The present study was designed to understand transcriptomic changes and the potential development of direct and cross-resistance in essential oil (EO)-adapted Escherichia coli O157:H7. The results demonstrated altered growth behaviors of E. coli O157:H7 during adaptation in sublethal thymol, carvacrol, and trans-cinnamaldehyde. Generally, EO-adapted bacteria showed enhanced resistance against subsequent lethal EO, heat, and oxidative stresses, with no induction of acid resistance in simulated gastric fluid. A transcriptomic analysis revealed the upregulation of related stress resistance genes and a downregulation of various virulence genes in EO-adapted cells. This study provides new insights into microbial EO adaptation behaviors and highlights the risk of resistance development in adapted bacteria.
KEYWORDS: E. coli O157:H7, RNA sequencing, essential oil adaptation, direct resistance, cross-resistance
ABSTRACT
Thymol, carvacrol, and trans-cinnamaldehyde are essential oil (EO) compounds with broad-spectrum antimicrobial activities against foodborne pathogens, including Escherichia coli O157:H7. However, little is known regarding direct resistance and cross-resistance development in E. coli O157:H7 after adaptation to sublethal levels of these compounds, and information is scarce on microbial adaptive responses at a molecular level. The present study demonstrated that E. coli O157:H7 was able to grow in the presence of sublethal thymol (1/2T), carvacrol (1/2C), or trans-cinnamaldehyde (1/2TC), displaying an extended lag phase duration and a lower maximum growth rate. EO-adapted cells developed direct resistance against lethal EO treatments and cross-resistance against heat (58°C) and oxidative (50 mM H2O2) stresses. However, no induction of acid resistance (simulated gastric fluid, pH 1.5) was observed. RNA sequencing revealed a large number (310 to 338) of differentially expressed (adjusted P value [Padj], <0.05; fold change, ≥5) genes in 1/2T and 1/2C cells, while 1/2TC cells only showed 27 genes with altered expression. In accordance with resistance phenotypes, the genes related to membrane, heat, and oxidative stress responses and genes related to iron uptake and metabolism were upregulated. Conversely, virulence genes associated with motility, biofilm formation, and efflux pumps were repressed. This study demonstrated the development of direct resistance and cross-resistance and characterized whole-genome transcriptional responses in E. coli O157:H7 adapted to sublethal thymol, carvacrol, or trans-cinnamaldehyde. The data suggested that caution should be exercised when using EO compounds as food antimicrobials, due to the potential stress resistance development in E. coli O157:H7.
IMPORTANCE The present study was designed to understand transcriptomic changes and the potential development of direct and cross-resistance in essential oil (EO)-adapted Escherichia coli O157:H7. The results demonstrated altered growth behaviors of E. coli O157:H7 during adaptation in sublethal thymol, carvacrol, and trans-cinnamaldehyde. Generally, EO-adapted bacteria showed enhanced resistance against subsequent lethal EO, heat, and oxidative stresses, with no induction of acid resistance in simulated gastric fluid. A transcriptomic analysis revealed the upregulation of related stress resistance genes and a downregulation of various virulence genes in EO-adapted cells. This study provides new insights into microbial EO adaptation behaviors and highlights the risk of resistance development in adapted bacteria.
INTRODUCTION
Escherichia coli O157:H7 is a Gram-negative facultative anaerobe with a low infectious dose (∼50 CFU). The Centers for Disease Control and Prevention (CDC) has estimated that E. coli O157:H7 infections cause approximately 73,000 illnesses, 2,200 hospitalizations, and 60 deaths annually in the United States, with an annual cost of 405 million dollars for these illnesses (1). Infection symptoms range from mild diarrhea to hemorrhagic colitis (HC) and the life-threatening hemolytic-uremic syndrome (HUS). The transmission of E. coli O157:H7 infections is mainly foodborne (52%), with ground beef, fresh produce, and unpasteurized dairy products being the main vehicles of outbreaks (1).
The food industry is constantly searching for safe and effective natural antimicrobials, due to consumers' demand for less synthetic preservatives in foods. Recently, plant-derived essential oils (EOs) have received strong research interest due to their historical use in foods (as flavorings), generally recognized as safe (GRAS) status, and wide-spectrum antimicrobial activities (2). Particularly, thymol (Thy), carvacrol (Car), and trans-cinnamaldehyde (TC), the major components of thyme, oregano, and cinnamon EOs, respectively, have demonstrated good antimicrobial activities against E. coli O157:H7, Listeria monocytogenes, and Salmonella enterica serovar Typhimurium (3).
Despite their promising antibacterial activities in vitro, evidences suggest that the hydrophobic EO compounds tend to interact with food components, including starch, proteins, and fats (4, 5). As a result of EO-food interaction, the actual amount of EO present in the food matrix might be at a sublethal level for food pathogens, which triggered the question whether it would induce stress adaptation and the formation of resistant bacteria. Previous studies have demonstrated that the exposure to sublethal stresses, such as mild acid or heat, induces adaptation responses in pathogens and makes them more resistant against subsequent lethal treatments (6, 7). However, limited information is available regarding the direct and cross-resistance development in pathogenic bacteria after exposure to sublethal EO stress. For example, Dubois-Brissonnet et al. (8) reported increased biocide resistance in S. Typhimurium after exposure to terpenes (Thy, Car, eugenol, and citral) at 0.25 to 0.9 MICs. A lack of direct and cross-tolerance (NaCl 5 g/100 ml, pH 5.2, 45°C) was reported in S. Typhimurium following exposure to EOs (rosemary and oregano) or their principal terpene compounds (1,8-cineole and carvacrol) at one-half and one-quarter MICs (9, 10). Hammer et al. (11) reported a lack of direct and antibiotic cross-resistance in Staphylococcus aureus and E. coli following exposure to tea tree EO and its major component terpinen-4-ol at a sublethal level. Chueca et al. (12) reported no increase in direct or cross-resistance (heat and pulsed electric fields) in E. coli following an overnight incubation with sublethal carvacrol or citral, but increased resistance was observed in bacteria after 10 days of consecutive adaptation.
Under stressful conditions, living cells alter their gene expression to maintain intracellular environments and cellular functions. Therefore, transcriptomic analyses, such as RNA sequencing (RNA-seq), enable us to visualize the changes in overall mRNA expression, which can facilitate the identification of microbial stress adaptation mechanisms. Compared to the traditional hybridization-based approach (microarray), RNA-seq has the advantages of not requiring prior knowledge of target gene sequences, low background noise, a large dynamic range (up to 9,000-fold change), and high accuracy (13). RNA-seq technology has been employed to identify genes and metabolic pathways involved in the adaptation of Listeria monocytogenes to growth on vacuum-packaged cold smoked salmon (14). However, limited RNA-seq studies have been carried out to understand the transcriptional landscape in foodborne pathogens adapting to EO stress. Therefore, the aims of the present study were to investigate the development of direct and cross-resistance in E. coli O157:H7 adapted to sublethal concentrations of EOs (Thy, Car, and TC) and to examine the genome-wide transcriptional responses using RNA-seq analysis.
RESULTS AND DISCUSSION
Effect of sublethal EOs on microbial growth.
Thy, Car, and TC were found to inhibit the growth of E. coli O157:H7 at the same MIC of 0.31 mg/ml, which was consistent with a previous study that showed the strong antimicrobial activities of the three compounds (15). Thus, in this study, EO-adapted cells were prepared by growing E. coli O157:H7 in the presence of 0.16 mg/ml (one-half MIC) Thy (1/2T), Car (1/2C), or TC (1/2TC), and microbial growth during EO adaptation was monitored over 24 h.
E. coli O157:H7 showed altered growth behaviors in the presence of sublethal EOs (Fig. 1), displaying significantly (P < 0.05) longer lag phase durations (6 to 8.5 h) and lower maximum growth rates (0.7 h−1) than control cells grown in tryptic soya broth (TSB; 1.4 h and 1.0 h−1, respectively). In addition, cells reached significantly (P < 0.05) lower final population densities (8.5 to 8.6 log CFU/ml) in medium containing sublethal Thy or Car, whereas sublethal TC did not significantly (P > 0.05) affect the final cell density (9.3 log CFU/ml) compared to that of the control (9.4 log CFU/ml) (Table 1). Similar to the present results, Kim et al. (16) reported an extension of the lag phase by 6 to 12 h in E. coli O157:H7 grown in the presence of 1/2MIC Car (0.5 mg/ml) or eugenol (0.5 mg/ml) compared to that of cells growing in TSB. Dubois-Brissonnet et al. (8) observed lower growth rates of S. Typhimurium in medium containing a sublethal level of terpenes, with growth rates negatively correlated with terpene concentrations. The lag phase indicates the time needed for microorganisms to repair injuries and return to a normal cellular condition that enables replication (6), while a lower growth rate correlates with stronger resistance against thermal (48°C), UVA, and solar disinfections in E. coli, due to the increasingly expressed stress response proteins (17).
