Phanerochaete chrysosporium is a white-rot fungus whose metabolism of lignin, aromatic pollutants, and lipids has been most extensively studied. This fungus harbors 154 cytochrome P450-encoding genes in the genome. As evidenced in this study, P. chrysosporium CYP505D6, a fused protein of P450 and its reductase, hydroxylates fatty alcohols (C9–15) and fatty acids (C9–15) at the ω-1 to ω-7 or ω-1 to ω-6 positions, respectively. Naphthalene and 1-naphthol were also hydroxylated, indicating that the substrate specificity of CYP505D6 is broader than those of the known fused proteins CYP102A1 and CYP505A1. The substrate versatility of CYP505D6 makes this enzyme an attractive candidate for biotechnological applications.
KEYWORDS: CYP505D6, CYP102A1, self-sufficient P450, Phanerochaete chrysosporium, white-rot fungus
ABSTRACT
The activity of a self-sufficient cytochrome P450 enzyme, CYP505D6, from the lignin-degrading basidiomycete Phanerochaete chrysosporium was characterized. Recombinant CYP505D6 was produced in Escherichia coli and purified. In the presence of NADPH, CYP505D6 used a series of saturated fatty alcohols with C9–18 carbon chain lengths as the substrates. Hydroxylation occurred at the ω-1 to ω-6 positions of such substrates with C9–15 carbon chain lengths, except for 1-dodecanol, which was hydroxylated at the ω-1 to ω-7 positions. Fatty acids were also substrates of CYP505D6. Based on the sequence alignment, the corresponding amino acid of Tyr51, which is located at the entrance to the active-site pocket in CYP102A1, was Val51 in CYP505D6. To understand the diverse hydroxylation mechanism, wild-type CYP505D6 and its V51Y variant and wild-type CYP102A1 and its Y51V variant were generated, and the products of their reaction with dodecanoic acid were analyzed. Compared with wild-type CYP505D6, its V51Y variant generated few products hydroxylated at the ω-4 to ω-6 positions. The products generated by wild-type CYP102A1 were hydroxylated at the ω-1 to ω-4 positions, whereas its Y51V variant generated ω-1 to ω-7 hydroxydodecanoic acids. These observations indicated that Val51 plays an important role in determining the regiospecificity of fatty acid hydroxylation, at least that at the ω-4 to ω-6 positions. Aromatic compounds, such as naphthalene and 1-naphthol, were also hydroxylated by CYP505D6. These findings highlight a unique broad substrate spectrum of CYP505D6, rendering it an attractive candidate enzyme for the biotechnological industry.
IMPORTANCE Phanerochaete chrysosporium is a white-rot fungus whose metabolism of lignin, aromatic pollutants, and lipids has been most extensively studied. This fungus harbors 154 cytochrome P450-encoding genes in the genome. As evidenced in this study, P. chrysosporium CYP505D6, a fused protein of P450 and its reductase, hydroxylates fatty alcohols (C9–15) and fatty acids (C9–15) at the ω-1 to ω-7 or ω-1 to ω-6 positions, respectively. Naphthalene and 1-naphthol were also hydroxylated, indicating that the substrate specificity of CYP505D6 is broader than those of the known fused proteins CYP102A1 and CYP505A1. The substrate versatility of CYP505D6 makes this enzyme an attractive candidate for biotechnological applications.
INTRODUCTION
White-rot fungi are capable of degrading a wide variety of recalcitrant aromatic compounds, including lignin and environmentally persistent pollutants (1–9). Fungal ligninolytic enzymes involved in the metabolism of aromatic compounds have been extensively studied (1, 2). Oxygenation reactions mediated by cytochrome P450 monooxygenase (P450) play important roles in fungal metabolism of recalcitrant xenobiotic compounds (3–9). Phanerochaete chrysosporium is a white-rot fungus whose ligninolytic and xenobiotic metabolism has been extensively studied (1–9). The sequence of the P. chrysosporium genome revealed the genetic diversity of fungal P450: as many as 154 P450 genes were identified therein (10–15). Among them, seven homologous genes encode fused proteins of cytochrome P450 and its reductase (11). P450BM3 (CYP102A1) of Bacillus megaterium and P450foxy (CYP505A1) of Fusarium oxysporum are also fusions of cytochrome P450 and its reductase and catalyze subterminal (ω-1 to ω-3) hydroxylation of fatty acids (16–18). Not only fatty acids but also fatty alcohols and amides are hydroxylated by CYP102A1 (19, 20). A CYP102 family enzyme isolated from Ktedonobacter racemifer, Krac9955, was recently reported to hydroxylate tetradecanoic acid at the ω-1 to ω-7 positions (21).
