In this study, an enzyme for imidazoline formation in pseudochelin biosynthesis was identified. Evidence for the involvement of this enzyme in the postulated reaction was obtained after in vitro reconstitution. Furthermore, the function of this enzyme was demonstrated in vivo by transferring the corresponding gene into the bacterium Myxococcus xanthus, which thereby became a producer of pseudochelin A. In addition to clarifying the molecular basis of imidazoline formation in siderophore biosynthesis, we describe the heterologous expression of a gene in a myxobacterium without chromosomal integration. Due to its metabolic proficiency, M. xanthus represents an interesting alternative to established host systems for the reconstitution and manipulation of biosynthetic pathways. Since the plasmid used in this study is easily adaptable for the expression of other enzymes as well, we expand the conventional expression strategy for myxobacteria, which is based on the integration of biosynthetic genes into the host genome.
KEYWORDS: Myxococcus xanthus, pseudochelin, myxochelin, expression plasmid, imidazoline synthase, biosynthesis, myxobacteria
ABSTRACT
Myxobacteria utilize the catechol natural products myxochelin A and B in order to maintain their iron homeostasis. Recently, the production of these siderophores, along with a new myxochelin derivative named pseudochelin A, was reported for the marine bacterium Pseudoalteromonas piscicida S2040. The latter derivative features a characteristic imidazoline moiety, which was proposed to originate from an intramolecular condensation reaction of the β-aminoethyl amide group in myxochelin B. To identify the enzyme catalyzing this conversion, we compared the myxochelin regulons of two myxobacterial strains that produce solely myxochelin A and B with those of P. piscicida S2040. This approach revealed a gene exclusive to the myxochelin regulon in P. piscicida S2040, coding for an enzyme of the amidohydrolase superfamily. To prove that this enzyme is indeed responsible for the postulated conversion, the reaction was reconstituted in vitro using a hexahistidine-tagged recombinant protein made in Escherichia coli, with myxochelin B as the substrate. To test the production of pseudochelin A under in vivo conditions, the amidohydrolase gene was cloned into the myxobacterial plasmid pZJY156 and placed under the control of a copper-inducible promoter. The resulting vector was introduced into the myxobacterium Myxococcus xanthus DSM 16526, a native producer of myxochelin A and B. Following induction with copper, the myxobacterial expression strain was found to synthesize small quantities of pseudochelin A. Replacement of the copper-inducible promoter with the constitutive pilA promoter led to increased production levels in M. xanthus, which facilitated the isolation and subsequent structural verification of the heterologously produced compound.
IMPORTANCE In this study, an enzyme for imidazoline formation in pseudochelin biosynthesis was identified. Evidence for the involvement of this enzyme in the postulated reaction was obtained after in vitro reconstitution. Furthermore, the function of this enzyme was demonstrated in vivo by transferring the corresponding gene into the bacterium Myxococcus xanthus, which thereby became a producer of pseudochelin A. In addition to clarifying the molecular basis of imidazoline formation in siderophore biosynthesis, we describe the heterologous expression of a gene in a myxobacterium without chromosomal integration. Due to its metabolic proficiency, M. xanthus represents an interesting alternative to established host systems for the reconstitution and manipulation of biosynthetic pathways. Since the plasmid used in this study is easily adaptable for the expression of other enzymes as well, we expand the conventional expression strategy for myxobacteria, which is based on the integration of biosynthetic genes into the host genome.
INTRODUCTION
Iron is indispensable for microbial viability. It acts as a component of several enzymes and enzymatic complexes and hence is essentially involved in various biological processes, such as respiration, amino acid synthesis, oxygen transport, and DNA biosynthesis. Although iron is abundant in nature, the metal exists predominantly in an insoluble and biologically inaccessible ferric (Fe3+) form under aerobic conditions and at physiological pH. To facilitate the uptake of ferric iron, many bacteria resort to low-molecular-weight iron-chelating compounds, so-called siderophores. These molecules are secreted from the cells in order to solubilize and to capture ferric iron. Subsequently, the ferric iron-siderophore complex is actively transported back into the bacterial cytoplasm and the coordinated ion is released by either reductive or hydrolytic mechanisms (1–3).
Myxobacteria are Gram-negative soil inhabitants. They possess an unusually complex life cycle, which involves differentiation from individual cells into multicellular fruiting bodies under starvation conditions, accompanied by sophisticated social behavior (4–6). In recent years, myxobacteria have gained increasing attention due to their enormous potential as producers of secondary metabolites with antifungal, antibacterial, and cytotoxic activities (7–9). To maintain their iron homeostasis, many myxobacteria produce catechol siderophores, such as the myxochelins (10–13) or the structurally related hyalachelins (14). Although the myxochelins are generally regarded as myxobacterial siderophores, they were occasionally also described for other bacteria. Examples include the actinomycete Nonomuraea pusilla (15) and the Chloroflexi bacterium Herpetosiphon aurantiacus (16).
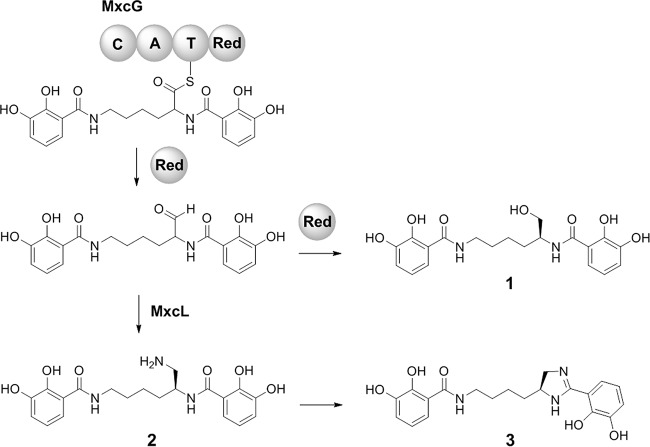
The biosynthesis of myxochelins has been thoroughly explored both on the genetic level (17) and on the biochemical level (11, 18–20). Briefly, a nonribosomal peptide synthetase (NRPS) termed MxcG links two 2,3-dihydroxybenzoate (DHBA) units with the amino groups of l-lysine. The resulting thioester intermediate is reduced to an aldehyde by the reductase domain of MxcG and thereby is released from the enzyme complex (Fig. 1). A subsequent reduction reaction forms myxochelin A (compound 1), whereas the transamination of the aldehyde, catalyzed by an aminotransferase (MxcL), leads to myxochelin B (compound 2). Recently, a novel myxochelin derivative termed pseudochelin A (compound 3) was isolated from the marine bacterium Pseudoalteromonas piscicida S2040 (21). Pseudochelin A features a characteristic 4,5-dihydroimidazole moiety, putatively emerging from an intramolecular cyclization reaction of myxochelin B (Fig. 1). In this study, the genetic basis of this biosynthetic reaction was determined by comparative genomics and heterologous reconstitution. Among others, Myxococcus xanthus strains that are able to produce pseudochelin A were engineered. For this, myxobacterial expression vectors were created.
FIG 1.
Late steps in the biosynthesis of myxochelin A (compound 1) and myxochelin B (compound 2), as well as pseudochelin A (compound 3). The NRPS MxcG consists of an N-terminal condensation (C) domain, followed by an adenylation (A), a thiolation (T), and a C-terminal reductase (Red) domain.
RESULTS
Organization of the myxochelin regulon in P. piscicida S2040.