FIG 1.
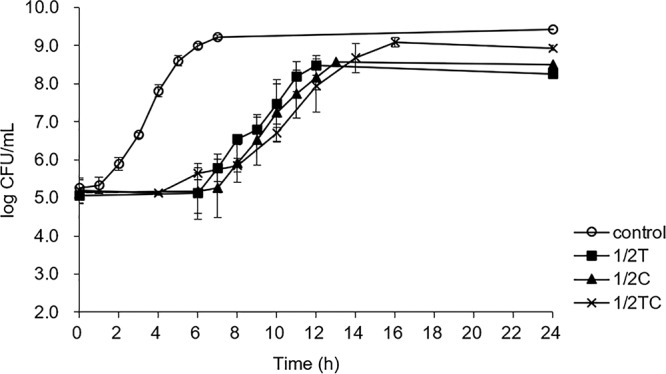
Growth curves of E. coli O157:H7 grown in TSB containing 1.6% (vol/vol) ethanol (control), 0.16 mg/ml thymol (1/2T), carvacrol (1/2C), or trans-cinnamaldehyde (1/2TC) for 24 h at 37°C.
TABLE 1.
Growth parameters of E. coli O157:H7 growing in TSB containing 1.6% (vol/vol) ethanol or 0.16 mg/ml thymol, carvacrol, or trans-cinnamaldehyde
| Treatmenta | Growth parameterb |
|||
|---|---|---|---|---|
| Initial cell density (log CFU/ml) | Lag phase duration (h) | Maximum growth rate (h−1) | Final cell density (log CFU/ml) | |
| Control | 5.2 ± 0.3 AB | 1.4 ± 0.4 A | 1.0 ± 0.1 A | 9.4 ± 0.1 A |
| 1/2T | 4.9 ± 0.2 A | 6.0 ± 0.4 B | 0.7 ± 0.1 B | 8.5 ± 0.2 B |
| 1/2C | 5.0 ± 0.2 A | 6.9 ± 0.9 B | 0.7 ± 0.1 B | 8.6 ± 0.3 B |
| 1/2TC | 5.5 ± 0.2 B | 8.5 ± 1.1 C | 0.7 ± 0.1 B | 9.3 ± 0.1 A |
Control, ethanol; 1/2T, 0.16 mg/ml thymol; 1/2C, 0.16 mg/ml carvacrol; 1/2TC, 0.16 mg/ml trans-cinnamaldehyde.
Different uppercase letters in the same column represent statistically significant differences (P < 0.05) between control and adapted cells.
Previous studies focused mainly on efficacies and mechanisms of EOs on microbial inactivation, but information is scarce regarding the microbial adaptive responses toward sublethal EOs, and less is understood about the underlying molecular mechanisms (18). Therefore, on the basis of the growth behaviors, nonadapted control (7 h) and 1/2T (12 h)-, 1/2C (13 h)-, and 1/2TC (17 h)-adapted cells were collected at early stationary phase and subjected to resistance assays and RNA-seq analysis.
Transcriptional responses of E. coli O157:H7 adapted to sublethal thymol, carvacrol, or trans-cinnamaldehyde.
The transcriptional profiles of EO-adapted cells were compared against those of a nonadapted control to identify the molecular mechanisms underlying microbial EO adaptation. Compared to that of the control, 1/2T and 1/2C cells showed drastic transcriptomic changes, with 338 and 310 genes, respectively, that were significantly (adjusted P value [Padj] of <0.05) differentially expressed by more than 5-fold. Conversely, 1/2TC cells only showed a small difference, with 27 differentially expressed genes (Fig. 2). Among the differentially expressed genes in 1/2T, 1/2C, and 1/2TC cells, 113 (33%), 120 (39%), and 0 genes, respectively, were downregulated, while 225 (67%), 190 (61%), and 27 (100%) genes, respectively, were upregulated.
FIG 2.
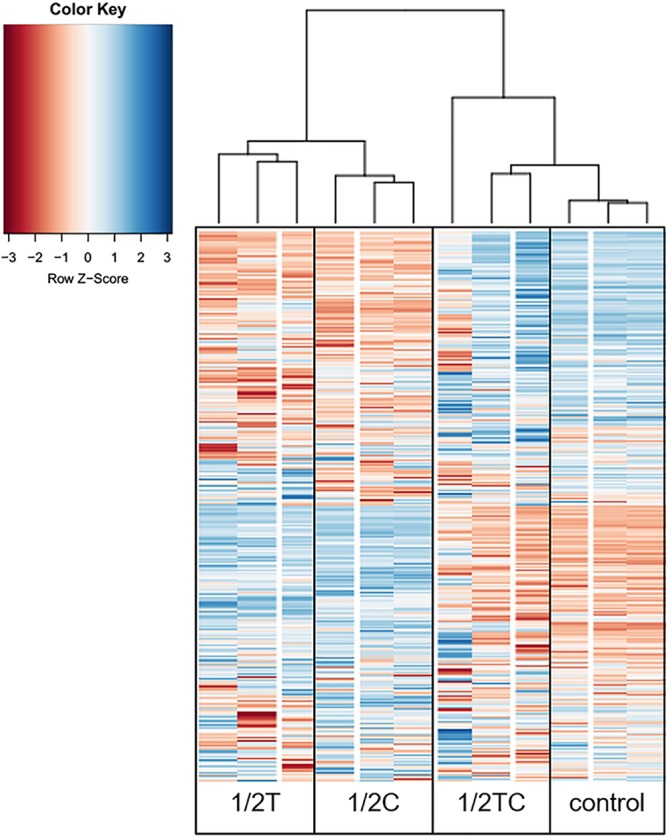
Heat map of genes whose mRNA level significantly changed in Escherichia coli O157:H7 growing in tryptic soya broth containing 1.6% (vol/vol) ethanol (control), 0.16 mg/ml thymol (1/2T), carvacrol (1/2C), or trans-cinnamaldehyde (1/2TC). Three biological replicates were included for each sample.
Genes induced in cells adapted to 1/2T and 1/2C were those related to stress responses, including the envelope stress phage shock operon (pspABCDG), heat shock protein (HSP) genes (σ32, hspQ, and ibpAB), and oxidative stress resistance genes (sodA, grxA, and trxC). In addition, genes encoding efflux pumps and iron transport were significantly (Padj < 0.05) upregulated in 1/2T- and 1/2C-adapted cells. Cells adapted to 1/2TC, conversely, mainly showed upregulated genes related to oxidative stress resistance and iron transport (see Table S1 in the supplemental material). Downregulated genes mainly consisted of those involved in motility (flagella and chemotaxis), virulence (shiga toxins and type III secretion system), and cell division (minCDE) (see Table S2).
Limited studies have been performed on transcriptomic changes in bacteria during EO adaptation. Rao et al. (19) reported that the exposure of Saccharomyces cerevisiae to Car at 0.005% (one-half MIC) or 0.01% (MIC) for 15 min induced 492 to 800 genes relating to stress response, drug efflux, and alternate metabolic pathways and repressed 430 to 603 genes involved in nucleic acid metabolism and cell division. Visvalingam et al. (20) observed a total of 195 and 466 differentially expressed genes in E. coli O157:H7 exposed to TC for 2 h and 4 h, respectively, with many stress-related genes upregulated at 2 h but not at 4 h. Kollanoor Johny et al. (21) found 566 and 483 differentially expressed genes in Salmonella enterica serotype Enteritidis exposed to sublethal TC and eugenol, respectively, with genes involved in motility, virulence, and biosynthetic pathways repressed and genes encoding heat shock response and efflux pumps induced. However, most of these reported studies examined transcriptomic changes after short EO exposures (15 min to 4 h), while resistance behaviors in EO-adapted cells were generally evaluated using overnight adapted cells, due to the known stronger resistance in stationary-phase cells than in their exponential-phase counterparts (22). Hence, the results from the present study offer new insights into the genetic responses of EO-adapted stationary-phase cells, and may be of relevance to the food industry where microbial cells are more likely to experience chronic exposure to sublethal levels of antimicrobial residues.
