In the past decade, various studies have described the effects of microbial volatiles on other (micro)organisms in vitro, but their broad-spectrum activity in vivo and the mechanisms underlying volatile-mediated plant growth promotion have not been addressed in detail. Here, we revealed that volatiles from root-associated bacteria of the genus Microbacterium can enhance the growth of different plant species and can prime plants for growth promotion without direct and prolonged contact between the bacterium and the plant. Collectively, these results provide new opportunities for sustainable agriculture and horticulture by exposing roots of plants only briefly to a specific blend of microbial volatile compounds prior to transplantation of the seedlings to the greenhouse or field. This strategy has no need for large-scale introduction or root colonization and survival of the microbial inoculant.
KEYWORDS: plant-microbe interactions, volatile organic compounds, VOCs, biostimulant
ABSTRACT
Volatile compounds produced by plant-associated microorganisms represent a diverse resource to promote plant growth and health. Here, we investigated the effect of volatiles from root-associated Microbacterium species on plant growth and development. Volatiles of eight strains induced significant increases in shoot and root biomass of Arabidopsis but differed in their effects on root architecture. Microbacterium strain EC8 also enhanced root and shoot biomass of lettuce and tomato. Biomass increases were also observed for plants exposed only briefly to volatiles from EC8 prior to transplantation of the seedlings to soil. These results indicate that volatiles from EC8 can prime plants for growth promotion without direct and prolonged contact. We further showed that the induction of plant growth promotion is tissue specific; that is, exposure of roots to volatiles from EC8 led to an increase in plant biomass, whereas shoot exposure resulted in no or less growth promotion. Gas chromatography–quadrupole time of flight mass spectometry (GC–QTOF-MS) analysis revealed that EC8 produces a wide array of sulfur-containing compounds, as well as ketones. Bioassays with synthetic sulfur volatile compounds revealed that the plant growth response to dimethyl trisulfide was concentration-dependent, with a significant increase in shoot weight at 1 μM and negative effects on plant biomass at concentrations higher than 1 mM. Genome-wide transcriptome analysis of volatile-exposed Arabidopsis seedlings showed upregulation of genes involved in assimilation and transport of sulfate and nitrate. Collectively, these results show that root-associated Microbacterium primes plants, via the roots, for growth promotion, most likely via modulation of sulfur and nitrogen metabolism.
IMPORTANCE In the past decade, various studies have described the effects of microbial volatiles on other (micro)organisms in vitro, but their broad-spectrum activity in vivo and the mechanisms underlying volatile-mediated plant growth promotion have not been addressed in detail. Here, we revealed that volatiles from root-associated bacteria of the genus Microbacterium can enhance the growth of different plant species and can prime plants for growth promotion without direct and prolonged contact between the bacterium and the plant. Collectively, these results provide new opportunities for sustainable agriculture and horticulture by exposing roots of plants only briefly to a specific blend of microbial volatile compounds prior to transplantation of the seedlings to the greenhouse or field. This strategy has no need for large-scale introduction or root colonization and survival of the microbial inoculant.
INTRODUCTION
Plant-associated bacteria produce an array of metabolites, including volatile organic compounds (VOCs). VOCs are low-molecular-weight molecules with a high vapor pressure that can disperse through the soil matrix, facilitating long-distance interactions between microorganisms and plants without direct contact (1). The production of VOCs by soil- and plant-associated microorganisms has long been recognized (2). Their effects on soilborne fungi have been reported since the early 1950s (3–7), but their impact on plant growth and health has only been recognized in the past decade. The VOCs 2,3-butanediol and 3-hydroxy-2-butanone, emitted by Bacillus species, enhanced the growth of Arabidopsis thaliana seedlings (8). Seedlings exposed to 2,3-butanediol also showed reduced symptoms of disease caused by a bacterial leaf pathogen (9). Since then, an increasing number of studies have shown the promising effects of bacterial VOCs in the inhibition of plant pathogens and the promotion of plant growth (10–15).
Previous studies on the plant growth-promoting effects by volatile-producing bacteria have been demonstrated mainly in vitro on nutrient-rich media in sealed petri dishes (16–18). Still, little is known about the potential of volatile compounds in agriculture and horticulture. The production of organic (such as 3-hydroxybutanone and dimethyl disulfide) and inorganic (such as NO and CO2) volatile compounds (here, volatiles) by bacteria in situ is not well-studied due to technical limitations. In soils, production levels of volatile compounds by bacteria are presumed to be low and strongly dependent on nutrient and oxygen availability, as well as on the physiological state of the bacteria (19). Variations in soil physicochemical characteristics can lead to a rapid and uneven evaporation of volatile compounds, resulting in inconsistent outcomes (20). Furthermore, plant exudates can affect bacterial densities and activity in the rhizosphere (21, 22), which in turn has impacts on the quantity and diversity of compounds produced in situ.
The genus Microbacterium represents Gram-positive bacteria of the Microbacteriaceae family within the Actinobacteria phylum (23). This genus currently comprises 97 species (http://www.bacterio.net/microbacterium.html), isolated from terrestrial and aquatic ecosystems and also from clinical and food samples (24–29). To date, the majority of functional studies on Microbacterium species relate to their ability to degrade hydrocarbons and complex polysaccharides of economic importance (30–34). Some Microbacterium strains have been shown to produce the plant growth hormone indoleacetic acid, solubilize phosphate, or exhibit 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity (35), but their effects on plant growth have received little attention. Microbacterium isolated from a soil suppressive to Rhizoctonia root rot of wheat, together with Pantoea and Exiguobacterium, enhanced the growth of wheat seedlings and reduced root infections by Rhizoctonia solani (36). However, the underlying mechanisms of volatile-mediated plant growth promotion by Microbacterium species have, to our knowledge, not yet been investigated.
In this study, we investigated the plant growth-promoting effects of the total volatile blend emitted by eight root-associated Microbacterium strains, encompassing organic and inorganic compounds. Using both in vitro and soil bioassays, we further investigated whether volatiles from the endophytic Microbacterium strain EC8 prime plants and whether the plants′ perception of these volatiles occurs via the root and/or shoot. Gas chromatography–time of flight mass spectometry (GC–QTOF-MS) analysis was performed to characterize the VOCs produced by Microbacterium. To further unravel the underlying molecular mechanisms of volatile-mediated growth promotion by Microbacterium, we conducted a genome-wide plant transcriptome analysis.
RESULTS
Volatile-mediated plant growth promotion by Microbacterium.
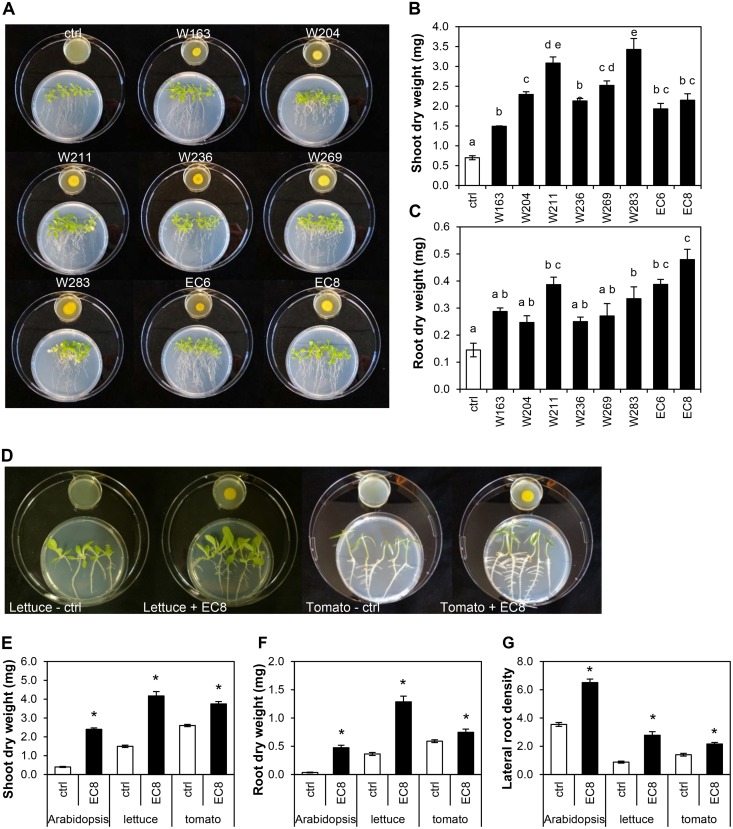
A total of 26 Microbacterium strains isolated from the rhizosphere and endosphere of sugar beet seedlings were phylogenetically characterized (see Fig. S1 in the supplemental material). To test the effects of volatile compounds from Microbacterium on growth of Arabidopsis, eight different strains were selected based on their phylogenetic distribution. Seven-day-old Arabidopsis seedlings were exposed to the total volatile blend, including organic and inorganic compounds such as CO2, emitted by each of the Microbacterium strains. Volatile blends from all eight strains promoted the growth of Arabidopsis seedlings in vitro with significant increases in shoot and root biomass relative to the untreated control (Fig. 1B and C). In addition, differences in root architecture induced by the eight Microbacterium strains were observed visually (Fig. 1A). With an increase of 230% in root biomass compared to control plants, volatiles from Microbacterium sp. strain EC8 induced the strongest increase in root biomass among the Microbacterium strains tested (Fig. 1C). Therefore, we decided to focus on this strain for its effects on growth of plant species other than Arabidopsis and to unravel the mechanisms underlying plant growth promotion. Upon in vitro exposure to the volatiles from EC8, lettuce seedlings showed increases of 178% in shoot biomass (t test, P < 0.001), 253% in root biomass (t test, P < 0.001), and 217% in lateral root density (t test, P < 0.001). Tomato seedlings showed increases of 44% in shoot biomass (t test, P < 0.001), 27% in root biomass (t test, P = 0.038), and 54% in lateral root density (t test, P < 0.001) compared to control seedlings (exposed to agar medium only) (Fig. 1D, E, F, and G). These results indicate that EC8 induces stronger growth-promoting effects on Arabidopsis and lettuce seedlings than on tomato seedlings.