Hydroxy fatty acids are industrially valuable compounds for the synthesis of various ceramides and additives, for example, for cosmetic formulations, lubricants, and adhesives (22–25). Moreover, these compounds are biomaterials of particular interest for food science and biomedical applications, for example, as drug and nutrient carriers for lipophilic compounds (26). Hydroxy fatty acids and their derivatives were also reported to play important roles in energy homeostasis, by controlling blood glucose and insulin levels, and exerting antidiabetic effects; in the immune system, exerting anti-inflammatory and antienteroviral effects; and in association with neurodegenerative diseases (27–34). Hydroxy fatty acids are chemically synthesized via a reductive cleavage of olefinic fatty acids and a reduction of dicarboxylic acids (35, 36). However, harsh reaction conditions and by-product formation make their production and purification processes costly (36). To overcome these limitations, development of biological processes has been attempted as an alternative strategy.
In a previous study, the 1-dodecanol metabolism of P. chrysosporium was analyzed, demonstrating that 1-dodecanol is hydroxylated at seven positions (ω to ω-6) (37). The current study aimed to characterize P. chrysosporium proteins involved in 1-dodecanol and dodecanoic acid metabolism. The expression of genes encoding seven CYP505 homologs, CYP505D1 to CYP505D7, was evaluated in fungi exposed to 1-dodecanol and dodecanoic acid. CYP505D6, which was the most strongly induced gene after a 2-h incubation with the substrates, was cloned. CYP505D6 was purified, and its activity was compared with those of other fused P. chrysosporium proteins.
RESULTS
Transcriptional regulation of class III cytochrome P450 in P. chrysosporium.
The genome of the white-rot basidiomycete P. chrysosporium encodes seven class III P450 proteins (CYP505D1, CYP505D2, CYP505D3, CYP505D4, CYP505D5, CYP505D6, and CYP505D7) (10, 11). Sequence alignment of the heme domains of CYP505D isozymes, CYP505A1 from F. oxysporum (UniProt identifier Q9Y8G7), Krac9955 from K. racemifer (D6TEN2), and CYP102A1 from B. megaterium (P14779), which is a well-characterized class III cytochrome P450, is shown in Fig. 1. CYP505D6 has 47%, 37%, and 39% amino acid sequence identities to CYP505A1, Krac9955, and CYP102A1, respectively. The heme-binding motifs of the P450 proteins are highly conserved (Fig. 1). Quantitative reverse transcription-PCR (RT-PCR) was used to investigate the transcriptional regulation of CYP505D genes in P. chrysosporium. All seven isoforms were expressed in fungi grown in high-carbon, low-nitrogen (HCLN) medium (Fig. 2), and each gene showed a different transcriptional response to exogenous 1-dodecanol and dodecanoic acid. CYP505D6 was most strongly induced by the two substrates (Fig. 2). Thus, CYP505D6 was chosen for biochemical characterization in this study.
FIG 1.