To confirm whether the increased expression of stress resistance genes would lead to measurable resistance behaviors, EO-adapted cells were challenged in lethal treatments, using EOs (Thy, Car, and TC), heat (58°C), acid (simulated gastric fluid, pH 1.5), and oxidative (H2O2, 50 mM) stresses.
Resistance of EO-adapted E. coli O157:H7 to thymol, carvacrol, and trans-cinnamaldehyde.
The stress resistance behaviors, as represented by D values, of nonadapted and EO-adapted E. coli O157:H7 against various lethal treatments are shown in Table 2. Cells adapted to 1/2T and 1/2C became more resistant against subsequent Thy (D value = 4.2 to 4.3 min), Car (4.5 to 5.1 min), and TC (160.7 to 161.7 min) treatments, whereas cells adapted to 1/2TC only developed direct resistance against TC treatment (243.7 min) compared to those of the control (D values of 2.3, 0.9, and 77.1 min, respectively) (Table 2). Exposure of E. coli strains to tea tree EO or terpinen-4-ol induced EO resistance, with a 2-fold increment in MIC in adapted cells compared to that of the nonadapted control (11). Similarly, Ultee et al. (23) reported enhanced survival of Bacillus cereus during lethal Car treatment after adaptation to sublethal (0.4 mM) Car.
TABLE 2.
D values of nonadapted and thymol-, carvacrol-, or trans-cinnamaldehyde-adapted Escherichia coli O157:H7 against thermal, acid, and hydrogen peroxide treatmentsa
| Treatment | D value (min)b |
|||
|---|---|---|---|---|
| Control | 1/2T | 1/2C | 1/2TC | |
| T (0.31 mg/ml) | 2.3 ± 0.3 A | 4.2 ± 0.1 B | 4.3 ± 0.6 B | 1.8 ± 0.4 A |
| C (0.31 mg/ml) | 0.9 ± 0.2 A | 4.5 ± 0.7 B | 5.1 ± 0.5 B | 0.8 ± 0.2 A |
| TC (0.63 mg/ml) | 77.1 ± 10.2 A | 161.7 ± 23.3 B | 160.7 ± 28.5 B | 243.7 ± 23.5 C |
| Thermal (58°C) | 1.5 ± 0.2 A | 2.4 ± 0.2 C | 2.4 ± 0.3 C | 1.9 ± 0.2 B |
| Acid (SGFc, pH 1.5) | 21.8 ± 2.3 A | 22.7 ± 1.6 A | 20.4 ± 3.1 A | 23.4 ± 1.1 A |
| Hydrogen peroxide (50 mM) | 4.1 ± 0.5 A | 5.4 ± 0.4 B | 5.5 ± 0.3 B | 5.1 ± 0.3 B |
Control, nonadapted; 1/2T, 0.16 mg/ml thymol; 1/2C, 0.16 mg/ml carvacrol; 1/2TC, 0.16 mg/ml trans-cinnamaldehyde.
Different uppercase letters in the same row represent statistically significant differences (P < 0.05) between control and adapted cells.
SGF, simulated gastric fluid.
A transcriptomic analysis revealed a strong upregulation of phage shock protein (psp) genes (416.9 to 1459.2-fold) in cells adapted to 1/2T and 1/2C but not in cells adapted to 1/2TC (Table 3). The Psp system is a conserved stress response system in E. coli that is activated by the disruption of cytoplasmic membrane (CM) integrity and a loss of proton motive force (pmf) (24), which is consistent with the known membrane-disintegrating properties of Thy and Car (15). Other membrane-altering stresses, such as osmotic shock (25) and hydrophobic solvents (26), also increased microbial psp gene expression. In contrast, the lack of psp induction in cells adapted to 1/2TC suggests that TC at a sublethal concentration probably did not affect membrane integrity, a phenomenon that was also reported by Helander et al. (15).
TABLE 3.
Selected upregulated stress response genes in Escherichia coli O157:H7 adapted to a sublethal level of thymol, carvacrol, or trans-cinnamaldehyde compared to nonadapted cellsa
| Gene identifier | Fold changeb |
Gene product description | ||
|---|---|---|---|---|
| 1/2T | 1/2C | 1/2TC | ||
| Cell envelope | ||||
| ECs1884 | 492.1 | 416.9 | —c | Phage shock protein, PspD |
| ECs1883 | 528.5 | 434.0 | — | PspC family transcriptional regulator |
| pspB | 615.4 | 465.0 | — | Phage shock protein, PspB |
| ECs1881 | 763.2 | 634.3 | — | Phage shock protein, PspA |
| pspG | 1459.2 | 1452.2 | — | Phage shock protein, PspG |
| cpxP | 566.9 | 374.6 | — | Cell envelope toxicity response protein, CpxP |
| Heat resistance | ||||
| ECs1050 | 8.4 | 9.8 | — | Heat shock protein, HspQ |
| ECs4310 | 8.5 | 6.0 | — | RNA polymerase factor sigma-32 |
| ECs4626 | 22.2 | 25.2 | — | Heat shock chaperone, IbpB |
| ECs4627 | 12.0 | 13.8 | — | Heat shock protein, IbpA |
| ECs0165 | 13.8 | 8.5 | — | Serine endoprotease, degrade damaged proteins |
| ECs2539 | 15.4 | 7.5 | — | Heat shock protein, HtpX |
| Acid resistance | ||||
| ECs5113 | 14.4 | — | Lysine decarboxylase 1, CadA | |
| cadB | 16.1 | — | Lysine/cadaverine antiporter, CadB | |
| ECs2303 | 37.4 | 42.6 | Acid-shock protein, resistance against moderate acid | |
| Oxidative stress resistance | ||||
| grxA | 9.3 | — | 8.8 | Glutaredoxin, GrxA |
| ECs4834 | 10.5 | — | — | Superoxide dismutase, SodA |
| ECs5045 | 10.4 | 9.0 | — | Redox-sensitive transcriptional activator, SoxR |
| ECs5044 | 17.1 | 17.1 | — | DNA-binding transcriptional regulator, SoxS |
| ECs2487 | 17.6 | 9.7 | — | Methionine sulfoxide reductase B |
| ECs3448 | 22.9 | 7.2 | 8.9 | Thioredoxin 2, TrxC |
| ECs3271 | 28.9 | 8.5 | 10.1 | Manganese transport protein, MntH |
| ECs0931 | 6.0 | 6.2 | — | Nitroreductase A |
| Iron transport | ||||
| ECs0635 | 9.8 | — | — | 2,3-Dihydro-2,3-dihydroxybenzoate dehydrogenase |
| entF | 7.9 | 5.5 | — | Enterobactin synthase subunit F, EntF |
| ECs0630 | 8.8 | — | — | Enterobactin exporter, EntS |
| ECs0634 | 11.3 | 5.8 | — | 2,3-Dihydro-2,3-dihydroxybenzoate synthetase, EntB |
| entE | 17.7 | 9.0 | — | 2,3-Dihydroxybenzoate-AMP ligase, EntE |
| ECs0632 | 23.8 | 10.3 | 7.9 | Isochorismate synthase, EntC |
| ECs0627 | 5.6 | 5.3 | 5.3 | Iron-enterobactin transporter ATP-binding protein |
| ECs0628 | 8.8 | — | 5.8 | Iron-enterobactin transporter permease |
| ECs0629 | 12.6 | 10.1 | 9.4 | Iron-enterobactin transporter membrane protein |
| ECs0631 | 11.1 | 7.3 | 6.8 | Iron-enterobactin transporter periplasmic binding protein |
| ECs1360 | 5.8 | — | — | Bifunctional enterobactin receptor/adhesin, Iha |
| ECs0624 | 17.8 | 14.3 | 6.0 | Enterobactin/ferric enterobactin esterase, Fes |
| ECs4380 | 5.0 | — | — | Heme utilization/transport protein, ChuA |
| ECs4382 | 16.0 | 9.7 | 7.3 | Hemin binding protein, ShuT |
| hemH | 9.4 | — | 7.4 | Ferrochelatase, HemH |
| ECs3917 | 5.8 | — | — | Ferrichrome iron receptor |
| ECs0154 | 7.2 | 6.9 | 8.3 | Ferrichrome outer membrane transporter |
| ECs1480 | 7.3 | — | — | Outer membrane receptor for Fe3+ uptake, FhuE |
| ECs5327 | 10.1 | 10.3 | 5.0 | Ferrioxamine B reductase, FhuF |
| ECs3890 | 11.2 | 8.2 | 5.2 | Biopolymer transport protein, ExbB |
| ECs3889 | 5.7 | — | — | Biopolymer transport protein, ExbD |
| feoA | 6.3 | 13.5 | 5.1 | Ferrous iron transport protein A, FeoA |
| feoB | 5.3 | 7.7 | — | Ferrous iron transport protein B, FeoB |
| ECs3916 | 5.3 | 6.2 | — | Iron-siderophore iron ABC transporter ATP-binding protein |
| ECs2055 | 5.2 | — | — | Outer membrane receptor for iron transport |
| ECs5531 | 8.8 | 10.3 | — | Bacterioferritin-associated ferredoxin, Bfd |
Cutoff criteria for upregulation were Padj of <0.05 and fold change ≥5.