FIG 1.
Plant growth promotion by volatiles emitted by Microbacterium strains. (A) Phenotypic changes of Arabidopsis seedlings exposed to volatiles from eight Microbacterium strains spot-inoculated on agar medium (10 μl at 109 CFU · ml−1) or from the agar medium only (ctrl). Pictures were taken 14 days after exposure. (B and C) Biomass (mean ± standard error [SE], n = 4 to 5) of shoots (B) and roots (C) of volatile-exposed and control seedlings. Different letters show statistically significant differences (one-way ANOVA, Tukey's HSD post hoc test, P < 0.05). (D) Phenotypic changes of Arabidopsis, lettuce, and tomato seedlings exposed to volatile compounds from Microbacterium strain EC8 or from the agar medium only (ctrl) for 12, 7, and 10 days, respectively. (E to G) Dry biomass (mean ± SE, n = 6 to 8) of shoots (E) and roots (F) and lateral root density (number of lateral roots/length [cm] of primary root) (G) of Arabidopsis, lettuce and tomato seedlings exposed to the volatiles from EC8. ctrl, control seedlings exposed to agar medium only; EC8, seedlings exposed to volatiles from EC8. Asterisks indicate statistically significant differences between volatile-exposed and control seedlings (independent samples t test, P < 0.05).
Volatile-mediated priming for plant growth promotion.
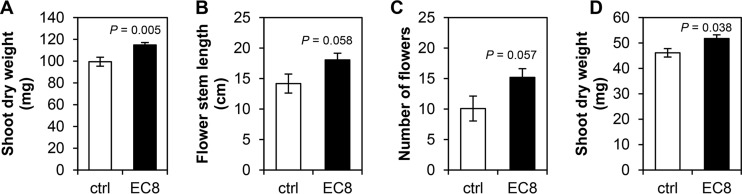
To test whether a relatively short exposure of Arabidopsis and lettuce seedlings to the bacterial volatiles could prime plant growth and development, seedlings were exposed in vitro to the volatiles from EC8 and then transplanted to soil without further exposure to the bacterial strain. The results showed that volatile exposure of 5 days for Arabidopsis or 4 days for lettuce seedlings (instead of 12 and 7 days, respectively) already promoted the growth of seedlings transplanted to and grown in soil for another 21 and 13 days, respectively. Arabidopsis plants preexposed to volatiles from EC8 showed a significant increase of 35% in shoot biomass (Fig. 2A; t test, P = 0.005). We also observed increases of 27% in the flower stem length (t test, P = 0.058) and 51% more flowers (t test, P = 0.057) (Fig. 2B and C). Lettuce plants showed a significant 12% increase in shoot biomass (Fig. 2D; t test, P = 0.038).
FIG 2.
Priming effects by volatiles from Microbacterium strain EC8 on the growth of Arabidopsis and lettuce seedlings. (A) Shoot dry biomass, (B) flower stem length, and (C) number of flowers of Arabidopsis plants (mean ± SE, n = 9); (D) shoot dry biomass of lettuce plants (mean ± SE, n = 4 to 5). ctrl, control plants exposed to agar medium only; EC8, seedlings exposed to volatiles from EC8. Statistically significant differences between volatile-exposed and control seedlings were determined with an independent samples t test.
Plant perception of volatiles from Microbacterium.
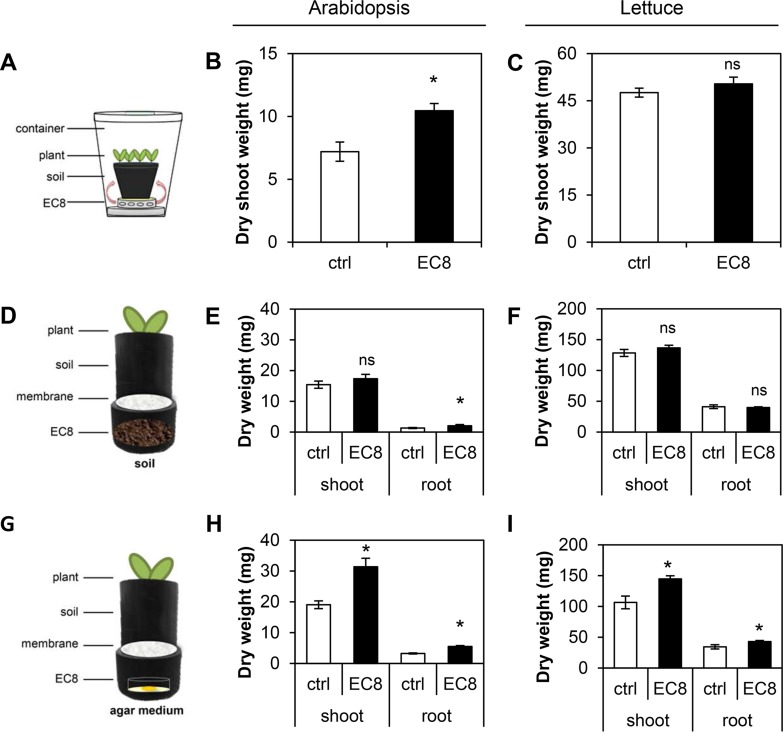
Two different experimental approaches were used to test the effects of volatiles from strain EC8 on plants grown in soil. These setups allowed us to test an “open” system, minimizing accumulation of bacterial CO2 as in the sealed plate assay described above. In the first setup, plants grown in potting soil were exposed to volatiles from EC8 grown on an agar plate inside a sterile closed container for 1 week, allowing exposure of the plant shoots to the bacterial volatiles (Fig. 3A). Exposure to the volatiles from EC8 resulted in a 45% increase of shoot biomass of Arabidopsis plants (Fig. 3B; t test, P = 0.002). However, no significant increases in shoot biomass were observed for lettuce plants (Fig. 3C; t test, P = 0.336). In the second experimental setup, plant roots were exposed to volatiles from strain EC8, either inoculated in a soil-sand mixture or inoculated onto agar medium. To expose only the roots to the bacterial volatiles, we used two-compartment pots separated by a membrane (Fig. 3D and G). The results showed that volatiles from EC8 inoculated into the soil-sand mixture promoted the growth of Arabidopsis roots (Fig. 3E; t test, P = 0.004) but not those of lettuce (Fig. 3F; t test, P = 0.694). Volatiles from EC8 grown on agar medium significantly enhanced the biomass of Arabidopsis and lettuce shoots (Fig. 3H and I; t test, P = 0.001 and P = 0.004, respectively) and roots (Fig. 3H and I; t test, P < 0.001 and P = 0.036, respectively).
FIG 3.
Exposure of plant shoots and roots to volatiles from Microbacterium strain EC8. (A) Experimental setup used to expose plant shoots to bacterial VOCs. (B and C) Shoot dry biomass (mean ± SE, n = 6) of volatile-exposed Arabidopsis (B) and lettuce (C) shoots. (D) Experimental setup used to expose plant roots to bacterial volatiles. Bacterial cells were inoculated in soil on the bottom compartment. (E and F) Dry biomass (mean ± SE, n = 9) of Arabidopsis (E) and lettuce (F) shoots and roots. (G) Experimental setup used to expose plant roots to bacterial volatiles. Bacterial cells were inoculated on agar medium on the bottom compartment. (H and I) Dry biomass (mean ± SE, n = 8 or 9) of Arabidopsis (H) and lettuce (I) shoots and roots. ctrl, control plants exposed to agar medium or soil only; EC8, plants exposed to volatiles from EC8; asterisks indicate a statistically significant difference between volatile-exposed and control seedlings; ns, no statistical differences (independent samples t test, P < 0.05).