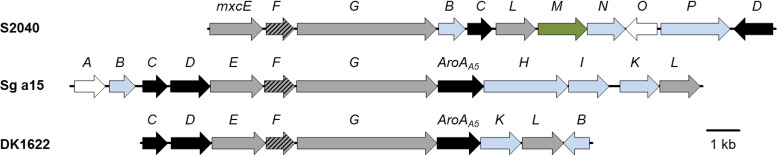
The myxobacteria Stigmatella aurantiaca Sg a15 and M. xanthus DK 1622 are known producers of myxochelin A and B (12, 17). Furthermore, the myxochelin regulon of S. aurantiaca Sg a15 has already been comprehensively analyzed (17); it comprises 12 genes, of which 3 (mxcE, mxcF, and mxcG) were shown previously to be sufficient for the assembly of myxochelin A from DHBA and l-lysine, while the aminotransferase gene mxcL is further needed for myxochelin B biosynthesis (11, 18). MxcF represents a bifunctional enzyme. It features an aryl carrier protein domain for tethering the activated DHBA building block during myxochelin assembly. Beyond that, its isochorismatase domain is involved in the conversion of chorismate into DHBA in conjunction with two other enzymes, MxcC and MxcD (17). The corresponding set of six core biosynthesis genes (mxcC to mxcG and mxcL) is also present in the myxochelin regulon of M. xanthus DK 1622, which consists of nine genes in total. Using the nucleotide sequences of the conserved biosynthesis genes as probes, a putative myxochelin locus was located in the genome of the pseudochelin producer P. piscicida S2040 by homology-driven alignment (Fig. 2). The corresponding regulon covers a contiguous DNA sequence of about 17 kb and includes 11 genes (GenBank accession numbers TW75_RS20420 to TW75_RS20470), which were renamed in this study according to the nomenclature used in previous publications (17–19). Seven of these genes have homologs in the myxochelin regulons of S. aurantiaca Sg a15 and M. xanthus DK 1622 (Table 1). In addition to the six myxochelin biosynthesis genes, P. piscicida S2040 contains a homolog of mxcB, coding for a NADPH-dependent ferric siderophore reductase. The latter removes the metal from an iron-bound myxochelin following its cellular uptake. Four genes (mxcM, mxcN, mxcO, and mxcP) are exclusively present in the regulon of P. piscicida S2040. The mxcN and mxcP open reading frames are predicted to be involved in siderophore transport and uptake, whereas mxcO encodes a regulator of the AraC family. Therefore, a role in the formation of pseudochelin is conceivable only for mxcM, the product of which was annotated as cytosine deaminase (22).
FIG 2.
Organization of the myxochelin regulons in P. piscicida S2040, S. aurantiaca Sg a15, and M. xanthus DK 1622. Genes involved in DHBA and myxochelin biosynthesis are highlighted in black and gray, respectively. The hatched mxcF is required during both processes. Transport genes are depicted in blue. Regulatory and accessory genes are marked in white. The candidate gene for pseudochelin formation is highlighted in green.
TABLE 1.
Distribution and putative function of myxochelin biosynthesis genes
| Gene | Putative function |
S. aurantiaca Sg a15 vs P. piscicida S2040a |
M. xanthus DK 1622 vs P. piscicida S2040a |
||
|---|---|---|---|---|---|
| % Id | % Qc | % Id | % Qc | ||
| mxcE | 2,3-DHBA-AMP ligase | 51 | 98 | 50 | 98 |
| mxcF | Isochorismatase/aryl carrier protein | 44 | 97 | 44 | 97 |
| mxcG | Nonribosomal peptide synthetase | 37 | 98 | 38 | 99 |
| mxcB | Ferric siderophore reductase | 37 | 90 | 36 | 86 |
| mxcC | 2,3-Dihydro-2,3-DHBA dehydrogenase | 50 | 96 | 50 | 95 |
| mxcL | Aldehyde aminotransferase | 58 | 93 | 62 | 93 |
| mxcM | Cytosine deaminase | — | — | — | — |
| mxcN | Major facilitator superfamily transporter | — | — | — | — |
| mxcO | AraC-type regulator | — | — | — | — |
| mxcP | TonB-dependent receptor | — | — | — | — |
| mxcD | Isochorismate synthase | 36 | 93 | 36 | 92 |
Similarities at the amino acid level for the corresponding enzymes in P. piscicida S2040, S. aurantiaca Sg a15, and M. xanthus DK 1622 are indicated. Id, identical amino acids; Qc, query cover; —, no homolog detected.
Bioinformatic analysis of MxcM.
A sequence analysis revealed that MxcM belongs to the amidohydrolase superfamily, whose members usually catalyze metal-dependent hydrolysis reactions (23, 24). One representative of this enzyme family, the imidazolonepropionase HutI, is known to cleave an imidazoline ring during histidine degradation (25). The reverse reaction, an intramolecular condensation, is reminiscent of the assumed formation of pseudochelin A. While sequence alignments of MxcM with different HutI homologs did not indicate significant similarities (see Table S1 in the supplemental material), a manual inspection showed that the active site residues of HutI (26, 27) are conserved in MxcM (Fig. S1A). However, the E252 residue of HutI, which seems necessary to initiate the hydrolysis reaction (26, 28), is replaced by a proline residue in MxcM. The closest homolog of MxcM with an assigned function is the amidohydrolase CbxE from Streptomyces sp. strain NTK937 (Fig. S1B and Table S2). This enzyme was found to be crucially involved in the biosynthesis of the benzoxazole antibiotic caboxamycin, and it was proposed to catalyze a two-step intramolecular cyclization and dehydration reaction, leading to the formation of an oxazoline ring (29). It is evident that the mechanism of this biosynthetic conversion is analogous to the formation of the imidazoline ring in pseudochelin A.
In vitro characterization of MxcM.
To verify the role of MxcM in pseudochelin A biosynthesis, the candidate gene was synthesized, cloned into pET28a, and overexpressed in Escherichia coli BL21(DE3) for the production of a 6×His-tagged protein. This recombinant enzyme was purified to homogeneity by immobilized metal affinity chromatography and was subsequently assayed with fermentation-derived myxochelin B (Fig. 3). High-performance liquid chromatography (HPLC) analysis showed the emergence of a new peak in the enzyme reaction, for which the high-resolution mass spectrometry (MS) and tandem mass spectrometry (MS/MS) data were consistent with pseudochelin A (21). When myxochelin B was incubated without 6×His-MxcM, no further peak was detected. Furthermore, the testing of synthetic myxochelin A (30) with 6×His-MxcM did not provide any evidence for an enzymatic conversion (Fig. 3).
FIG 3.
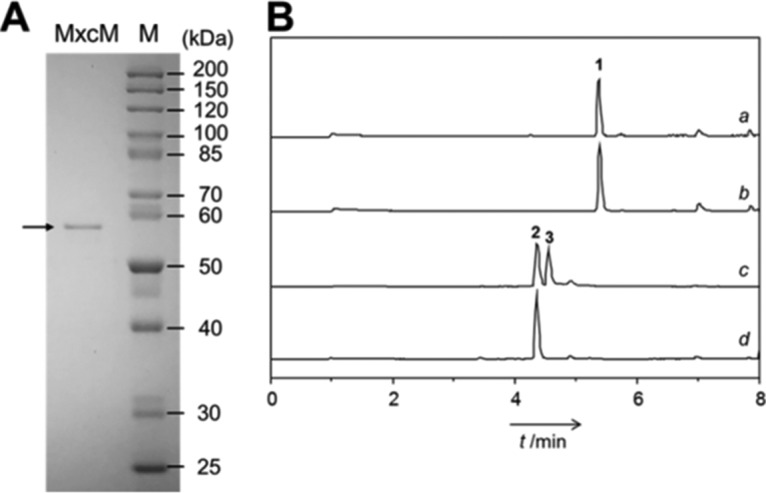
(A) SDS-PAGE of purified 6×His-MxcM. The calculated molecular mass of the recombinant protein is 58.786 kDa. Lane M, protein ladder. (B) Results of the in vitro testing of 6×His-MxcM. Total ion chromatograms of myxochelin A (compound 1) with 6×His-MxcM (profile a), myxochelin A (compound 1) without 6×His-MxcM (profile b), myxochelin B (compound 2) with 6×His-MxcM (profile c), and myxochelin B (compound 2) without 6×His-MxcM (profile d) are shown. The additional peak in profile c corresponds to pseudochelin A (compound 3).