1/2T, 0.16 mg/ml thymol; 1/2C, 0.16 mg/ml carvacrol; 1/2TC, 0.16 mg/ml trans-cinnamaldehyde.
—, no detection of differentially expressed gene at the set criteria.
The upregulated psp gene possibly contributed to the observed EO resistance in adapted cells by maintaining membrane structure and functions during stress exposure. PspA was reported to reduce lipid bilayer deformation (27) and suppress proton leakage from damaged CMs (28) by interacting with phosphatidylserine (PS) and phosphatidylglycerol (PG) and reducing the fluidity of the distorted phospholipid bilayer. In addition, PspB and PspC prevent lethal cytoplasmic membrane permeability induced by secretin (29). An alteration of the membrane fatty acid composition and a reduction of membrane fluidity counter the measures of microbial cells against EO damages (23), which are known to cause membrane fluidization and permeabilization (18). These results suggested that the use of EOs at sublethal concentrations should be avoided, due to the potential development of EO-resistant bacteria.
Heat resistance of EO-adapted E. coli O157:H7.
All EO-adapted E. coli O157:H7 groups showed significantly (P < 0.05) increased heat resistance (D value = 1.9 to 2.4 min) compared to that of the nonadapted control (1.5 min) (Table 2). Unlike the present results, studies evaluating other Gram-negative bacteria such as Pseudomonas aeruginosa and S. Typhimurium reported no change in resistance toward mild heat (45°C) treatment (9, 30), which might be attributed to the different microbial strains and lethal conditions used.
In accordance with the present experimental results, RNA-seq revealed the upregulation of various heat shock protein (HSP) genes in cells adapted to 1/2T and 1/2C but not in those adapted to 1/2TC. These include the RNA polymerase sigma factor σ32, which regulates the transcription of approximately 30 HSPs involved in microbial heat survival (31), molecular chaperons (IbpA and IbpB) that stabilize and protect proteins against irreversible denaturation (32), and proteases (HtpX, HspQ, and ECs0165) that degrade damaged proteins (33) (Table 3). Burt et al. (34) demonstrated that an overnight incubation of E. coli O157:H7 with sublethal Car (1 mM) inhibited flagellin synthesis and induced a significant production of HSP60. By using proteomic analysis, Pasqua et al. (35) demonstrated the upregulation of HSPs and the outer membrane stress response in S. enterica serovar Thompson exposed to sublethal (0.01%) Thy. By ensuring the quality of proteins, the HSPs exert homeostatic control of the biological membrane and cellular functions during a time of stress, thereby enhancing the survival of E. coli at elevated temperatures (36).
The induction of HSPs during EO adaptation might be attributed to a microbial regulatory response to abnormalities in membrane structures, induced by EO insertion into the phospholipid bilayer. Hydrostatic pressure (30 to 50 MPa), which is known to solidify cellular membrane lipids and decrease membrane fluidity (37), and adaptation to lettuce leaf surface, which causes microbial membrane damage by desiccation stress (25), were reported to induce HSPs in E. coli. In addition, direct EO-protein interaction might also elicit an HSP response by creating misfolded and damaged proteins in the cytoplasm. Nobre et al. (38) reported that the treatment of E. coli with the carbon monoxide (CO)-releasing compound CORM-2, whose antimicrobial activity was attributed to the binding and inhibition of respiratory chain enzymes, induced a range of HSPs in bacteria.
The development of direct resistance against TC treatment and cross-resistance against heat treatment in E. coli O157:H7 adapted to 1/2TC was not explained by the RNA-seq results. Previous studies have demonstrated that transcriptional responses diminish with time. For example, the induction of stress response genes was observed to peak at 10 to 15 min after drug exposure in Candida albicans, followed by rapid decay by 30 min postexposure (39). Similarly, for E. coli grown under a high pressure stress condition, the levels of HSPs induced were much higher in early-log-phase cells (3 to 5 h) than in late-log-phase cells (7 to 11 h) (37). It seems that mRNAs were transiently induced in response to stress exposure and subsequently degraded for the bacterial cells to readjust to the new environment. Hence, judging from the long duration (17 h) (Fig. 1) required for E. coli O157:H7 to reach stationary phase when grown in the presence of sublethal TC compared to that when grown in Thy (12 h) and Car (13 h), we suspect that the necessary stress-related transcriptional changes were already completed by the time we collected the bacterial cells for RNA-seq analysis. To confirm this hypothesis, a proteomic analysis should be performed in future investigations to determine if stress-resistant proteins are synthesized in 1/2TC-adapted cells. Nevertheless, the results from the present study demonstrated an enhanced heat resistance in all adapted E. coli O157:H7 cells, suggesting that the use of sublethal EOs should be avoided during industrial application.
Acid resistance of EO-adapted E. coli O157:H7.
The low acidity in the human stomach (pH 1.5 to 3.0) acts as the first defense line against microbial infection; hence, the ability of enteric pathogens, such as E. coli O157:H7, to survive extreme acidic conditions is an important virulence property that is inversely correlated with the infectious dose (1). No significant difference was detected for acid resistance when nonadapted and EO-adapted E. coli O157:H7 cells were exposed to simulated gastric fluid (SGF) at pH 1.5 (Table 2). A similar lack of acid tolerance induction was demonstrated in S. Typhimurium and P. aeruginosa following adaptation to sublethal Car (9, 30); however, most of these studies were conducted using mild acidic treatment (lactic acid, pH 5.2) that did not cause significant microbial inactivation. On the other hand, the present study evaluated the acid resistance of EO-adapted E. coli O157:H7 under a strong acid condition (pH 1.5) in simulated gastric fluid, which is more relevant to their clinical impacts.
Data for the downregulation and upregulation of acid resistance (AR) genes in EO-adapted cells are presented in Table S2 in the supplemental material and Table 3. The downregulated arginine-dependent AR system, consisting of arginine decarboxylase (aidA), arginine:agmatine antiporter (adiC), and a transcription regulator (adiY), functions by consuming intracellular protons, thereby maintaining the internal pH within a physiological range conducive to E. coli survival in an acidic environment (1). In addition, the downregulated hdeA and hdeB encoding chaperone proteins are known to protect periplasmic proteins from acid damage by binding to acid-denatured proteins and preventing their irreversible aggregation (40). Conversely, the upregulated lysine-dependent AR, consisting of lysine decarboxylase (cadA) and lysine-cadaverine exchanger (cadB), offers a weaker acid protection for E. coli at pH 2.5 (41). Previous studies demonstrated the upregulation of AR genes in E. coli as a direct stress response following exposure to organic/inorganic acids (42) or as a general stress response when grown in a food matrix, such as ground beef extract (43) and lettuce (25).
One possible explanation for the lack of acid resistance in the SGF assay is that E. coli O157:H7 possesses multiple AR systems, namely, the oxidative AR, and the arginine-, glutamine-, and lysine-dependent AR systems (40). Hence, the up- or downregulation of one or two AR systems might not result in a physiological impact. For example, Carter et al. (44) reported that the deletion of the hdeB gene in E. coli O157:H7 did not affect its survival at pH 2.0, while Price et al. (45) found that the arginine-dependent AR system was not essential for survival in both apple cider (pH 3.5) and bovine gastrointestinal tract (pH 2.0 to 2.5), due to the activities of other AR systems. Moreover, the glutamine-dependent AR genes, which are associated with protection in a strong acid environment (< pH 2.0), were not significantly induced in EO-adapted cells. The other AR systems, such as arginine-dependent AR (pH ≥ 2.5) (40) and HdeA and HdeB (pH 2 to 3) (46), are known to be the most efficient under milder acid conditions. Therefore, the testing condition in the present study (pH 1.5) might be too harsh for the up- or downregulated ARs to show a physiological impact.