Characterization and activity of VOCs from Microbacterium strain EC8.
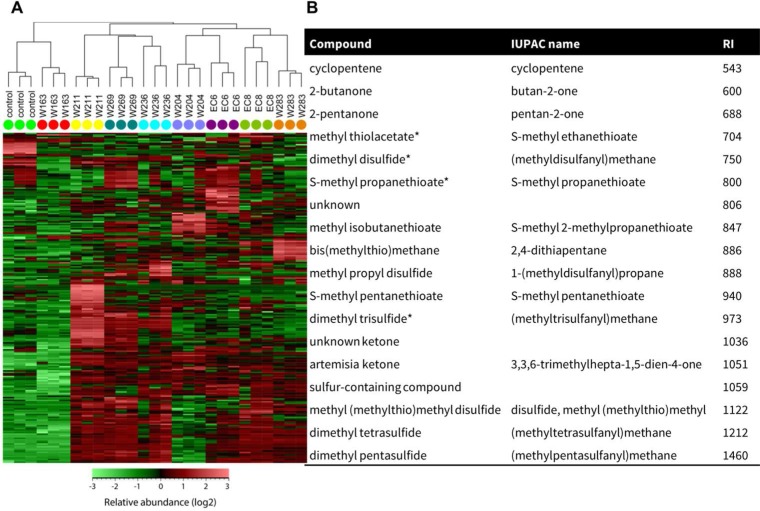
Analysis of the headspace of cultures of the eight Microbacterium strains provided a global profile of their VOCs. Hierarchical cluster analysis showed that the VOC profiles of the eight strains were diverse and different (Fig. 4A). To study this diversity in more detail, headspace VOCs of cultures of EC8 were collected for 6 days and analyzed by GC–QTOF-MS. A total of 18 VOCs were detected that were not found in the control (agar medium only) or were detected with peak areas at least 2-fold larger and significantly different (t test, P < 0.05) from the VOCs in the control. The vast majority of VOCs that met these criteria were identified as sulfur-containing compounds (Fig. 4B). The sulfur-containing compounds detected in our study included dimethyl disulfide and dimethyl trisulfide, commonly found for other bacterial genera, but also rarer compounds such as S-methyl 2-methylpropanethioate and S-methyl pentanethioate and four ketones.
FIG 4.
Volatile organic compounds (VOCs) from Microbacterium strains. (A) Hierarchical cluster and heat map analyses of VOC profiles of Microbacterium. Columns represent three replicate VOC measurements of each of the 8 isolates and the medium alone (control). Rows represent the different VOCs (green, low abundance; red, high abundance). (B) List of VOCs from Microbacterium strain EC8. VOCs displayed were detected only for EC8 or were significantly different (Student's t test, P < 0.05, n = 3) and detected at peak intensities at least twice as high as those in the control (medium only). Compounds were putatively annotated by comparing their mass spectra and calculated linear retention indices (RI) with those of NIST and in-house mass spectral libraries and standard (*).
To determine if the sulfur VOCs detected for EC8 contribute to plant growth promotion, dimethyl disulfide and dimethyl trisulfide were tested as single compounds and as a blend for their effects on growth of Arabidopsis seedlings. Seedlings were exposed to 20 μl of the single compounds at 6 different concentrations ranging from 1 nM to 100 mM, including concentrations previously described for different bacteria (12, 37). In addition, seedlings were also exposed to mixtures (1:1) of these compounds at concentrations of 100 nM, 100 μM, and 100 mM. The results showed that dimethyl disulfide had no effect on Arabidopsis shoot or root biomass. In fact, a slight growth reduction was observed relative to the control (dichloromethane [DCM] solvent alone) for plants exposed to 20 μl at 100 nM and 100 mM dimethyl disulfide (Fig. 5B). A reduction in shoot and root weight was also observed when seedlings were exposed to a mixture of the two sulfur compounds at 100 mM (Fig. 5D). Plants exposed to dimethyl trisulfide at a concentration of 1 μM showed a significant increase in shoot weight (Fig. 5C), whereas negative effects on plant growth were observed at concentrations of 1 mM and higher.
FIG 5.
Effects of synthetic sulfur volatile compounds on shoot and roots of Arabidopsis seedlings. (A) Experimental setup for exposing seedlings to volatile synthetic compounds in vitro. Shoot and dry weight (mean ± SE, n = 5) of Arabidopsis exposed to 20 μl of (B) dimethyl disulfide, (C) dimethyl trisulfide, and (D) mixture (1:1) of dimethyl disulfide and dimethyl trisulfide at different concentrations. Dichloromethane (DCM) was used as the solvent. Control plants were not exposed to volatile compounds. Different letters indicate statistically significant differences (one-way ANOVA, P < 0.05).
Plant transcriptional changes induced by volatiles from Microbacterium strain EC8.
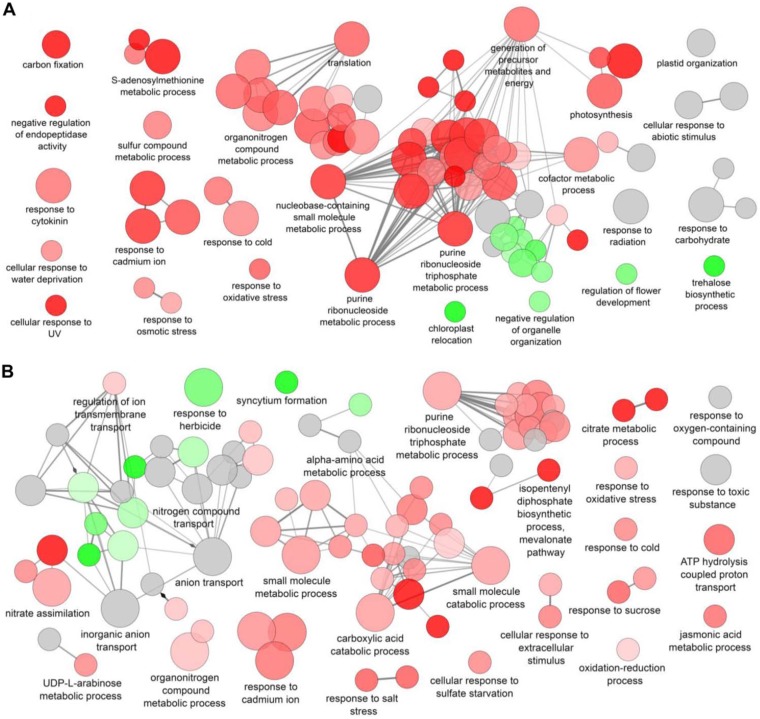
To begin to understand the molecular mechanisms underlying volatile-mediated plant growth promotion by EC8, RNA-seq analysis was performed for Arabidopsis seedlings exposed for 1 week to the bacterial volatiles. Genes of shoot and root tissues with an adjusted P value of <0.05 and with a log2 ratio of ≥0.585 or ≤−0.585 (1.5-fold change) were considered differentially expressed from the nonexposed (control) seedlings. A total of 946 (545 upregulated and 401 downregulated) and 1,361 (698 upregulated and 663 downregulated) differentially expressed genes (DEGs) were identified in shoot and root tissues, respectively. Gene Ontology (GO) terms associated with shoot DEGs were grouped into 23 functional clusters, including “purine ribonucleoside metabolic process,” “response to cytokinin,” “response to ethylene,” “response to oxidative stress.” and several processes related to sulfur metabolism, including “sulfate assimilation,” “sulfur compound catabolic process,” “sulfur compound metabolic process,” and “S-adenosylmethionine metabolic process” (Fig. 6A; see also Fig. S2 in the supplemental material). Downregulated shoot DEGs were grouped into 13 functional clusters and included “cellular carbohydrate metabolic process,” “regulation of postembryonic development,” “plastid organization,” and “movement of cell or subcellular component” (Fig. 6A and S2). GO terms associated with upregulated DEGs in root tissue were grouped into 31 functional clusters, including “nitrate assimilation,” “small molecule catabolic process,” “jasmonic acid metabolic process,” “regulation of actin filament polymerization,” “acetyl-CoA metabolic process,” and “response to oxidative stress” (Fig. 6B; see also Fig. S3 in the supplemental material). Downregulated root DEGs were grouped into 6 functional clusters, including “anion transport,” “response to herbicide,” “transmembrane transport,” and “syncytium formation” (Fig. 6B and S3).
FIG 6.