Construction of a vector for inducible expression of mxcM in M. xanthus.
To demonstrate the functionality of MxcM under in vivo conditions, the corresponding gene was expressed in M. xanthus DSM 16526. This myxobacterial strain had been confirmed previously to synthesize myxochelin B and showed no signs of pseudochelin A production. For the gene transfer, a replicative expression plasmid was created. The respective vector was derived from pZJY156, which had been demonstrated to replicate stably in M. xanthus as well as in E. coli (31, 32). A codon-optimized version of the target gene mxcM from P. piscicida S2040 was cloned into the vector and placed under the control of the copper-inducible promoter PcuoA. The expression of this promoter is known to correlate linearly with the copper concentration in the culture medium (33, 34). The final expression vector, pJK3 (Fig. 4A), was introduced into M. xanthus DSM 16526 via electroporation (35, 36). Transformants were selected by their acquired kanamycin resistance and, after several rounds of subcultivation, were analyzed by colony PCR to validate the presence of free, nonintegrated plasmid. The annealing sites of the primers used were located adjacent to each other inside the kanamycin resistance gene, thus leading to amplification of the entire pJK3 plasmid. Separation of the PCR products by horizontal electrophoresis revealed the expected product size of 8.9 kb (Fig. 4B). If any vector linearization had occurred (e.g., by integration of pJK3 into the host genome), then no PCR products would have been obtained. Ultimate evidence for the existence of autonomous plasmid DNA was obtained by sequencing (see Table S3 in the supplemental material).
FIG 4.
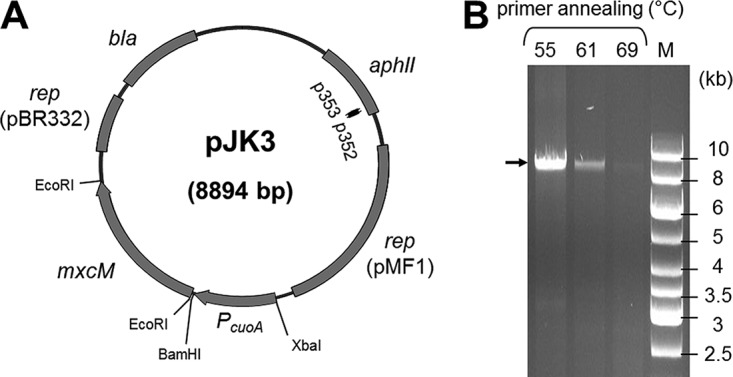
(A) Schematic map of the expression plasmid pJK3. Binding sites and the orientation of primers p352 and p353 are displayed as black arrows. (B) PCR-based confirmation of pJK3. A gradient PCR using biomass from the M. xanthus expression strain as the template was performed in order to amplify pJK3 with primers p352 and p353. The expected amplicon is marked with an arrow. Lane M, DNA ladder.
Copper-induced expression of the mxcM gene in M. xanthus DSM 16526.
Prior to the expression experiment, we evaluated the effect of the inducing agent (CuSO4) on myxochelin production in the M. xanthus DSM 16526 wild-type strain. Since the liquid chromatography (LC)-MS profiles of supplemented and nonsupplemented cultures showed no differences, an impact of CuSO4 on myxochelin production could be ruled out. Furthermore, it was verified that myxochelin production was not affected in an M. xanthus DSM 16526 strain carrying the empty expression vector (data not shown). Comparison of the extracts of wild-type and expression strains, after induction with 115 μM CuSO4, by LC-MS measurements combined with MS/MS analyses indicated the presence of pseudochelin A exclusively in the extract of the expression strain (Fig. 5). MS/MS spectra of the putative pseudochelin A peak and an authentic myxochelin A standard both showed a Δ136 fragment (myxochelin A, m/z 405 → m/z 269; pseudochelin A, m/z 386 → m/z 250), which can be attributed to the loss of a 2,3-dihydroxybenzaldehyde moiety. The elimination of a 2,3-dihydroxybenzamide moiety was also observed (pseudochelin A, m/z 386 → m/z 233). These observations are in accordance with previous reports (21, 37). A distinctive Δ18 fragment of myxochelin A arises from the dehydration of its primary alcohol function in the lysinol residue. The same structural motif is not present in pseudochelin A, which supports the MS-based assignment.
FIG 5.
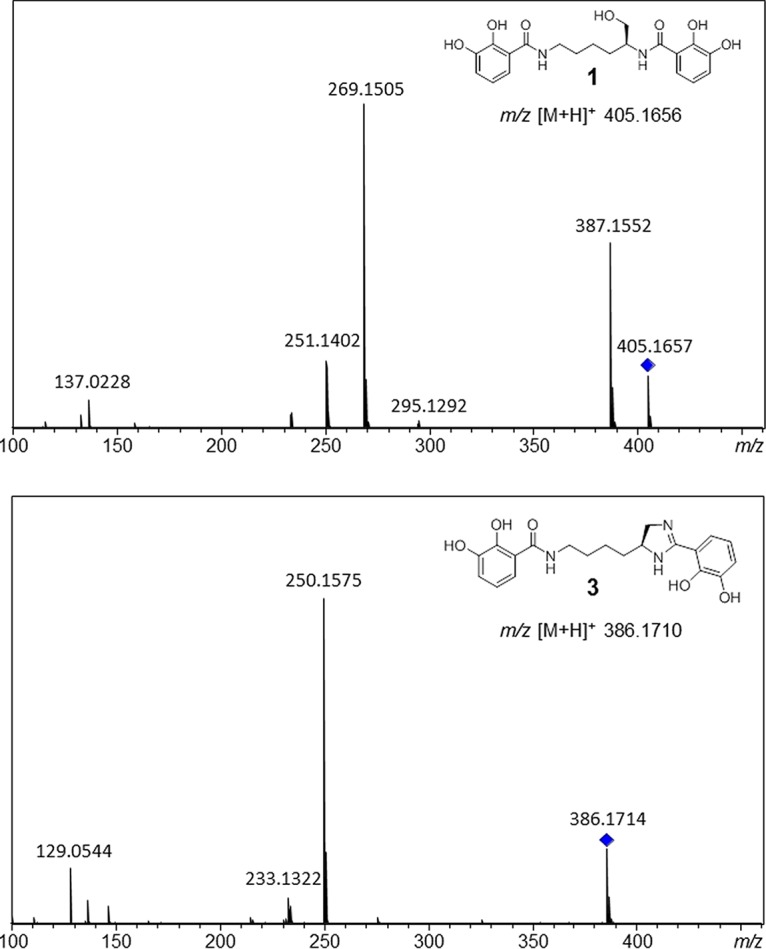
MS/MS fragmentation patterns of myxochelin A (compound 1) and pseudochelin A (compound 3). Parent ions are indicated with diamonds.