Amino acid sequence alignment of heme domains of the CYP505D isozymes, CYP102A1, CYP505A1, and Krac9955. Sequences of CYP505D1 (protein identification number ug.73.17.1 [https://drnelson.uthsc.edu/Phanerochaete.P450s.htm]), CYP505D2 (JGI MycoCosm protein ID 3035819), CYP505D3 (JGI MycoCosm protein ID 137435), CYP505D4 (JGI MycoCosm protein ID 3002540), CYP505D5 (JGI MycoCosm protein ID 2900820), CYP505D6 (JGI MycoCosm protein ID 2932311), and CYP505D7 (JGI MycoCosm protein ID 2925787) from P. chrysosporium; CYP102A1 from B. megaterium (UniProt identifier P14779); CYP505A1 from F. oxysporum (UniProt identifier Q9Y8G7); and Krac9955 from K. racemifer (UniProt identifier D6TEN2) are shown. All protein identifications except that of CYP505D1 are from P. chrysosporium JGI genome version 2.2. The protein identification of CYP505D1 is from P. chrysosporium JGI genome version 1.0 because it is not annotated in the updated genome database. The heme-binding motif is underlined. The Tyr51 residue located at the entrance of the substrate-accessing channel of CYP102A1 and the corresponding amino acids in CYP505Ds are boxed. The sequences were aligned using CLUSTAL W.
FIG 2.
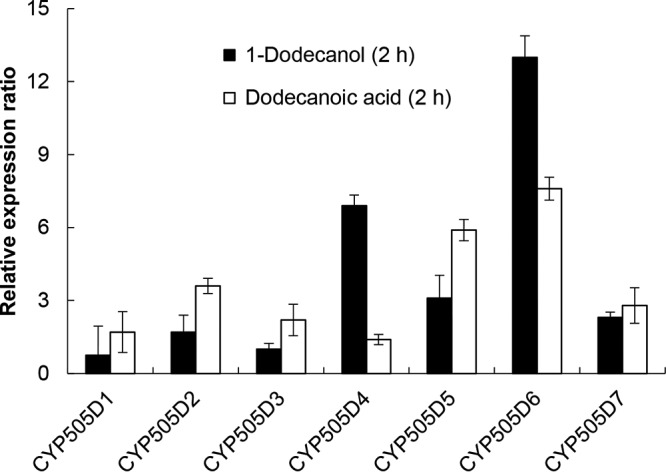
Gene expression profiles of class III P450 proteins in response to exogenous 1-dodecanol and dodecanoic acid (lauric acid). The abundances of amplified cDNA fragments of CYP505D1, CYP505D2, CYP505D3, CYP505D4, CYP505D5, CYP505D6, and CYP505D7 transcripts were normalized by comparison with that of ACT1. The relative normalized expression for each gene in fungi upon a 2-h exposure to 1-dodecanol or dodecanoic acid is shown as a ratio to the normalized expression in the absence of the substrate. Data are presented as mean values ± standard deviations (error bars) from four independent experiments.
Production and purification of recombinant CYP505D6.
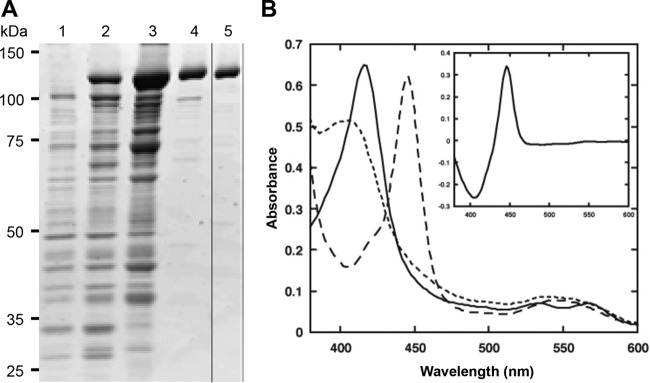
Heterologous expression of several class III P450 proteins from bacteria and fungi in Escherichia coli expression systems was previously reported (17, 19, 38, 39). When the production of CYP505D6 was induced in the pBAD/TOPO vector system, a new protein band in the soluble fraction was apparent during SDS-PAGE analysis (Fig. 3A). The molecular mass of the band (120 kDa) was consistent with the molecular mass of CYP505D6 predicted from its deduced amino acid sequence (118.0 kDa). Recently, CYP505D6 was produced by using the pET expression system (40). After 48 h of incubation with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the expression yield of CYP505D6 was 665 nmol/liter (40). In the present study, the expression yield of soluble CYP505D6 produced in E. coli was slightly improved (955 nmol/liter) by using the pBAD/TOPO vector system. The purified recombinant protein showed a typical P450 CO difference spectrum, with the absorption maximum at 448 nm and no maximum at 420 nm (Fig. 3B). The heme content in CYP505D6 was 0.45 to 0.55 mol of protoheme per mol of protein. In our previous study, the absorption spectra of native and recombinant CYP505A1 enzymes were determined. However, a broad shoulder in the 440- to 490-nm region derived from flavin prosthetic groups (16) was not prominent because of low flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) contents (17, 41). The absorption