Global visualization of GO terms assigned for differentially expressed genes (DEGs) of Arabidopsis exposed to volatiles from Microbacterium strain EC8. Functional groups of upregulated DEGs of shoots (A) and roots (B) are shown in red, whereas downregulated DEGs are shown in green. Functional groups with upregulated and downregulated DEGs are shown in gray. Single cluster analysis was performed using Cytoscape software with the ClueGO plugin. The fusion option was used to reduce redundancy of GO terms. Networks with terms functionally grouped with GO pathways are indicated as nodes (two-sided hypergeometric test corrected with the Benjamini-Hochberg procedure; P < 0.05) linked by their kappa score levels (≥0.4), with only the label of the most significant term per group shown. The node size represents the term enrichment significance; smaller nodes indicate larger P values, while larger nodes indicate smaller P values.
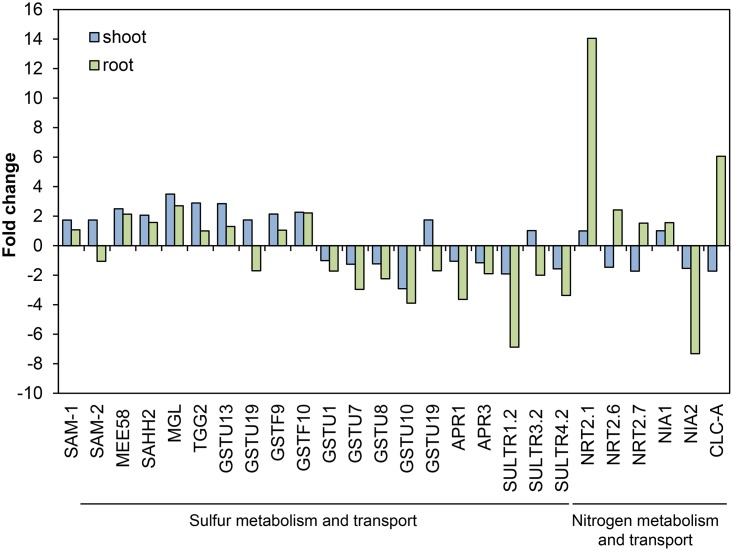
A total of 20 genes involved in sulfur metabolism and transport were found to be differentially expressed in shoot and root tissues upon exposure to volatiles from EC8 (Fig. 7). DEGs involved in sulfur metabolism were mostly upregulated in shoots but downregulated in root tissue. Genes encoding the S-adenosylmethionine synthases SAM-1 (AT1G02500.1) and SAM-2 (AT4G01850.2) and the adenosylhomocysteinases MEE58 (AT4G13940.1) and SAHH2 (AT3G23810.1) were specifically upregulated in shoot tissue, whereas genes encoding the phosphosulfate reductases APR1 (AT4G04610.1) and APR3 (AT4G21990.1) were specifically downregulated in root tissue. DEGs involved in sulfur transport, such as the genes encoding the sulfate transporters SULTR1.2 (AT1G78000.2), SULTR3.2 (AT4G02700.1), and SULTR4.2 (AT3G12520.2), were downregulated in root tissue. Furthermore, several genes encoding glutathione S-transferases (GSTs) were upregulated in shoot tissue, such as GSTU13 (AT1G27130.1), GSTU19 (AT1G78380.1), GSTF9 (AT2G30860.1), and GSTF10 (AT2G30870.1), whereas GSTU1 (AT2G29490.1), GSTU7 (AT2G29420.1), GSTU8 (AT3G09270.1), GSTU10 (AT1G74590.1), and GSTU19 (AT1G78380.1) were downregulated in root tissue. GSTs are ubiquitous in plants and have been suggested to be involved in herbicide detoxification and stress responses (38). However, little is known about their roles in normal plant physiology, during biotic and abiotic stress responses (39), and in bacterium-plant interactions. The methionine gamma-lyase (MGL) (AT1G64660.1), which is involved in methionine homeostasis, and the beta-thioglucoside glucohydrolase TGG2 (AT5G25980.2), which catalyzes the hydrolysis of glucosinolates, were 3.5- and 2.9-fold upregulated in shoot tissue, respectively. Collectively these results indicated that volatiles from EC8 have a significant impact on sulfur metabolism and transport in Arabidopsis seedlings.
FIG 7.
Differentially expressed genes (DEGs) involved in sulfur and nitrogen transport and metabolism of Arabidopsis seedlings exposed to volatiles from Microbacterium strain EC8. One-week-old seedlings were exposed to the bacterial volatiles for 1 week. Shoot DEGs are shown in blue and root DEGs are shown in green. Fold change (FC) was calculated using the log2FC (volatile-exposed seedlings/control).
Furthermore, our transcriptome analysis showed an enrichment of genes involved in nitrate-related processes in volatile-exposed root tissue (Fig. 7). DEGs of root tissue involved in nitrate assimilation were upregulated, whereas DEGs involved in nitrate reduction were downregulated. Expression of genes encoding the three nitrate transporters NRT2.1 (AT1G08090.1), NRT2.6 (AT3G45060.1), and NRT2.7 (AT5G14570.1) and a chlorine channel, CLC-A, was upregulated. Other genes involved in nitrate-related processes, such as the genes for the nitrate reductases NIA1 (AT1G77760.1) and NIA2 (AT1G37130.1), were downregulated. Shoot genes involved in nitrate-related processes were not found to be differentially expressed. Nitrate has been reported to not only serve as a nutrient for plants but also to act as a signal in the regulation of carbon and nitrogen metabolism (40).
DISCUSSION
Members of the Microbacterium genus are widespread in nature; however, their VOC-mediated effects on plant growth and development are not well studied. Here, we described the effects of volatiles from the endophytic Microbacterium strain EC8 on plant growth and on priming seedlings for growth promotion. In addition, we showed how this strain induced specific transcriptional changes in seedlings exposed to the volatile compounds.
To date, most studies on the effects of bacterial volatiles on plant growth promotion have been described in vitro and on strains belonging to the bacterial genera Pseudomonas and Bacillus (11, 12, 18, 20, 39). We show that volatiles from EC8 induce an increase in shoot and root biomass, as well as in lateral root density. These effects were observed not only for the model plant, Arabidopsis, but also for crop plants, such as lettuce and tomato. In in vitro bioassays, plants were exposed to the total volatile blend, i.e., to organic and inorganic volatile compounds, including CO2. Therefore, CO2 could possibly contribute to plant growth-promoting effects (41). However, in previous studies with fungi and bacteria, we showed that CO2 has a role, but a minor one, as several microorganisms with similar densities/biomass failed to promote plant growth in the sealed plate assays (17, 42). Furthermore, plants exposed to CO2 at levels 3-fold higher than ambient levels did not show a significant biomass increase (42). Additionally, earlier studies showed that Bacillus strains with mutations in specific bacterial genes involved in the production of different VOCs had a reduced ability to induce growth promotion (8). Collectively, these results further demonstrate the role of bacterial volatile organic compounds in plant growth promotion.
Several previous studies have described the growth-promoting effects of bacterial volatiles. In these studies, plants were exposed to the volatiles for a long period of time. Volatiles from Bacillus subtilis GB03 promoted the growth of Arabidopsis and sustained the growth for 12 weeks under constant exposure to the bacterial compounds (43). Here, we tested the effects of temporary exposure. Our results demonstrated that short exposure to the volatiles from EC8 prime seedlings of Arabidopsis and lettuce for growth promotion. Priming is defined here as a mechanism and/or substance, in this case volatile compounds, that prepares plants for subsequent/future action, i.e., enhanced growth. Therefore, a prolonged VOC exposure is not necessary to sustain plant growth promotion by the bacterial volatiles. These findings hold a promising tool for improving plant growth, as it does not require a long exposure period or environmental introduction of the bacterial strain in soil.
Currently, knowledge on plant perception of volatiles from root-associated microorganisms is lacking. Most results on plant perception of volatiles originate from studies on plant-plant communication aboveground. Plants can sense volatiles emitted by neighboring plants under herbivore attack and subsequently enhance resistance (44, 45). Our results showed that roots sense and respond to volatiles from EC8 in a context-dependent manner. Volatiles from EC8 inoculated on agar medium promoted the growth of both Arabidopsis and lettuce grown in a soil-sand mixture, but when EC8 was inoculated in soil, only Arabidopsis roots showed growth promotion. Variations in nutrient composition may considerably change the type and the amounts of volatiles produced in soil (46). This may explain why EC8 enhanced plant growth to a larger extent on agar medium than in a soil-sand mixture. In addition, the involvement of different perception mechanisms or nutrient absorption/degradation pathways in lettuce seedlings may explain the different phenotypes observed between Arabidopsis and lettuce. Another mechanism may be that the effects are concentration-dependent and that lettuce shoots need a higher concentration of the specific volatiles for triggering growth promotion.