Constitutive expression of the mxcM gene in M. xanthus DSM 16526.
To obtain unequivocal evidence that the mxcM gene is involved in the biosynthesis of pseudochelin A, we set out to confirm the identity of the heterologously produced compound by nuclear magnetic resonance (NMR) spectroscopy. However, only trace amounts of pseudochelin A could be recovered from the copper-inducible expression strain (data not shown). To evaluate whether the low production level could be overcome, PcuoA was replaced with the constitutive promoter PpilA (38), resulting in the generation of the expression vector pJK5 (Fig. S2). Transformation of M. xanthus DSM 16526 with this plasmid led to a strain exhibiting increased pseudochelin A titers. A total of 95 mg of pseudochelin A could be isolated from a 6-liter culture. NMR spectroscopic analyses of this material verified the identity of pseudochelin A (Fig. S3 to S7).
DISCUSSION
Pseudochelin A is a catechol siderophore that is structurally closely related to the myxochelins (10, 13) and hyalachelins (14). In this study, we identified a gene cluster for pseudochelin A biosynthesis in P. piscicida S2040 and we demonstrated that a single enzyme, MxcM, is needed to provide a myxochelin-producing bacterium the potential for pseudochelin biosynthesis.
From a biological perspective, the chemical diversity of myxochelin-type siderophores is intriguing. To date, however, it is unclear whether the minor structural differences between these compounds have important consequences, e.g., in terms of iron affinity or siderophore uptake. Biosynthetic genes for the production of myxochelin-type molecules are found in several bacterial lineages, according to an analysis of genomic data. The occurrence of siderophore pathways in taxonomically distinct bacteria is not without precedence, as exemplified by the yersiniabactin locus (39). This wide distribution is usually attributed to lateral gene transfer and a strong evolutionary driving force to maintain the newly acquired biosynthesis genes (39). As expected, myxobacteria predominate among the myxochelin producers (see Fig. S8 in the supplemental material). The myxobacterial producer strains were isolated from different locations around the globe, which suggests that myxochelin biosynthesis arose quite early in this bacterial group. In contrast, very few actinomycetes were identified as myxochelin producers. Since myxobacteria and actinomycetes are both soil-inhabiting microorganisms, it is possible that some actinomycetes acquired the myxochelin biosynthesis genes by horizontal gene transfer. This would explain the restricted occurrence of the myxochelin regulon in this taxon.
Most myxobacteria that were analyzed possess the mxcL gene, but they lack an imidazoline synthase gene. Therefore, they are assumed to synthesize myxochelin B. Few bacteria (including 4 myxobacterial strains) were predicted to produce myxochelin A exclusively. The capacity for pseudochelin formation, specified by the presence of the amidohydrolase-encoding gene mxcM, seems to be restricted to members of the Gammaproteobacteria, particularly to members of the orders Alteromonadales, Oceanospirillales, and Vibrionales (Fig. S8). Considering the marine origin of the corresponding species (21, 40–43), the production of pseudochelin, compared to the myxochelins, might have an ecological advantage in aqueous environments. It is noteworthy in this context that the genomes of marine myxobacteria (44) are devoid of myxochelin biosynthesis genes.
Interestingly, homologs of MxcM are encoded not only in myxochelin-type pathways. The amidohydrolase CbxE from Streptomyces sp. strain NTK937 is involved in the biosynthesis of the antibiotic caboxamycin (29). While CbxE catalyzes the formation of an oxazoline ring, MxcM is responsible for imidazoline ring formation, putatively following an analogous reaction mechanism. Another homolog of MxcM was found in Streptomyces megasporus strain NRRL B-16372 (NCBI Protein accession number WP_031508998). The corresponding open reading frame is embedded in a gene cluster bearing remarkable similarity to the loci for antibiotic A33853 (45, 46) and nataxazole (47, 48) biosynthesis, which are found in the genomes of Streptomyces sp. strain NRRL 12068 and Streptomyces sp. strain Tü 6176, respectively.
In this study, the function of MxcM was probed in vitro as well as in vivo by heterologous expression in M. xanthus. This myxobacterium not only is a model organism for the analysis of bacterial motility, development, and predation (6, 49, 50) but also represents an attractive host for the reconstitution of biosynthetic pathways and genes. Of particular note in this context is its inherent proficiency in natural product assembly, including the allocation of a variety of biosynthetic building blocks (51). Although these properties are common to many fruiting myxobacteria, M. xanthus is further distinguished by its comparatively short generation time (52, 53) and its susceptibility to genetic manipulations (35, 36). Therefore, it is no surprise that M. xanthus has already served as host for the production of several secondary metabolites, including the myxobacterial compounds epothilone (53), myxothiazol (54), myxochromid (55), myxopyronin (56), corallopyronin (56), and vioprolide (57) and the actinobacterial polyketide oxytetracycline from Streptomyces rimosus (58). Hitherto, chromosomal integration of target genes was considered the method of choice for the engineering of M. xanthus expression strains. To this end, selected genes or entire biosynthetic gene clusters were incorporated into the chromosome, either by targeted homologous recombination or by random transpositional integration (53–58). The discovery of the first myxobacterial plasmid, pMF1, and the subsequent availability of a stable shuttle vector (31) set the stage for a plasmid-based expression system applicable in M. xanthus. Until now, however, pZJY156-derived vectors were utilized only during complementation studies, in order to restore certain gene functions in null mutants (31, 59).
In contrast to chromosomal integration, multicopy plasmids in general favor higher expression rates, due to manifold availability of the gene sequence. Therefore, it was expected that the pJK3-based expression of mxcM in M. xanthus would provide sufficient protein for the conversion of myxochelin B to pseudochelin A, especially since the wild-type strain had been identified as an efficient producer of myxochelin B. Nevertheless, only traces of pseudochelin A were detected in the corresponding strain. To test whether the copper-inducible expression affected the production of pseudochelin A, a promoter exchange was carried out. The resulting pJK5 strain, in which mxcM was constitutively expressed under the control of the PpilA promoter, produced appreciable quantities of pseudochelin A. A possible explanation for the low productivity of the pJK3 strain might be that the expression of mxcM started too late. In that case, most of the biosynthesized myxochelin B would already have been secreted and the enzymatic conversion would have suffered from a lack of substrate in the myxobacterial cells. Although the tight copper dependency and functionality of the PcuoA promoter had been demonstrated previously in M. xanthus, it had also been noted that the promoter reached its greatest activity 24 to 48 h after copper addition (34). The delayed response of the PcuoA promoter and consequently the delayed formation of active enzyme inside the host, combined with the active efflux of the siderophore, would also provide an explanation for the observed accumulation of myxochelin B in the culture broth of the pJK3 strain.
In summary, the previously proposed enzymatic conversion of myxochelin B into pseudochelin A could be verified, and the amidohydrolase involved, MxcM, was identified. Furthermore, heterologous production of pseudochelin A in the nonnative host M. xanthus was achieved using a plasmid-based expression system. The capacity for myxochelin biosynthesis was shown to be much more widespread than reported previously, and pseudochelin A biosynthesis could be traced back uniquely to members of Gammaproteobacteria.
MATERIALS AND METHODS
Strains and growth conditions.