Overall, the results from the present study suggest that EO adaptation did not induce acid resistance in E. coli O157:H7 at pH 1.5 in SGF. Nevertheless, future works should also evaluate the survival of adapted bacteria under milder conditions (pH 2.0 to 3.5), which are commonly encountered in acidic foods (e.g., apple cider) (45), to assess the virulence of these EO-adapted pathogens.
Oxidative stress resistance of EO-adapted E. coli O157:H7.
During host-pathogen interactions, microorganisms usually encounter a high level of O2−, generated by the mammalian immune systems. Thus, the ability of pathogens to resist oxidative stress would enhance their survival during infection and contribute to their virulence (47). All EO-adapted E. coli O157:H7 cells displayed a significantly (P < 0.05) stronger resistance (D value, 5.1 to 5.5 min) against oxidative stress (50 mM H2O2) than the nonadapted control (D value, 4.1 min) (Table 2). Similar to this observation, Dubois-Brissonnet et al. (8) observed an increased resistance in sublethal terpene-adapted S. Typhimurium against the biocide peracetic acid (PA), an oxidizing agent that interacts with and causes damage to microbial macromolecules.
In agreement with the observed phenotype, genes related to oxidative stress defense were significantly (P < 0.05) upregulated in EO-adapted cells. These include transcription regulators (soxRS) (48), a reactive oxygen species (ROS) scavenger (sodA) and its manganese cofactor transporter (mntH) (49), and several oxidative damage repair enzymes (msrB, trxC, and grxA), which help to restore lost structure and functions in oxidized proteins, thereby maintaining normal cellular activities and enhancing microbial survival under oxidative stress (50) (Table 3). A similar induction of oxidative stress response genes was observed in E. coli upon exposure to epigallocatechin gallate (EGCG) (soxR, sodA) (51) and cinnamaldehyde (20). The induction of these genes in EO-adapted cells suggests that EOs might impose oxidative stress on E. coli O157:H7. Chueca et al. (52, 53) demonstrated that Car, citral, and (+)-limonene induce ROS formation and result in oxidative DNA damage in E. coli. Similarly, Xiong et al. (51) reported that the bactericidal activity of polyphenol EGCG against E. coli was due to its ability to induce intracellular superoxide formation. The mechanism by which EOs triggered ROS production, however, remains to be elucidated. Xiong et al. (51) suggested that the ROS production was a result of metabolic instability when microbial cells were challenged with bactericidal agents.
Apart from the induction of the oxidative stress response, a large number of genes related to iron transport and metabolism, such as iron-enterobactin transporters, Fe2+ transporters (feoAB), and heme utilization (chuA) (54), were upregulated in EO-adapted cells (Table 3). Iron is an essential nutrient for bacteria, as it is contained within the redox center of various metabolic enzymes. Hence, an induced uptake and metabolism of iron may enhance microbial survival under stress conditions (54). The overexpression of iron acquisition genes was reported in S. cerevisiae exposed to Thy (55) and in E. coli exposed to n-butanol (56) and was found to contribute to microbial oxidative resistance (by sequestering free irons, which generate ROS via the Fenton reaction) and host colonization, where the concentration of free iron is extremely low (57).
The results obtained demonstrated that EO adaptation enhanced oxidative stress resistance in E. coli O157:H7, which possibly contributes to a better survival of these pathogens during host infection. Hence, the use of sublethal EOs should be avoided.
RT-qPCR validation.
Reverse transcription-quantitative PCR (RT-qPCR) results confirmed the increased expression of genes related to membrane (pspD and pspG), heat (ibpB), and oxidative (grxA and soxS) stress responses and iron transport (ECs0632 and feoA) in EO-adapted E. coli O157:H7 cells compared to that in the nonadapted control (Fig. 3). The numerical values of fold change obtained from RNA-seq were generally higher than those from PCR, suggesting a wider dynamic range of RNA-seq analysis. Different from Bi et al. (55), who reported a strong positive correlation (r = 0.96) between RT-PCR and microarray results in Thy-treated S. cerevisiae, we did not observe a good correlation between these two methods (data not shown), possibly due to the different batches of RNA used. Similarly, King et al. (42) reported a similar trend of up- and downregulation of genes in acid-stressed E. coli by RT-qPCR and microarray, but with a poor correlation of the numerical values of fold changes.
FIG 3.
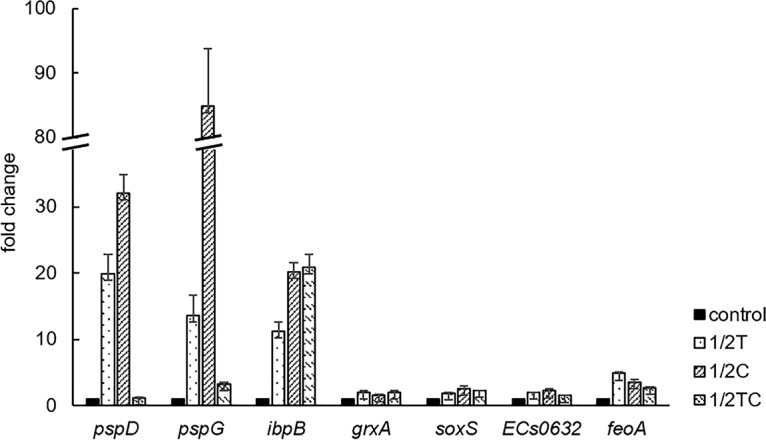
Expression levels of selected genes related to membrane (pspD, pspG), heat (ibpB), and oxidative (grxA, soxS) stress responses and iron uptake (ECs0632, feoA), in E. coli O157:H7 nonadapted (control) or adapted to a sublethal concentration of thymol (1/2T), carvacrol (1/2C), or trans-cinnamaldehyde (1/2TC), determined by RT-qPCR.
Conclusion.
In light of these findings, it can be concluded that the adaptation of E. coli O157:H7 to a sublethal level of thymol, carvacrol, or trans-cinnamaldehyde induced both direct and cross-resistance against subsequent lethal treatments using essential oils (EO), heat, acid, and oxidative stresses. A transcriptomic analysis by RNA-seq revealed differentially expressed genes that were generally consistent with the resistance phenotypes. Thus, the results from the present work suggest that caution should be exercised regarding the use of EO as a food antimicrobial, due to the potential development of resistance.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
A frozen stock of E. coli O157:H7 (ATCC 35150) was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and stored at −80°C in Cryoinstant vials with porous beads (DeltaLab, Barcelona, Spain). Frozen cultures were activated by two consecutive 24-h transfers at 37°C in 10 ml sterile TSB (Oxoid, Hampshire, England), and daily transfer was performed to maintain cell viability. Cells were washed twice in 1× phosphate-buffered saline (PBS) (Vivantis Inc., Oceanside, CA, USA) with centrifugation (5000 × g at 4°C for 5 min) and diluted in TSB to appropriate concentrations before use.
Determination of MIC.
Thymol (Thy), carvacrol (Car), and trans-cinnamaldehyde (TC) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Stock solutions of EOs were prepared in 95% ethanol at 100 mg/ml and stored at 4°C in the dark until use. The MICs of EOs were determined using the Clinical and Laboratory Standards Institute (CLSI) microdilution method (58). Serial 2-fold dilutions of EO compounds were prepared in TSB in a sterile 96-well plate (Thermo Fisher Scientific, Waltham, MA, USA), and equal volumes (100 μl) of EO and the bacterial solution were mixed. The final microbial inoculum level was 105 CFU/ml and final EO concentration range was 0.08 to 10.0 mg/ml. After a 24-h incubation at 37°C, 10 μl of 0.01% (wt/vol) resazurin (Sigma-Aldrich) solution was added to each well and incubated for 2 h at 37°C before visual examination of the color change. The MIC in this study was defined as the minimum EO concentration that inhibited microbial growth, as determined by a lack of color change in resazurin indicator. The highest ethanol concentration (2.5% [vol/vol]) was determined to have no impact on microbial survival (data not shown). TSB and E. coli O157:H7 inoculated in TSB without EO were used as negative and positive growth controls, respectively.