VOC profiling by GC–QTOF-MS showed an enrichment of sulfur-containing compounds in the headspace of EC8, including dimethyl disulfide and dimethyl trisulfide, frequently found for other bacterial genera, and also rarer compounds, such as S-methyl 2-methylpropanethioate and S-methyl pentanethioate. Dimethyl disulfide from Bacillus sp. strain BG55 has been described to promote the growth of Nicotiana attenuata plants grown under sulfur-limiting conditions. This effect was attributed, in part, to absorption and assimilation of this VOC (12). In our study, exposure of Arabidopsis seedlings, grown under nonlimiting sulfur conditions, to dimethyl disulfide did not promote plant growth, whereas exposure to dimethyl trisulfide affected plant growth in a concentration-dependent manner. How this specific sulfur VOC from EC8 is perceived by the plant roots and which signal transduction pathways are induced, leading to growth promotion, will be addressed in future studies.
Our genome-wide transcriptome analyses further revealed that volatiles from EC8 differently regulate plant genes involved in sulfur assimilation and biosynthesis, as well as in nitrogen transport and assimilation. The processes of assimilation of nitrogen and sulfur by plants are well-coordinated and are involved in the synthesis of cysteine, an important structural and functional component of proteins and enzymes. However, the molecular mechanisms, sensors, and signals involved in this regulation are largely unknown (47). The nitrogen transporter NTR2.1, which showed a 14-fold upregulation in Arabidopsis exposed to volatiles from EC8, has been reported to be regulated by nitrate and to function as a negative regulator of lateral root initiation under high-sucrose and low-nitrate condition, whereas NRT2.6 has been reported to be involved in growth promotion of Arabidopsis by the rhizobacterium Phyllobacterium brassicacearum STM196 (48, 49). The transporter NRT2.6, together with NRT2.5, was found to be upregulated in Arabidopsis leaves inoculated with the bacteria, suggesting that these genes might be part of the regulation of the nitrogen control of root development (49). Among the genes involved in sulfur metabolism and transport, different members of the glutathione S-transferase (GST) family were found to be differentially expressed in shoot and root tissues. GSTs are ubiquitous in plants and have been suggested to be involved in herbicide detoxification and stress responses (38). However, little is known about their roles in normal plant physiology, during biotic and abiotic stress response (39), and in bacterium-plant interactions.
Previous studies have shown that microbial volatiles promote plant growth and alter plant development by modulating auxin signaling and transport in the plant (11, 15, 42, 50). Our results showed that exposure of Arabidopsis to volatiles from EC8 upregulated the expression of the auxin receptor TIR1 in both shoot and root tissues. This receptor mediates the degradation of Aux/indole-3-acetic acid (IAA) proteins and auxin-regulated transcription and, together with the Skp-, cullin-, and F-box-containing (SCF) complex′s ubiquitin ligase proteins, regulates root and hypocotyl growth, lateral root formation, and cell elongation (51). Here, we found an upregulation of nitrilase 2 in roots exposed to volatiles from EC8. However, we did not identify an enrichment of other auxin-related genes, suggesting that EC8-mediated plant growth promotion may involve other mechanisms.
Coupling the results from the VOC profiling and the bioassays with the synthetic sulfur volatile compound dimethyl trisulfide to the results from the plant transcriptome analyses suggests a modulation of sulfur metabolism and transport by EC8. In nature, inorganic sulfur is taken up by roots in the form of sulfate. However, 95% of the sulfur present in soils is bound to organic molecules (organosulfur) and is not directly available to plants. Soil microorganisms play a critical role in sulfur acquisition by catalyzing organosulfur compounds, allowing uptake by the plants (52). Although volatiles from EC8 were also found to differently regulate genes involved in nitrogen-related processes, we did not detect an enrichment of nitrogen-containing compounds in the headspace of EC8 in our analysis. However, it may be that inorganic compounds, including nitrogen compounds, which were not detected by the method used here, are produced in the headspace of strain EC8 and might contribute to plant growth promotion. An example is nitrogen oxide (NO), which has been demonstrated to interact with plant hormones, influencing several plant developmental processes, including root growth and lateral root formation (53, 54). Of note, NO produced by Azospirillum brasilense has been associated with growth promotion and induction of lateral root formation in tomato plants (55, 56).
In conclusion, volatiles produced by Microbacterium represent a new source of natural compounds for stimulation of plant growth. Priming seedlings by a short exposure to Microbacterium volatiles provides an exciting new strategy for plant growth promotion. Our analysis of the transcriptional changes induced in Arabidopsis by Microbacterium volatiles identified several differentially expressed genes. This knowledge advances our understanding of the underlying molecular mechanisms of volatile-mediated plant growth promotion and provides a basis for future experiments to validate the role of specific pathways in volatile-mediated growth promotion. Further identification of the bioactive volatiles in lab and open field conditions and characterization of their ecological functions will contribute to reveal novel mechanisms for improving crop production for sustainable agriculture and, ultimately, to minimize fertilizer inputs.
MATERIALS AND METHODS
Isolation of Microbacterium.
Microbacterium isolates were obtained from the rhizosphere (roots with adhering soil) of sugar beet plants (Beta vulgaris cv. Aligator), as previously described (17). An additional isolation method with modifications was used to isolate endophytic Microbacterium (57). For that, roots of sugar beet seedlings were rinsed with 10 ml of 10 mM MgSO4 · 7H2O at pH 7.0 (here, buffer) to remove the rhizospheric soil and washed three times with buffer supplemented with 0.01% Tween 20 (vol/vol). Subsequently, roots were surface sterilized for 2 min under slow agitation in 1% (vol/vol) sodium hypochlorite solution supplemented with 0.01% (vol/vol) Tween 20 and then rinsed five times with buffer. To confirm that the roots were sterile, treated roots were spread on Luria-Bertani (LB; Oxoid Thermo Scientific, Lenexa, KS) and 1/10th-strength tryptic soy agar (1/10th TSA; Difco, BD Laboratories, Houston, TX) plates. In addition, 100 μl of the last rinsing step solution was also plated. To separate plant from microbial cells, the surface-sterilized root tissue was disrupted using a blender. The homogenate was filtered consecutively through 25-μm and 10-μm mesh cheesecloth to remove plant tissue. The flowthrough was further cleaned by centrifugation at 500 rpm for 1 min, and the supernatant was transferred to a new tube. Bacterial cells were collected by centrifuging the supernatant at 9,500 rpm for 15 min. The pellet, consisting mainly of endophytic microorganisms, was suspended in 3.5 ml of buffer supplemented with Nycodenz resin (Progen Biotechnik, Germany) to a final concentration of 50% (wt/vol). A Nycodenz density gradient was mounted above the sample by slowly depositing various layers of Nycodenz (3 ml of 35% Nycodenz, 2 ml of 20% Nycodenz, and 2 ml of 10% Nycodenz), and the gradient was centrifuged for 45 min at 8,500 rpm (Sorvall HB-6). Endophytic bacteria, visualized as a whitish band, were recovered by pipetting. The recovered cells were washed five times with buffer and centrifuged at 13,000 rpm for 5 min in order to remove the Nycodenz resin. Finally, bacterial cells were suspended in 500 μl of buffer and recovered by quick centrifugation at 16,000 rpm. Samples were frozen in liquid nitrogen and stored at −80°C. For the isolation of single cells, 100 μl was plated on 1/10th TSA medium.
Growth conditions of the Microbacterium strains.
Microbacterium strains were grown on tryptone soy broth (Oxoid Thermo Scientific, Lenexa, KS) supplemented with 18 g of technical agar (Oxoid Thermo Scientific, Lenexa, KS) for 3 days at 21°C. Cells were obtained from the agar plates and mixed with buffer. Cell density was measured and adjusted to an optical density at 600 nm (OD600) of 1 (∼109 CFU · ml−1).
Phylogeny of Microbacterium.
To phylogenetically characterize the Microbacterium isolates, 16S rRNA genes were amplified by PCR. Amplifications were conducted using primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1392R (5′-ACGGGCGGTGTGTACA-3′) or 27F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-ACCTTGTTACGACGACTT-3′) (58, 59). For obtaining DNA, cell suspensions were prepared in Tris-EDTA (TE) buffer and centrifuged at 13,000 rpm for 10 min. After centrifugation, 2-μl volumes of the supernatants were used for the PCRs. PCR products were purified and sequenced at Macrogen, Inc. The amplified 16S rRNA gene sequences (700 to 800 bp) of the Microbacterium isolates were compared with the 16S rRNA gene sequences of Microbacterium type strains. Phylogenetic analysis using partial sequences of 16S rRNA gene, resulting from alignment of 732 sites, was performed with Muscle (60) in MEGA6 (61) (Fig. S1). A neighbor-joining consensus tree was constructed using the Tamura 3-parameter model with the optimal model parameters and the option of complete deletion of gaps and gamma distribution (62). Confidence levels for the branching points were determined using 1,000 bootstrap replicates. A total of eight Microbacterium strains were selected based on the phylogenetic distribution for testing the effects on plant growth via the production of VOCs.
Plant material.