M. xanthus DSM 16526 wild-type and expression strains were routinely cultured in modified CTPM medium (1.0% [wt/vol] casitone, 0.12% [wt/vol] Tris, 0.02% [wt/vol] KH2PO4, 0.06% [wt/vol] MgSO4·7H2O, and 0.00005% [wt/vol] vitamin B12 [pH 7.5]) (60). Cultivation was conducted at 30°C, and liquid cultures were shaken at 130 rpm. Escherichia coli DH5α and E. coli BL21(DE3) were grown in standard lysogeny broth (LB) (Carl Roth) at 37°C, with agitation at 200 rpm. Antibiotics were used, when necessary, at the following concentrations: 100 μg/ml ampicillin, and 50 μg/ml kanamycin.
Analytical methods.
HPLC experiments were conducted with a Shimadzu HPLC system (LC-20AD system with SPD-M20A detector). LC-MS and LC-MS/MS analyses were performed with an Agilent 1260 Infinity HPLC system combined with a Compact quadrupole-time of flight (Q-TOF) mass spectrometer (Bruker Daltonics). The Q-TOF mass spectrometer was interfaced with an electrospray ionization source. All analyses were performed in positive ion mode using a capillary potential of 4.5 kV. The desolvation gas (N2) temperature was 220°C, with a flow rate of 12 liters/min. Compounds were infused into the electrospray source by the 1260 Infinity HPLC system at a flow rate of 0.4 ml/min. The protonated molecular ions of test compounds were generated at an end plate offset of 500 V. Nitrogen was used as the collision gas, at a pressure of 5 mPa. MS/MS experiments were performed in multiple reaction monitoring (MRM) mode, with a collision gas energy of 23 eV. NMR spectra were recorded on a Bruker AV 600-MHz Avance III HD system (CryoProbe), with methanol-d4 as the solvent and internal standard. The solvent signals were referenced to δH 3.31 ppm and δC 49.0 ppm.
Production and purification of myxochelin B.
For the production of myxochelin B, M. xanthus DSM 16526 was grown for 5 days in modified CTPM medium supplemented with DHBA. The biosynthetic precursor was added as a filter-sterilized solution (final concentration, 30 mg/liter) prior to the inoculation of M. xanthus. To facilitate the recovery of the siderophore, the myxobacteria were grown in the presence of 2% (wt/vol) Amberlite XAD7HP resin (Sigma-Aldrich). At the end of cultivation, the adsorber resin was collected by filtration, washed with water, and extracted exhaustively with methanol. Isolation of myxochelin B from the methanol extract was accomplished with two consecutive reverse-phase HPLC steps. The initial separation was carried out on a Nucleodur C18 gravity column (250 by 10 mm, 3 μm; Macherey-Nagel), using a linear gradient of methanol in water supplemented with 0.1% (vol/vol) trifluoroacetic acid (10% to 100% methanol within 15 min, followed by 100% methanol for an additional 10 min), at a flow rate of 2.5 ml/min. Final purification of myxochelin B was achieved on a Nucleodur Sphinx RP column (250 by 10 mm, 5 μm; Macherey-Nagel), with identical eluents and flow rate but a different gradient (25% to 48% methanol over 27 min, 48% to 75% methanol within 8 min, and 75% methanol for 5 min). The elution of compounds was monitored with a diode array detector.
Plasmids, nucleic acids, and protein sequences.
Plasmids used in this study are listed in Table 2. The native mxcM gene can be found in GenBank under the locus tag TW75_RS20450 (GenBank protein accession number WP_045965770). The shuttle vector pZJY156 and the codon-adapted version of the P. piscicida mxcM gene were synthesized by ATG:biosynthetics. The codon adaptation of mxcM was carried out manually, referring to the codon usage of M. xanthus deposited in the Kazusa database (61). Nucleotide sequences of P. piscicida S2040, Stigmatella aurantiaca Sg a15, and Myxococcus xanthus DK 1622 were retrieved from GenBank (accession numbers NZ_JXXW00000000, AF299336, and NC_008095, respectively) . Homologies of nucleotide sequences were evaluated using the Basic Local Alignment Search Tool (BLAST) (62). Protein sequences of imidazolonepropionases were also obtained from GenBank. Alignments were performed with the open access software GeneStudio Pro.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| pET28a(+) | E. coli expression vector; Kmr | Novagen |
| pET28a(+)-mxcM | pET28a(+) carrying synthetic mxcM gene; Kmr | This study |
| pJET1.2 | E. coli cloning vector; Ampr | Thermo Fisher Scientific |
| pJK1 | pJET1.2 carrying 825-bp promoter region PcuoA from DSM 16526; Ampr | This study |
| pJK2 | Subcloning vector, pZJY156-mxcM; Kmr | This study |
| pJK3 | M. xanthus expression vector, pZJY156-PcuoA-mxcM; Kmr | This study |
| pJK4 | pJET1.2 carrying 207-bp promoter region PpilA from DSM 16526; Ampr | This study |
| pJK5 | M. xanthus expression vector, pZJY156-PpilA-mxcM; Kmr | This study |
| pZJY156 | M. xanthus-E. coli shuttle vector; Ampr, Kmr | 31, 32 |
Kmr, kanamycin resistance; Ampr, ampicillin resistance.
General DNA methods.
Genomic DNA was extracted from M. xanthus DSM 16526 following a previously published protocol (63). Plasmids from E. coli DH5α were isolated using a Roti-Prep plasmid minikit (Carl Roth). DNA fragments from agarose electrophoresis gels were extracted with the QIAquick gel extraction kit (Qiagen). Restriction digestions were performed using FastDigest restriction enzymes (Thermo Fisher Scientific). Ligation of DNA fragments was achieved using a T4 DNA ligase (Thermo Fisher Scientific). If required, plasmid DNA was dephosphorylated, using the FastAP thermosensitive alkaline phosphatase (Thermo Fisher Scientific), prior to ligation. Subcloning was performed in E. coli DH5α using standard protocols.
Production and purification of recombinant 6×His-MxcM.
The synthetic mxcM gene was cloned into the EcoRI restriction site of pET28a(+). The resulting expression plasmid, pET28a(+)-mxcM, was transferred into chemically competent E. coli BL21(DE3). For the production of 6×His-tagged MxcM, the expression strain was cultivated at 37°C in terrific broth medium (12.0% [wt/vol] tryptone/peptone, 24.0% [wt/vol] yeast extract, 0.4% [vol/vol] glycerol, 2.3% [wt/vol] KH2PO4, and 12.5% [wt/vol] K2HPO4), to an optical density at 600 nm (OD600) of 0.6. After induction with isopropyl-β-d-1-thiogalactopyranoside (IPTG) (final concentration, 1 mM), the incubation was continued at 16°C for 20 h. Cells were harvested by centrifugation (4,000 rpm for 15 min at 4°C) and resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 10 mM imidazole, 300 mM NaCl, and 10% glycerol). After cell disruption by ultrasonic treatment, the cell debris was removed by centrifugation (4,500 rpm for 30 min at 4°C). Purification of 6×His-labeled MxcM from the supernatant was achieved by affinity chromatography using Protino Ni-nitrilotriacetic acid (NTA)-agarose (Macherey-Nagel) as the stationary phase, in a polypropylene column (Qiagen). The recombinant protein was eluted using lysis buffer containing 250 mM imidazole. After desalting with a PD-10 desalting column (GE Healthcare Life Sciences) and concentration with an Amicon centrifugal filter unit (Ultra-15, with a molecular weight cutoff value of 10 kDa; Merck Millipore), 6×His-MxcM was kept in 0.1 M phosphate buffer (pH 7.0).
In vitro testing of 6×His-MxcM.