Growth kinetics of E. coli O157:H7 in sublethal EOs.
EO-adapted bacterial cells were prepared by inoculating overnight E. coli O157:H7 into TSB containing Thy, Car, or TC at one-half MIC and incubating at 37°C. Cell growth was monitored by sampling at appropriate time intervals, diluting in 0.1% (wt/vol) peptone water (PW; Oxoid), and plating on tryptic soya agar (TSA). The number of viable cells, expressed as log CFU/ml, was plotted against time. The growth curves and growth parameters were generated by fitting the data to the equation of Baranyi and Roberts (59) using DMFit (https://browser.combase.cc/DMFit.aspx). On the basis of the growth curve, EO-adapted and nonadapted (control) E. coli O157:H7 cells were collected at early stationary phase for resistance assays and RNA-seq analysis.
Determination of EO resistance.
EO-adapted and nonadapted (control) E. coli O157:H7 cells were inoculated (final inoculum, 105 CFU/ml) in 10 ml TSB containing a lethal concentration of Thy, Car, or TC. Aliquots were withdrawn at appropriate time intervals, diluted in 0.1% PW, and plated on TSA to determine the viable cell count. Survival curves were constructed by plotting viable cell counts (log CFU/ml) against treatment time. The best fit line of the survival curves was determined using linear regression with Microsoft Excel (Microsoft Corp., Redmond, WA), and the D value (min) was calculated as the negative reciprocal of the slope.
Determination of heat resistance.
EO-adapted and nonadapted (control) E. coli O157:H7 cells were inoculated (final inoculum, 105 CFU/ml) in 10 ml TSB prewarmed at 58.0°C in a stirring water bath (Valchim, Milan, Italy). At appropriate time intervals, aliquots were withdrawn from the test tube and immediately cooled in iced water for 5 min, diluted in 0.1% PW, and plated on TSA to count viable cells. The D values (min) of EO-adapted and nonadapted cells were calculated as described above.
Determination of acid resistance.
The acid resistance of E. coli O157:H7 was determined using simulated gastric fluid (SGF) at pH 1.5. SGF was prepared with the following composition: 8.3 g/liter proteose-peptone (Oxoid), 3.5 g/liter d-glucose (Goodrich Chemical Enterprise, Singapore), 2.05 g/liter NaCl (Goodrich Chemical Enterprise), 0.6 g/liter KH2PO4 (Sigma-Aldrich), 0.11 g/liter CaCl2 (Goodrich Chemical Enterprise), 0.37 g/liter KCl (Goodrich Chemical Enterprise), 0.1 g/liter lysozyme (Sigma-Aldrich), and 13.3 mg/liter pepsin (Sigma-Aldrich). All compounds were dissolved in distilled water and autoclaved, except for lysozyme and pepsin, which were filter sterilized (0.2 μm). The mixing of sterile components was performed inside a biosafety cabinet (BSC; ESCO, Marietta, OH, USA), and the final pH was adjusted to 1.5 using 5.0 N HCl (Merck, Darmstadt, Germany) (7). Adapted and nonadapted (control) E. coli O157:H7 cells were inoculated (final inoculum, 105 CFU/ml) in 10 ml SGF solution, which was prewarmed to 37°C in a stirring water bath (Valchim). Microbial survival was monitored by sampling at appropriate time intervals, and D values (min) of EO-adapted and nonadapted cells were calculated as described above.
Determination of oxidative stress resistance.
EO-adapted and nonadapted (control) E. coli O157:H7 cells were inoculated (final inoculum, 105 CFU/ml) in 10 ml 1× PBS containing 50 mM hydrogen peroxide (H2O2; Sigma-Aldrich) and incubated at room temperature (ca. 23°C). Samples were removed periodically to determine the number of survivors, and D values were calculated as described above.
Transcriptomic analysis.
To understand transcriptional response of E. coli O157:H7 (ATCC 35150) adapting to EO stress, a comparative transcriptomic analysis was performed between EO-adapted E. coli O157:H7 cells and the nonadapted control. The experiment was conducted with three biological replicates for each sample type. Early stationary-phase nonadapted and EO-adapted E. coli O157:H7 cultures were stabilized with RNAprotect Bacteria reagent (Qiagen, Hilden, Germany), and RNA extraction was performed using an RNeasy minikit (Qiagen) with on-column DNase digestion, according to the manufacturer's protocols. The quality and concentration of extracted RNA were verified using a Nanodrop 1000 (Thermo Scientific Inc., Wilmington, DE, USA), and the integrity was checked by agarose gel electrophoresis. The total RNA was treated again with a Turbo DNase (Life Technologies, USA) kit and purified with Agencourt RNAClean XP magnetic beads (Beckman Coulter). Agilent 2200 TapeStation (Agilent Technologies, USA) was employed to determine the quality and integrity of the total RNA. The quantification of the total RNA and DNA contamination was performed with Qubit 2.0 fluorometer assays (Life Technologies, CA, USA). Total RNA (5 μg) was rRNA depleted with a Ribo-Zero bacterial rRNA removal kit (Illumina, USA), and NEBNext RNA first strand synthesis module and NEBNext Ultra Directional RNA second strand synthesis module kits (New England BioLabs, USA) were used to synthesize double-stranded cDNA from up to 100 ng of ribosome-depleted RNA. The cDNA purification was performed with Agencourt AMPure XP magnetic beads (Beckman Coulter). Double-stranded cDNA was quantified with Qubit 2.0 fluorometer assays (Life Technologies, CA, USA), and samples were submitted to SCELSE's sequencing facility for TruSeq Stranded mRNA library preparation (Illumina, USA).
The library was sequenced on an Illumina HiSeq 2500 platform (Illumina) using a paired-end protocol and a read length of 2 × 100 bp. The raw reads were quality filtered, adaptors were removed, and any reads below 50 bp were discarded. The reads were then mapped onto the E. coli O157:H7 strain Sakai reference genome (NC_002695), and the raw read count table was generated using CLC genomics Workbench 10.0 (CLC Bio, Arhus, Denmark). Statistical analysis was performed using the DESeq2 package (60), with transcript counts normalized to effective library size. A negative binomial test was used to identify differentially expressed genes, with cutoff criteria set at an adjusted P value (Padj) of <0.05 and a fold change of ≥5.
RT-qPCR.
Seven genes that showed significant (Padj < 0.05) differential expression in RNA-seq were validated by RT-qPCR. The 16S rRNA gene was used as an endogenous control for normalization within samples. Forward and reverse PCR primers for the target genes were designed on the basis of the NCBI E. coli O157:H5 strain Sakai genomic sequence (NC_002695) using Prime3 software (http://bioinfo.ut.ee/primer3-0.4.0/) with the following criteria: amplicon size, 100 to150 bp; calculated primer melting temperature, 57 to 62°C; GC content, 40 to 60%; and probabilities of primer-dimer/hairpin formation were minimized (Table 4).
TABLE 4.
Primer sequences
| Function | Gene | Primer sequence (5′→3′)a | Size (bp) |
|---|---|---|---|
| Housekeeping | 16S rRNA | F: AGAGGATGACCAGCCACAC | 194 |
| R: CGGGTAACGTCAATGAGCAAAG | |||
| Membrane stress response | pspD | F: GGCAAAAGGTAAAGCCTGGT | 112 |
| R: CGGCGAGCAACTGATTTTAT | |||
| pspG | F: GTTTCGTTGCTGGGCATTAT | 101 | |
| R: AGTAACCACGGCAGCAACTT | |||
| Heat stress resistance | ibpB | F: CAAGGGTAATGCGGTAGTGG | 116 |
| R: TCAATGGATCGGTTTTGACA | |||
| Oxidative stress resistance | grxA | F: TCAGGCGTCCAGATTCTCTT | 143 |
| R: GTGCGGAAGGGATCACTAAA | |||
| soxS | F: ACGGCGTTGACGAATGTAAT | 107 | |
| R: ACATTGATGTCGTCGCAAAA | |||
| Iron transport | ECs0632 | F: GACTCAGGCGATGAAAGAGG | 112 |
| R: TTCAAAGGGAGTTGCGAGAT | |||
| feoA | F: CGCCAAAAACTGCTTTCTCT | 118 | |
| R: TGCGTAATACCAGGCTCACA |
F, forward; R, reverse.