For the in vitro assays, seeds of Arabidopsis (Arabidopsis thaliana Col-0) were surface sterilized for 3 h by placing seeds in Eppendorf tubes open in a desiccator jar. Two beakers, each containing 50 ml of sodium hypochlorite solution, were placed inside, and 1.5 ml of 37% hydrochloric acid was added to each beaker. The desiccator jar was closed, and the seeds were sterilized by chlorine gas. Eppendorf tubes containing the sterile seeds were kept open in the flow cabinet for 30 min and after that placed on a wet paper filter in a petri dish. The petri dish was sealed and wrapped in tin foil and kept at 4°C for 3 to 4 days. Seeds of lettuce (Lactuca sativa) and tomato (Solanum lycopersicum L.) were surface sterilized by soaking in 70% ethanol for 2 min, followed by soaking in 1% sodium hypochlorite solution for 20 min. After soaking, seeds were rinsed three times in sterile demineralized water. Plants were kept in climate cabinets at 21°C (180 μmol light m–2 · s–1 at plant level; 16 h:8 h, light:dark; and 70% rH).
In vitro plant growth promotion assay.
Sterile seeds of Arabidopsis, lettuce, and tomato were sown on petri dishes (diameter, 90 mm) containing 25 ml of 0.5× Murashige and Skoog (MS) medium (63) supplemented with 0.5% sucrose. These petri dishes (without lids) were kept inside a larger petri dish (diameter, 145 mm) which was sealed and kept in the climate cabinet. After 4 days, seedlings were exposed to the bacterial volatiles or to agar medium by introducing a small petri dish (diameter, 35 mm) containing a 3-day-old bacterial culture or the agar medium (control). Petri dishes (diameter, 145 mm) were sealed and kept in the climate cabinet. Plant shoot and root biomass was determined after 12 to 14, 10, and 7 days for Arabidopsis, tomato, and lettuce seedlings, respectively. To test if a short exposure to the bacterial volatiles had an effect on plant growth, seedlings were exposed in vitro using the three-compartment setup described above. Seven-day-old Arabidopsis and 3-day-old lettuce seedlings were exposed for five and 3 days, respectively, and then transplanted to soil. Plants were kept in plastic pots containing 130 g of potting soil with 40% moisture. A total of 5 to 9 replicates were used per treatment. Arabidopsis shoot biomass, number of flowers, and length of flower stems were determined 21 days after soil transplantation. Lettuce biomass was determined 13 days after soil transplantation. Data were analyzed by independent samples t test and one-way analysis of variance (ANOVA) with a Tukey's honestly significant difference (HSD) test (P < 0.05).
To test the effects of synthetic sulfur volatile compounds, 2-compartment petri dishes (diameter, 90 mm) were used (Fig. 5A). Five sterile Arabidopsis seeds were grown in one compartment containing 0.5× MS medium supplemented with 0.5% sucrose. In the second compartment, 20 μl of each different dilution (1 nM, 100 nM, 1 μM, 100 μM, 1 mM, and 100 mM) of the synthetic compounds and of the mixture was applied to a sterile filter paper (1.5 × 1.5 cm). These concentrations included those previously detected in the headspaces of different bacteria (12, 37). Compounds were diluted with dichloromethane (DCM). For controls, the second compartment was left empty or 20 μl of DCM was applied to the filter paper. Petri dishes were immediately sealed and incubated in a growth cabinet. Shoot and root biomass was determined after 2 weeks. Five biological replicates were prepared and statistical differences were determined by one-way ANOVA with Tukey's HSD test (P < 0.05).
Shoot and root exposure to bacterial volatiles.
To expose plant shoots and roots to the bacterial volatiles, two different experimental setups were used. For the exposure of plant shoots, a closed sterile container (OS140box; Duchefa Biochemie, Haarlem, the Netherlands) was used. Seedlings were sown in pots (inner diameter, 6.5 cm; height, 5 cm) containing potting soil and kept in the climate chamber. Microbacterium strain EC8 was inoculated on petri dishes containing TSA medium and incubated for 6 days at 21°C. Ten holes were made in the walls of these petri dishes to allow diffusion of the bacterial volatiles as displayed in Fig. 3A. Arabidopsis, lettuce, and tomato seedlings were exposed to the bacterial volatiles 7, 4, and 6 days after sowing, respectively. After 1 week of cocultivation, pots were kept open in the flow cabinet for 30 min to remove excess condensation on the pot walls. Plants were exposed three more days and allowed to grow for 4 days in the absence of the bacterial volatile compounds. After that, shoot biomass was determined. For the exposure of plant roots, two-compartment pots were used. Top and bottom compartments were separated by a polyester membrane (5 μm, Nedfilter, Lelystad, The Netherlands). The upper compartment (inner diameter: 5.5 cm, height: 8 cm) was filled with a potting soil-sand mixture (1:2, vol/vol; 25% moisture), where one Arabidopsis or lettuce seed was sown. The bottom compartment (inner diameter: 6.5 cm, height: 4.5 cm) was filled with the soil-sand mixture mixed with the bacterial culture (107 CFU · g−1 soil) or a petri dish (diameter, 35 mm) containing a 3-day-old bacterial culture on TSA medium (initial concentration, 109 CFU · ml−1) previously incubated at 21°C. Shoot and root biomass was determined 3 weeks after sowing. Data were analyzed by independent samples t test (P < 0.05).
VOC profiling of Microbacterium.
For profiling the VOCs produced by the Microbacterium isolates, solid-phase microextraction (SPME) with a 65-mm polydimethylsiloxane-divinylbenzene fiber (Supelco, Bellefonte, PA) was used. Isolates were inoculated (10 µl at an OD600 of 1) individually in 10-ml sterile glass vials containing 2.5 ml of TSA medium. A total of 3 replicates per treatment were used, and vials containing medium only served as the control. All vials were closed and incubated at 30°C. VOCs from the headspace of each vial were collected after 7 days. VOCs were analyzed by gas chromatography-mass spectrometry (GC-MS), and raw data were processed as previously described (17). Hierarchical cluster analysis (HCA) using Pearson's correlation coefficient with the unweighted pair group method with arithmetic mean (UPGMA) algorithm was performed with GeneMaths XT version 2.11 (Applied Maths, Belgium).
Based on the results of the plant growth-promotion assays, we decided to study the VOC profile of Microbacterium strain EC8 in detail. Bacterial cells (100 μl; OD600 = 0.1) were plated on sterile glass petri dishes (diameter, 90 mm) containing 20 μl of TSA medium. Petri dishes were sealed and incubated at 30°C. VOC collection started right after plating, and for that, the lids of these petri dishes were designed with an outlet where the Tenax tubes were connected and kept for 6 consecutive days. Trapped compounds were subjected to gas chromatography–quadrupole time of flight mass spectrometry (GC–QTOF-MS). Compounds were desorbed from the Tenax tubes in a thermodesorption unit (model UnityTD-100; Markes International Ltd., Llantrisant, UK) at 210°C for 12 min (helium flow, 50 ml · min−1) using a 1:20 split ratio. Released compounds were focused on a cold trap at −10°C and introduced into the GC–QTOF-MS apparatus (7890B GC and 7200A QTOF; Agilent, Santa Clara, CA). Compounds were transferred to the analytical column (30 m × 0.25 mm internal diameter; film thickness, 0.25 μm; RXI-5MS 13424-6850; Restek, Bellefonte, PA) by heating the cold trap to 250°C for 12 min. The temperature program of the GC oven was 39°C for 2 min, from 39°C to 95°C at 3.5°C · min−1, from 95°C to 165°C at 6°C · min−1, 165°C to 250°C at 15°C · min−1, and finally from 250°C to 300°C at 40°C · min−1, with a 20-min hold at a constant gas flow of 1.2 ml · min−1. Mass spectra were acquired by electron impact ionization (70 eV) scanning from m/z 30 to 400 with a scan rate of 4 scans · s−1.
Mass spectra were analyzed with MassHunter qualitative analysis software B.07.00 (Agilent Technologies, Santa Clara, CA) using the GC–QTOF-MS qualitative analysis module. VOCs were selected based on three criteria, namely, peak intensity of at least 104 arbitrary units (a.u.), P < 0.05 (Student's t test), and a fold change (FC) of >2. Selected VOCs were tentatively identified by comparison of the mass spectra with those of NIST (National Institute of Standards and Technology, USA) and Wiley libraries and by comparing the experimentally calculated linear retention indices (LRI) with the literature values.
Plant transcriptome analysis.