Hexahistidine-tagged MxcM (final concentration, 2 μM) was incubated at 30°C with myxochelin A or B (0.5 mM) and 50 mM Tris-HCl buffer (pH 7.0), in a total reaction volume of 0.5 ml. As controls, the substrate and/or the recombinant enzyme was omitted. After 1 h, the reaction was stopped by the addition of 15% (vol/vol) acetonitrile. Product formation was analyzed by LC-MS measurements.
Construction of the myxobacterial expression vectors pJK3 and pJK5.
The promoter regions PcuoA and PpilA were amplified from M. xanthus DSM 16526 genomic DNA using the Phusion Flash high-fidelity PCR master mix (Thermo Fisher Scientific). The primers p357 (5′-GCATGGATCCGAAGCCTCTTCACGAATGGATGG-3′) and p358 (5′-GAGCTCTAGATGTCGGCCATGAACGGCACTTCACG-3′), carrying BamHI and XbaI restriction sites, respectively, were used to amplify the region 825 bp upstream of the cuoA gene (locus tag MXAN_3420). The PCR was conducted with the following conditions: 98°C for 10 s; 30 cycles of 98°C for 1 s, 64°C for 5 s, and 72°C for 12 s; and 72°C for 1 min. The amplicon was ligated into pJET1.2 to yield pJK1. Subsequently, the synthetic gene mxcM, in its codon-adapted form, was ligated into the EcoRI site of pZJY156 in order to create pJK2. The PcuoA sequence was excised from pJK1 using BamHI and XbaI. The vector pJK2 was also digested with BamHI and XbaI and the backbone fragment was ligated with the promoter sequence to generate the expression vector pJK3. To amplify the region 207 bp upstream of the pilA gene (locus tag MXAN_5783), we used the primers p493 (5′-GGATCCGGGAGCGCTTCGGATGCGTAG-3′) and p494 (5′-GGTACCGGGGGTCCTCAGAGAAGG-3′), which introduced BamHI and KpnI restriction sites into the PCR product. The PCR was conducted as described before. The resulting amplicon was subcloned into pJET1.2 to yield pJK4; after digestion, the corresponding DNA fragment was ligated into the BamHI-KpnI site of pJK2 to generate the expression vector pJK5.
Generation of the M. xanthus expression strains.
Transformation of M. xanthus DSM 16526 was achieved by electroporation, according to previously published protocols (35, 36). To verify the successful uptake of the plasmids pJK3 and pJK5, the M. xanthus transformants were grown for 4 to 5 days at 30°C on solid CTPM medium supplemented with kanamycin. Biomass was collected and used as the template for colony PCR. For this purpose, cells were suspended in MilliQ water and incubated for 3 min at 99°C. The PCR was performed using DreamTaq DNA polymerase (Thermo Fisher Scientific). The optimum primer annealing was examined using a temperature gradient from 55°C to 69°C. The PCR was conducted under the following conditions: 95°C for 3 min; 30 cycles of 95°C for 45 s, 55°C/61°C/69°C for 30 s, and 72°C for 8 min; and final extension at 72°C for 10 min. Primers p352 (5′-GCTGACCGCTTCCTCGTGCTTTACG-3′) and p353 (5′-CCGCCAAGCTCTTCAGCAATATCACGGG-3′) were used in order to amplify the entire pJK3 (size, 8.9 kb) or pJK5 (size, 8.3 kb) plasmid. The corresponding PCR products were purified and used as the templates for subsequent PCRs in order to amplify the kanamycin resistance gene aphII with primers p268 (5′-GGATTGCACGCAGGTTCTCCGG-3′) and p269 (5′-CGATAGAAGGCGATGCGCTGCG-3′) and the pMF1-based origin of replication with primers p342 (5′-CTGGTGCCTGTCGTCATGGAGAC-3′) and p343 (5′-CACGCACACCGACGAGTCATAG-3′). The pBR332-based origin of replication was amplified together with the ampicillin resistance gene bla using the primers p344 (5′-GCTCAAGTCAGAGGTGGCGAAACC-3′) and p345 (5′-GCGAGACGAAAGGGCCTCGTGATACGCC-3′). The PCR products were purified and used as the templates for sequencing reactions (Microsynth Seqlab).
Heterologous production and isolation of pseudochelin A from recombinant M. xanthus.
In order to maintain the pJK3 and pJK5 vectors, cultures of the M. xanthus expression strains were grown in the presence of kanamycin. Expression of the target gene in pJK3 was induced by the addition of copper(II) sulfate (115 μM). After 4 days of incubation, the culture broths were extracted with ethyl acetate, and pseudochelin A production was verified by high-resolution MS and MS/MS analyses. Myxochelin A served as a control during these experiments. For the isolation of pseudochelin A, the M. xanthus strain harboring pJK5 was incubated for 4 days in modified CTPM medium supplemented with 2,3-DHBA. To assist recovery of the metabolite, cultivation was carried out in the presence of 2% (wt/vol) Amberlite XAD7HP resin (Sigma-Aldrich). At the end of fermentation, the adsorber resin was collected by filtration, washed with water, and exhaustively eluted with methanol. Isolation of pseudochelin A from the methanol extract was accomplished by two consecutive reverse-phase HPLC steps. The first separation was achieved on a Nucleodur 100-5 C18 ec VarioPrep column (125 by 21 mm, 5 μm; Macherey-Nagel), using a linear gradient of methanol in water supplemented with 0.1% (vol/vol) trifluoroacetic acid (15% to 60% methanol within 20 min, 60% to 100% over 5 min, and 100% methanol for an additional 7 min), at a flow rate of 8 ml/min. Final purification of pseudochelin A was achieved on the same column with identical eluents and flow rate but under isocratic conditions, using a mobile phase consisting of 32% methanol. The elution of compounds was monitored with a diode array detector.
Accession number(s).
The nucleotide sequence of the synthetic mxcM gene was deposited in GenBank under accession number MH248775.
Supplementary Material
ACKNOWLEDGMENTS
Financial support from the Bundesministerium für Bildung und Forschung within the program InfectControl 2020 (grant 03ZZ0808A) is gratefully acknowledged.
We thank Anna Tippelt for valuable discussions and Katharina Kuhr and Chantale Zammarelli (TU Dortmund University) for conducting MS and MS/MS measurements. We thank Karin Martin and Florian Baldeweg (Leibniz Institute for Natural Product Research and Infection Biology) for technical support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01789-18.
