. 2001;45:723–726. doi: 10.1128/AAC.45.3.723-726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kor S.B., Choo Q.C., Chew C.H. New integron gene arrays from multiresistant clinical isolates of members of the Enterobacteriaceae and Pseudomonas aeruginosa from hospitals in Malaysia. J. Med. Microbiol. 2013;62:412–420. doi: 10.1099/jmm.0.053645-0. [DOI] [PubMed] [Google Scholar]
- 44.Ali T., Ur Rahman S., Zhang L., Shahid M., Zhang S., Liu G., Gao J., Han B. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to ISCR1. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J., Bae I.K., Jeong S.H., Chang C.L., Lee C.H., Lee K. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J. Antimicrob. Chemother. 2011;66:1263–1268. doi: 10.1093/jac/dkr106. [DOI] [PubMed] [Google Scholar]
- 46.Liao X.P., Xia J., Yang L., Li L., Sun J., Liu Y.H., Jiang H.X. Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bae I.K., Lee Y.H., Jeong H.J., Hong S.G., Lee S.H., Jeong S.H. A novel blaCTX-M-14 gene-harboring complex class 1 integron with an In4-like backbone structure from a clinical isolate of Escherichia coli. Diagn. Microbiol. Infect. Dis. 2008;62:340–342. doi: 10.1016/j.diagmicrobio.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Wang J., Stephan R., Zurfluh K., Hächler H., Fanning S. Characterization of the genetic environment of blaESBLgenes, integrons and toxin-antitoxin systems identified on large transferrable plasmids in multi-drug resistant Escherichia coli. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2014.00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Leverstein Hall M.A., Dierikx C.M., Cohen Stuart J., Voets G.M., van den Munckhof M.P., van Essen-Zandbergen A., Platteel T., Fluit A.C., van de Sande-Bruinsma N., Scharinga J., et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011;17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 50.Kiiru J., Butaye P., Goddeeris B.M., Kariuki S. Analysis for prevalence and physical linkages amongst integrons, ISEcp 1, ISCR 1, Tn 21 and Tn 7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011) BMC Microbiol. 2013;13 doi: 10.1186/1471-2180-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porse A., Schønning K., Munck C., Sommer M.O.A. Survival and Evolution of a Large Multidrug Resistance Plasmid in New Clinical Bacterial Hosts. Mol. Biol. Evol. 2016;33:2860–2873. doi: 10.1093/molbev/msw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Billard-Pomares T., Fouteau S., Jacquet M.E., Roche D., Barbe V., Castellanos M., Bouet J.Y., Cruveiller S., Médigue C., Blanco J., et al. Characterization of a P1-like bacteriophage carrying an SHV-2 extended-spectrum β-lactamase from an Escherichia coli strain. Antimicrob. Agents Chemother. 2014;58:6550–6557. doi: 10.1128/AAC.03183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doi Y., Hazen T.H., Boitano M., Tsai Y.C., Clark T.A., Korlach J., Rasko D.A. Whole-genome assembly of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 carbapenemases using single-molecule, real-time sequencing. Antimicrob. Agents Chemother. 2014;58:5947–5953. doi: 10.1128/AAC.03180-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y., Zeng Z., Chen S., Ma J., He L., Liu Y., Deng Y., Lei T., Zhao J., Liu J.H. High prevalence of blaCTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 2010;16:1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x. [DOI] [PubMed] [Google Scholar]
- 55.Tamang M.D., Nam H.M., Gurung M., Jang G.C., Kim S.R., Jung S.C., Park Y.H., Lim S.K. Molecular characterization of CTX-M β-lactamase and associated addiction systems in Escherichia coli circulating among cattle, farm workers, and the farm environment. Appl. Environ. Microbiol. 2013;79:3898–3905. doi: 10.1128/AEM.00522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez-Martinez J.M., Poirel L., Canton R., Nordmann P. Common region CR1 for expression of antibiotic resistance genes. Antimicrob. Agents Chemother. 2006;50:2544–2546. doi: 10.1128/AAC.00609-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toleman M.A., Bennett P.M., Walsh T.R. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 2006;58 doi: 10.1093/jac/dkl204. [DOI] [PubMed] [Google Scholar]
- 58.Toleman M.A., Bennett P.M., Walsh T.R. ISCR Elements: Novel Gene-Capturing Systems of the 21st Century? Microbiol. Mol. Biol. Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Millan B., Castro D., Araque M., Ghiglione B., Gutkind G. ISCR1 associated with blaCTX-M-1 y blaCTX-M-2 genes in IncN and IncFIIA plasmids isolated from Klebsiella pneumoniae of nosocomial origin in Mérida, Venezuela. Biomédica. 2012;33:268–275. doi: 10.7705/biomedica.v33i2.774. [DOI] [PubMed] [Google Scholar]
- 60.Woodford N., Carattoli A., Karisik E., Underwood A., Ellington M.J., Livermore D.M. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 2009;53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.