Total RNA was extracted from shoot and root tissues of Arabidopsis seedlings exposed for 1 week to the volatiles, including organic and inorganic compounds, from Microbacterium strain EC8. Seedlings exposed to TSA medium only were used as the control. For plant RNA sequencing, total RNA was extracted from roots and shoots. For each treatment, 4 replicates were used; each replicate consisted of 4 plates with 6 seedlings each in order to obtain enough biomass. RNA was obtained from frozen tissues with Trizol reagent (Invitrogen). The RNA samples were further purified using the NucleoSpin RNA II kit (Macherey-Nagel) and kept at −80°C until sequencing. For RNA sequencing, samples were processed using the NebNext Ultra directional RNA library prep kit for Illumina at ServiceXS (GenomeScan B.V., Leiden, the Netherlands). Briefly, mRNA was isolated from the total RNA using oligo(dT) magnetic beads. After fragmentation of the mRNA, cDNA was synthesized, ligated with sequencing adapters, and amplified by PCR in order to obtain cDNA libraries. Each cDNA library was individually analyzed for quality and yield using a fragment analyzer. cDNA was then clustered and a concentration of 1.6 pM was sequenced with an Illumina NextSeq 500 sequencer.
Illumina sequences were trimmed and filtered with FastQC using a threshold of 25 (quality value [Q] > 25). Quality-trimmed reads were counted using the RSEM software package (64) and transformed into reads per kilobase per million reads (RPKM). Reads were mapped to the Arabidopsis reference genes using the software Bowtie 2 v.2.1.0 (65). The Bioconductor package DESeq2 (66) was used for normalization and differential expression analyses. The P value was obtained from the differential gene expression test. False discovery rate (FDR) manipulation was used to determine the P value threshold in multiple tests and analyses. Significant differentially expressed genes (DEGs) were selected using an FDR of <0.05 and an absolute value of the log2 ratio of ≥0.585 (at least 1.5-fold higher than the expression level in the control) or ≤−0.585 (at least 1.5-fold lower than the expression level in the control) as thresholds. Biological interpretation of the DEGs was performed using Cytoscape software with the ClueGO plugin (67).
Accession number(s).
Raw RNA-seq data are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) and assigned to BioProject accession no. PRJNA492842 and BioSample accession numbers SAMN09711604 to SAMN09711619.
Supplementary Material
ACKNOWLEDGMENTS
We thank Victor de Jager for the quality check of the transcriptome data and Hans Zweers for running the samples with GC–QTOF-MS.
This is publication number 6582 from the Netherlands Institute of Ecology, NIOO-KNAW.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01865-18.
REFERENCES
- 1.Wheatley RE. 2002. The consequences of volatile organic compound mediated bacterial and fungal interactions. Antonie Van Leeuwenhoek 81:357–364. doi: 10.1023/A:1020592802234. [DOI] [PubMed] [Google Scholar]
- 2.Zoller HF, Clark WM. 1921. The production of volatile fatty acids by bacteria of the dysentery group. J Gen Physiol 3:325–330. doi: 10.1085/jgp.3.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernando WGD, Ramarathnam R, Krishnamoorthy AS, Savchuk SC. 2005. Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol Biochem 37:955–964. doi: 10.1016/j.soilbio.2004.10.021. [DOI] [Google Scholar]
- 4.Kai M, Effmert U, Berg G, Piechulla B. 2007. Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch Microbiol 187:351–360. doi: 10.1007/s00203-006-0199-0. [DOI] [PubMed] [Google Scholar]
- 5.Epstein L, Lockwood JL. 1984. Effect of soil microbiota on germination of Bipolaris victoriae conidia. Trans Br Mycol Soc 82:63–69. doi: 10.1016/S0007-1536(84)80212-0. [DOI] [Google Scholar]
- 6.Hora TS, Baker R. 1972. Extraction of a volatile factor from soil-inducing fungistasis. Phytopathology 62:1475–1476. doi: 10.1094/Phyto-62-1475. [DOI] [Google Scholar]
- 7.Dobbs CG, Hinson WH. 1953. A widespread fungistasis in soils. Nature 172:197–199. doi: 10.1038/172197a0. [DOI] [PubMed] [Google Scholar]
- 8.Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW. 2003. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci U S A 100:4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Pare PW. 2004. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026. doi: 10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vespermann A, Kai M, Piechulla B. 2007. Rhizobacterial volatiles affect the growth of fungi and Arabidopsis thaliana. Appl Environ Microbiol 73:5639–5641. doi: 10.1128/AEM.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS, Pare PW. 2007. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839–851. doi: 10.1007/s00425-007-0530-2. [DOI] [PubMed] [Google Scholar]
- 12.Meldau DG, Meldau S, Hoang LH, Underberg S, Wunsche H, Baldwin IT. 2013. Dimethyl disulfide produced by the naturally associated bacterium Bacillus sp B55 promotes Nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell 25:2731–2747. doi: 10.1105/tpc.113.114744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garbeva P, Hordijk C, Gerards S, de Boer W. 2014. Volatiles produced by the mycophagous soil bacterium Collimonas. FEMS Microbiol Ecol 87:639–649. doi: 10.1111/1574-6941.12252. [DOI] [PubMed] [Google Scholar]
- 14.Cordero P, Principe A, Jofre E, Mori G, Fischer S. 2014. Inhibition of the phytopathogenic fungus Fusarium proliferatum by volatile compounds produced by Pseudomonas. Arch Microbiol 196:803–809. doi: 10.1007/s00203-014-1019-6. [DOI] [PubMed] [Google Scholar]
- 15.Bailly A, Groenhagen U, Schulz S, Geisler M, Eberl L, Weisskopf L. 2014. The inter-kingdom volatile signal indole promotes root development by interfering with auxin signalling. Plant J 80:758–771. doi: 10.1111/tpj.12666. [DOI] [PubMed] [Google Scholar]
- 16.Blom D, Fabbri C, Connor EC, Schiestl FP, Klauser DR, Boller T, Eberl L, Weisskopf L. 2011. Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ Microbiol 13:3047–3058. doi: 10.1111/j.1462-2920.2011.02582.x. [DOI] [PubMed] [Google Scholar]
- 17.Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, van Wezel GP, Raaijmakers JM. 2015. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front Microbiol 6:1081. doi: 10.3389/fmicb.2015.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernández-León R, Rojas-Solís D, Contreras-Pérez M, Orozco-Mosqueda MDC, Macías-Rodríguez LI, Reyes-de la Cruz H, Valencia-Cantero E, Santoyo G. 2015. Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol Control 81:83–92. doi: 10.1016/j.biocontrol.2014.11.011. [DOI] [Google Scholar]
- 19.Insam H, Seewald MSA. 2010. Volatile organic compounds (VOCs) in soils. Biol Fertil Soils 46:199–213. doi: 10.1007/s00374-010-0442-3. [DOI] [Google Scholar]
- 20.Ryu C-M. 2015. Bacterial volatiles as airborne signals for plants and bacteria, p 53–61. In Lugtenberg B. (ed), Principles of plant-microbe interactions: microbes for sustainable agriculture. Springer International Publishing, Cham, Switzerland [Google Scholar]
- 21.Lemanceau P, Corberand T, Gardan L, Latour X, Laguerre G, Boeufgras J, Alabouvette C. 1995. Effect of two plant species, flax (Linum usitatissinum L.) and tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl Environ Microbiol 61:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grayston SJ, Wang S, Campbell CD, Edwards AD. 1998. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem 30:369–378. doi: 10.1016/S0038-0717(97)00124-7. [DOI] [Google Scholar]
- 23.Stackebrandt E, Rainey FA, Ward-Rainey NL. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Evol Microbiol 47:479–491. [Google Scholar]
- 24.Lee JS, Lee KC, Park YH. 2006. Microbacterium koreense sp. nov., from sea water in the South Sea of Korea. Int J Syst Evol Microbiol 56:423–427. doi: 10.1099/ijs.0.63854-0. [DOI] [PubMed] [Google Scholar]
- 25.Anand S, Bala K, Saxena A, Schumann P, Lal R. 2012. Microbacterium amylolyticum sp. nov., isolated from soil from an industrial waste site. Int J Syst Evol Microbiol 62:2114–2120. doi: 10.1099/ijs.0.034439-0. [DOI] [PubMed] [Google Scholar]
- 26.Karojet S, Kunz S, van Dongen JT. 2012. Microbacterium yannicii sp. nov., isolated from Arabidopsis thaliana roots. Int J Syst Evol Microbiol 62:822–826. doi: 10.1099/ijs.0.026955-0. [DOI] [PubMed] [Google Scholar]