REFERENCES
- 1.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Saha R, Saha N, Donofrio RS, Bestervelt LL. 2013. Microbial siderophores: a mini review. J Basic Microbiol 53:303–317. doi: 10.1002/jobm.201100552. [DOI] [PubMed] [Google Scholar]
- 3.Kurth C, Kage H, Nett M. 2016. Siderophores as molecular tools in medical and environmental applications. Org Biomol Chem 14:8212–8227. doi: 10.1039/C6OB01400C. [DOI] [PubMed] [Google Scholar]
- 4.Velicer GJ, Vos M. 2009. Sociobiology of the myxobacteria. Annu Rev Microbiol 63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 5.Cao P, Dey A, Vassallo CN, Wall D. 2015. How myxobacteria cooperate. J Mol Biol 427:3709–3721. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bretl DJ, Kirby JR. 2016. Molecular mechanisms of signaling in Myxococcus xanthus development. J Mol Biol 428:3805–3830. doi: 10.1016/j.jmb.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Schäberle TF, Lohr F, Schmitz A, König GM. 2014. Antibiotics from myxobacteria. Nat Prod Rep 31:953–972. doi: 10.1039/c4np00011k. [DOI] [PubMed] [Google Scholar]
- 8.Korp J, Vela Gurovic MS, Nett M. 2016. Antibiotics from predatory bacteria. Beilstein J Org Chem 12:594–607. doi: 10.3762/bjoc.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann J, Fayad AA, Müller R. 2017. Natural products from myxobacteria: novel metabolites and bioactivities. Nat Prod Rep 34:135–160. doi: 10.1039/C6NP00106H. [DOI] [PubMed] [Google Scholar]
- 10.Kunze B, Bedorf N, Kohl W, Höfle G, Reichenbach H. 1989. Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales): production, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 42:14–17. doi: 10.7164/antibiotics.42.14. [DOI] [PubMed] [Google Scholar]
- 11.Gaitatzis N, Kunze B, Müller R. 2005. Novel insights into siderophore formation in myxobacteria. ChemBioChem 6:365–374. doi: 10.1002/cbic.200400206. [DOI] [PubMed] [Google Scholar]
- 12.Krug D, Zurek G, Revermann O, Vos M, Velicer GJ, Müller R. 2008. Discovering the hidden secondary metabolome of Myxococcus xanthus: a study of intraspecific diversity. Appl Environ Microbiol 74:3058–3068. doi: 10.1128/AEM.02863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schieferdecker S, König S, Koeberle A, Dahse HM, Werz O, Nett M. 2015. Myxochelins target human 5-lipoxygenase. J Nat Prod 78:335–338. doi: 10.1021/np500909b. [DOI] [PubMed] [Google Scholar]
- 14.Nadmid S, Plaza A, Lauro G, Garcia R, Bifulco G, Müller R. 2014. Hyalachelins A–C, unusual siderophores isolated from the terrestrial myxobacterium Hyalangium minutum. Org Lett 16:4130–4133. doi: 10.1021/ol501826a. [DOI] [PubMed] [Google Scholar]
- 15.Miyanaga S, Obata T, Onaka H, Fujita T, Saito N, Sakurai H, Saiki I, Furumai T, Igarashi Y. 2006. Absolute configuration and antitumor activity of myxochelin A produced by Nonomuraea pusilla TP-A0861. J Antibiot (Tokyo) 59:698–703. doi: 10.1038/ja.2006.93. [DOI] [PubMed] [Google Scholar]
- 16.Kiss H, Nett M, Domin N, Martin K, Maresca JA, Copeland A, Lapidus A, Lucas S, Berry KW, Glavina Del Rio T, Dalin E, Tice H, Pitluck S, Richardson P, Bruce D, Goodwin L, Han C, Detter JC, Schmutz J, Brettin T, Land M, Hauser L, Kyrpides NC, Ivanova N, Göker M, Woyke T, Klenk HP, Bryant DA. 2011. Complete genome sequence of the filamentous gliding predatory bacterium Herpetosiphon aurantiacus type strain (114-95T). Stand Genomic Sci 5:356–370. doi: 10.4056/sigs.2194987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silakowski B, Kunze B, Nordsiek G, Blöcker H, Höfle G, Müller R. 2000. The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur J Biochem 267:6476–6485. doi: 10.1046/j.1432-1327.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 18.Gaitatzis N, Kunze B, Müller R. 2001. In vitro reconstitution of the myxochelin biosynthesis machinery of Stigmatella aurantiaca Sg a15: biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc Natl Acad Sci U S A 98:11136–11141. doi: 10.1073/pnas.201167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Weissman KJ, Müller R. 2008. Myxochelin biosynthesis: direct evidence for two- and four-electron reduction of a carrier protein-bound thioester. J Am Chem Soc 130:7554–7555. doi: 10.1021/ja8025278. [DOI] [PubMed] [Google Scholar]
- 20.Korp J, König S, Schieferdecker S, Dahse H-M, König GM, Werz O, Nett M. 2015. Harnessing enzymatic promiscuity in myxochelin biosynthesis for the production of 5-lipoxygenase inhibitors. ChemBioChem 16:2445–2450. doi: 10.1002/cbic.201500446. [DOI] [PubMed] [Google Scholar]
- 21.Sonnenschein EC, Stierhof M, Goralczyk S, Vabre FM, Pellissier L, Hanssen KØ, de la Cruz M, Díaz C, de Witte P, Copmans D, Andersen JH, Hansen E, Kristoffersen V, Tormo JR, Ebel R, Milne BF, Deng H, Gram L, Jaspars M, Tabudravu JN. 2017. Pseudochelin A, a siderophore of Pseudoalteromonas piscicida S2040. Tetrahedron 73:2633–2637. doi: 10.1016/j.tet.2017.03.051. [DOI] [Google Scholar]
- 22.Machado H, Sonnenschein EC, Melchiorsen J, Gram L. 2015. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genomics 16:158. doi: 10.1186/s12864-015-1365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm H, Sander C. 1997. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins 28:72–82. doi:. [DOI] [PubMed] [Google Scholar]
- 24.Seibert CM, Raushel FM. 2005. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 25.Michal G. 1999. Biochemical pathways: an atlas of biochemistry and molecular biology. John Wiley & Sons, New York, NY. [Google Scholar]
- 26.Yu Y, Liang YH, Brostromer E, Quan JM, Panjikar S, Dong YH, Su XD. 2006. A catalytic mechanism revealed by the crystal structures of the imidazolonepropionase from Bacillus subtilis. J Biol Chem 281:36929–36936. doi: 10.1074/jbc.M607703200. [DOI] [PubMed] [Google Scholar]
- 27.Tyagi R, Eswaramoorthy S, Burley SK, Raushel FM, Swaminathan S. 2008. A common catalytic mechanism for proteins of HutI family. Biochemistry 47:5608–5615. doi: 10.1021/bi800180g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su H, Sheng X, Liu Y. 2016. Exploring the substrate specificity and catalytic mechanism of imidazolonepropionase (HutI) from Bacillus subtilis. Phys Chem Chem Phys 18:27928–27938. doi: 10.1039/C6CP04918D. [DOI] [PubMed] [Google Scholar]
- 29.Losada AA, Cano-Prieto C, García-Salcedo R, Braña AF, Méndez C, Salas JA, Olano C. 2017. Caboxamycin biosynthesis pathway and identification of novel benzoxazoles produced by cross-talk in Streptomyces sp. NTK 937. Microb Biotechnol 10:873–885. doi: 10.1111/1751-7915.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schieferdecker S, Nett M. 2016. A fast and efficient method for the preparation of the 5-lipoxygenase inhibitor myxochelin A. Tetrahedron Lett 57:1359–1360. doi: 10.1016/j.tetlet.2016.02.047. [DOI] [Google Scholar]