The template cDNA was synthesized from 1 μg of extracted total RNA by reverse transcription using a GoScript reverse transcription system (Promega, Fitchburg, WI, USA) with random primers according to the manufacturer's instructions. The cDNA was used as a template for qPCR with SYBR Select master mix kit (catalog no. 4472953; Applied Biosystems, Carlsbad, CA, USA), with each qPCR mix (total volume 25 μl) consisting of 1 μl cDNA, 1 μl each forward and reverse primer (25 μM stock), 10 μl SYBR green supermix, and 7 μl nuclease-free water. Real-time PCR was performed on a StepOnePlus real-time PCR system (Applied Biosystems) with the following thermal cycling conditions: 95°C for 30 s (denaturation and polymerase activation) and 40 cycles of 95°C for 3 s and 60°C for 30 s (amplification). The specificity of the PCR was determined with a melting curve analysis (60 to 95°C with a heating rate of 0.3°C/s) and agarose gel electrophoresis. The relative changes in gene expression in EO-adapted cells compared to that of the nonadapted control were calculated using the 2−ΔΔCT method (61).
Statistical analysis.
All experiments were performed in duplicates and repeated at least three times (n = 6). The results are represented as means ± standard deviations, and significant differences (P < 0.05) were determined by one-way analysis of variance (ANOVA) and a Tukey's post hoc test with the IBM SPSS statistical software (version 20; SPSS Inc., IBM Corporation, Armonk, NY, USA).
Accession number(s).
Raw reads obtained during the RNA-Seq analysis were submitted to GenBank under BioProject number PRJNA476677.
Supplementary Material
ACKNOWLEDGMENT
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01616-18.
REFERENCES
- 1.Lim JY, Yoon JW, Hovde CJ. 2010. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J Microbiol Biotechnol 20:5–14. [PMC free article] [PubMed] [Google Scholar]
- 2.Burt S. 2004. Essential oils: their antibacterial properties and potential applications in foods—a review. Int J Food Microbiol 94:223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Hyldgaard M, Mygind T, Meyer RL. 2012. Essential oils in food preservation: mode of action, synergies, and interactions with food matrix components. Front Microbiol 3:12. doi: 10.3389/fmicb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cava-Roda R, Taboada-Rodriguez A, Valverde-Franco M, Marin-Iniesta F. 2010. Antimicrobial activity of vanillin and mixtures with cinnamon and clove essential oils in controlling Listeria monocytogenes and Escherichia coli O157:H7 in milk. Food Bioproc Tech 5:2120–2131. doi: 10.1007/s11947-010-0484-4. [DOI] [Google Scholar]
- 5.Gutierrez J, Barry-Ryan C, Bourke P. 2008. The antimicrobial efficacy of plant essential oil combinations and interactions with food ingredients. Int J Food Microbiol 124:91–97. doi: 10.1016/j.ijfoodmicro.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 6.Wesche AM, Gurtler JB, Marks BP, Ryser ET. 2009. Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J Food Prot 72:1121–1138. doi: 10.4315/0362-028X-72.5.1121. [DOI] [PubMed] [Google Scholar]
- 7.Yuk HG, Marshall DL. 2004. Adaptation of Escherichia coli O157:H7 to pH alters membrane lipid composition, verotoxin secretion, and resistance to simulated gastric fluid acid. Appl Environ Microbiol 70:3500–3505. doi: 10.1128/AEM.70.6.3500-3505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubois-Brissonnet F, Naitali M, Mafu AA, Briandet R. 2011. Induction of fatty acid composition modifications and tolerance to biocides in Salmonella enterica serovar Typhimurium by plant-derived terpenes. Appl Environ Microbiol 77:906–910. doi: 10.1128/AEM.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luz IS, Gomes-Neto NJ, Tavares AG, Nunes PC, Magnani M, de Souza EL. 2012. Evidence for lack of acquisition of tolerance in Salmonella enterica serovar Typhimurium ATCC 14028 after exposure to subinhibitory amounts of Origanum vulgare L. essential oil and carvacrol. Appl Environ Microbiol 78:5021–5024. doi: 10.1128/AEM.00605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes-Neto NJ, Luz IS, Franco OL, Magnani M, Souza EL. 2014. Tolerance evaluation in Salmonella enterica serovar Typhimurium challenged with sublethal amounts of Rosmarinus officinalis L. essential oil or 1,8-cineole in meat model. Int J Food Sci Technol 49:1912–1917. doi: 10.1111/ijfs.12522. [DOI] [Google Scholar]
- 11.Hammer KA, Carson CF, Riley TV. 2012. Effect of Melaleuca alternifolia (tea tree) essential oil and the major monoterpene component terpinen-4-ol on the development of single- and multistep antibiotic resistance and antimicrobial susceptibility. Antimicrob Agents Chemother 56:909–915. doi: 10.1128/AAC.05741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chueca B, Berdejo D, Gomes-Neto NJ, Pagán R, García-Gonzalo D. 2016. Emergence of hyper-resistant Escherichia coli MG1655 derivative strains after applying sub-inhibitory doses of individual constituents of essential oils. Front Microbiol 7:273. doi: 10.3389/fmicb.2016.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang S, Orsi RH, den Bakker HC, Wiedmann M, Boor KJ, Bergholz TM. 2015. Transcriptomic analysis of the adaptation of Listeria monocytogenes to growth on vacuum-packed cold smoked salmon. Appl Environ Microbiol 81:6812–6824. doi: 10.1128/AEM.01752-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helander IM, Alakomi HL, Latva-Kala K, Mattila-Sandholm T, Pol I, Smid EJ, Gorris LGM, von Wright A. 1998. Characterization of the action of selected essential oil components on Gram-negative bacteria. J Agric Food Chem 46:3590–3595. doi: 10.1021/jf980154m. [DOI] [Google Scholar]
- 16.Kim J, Marshall MR, Wei C. 1995. Antibacterial activity of some essential oil components against five foodborne pathogens. J Agric Food Chem 43:2839–2845. doi: 10.1021/jf00059a013. [DOI] [Google Scholar]
- 17.Berney M, Weilenmann HU, Ihssen J, Bassin C, Egli T. 2006. Specific growth rate determines the sensitivity of Escherichia coli to thermal, UVA, and solar disinfection. Appl Environ Microbiol 72:2586–2593. doi: 10.1128/AEM.72.4.2586-2593.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Souza EL. 2016. The effects of sublethal doses of essential oils and their constituents on antimicrobial susceptibility and antibiotic resistance among food-related bacteria: a review. Trends Food Sci Technol 56:1–12. doi: 10.1016/j.tifs.2016.07.012. [DOI] [Google Scholar]
- 19.Rao A, Zhang Y, Muend S, Rao R. 2010. Mechanism of antifungal activity of terpenoid phenols resembles calcium stress and inhibition of the TOR pathway. Antimicrob Agents Chemother 54:5062–5069. doi: 10.1128/AAC.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visvalingam J, Hernandez-Doria JD, Holley RA. 2013. Examination of the genome-wide transcriptional response of Escherichia coli O157:H7 to cinnamaldehyde exposure. Appl Environ Microbiol 79:942–950. doi: 10.1128/AEM.02767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kollanoor Johny A, Frye JG, Donoghue A, Donoghue DJ, Porwollik S, McClelland M, Venkitanarayanan K. 2017. Gene expression response of Salmonella enterica serotype Enteritidis phage type 8 to subinhibitory concentrations of the plant-derived compounds trans-cinnamaldehyde and eugenol. Front Microbiol 8:1828. doi: 10.3389/fmicb.2017.01828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold KW, Kaspar CW. 1995. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol 61:2037–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ultee A, Kets EPW, Alberda M, Hoekstra FA, Smid EJ. 2000. Adaptation of the food-borne pathogen Bacillus cereus to carvacrol. Arch Microbiol 174:233–238. doi: 10.1007/s002030000199. [DOI] [PubMed] [Google Scholar]
- 24.Flores-Kim J, Darwin AJ. 2016. The phage shock protein response. Annu Rev Microbiol 70:83–101. doi: 10.1146/annurev-micro-102215-095359. [DOI] [PubMed] [Google Scholar]
- 25.Fink RC, Black EP, Hou Z, Sugawara M, Sadowsky MJ, Diez-Gonzalez F. 2012. Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl Environ Microbiol 78:1752–1764. doi: 10.1128/AEM.07454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi H, Yamamoto M, Aono R. 1998. Appearance of a stress-response protein, phage-shock protein A, in Escherichia coli exposed to hydrophobic organic solvents. Microbiology 144:353–359. doi: 10.1099/00221287-144-2-353. [DOI] [PubMed] [Google Scholar]