- 27.Sharma P, Diene SM, Thibeaut S, Bittar F, Roux V, Gomez C, Reynaud-Gaubert M, Rolain JM. 2013. Phenotypic and genotypic properties of Microbacterium yannicii, a recently described multidrug resistant bacterium isolated from a lung transplanted patient with cystic fibrosis in France. BMC Microbiol 13:97. doi: 10.1186/1471-2180-13-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soto-Rodriguez SA, Cabanillas-Ramos J, Alcaraz U, Gomez-Gil B, Romalde JL. 2013. Identification and virulence of Aeromonas dhakensis, Pseudomonas mosselii and Microbacterium paraoxydans isolated from Nile tilapia, Oreochromis niloticus, cultivated in Mexico. J Appl Microbiol 115:654–662. doi: 10.1111/jam.12280. [DOI] [PubMed] [Google Scholar]
- 29.Cogan TM, Goerges S, Gelsomino R, Larpin S, Hohenegger M, Bora N, Jamet E, Rea MC, Mounier J, Vancanneyt M, Gueguen M, Desmasures N, Swings J, Goodfellow M, Ward AC, Sebastiani H, Irlinger F, Chamba JF, Beduhn R, Scherer S. 2014. Biodiversity of the surface microbial consortia from Limburger, Reblochon, Livarot, Tilsit, and Gubbeen cheeses. Microbiol Spectr 2:Cm-0010–2012. doi: 10.1128/microbiolspec.CM-0010-2012. [DOI] [PubMed] [Google Scholar]
- 30.Qian F, An L, Wang M, Li C, Li X. 2007. Isolation and characterization of a xanthan-degrading Microbacterium sp. strain XT11 from garden soil. J Appl Microbiol 102:1362–1371. doi: 10.1111/j.1365-2672.2006.03215.x. [DOI] [PubMed] [Google Scholar]
- 31.Sheng XF, He LY, Zhou L, Shen YY. 2009. Characterization of Microbacterium sp. F10a and its role in polycyclic aromatic hydrocarbon removal in low-temperature soil. Can J Microbiol 55:529–535. doi: 10.1139/w09-005. [DOI] [PubMed] [Google Scholar]
- 32.Corretto E, Antonielli L, Sessitsch A, Kidd P, Weyens N, Brader G. 2015. Draft genome sequences of 10 Microbacterium spp., with emphasis on heavy metal-contaminated environments. Genome Announc 3:e00432-15. doi: 10.1128/genomeA.00432-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim EJ, Fathoni A, Jeong GT, Jeong HD, Nam TJ, Kong IS, Kim JK. 2013. Microbacterium oxydans, a novel alginate- and laminarin-degrading bacterium for the reutilization of brown-seaweed waste. J Environ Manage 130:153–159. doi: 10.1016/j.jenvman.2013.08.064. [DOI] [PubMed] [Google Scholar]
- 34.Kim KK, Park HY, Park W, Kim IS, Lee ST. 2005. Microbacterium xylanilyticum sp. nov., a xylan-degrading bacterium isolated from a biofilm. Int J Syst Evol Microbiol 55:2075–2079. doi: 10.1099/ijs.0.63706-0. [DOI] [PubMed] [Google Scholar]
- 35.Madhaiyan M, Poonguzhali S, Lee JS, Lee KC, Saravanan VS, Santhanakrishnan P. 2010. Microbacterium azadirachtae sp. nov., a plant-growth-promoting actinobacterium isolated from the rhizoplane of neem seedlings. Int J Syst Evol Microbiol 60:1687–1692. doi: 10.1099/ijs.0.015800-0. [DOI] [PubMed] [Google Scholar]
- 36.Barnett SJ, Roget DK, Ryder MH. 2006. Suppression of Rhizoctonia solani AG-8 induced disease on wheat by the interaction between Pantoea, Exiguobacterium, and Microbacteria. Aust J Soil Res 44:331. doi: 10.1071/SR05113. [DOI] [Google Scholar]
- 37.Farag MA, Ryu CM, Sumner LW, Pare PW. 2006. GC-MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochemistry 67:2262–2268. doi: 10.1016/j.phytochem.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 38.Wagner U, Edwards R, Dixon DP, Mauch F. 2002. Probing the diversity of the arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49:515–532. doi: 10.1023/A:1015557300450. [DOI] [PubMed] [Google Scholar]
- 39.Nutricati E, Miceli A, Blando F, De Bellis L. 2006. Characterization of two Arabidopsis thaliana glutathione S-transferases. Plant Cell Rep 25:997–1005. doi: 10.1007/s00299-006-0146-1. [DOI] [PubMed] [Google Scholar]
- 40.Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M. 1997. Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9:783–798. doi: 10.1105/tpc.9.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kai M, Piechulla B. 2009. Plant growth promotion due to rhizobacterial volatiles—an effect of CO2? FEBS Lett 583:3473–3477. doi: 10.1016/j.febslet.2009.09.053. [DOI] [PubMed] [Google Scholar]
- 42.Cordovez V, Mommer L, Moisan K, Lucas-Barbosa D, Pierik R, Mumm R, Carrion VJ, Raaijmakers JM. 2017. Plant phenotypic and transcriptional changes induced by volatiles from the fungal root pathogen Rhizoctonia solani. Front Plant Sci 8:1262. doi: 10.3389/fpls.2017.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie X, Zhang H, Pare PW. 2009. Sustained growth promotion in Arabidopsis with long-term exposure to the beneficial soil bacterium Bacillus subtilis (GB03). Plant Signal Behav 4:948–953. doi: 10.4161/psb.4.10.9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelberth J, Alborn HT, Schmelz EA, Tumlinson JH. 2004. Airborne signals prime plants against insect herbivore attack. Proc Natl Acad Sci U S A 101:1781–1785. doi: 10.1073/pnas.0308037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heil M, Silva Bueno JC. 2007. Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc Natl Acad Sci U S A 104:5467–5472. doi: 10.1073/pnas.0610266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheatley RE, Millar SE, Griffiths DW. 1996. The production of volatile organic compounds during nitrogen transformations in soils. Plant and Soil 181:163–167. doi: 10.1007/BF00011303. [DOI] [Google Scholar]
- 47.Kruse C, Jost R, Lipschis M, Kopp B, Hartmann M, Hell R. 2007. Sulfur-enhanced defence: effects of sulfur metabolism, nitrogen supply, and pathogen lifestyle. Plant Biol (Stuttg) 9:608–619. doi: 10.1055/s-2007-965432. [DOI] [PubMed] [Google Scholar]
- 48.Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. 2005. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci U S A 102:13693–13698. doi: 10.1073/pnas.0504219102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kechid M, Desbrosses G, Rokhsi W, Varoquaux F, Djekoun A, Touraine B. 2013. The NRT2.5 and NRT2.6 genes are involved in growth promotion of Arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain Phyllobacterium brassicacearum STM196. New Phytol 198:514–524. doi: 10.1111/nph.12158. [DOI] [PubMed] [Google Scholar]
- 50.Bitas V, McCartney N, Li N, Demers J, Kim JE, Kim HS, Brown KM, Kang S. 2015. Fusarium oxysporum volatiles enhance plant growth via affecting auxin transport and signaling. Front Microbiol 6:1248. doi: 10.3389/fmicb.2015.01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 52.Kertesz MA, Mirleau P. 2004. The role of soil microbes in plant sulphur nutrition. J Exp Bot 55:1939–1945. doi: 10.1093/jxb/erh176. [DOI] [PubMed] [Google Scholar]
- 53.Stöhr C, Stremlau S. 2006. Formation and possible roles of nitric oxide in plant roots. J Exp Bot 57:463–470. doi: 10.1093/jxb/erj058. [DOI] [PubMed] [Google Scholar]
- 54.Freschi L. 2013. Nitric oxide and phytohormone interactions: current status and perspectives. Front Plant Sci 4:398. doi: 10.3389/fpls.2013.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Creus CM, Graziano M, Casanovas EM, Pereyra MA, Simontacchi M, Puntarulo S, Barassi CA, Lamattina L. 2005. Nitric oxide is involved in the Azospirillum brasilense-induced lateral root formation in tomato. Planta 221(2):297–303. doi: 10.1007/s00425-005-1523-7. [DOI] [PubMed] [Google Scholar]
- 56.Molina-Favero C, Creus CM, Simontacchi M, Puntarulo S, Lamattina L. 2008. Aerobic nitric oxide production by Azospirillum brasilense Sp245 and its influence on root architecture in tomato. Mol Plant Microbe Interact 21:1001–1009. doi: 10.1094/MPMI-21-7-1001. [DOI] [PubMed] [Google Scholar]
- 57.Ikeda S, Kaneko T, Okubo T, Rallos LE, Eda S, Mitsui H, Sato S, Nakamura Y, Tabata S, Minamisawa K. 2009. Development of a bacterial cell enrichment method and its application to the community analysis in soybean stems. Microb Ecol 58:703–714. doi: 10.1007/s00248-009-9566-0. [DOI] [PubMed] [Google Scholar]
- 58.Lane DJ. 1991. 16S/23S rRNA sequencing. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 59.DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK. 2009. Selective progressive response of soil microbial community to wild oat roots. ISME J 3:168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 60.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 63.Murashige T, Skoog F. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 64.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:1–16. doi: 10.1186/1471-2105-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.