- 31.Zhao JY, Zhong L, Shen MJ, Xia ZJ, Cheng QX, Sun X, Zhao GP, Li YZ, Qin ZJ. 2008. Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl Environ Microbiol 74:1980–1987. doi: 10.1128/AEM.02143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng J, Chen XJ, Sun X, Wang N, Li YZ. 2012. Characterization of the replication origin of the myxobacterial self-replicative plasmid pMF1. Plasmid 68:105–112. doi: 10.1016/j.plasmid.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Sánchez-Sutil MC, Gómez-Santos N, Moraleda-Muñoz A, Martins LO, Pérez J, Muñoz-Dorado J. 2007. Differential expression of the three multicopper oxidases from Myxococcus xanthus. J Bacteriol 189:4887–4898. doi: 10.1128/JB.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez-Santos N, Treuner-Lange A, Moraleda-Muñoz A, García-Bravo E, García-Hernández R, Martínez-Cayuela M, Pérez J, Søgaard-Andersen L, Muñor-Dorado J. 2012. Comprehensive set of integrative plasmid vectors for copper-inducible gene expression in Myxococcus xanthus. Appl Environ Microbiol 78:2515–2521. doi: 10.1128/AEM.07502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashefi K, Hartzell PL. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol Microbiol 15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 36.Youderian P, Burke N, White DJ, Hartzell PL. 2003. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol 49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Choi JN, Kim P, Sok DE, Nam SW, Lee CH. 2009. LC-MS/MS profiling-based secondary metabolite screening of Myxococcus xanthus. J Microbiol Biotechnol 19:51–54. [PubMed] [Google Scholar]
- 38.Wu SS, Kaiser D. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol 179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greule A, Marolt M, Deubel D, Peintner I, Zhang S, Jessen-Trefzer C, De Ford C, Burschel S, Li S-M, Friedrich T, Merfort I, Lüdeke S, Bisel P, Müller M, Paululat T, Bechthold A. 2017. Wide distribution of foxicin biosynthetic gene clusters in Streptomyces strains: an unusual secondary metabolite with various properties. Front Microbiol 8:221. doi: 10.3389/fmicb.2017.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmström C, Kjelleberg S. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol 30:285–293. doi: 10.1016/S0168-6496(99)00063-X. [DOI] [PubMed] [Google Scholar]
- 41.Lee HK, Chun J, Moon EY, Ko SH, Lee DS, Lee HS, Bae KS. 2001. Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int J Syst Evol Microbiol 51:661–666. doi: 10.1099/00207713-51-2-661. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Zhang Z, Wang Y, Zheng Y, Zhang XH. 2016. Enterovibrio pacificus sp. nov., isolated from seawater, and emended descriptions of Enterovibrio coralii and the genus Enterovibrio. Int J Syst Evol Microbiol 66:319–325. doi: 10.1099/ijsem.0.000721. [DOI] [PubMed] [Google Scholar]
- 43.Amin AKMR, Tanaka M, Al-Saari N, Feng G, Mino S, Ogura Y, Hayashi T, Meirelles PM, Thompson FL, Gomez-Gil B, Sawabe T, Sawabe T. 2017. Thaumasiovibrio occultus gen. nov. sp. nov. and Thaumasiovibrio subtropicus sp. nov. within the family Vibrionaseae, isolated from coral reed seawater off Ishigaki Island, Japan. Syst Appl Microbiol 40:290–296. doi: 10.1016/j.syapm.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Dávila-Céspedes A, Hufendiek P, Crüsemann M, Schäberle TF, König GM. 2016. Marine-derived myxobacteria of the suborder Nannocystineae: an underexplored source of structurally intriguing and biologically active metabolites. Beilstein J Org Chem 12:969–984. doi: 10.3762/bjoc.12.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michel KH, Boeck LD, Hoehn MM, Jones ND, Chaney MO. 1984. The discovery, fermentation, and structure of antibiotic A33853 and its tetraacetyl derivative. J Antibiot (Tokyo) 37:441–445. doi: 10.7164/antibiotics.37.441. [DOI] [PubMed] [Google Scholar]
- 46.Lv M, Zhao J, Deng Z, Yu Y. 2015. Characterization of the biosynthetic gene cluster of benzoxazole antibiotic A33853 reveals unusual assembly logic. Chem Biol 22:1313–1324. doi: 10.1016/j.chembiol.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Sommer PS, Almeida RC, Schneider K, Beil W, Süssmuth RD, Fiedler HP. 2008. Nataxazole, a new benzoxazole derivative with antitumor activity produced by Streptomyces sp. Tü 6176. J Antibiot (Tokyo) 61:683–686. doi: 10.1038/ja.2008.97. [DOI] [PubMed] [Google Scholar]
- 48.Cano-Prieto C, García-Salcedo R, Sánchez-Hidalgo M, Braña AF, Fiedler HP, Méndez C, Salas JA, Olano C. 2015. Genome mining of Streptomyces sp. Tü 6176: characterization of the nataxazole biosynthetic pathway. ChemBioChem 16:1461–1473. doi: 10.1002/cbic.201500153. [DOI] [PubMed] [Google Scholar]
- 49.Zusman DR, Scott AE, Yang Z, Kirby JR. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol 5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 50.Keane R, Berleman J. 2016. The predatory life cycle of Myxococcus xanthus. Microbiology 162:1–11. doi: 10.1099/mic.0.000208. [DOI] [PubMed] [Google Scholar]
- 51.Wenzel SC, Müller R. 2009. Myxobacteria: ‘microbial factories’ for the production of bioactive secondary bioactive secondary metabolites. Mol Biosyst 5:567–574. doi: 10.1039/b901287g. [DOI] [PubMed] [Google Scholar]
- 52.Dworkin M. 1962. Nutritional requirements for vegetative growth of Myxococcus xanthus. J Bacteriol 84:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Julien B, Shah S. 2002. Heterologous expression of epothilone biosynthetic genes in Myxococcus xanthus. Antimicrob Agents Chemother 46:2772–2778. doi: 10.1128/AAC.46.9.2772-2778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perlova O, Fu J, Kuhlmann S, Krug D, Stewart AF, Zhang Y, Müller R. 2006. Reconstitution of the myxothiazol biosynthetic gene cluster by Red/ET recombination and heterologous expression in Myxococcus xanthus. Appl Environ Microbiol 72:7485–7494. doi: 10.1128/AEM.01503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu J, Wenzel SC, Perlova O, Wang J, Gross F, Tang Z, Yin Y, Stewart AF, Müller R, Zhang Y. 2008. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Res 36:e113. doi: 10.1093/nar/gkn499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sucipto H, Pogorev D, Luxenberger E, Wenzel SC, Müller R. 2017. Heterologous production of myxobacterial α-pyrone antibiotics in Myxococcus xanthus. Metab Eng 44:160–170. doi: 10.1016/j.ymben.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Yan F, Auerbach D, Chai Y, Keller L, Tu Q, Hüttel S, Glemser A, Grab HA, Bach T, Zhang Y, Müller R. 2018. Biosynthesis and heterologous production of vioprolides: rational biosynthetic engineering and unprecedented 4-methylazetidine-carboxylic acid formation. Angew Chem Int Ed Engl 57:8754–8759. doi: 10.1002/anie.201802479. [DOI] [PubMed] [Google Scholar]
- 58.Stevens DC, Henry MR, Murphy KA, Boddy CN. 2010. Heterologous expression of the oxytetracycline biosynthetic pathway in Myxococcus xanthus. Appl Environ Microbiol 76:2681–2683. doi: 10.1128/AEM.02841-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhat S, Zhu X, Patel RP, Orlando R, Shimkets LJ. 2011. Identification and localization of Myxococcus xanthus porins and lipoproteins. PLoS One 6:e27475. doi: 10.1371/journal.pone.0027475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hartzell PL. 1997. Complementation of sporulation and motility defects in prokaryote by a eukaryotic GTPase. Proc Natl Acad Sci U S A 94:9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakamura Y, Gojobori T, Ikemura T. 1997. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res 25:244–245. doi: 10.1093/nar/25.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 63.Neumann B, Pospiech A, Schairer HU. 1992. Rapid isolation of genomic DNA from Gram-negative bacteria. Trends Genet 8:332–333. doi: 10.1016/0168-9525(92)90269-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.