- 27.McDonald C, Jovanovic G, Ces O, Buck M. 2015. Membrane stored curvature elastic stress modulates recruitment of maintenance proteins PspA and Vipp1. mBio 6:e01188-. doi: 10.1128/mBio.01188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi R, Suzuki T, Yoshida M. 2007. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol 66:100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- 29.Horstman NK, Darwin AJ. 2012. Phage shock proteins B and C prevent lethal cytoplasmic membrane permeability in Yersinia enterocolitica. Mol Microbiol 85:445–460. doi: 10.1111/j.1365-2958.2012.08120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da Silva Luz I, Gomes-Neto NJ, Magnani M, de Souza EL. 2014. Assessment of tolerance induction by Origanum vulgare L. essential oil or carvacrol in Pseudomonas aeruginosa cultivated in a meat-base broth and in a meat model. Food Sci Technol Int 21:571–580. doi: 10.1177/1082013214554467. [DOI] [PubMed] [Google Scholar]
- 31.Arsène F, Tomoyasu T, Bukau B. 2000. The heat shock response of Escherichia coli. Int J Food Microbiol 55:3–9. doi: 10.1016/S0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 32.Ratajczak E, Zietkiewicz S, Liberek K. 2009. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J Mol Biol 386:178–189. doi: 10.1016/j.jmb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Shimohata N, Chiba S, Saikawa N, Ito K, Akiyama Y. 2002. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7:653–662. doi: 10.1046/j.1365-2443.2002.00554.x. [DOI] [PubMed] [Google Scholar]
- 34.Burt SA, van der Zee R, Koets AP, de Graaff AM, van Knapen F, Gaastra W, Haagsman HP, Veldhuizen EJA. 2007. Carvacrol induces heat shock protein 60 and inhibits synthesis of flagellin in Escherichia coli O157:H7. Appl Environ Microbiol 73:4484–4490. doi: 10.1128/AEM.00340-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Pasqua R, Mamone G, Ferranti P, Ercolini D, Mauriello G. 2010. Changes in the proteome of Salmonella enterica serovar Thompson as stress adaptation to sublethal concentrations of thymol. Proteomics 10:1040–1049. [DOI] [PubMed] [Google Scholar]
- 36.Shimuta T, Nakano K, Yamaguchi Y, Ozaki S, Fujimitsu K, Matsunaga C, Noguchi K, Emoto A, Katayama T. 2004. Novel heat shock protein HspQ stimulates the degradation of mutant DnaA protein in Escherichia coli. Genes Cells 9:1151–1166. doi: 10.1111/j.1365-2443.2004.00800.x. [DOI] [PubMed] [Google Scholar]
- 37.Ishii A, Oshima T, Sato T, Nakasone K, Mori H, Kato C. 2005. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles 9:65–73. doi: 10.1007/s00792-004-0414-3. [DOI] [PubMed] [Google Scholar]
- 38.Nobre LS, Al-Shahrour F, Dopazo J, Saraiva LM. 2009. Exploring the antimicrobial action of a carbon monoxide-releasing compound through whole-genome transcription profiling of Escherichia coli. Microbiology 155:813–824. doi: 10.1099/mic.0.023911-0. [DOI] [PubMed] [Google Scholar]
- 39.Gamarra S, Rocha EMF, Zhang YQ, Park S, Rao R, Perlin DS. 2010. Mechanism of the synergistic effect of amiodarone and fluconazole in Candida albicans. Antimicrob Agents Chemother 54:1753–1761. doi: 10.1128/AAC.01728-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richard HT, Foster JW. 2003. Acid resistance in Escherichia coli. Adv Appl Microbiol 52:167–186. doi: 10.1016/S0065-2164(03)01007-4. [DOI] [PubMed] [Google Scholar]
- 41.Iyer R, Williams C, Miller C. 2003. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol 185:6556–6561. doi: 10.1128/JB.185.22.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King T, Lucchini S, Hinton JCD, Gobius K. 2010. Transcriptomic analysis of Escherichia coli O157:H7 and K-12 cultures exposed to inorganic and organic acids in stationary phase reveals acidulant- and strain-specific acid tolerance responses. Appl Environ Microbiol 76:6514–6528. doi: 10.1128/AEM.02392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fratamico PM, Wang S, Yan X, Zhang W, Li Y. 2011. Differential gene expression of E. coli O157:H7 in ground beef extract compared to tryptic soy broth. J Food Sci 76:M79–M87. doi: 10.1111/j.1750-3841.2010.01952.x. [DOI] [PubMed] [Google Scholar]
- 44.Carter MQ, Louie JW, Fagerquist CK, Sultan O, Miller WG, Mandrell RE. 2012. Evolutionary silence of the acid chaperone protein HdeB in enterohemorrhagic Escherichia coli O1517:H7. Appl Environ Microbiol 78:1004–1014. doi: 10.1128/AEM.07033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price SB, Wright JC, DeGraves FJ, Castanie-Cornet MP, Foster JW. 2004. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl Environ Microbiol 70:4792–4799. doi: 10.1128/AEM.70.8.4792-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kern R, Malki A, Abdallah J, Tagourti J, Richarme G. 2007. Escherichia coli HdeB is an acid stress chaperone. J Bacteriol 189:603–610. doi: 10.1128/JB.01522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi K, Fujikawa M, Kozawa T. 2014. Oxidative stress sensing by the iron-sulfur cluster in the transcription factor, SoxR. J Inorg Biochem 133:87–91. doi: 10.1016/j.jinorgbio.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 48.Pomposiello P, Bennik MHJ, Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli response to superoxide stress and sodium salicylate. J Bacteriol 183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabri M, Caza M, Proulx J, Lymberopoulos MH, Bree A, Moulin-Schouleur M, Curtis R III, Dozois CM. 2008. Contribution of the SitABCD, MntH, and FeoB metal transporters to the virulence of avian pathogenic Escherichia coli O78 strain χ7122. Infect Immun 76:601–611. doi: 10.1128/IAI.00789-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J, Holmgren A. 2014. The thioredoxin antioxidant system. Free Radic Biol Med 66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 51.Xiong LG, Chen YJ, Tong JW, Huang JA, Li J, Gong YS, Liu ZH. 2017. Tea polyphenol epigallocatechin gallate inhibits Escherichia coli by increasing endogenous oxidative stress. Food Chem 217:196–204. doi: 10.1016/j.foodchem.2016.08.098. [DOI] [PubMed] [Google Scholar]
- 52.Chueca B, Pagan R, Garcia-Gonzalo D. 2014. Differential mechanism of Escherichia coli inactivation by (+)-limonene as a function of cell physiological state and drug's concentration. PLoS One 9:e94072. doi: 10.1371/journal.pone.0094072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chueca B, Pagan R, Garcia-Gonzalo D. 2014. Oxygenated monoterpenes citral and carvacrol cause oxidative damage in Escherichia coli without the involvement of tricarboxylic acid cycle and Fenton reaction. Int J Food Microbiol 189:126–131. doi: 10.1016/j.ijfoodmicro.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Braun V. 2003. Iron uptake by Escherichia coli. Front Biosci (Landmark Ed) 8:s1409–s1421. doi: 10.2741/1232. [DOI] [PubMed] [Google Scholar]
- 55.Bi X, Guo N, Jin J, Liu J, Feng H, Shi J, Xiang H, Wu X, Dong J, Hu H, Yan S, Yu C, Wang X, Deng X, Yu L. 2010. The global gene expression profile of the model fungus Saccharomyces cerevisiae induced by thymol. J Appl Microbiol 108:712–722. doi: 10.1111/j.1365-2672.2009.04470.x. [DOI] [PubMed] [Google Scholar]
- 56.Reyes LH, Almario MP, Winkler J, Orozco MM, Kao KC. 2012. Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab Eng 14:579–590. doi: 10.1016/j.ymben.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 57.Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, Kaper JB. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun 70:5416–5427. doi: 10.1128/IAI.70.10.5416-5427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Othman M, San Loh H, Wiart C, Khoo TJ, Lim KH, Ting KN. 2011. Optimal methods for evaluating antimicrobial activities from plant extracts. J Microbiol Methods 84:161–166. doi: 10.1016/j.mimet.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 59.Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23:277–294. doi: 10.1016/0168-1605(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