spectra of recombinant CYP505D6 were similar to those of CYP505A1. The contents of FAD and FMN in CYP505D6 were determined by a fluorometric method (42) to be 0.22 and 0.41 mol (mol protein)−1, respectively, indicating that CYP505D6 also has low contents of these cofactors. Upon titration with 1-dodecanol and dodecanoic acid, a type I substrate binding spectrum of CYP505D6 was observed (see Fig. S1 in the supplemental material), indicating a spin-state transition of the heme from the low to the high spin state upon substrate binding. Apparent Kd (dissociation constant) values of 81.5 μM and 155.8 μM were determined for the binding of 1-dodecanol and dodecanoic acid, respectively (Fig. S1). These results indicated that CYP505D6 was successfully produced, highly purified, and active.
FIG 3.
SDS-PAGE analysis and absorption spectra of recombinant CYP505D6. (A) SDS-PAGE analysis of purified CYP505D6. The expression of CYP505D6 was induced by the addition of arabinose (final concentration of 0.2%, wt/vol), and the culture was allowed to continue for up to 48 h at 30°C. Protein profiles before induction (time zero) (lane 1) and after a 48-h induction (lane 2) are shown. The crude extract was loaded onto a DEAE-Sepharose column. The protein fraction was continuously eluted off the DEAE-Sepharose column (lane 3) and the 2′,5′-ADP-Sepharose column (lane 4), followed by gel filtration chromatography (lane 5) and analysis of the eluates. (B) Absorption spectra of CYP505D6. Solid line, resting (oxidized); dotted line, dithionite reduced; dashed line, dithionite reduced plus CO; inset, CO difference spectrum (CO bound minus dithionite reduced).
Catalytic properties of CYP505D6.
To evaluate the enzymatic activity of the recombinant CYP505D6, its catalytic conversions of 1-dodecanol and dodecanoic acid were analyzed. CYP505D6 has low contents of the FAD and FMN cofactors. In our previous study, prior incubation with free FAD and FMN increased the specific activity of native and recombinant CYP505A1 enzymes, which have low flavin contents (17, 41). A similar phenomenon was also observed in CYP505D6. The enzyme activity with 1.0 μM FAD and 1.0 μM FMN, which were added 30 min before the reaction, reached levels 1.5-fold higher than that without FAD and FMN. Hydroxylated products of 1-dodecanol conversions were determined by gas chromatography-mass spectrometry (GC-MS) analysis (Fig. 4A; see also Fig. S1 in the supplemental material). The substrate was efficiently converted, with seven products generated. The mass spectra of the trimethylsilyl (TMS) derivatives of the reaction products had the same fragmentation patterns as those previously reported for dodecanediols hydroxylated at positions ω-1 to ω-6 (37). An additional peak with a slightly shorter retention time exhibited a similar fragmentation pattern, with an ion fragment at m/z 201, [C7H15CHOSi(CH3)3]+, characteristic for dodecanediols hydroxylated at the ω-7 position (Fig. S2). Thus, CYP505D6 converted 1-dodecanol to a series of dodecanediol isomers by incorporating an oxygen atom at the ω-1, -2, -3, -4, -5, -6, and -7 positions. During 1-dodecanol conversion, ω-1-, ω-2-, and ω-3-hydroxylated products were the dominant products (Fig. 4A); the yields of products hydroxylated at the ω-4, ω-5, ω-6, and ω-7 positions were lower (Fig. 4A).
FIG 4.
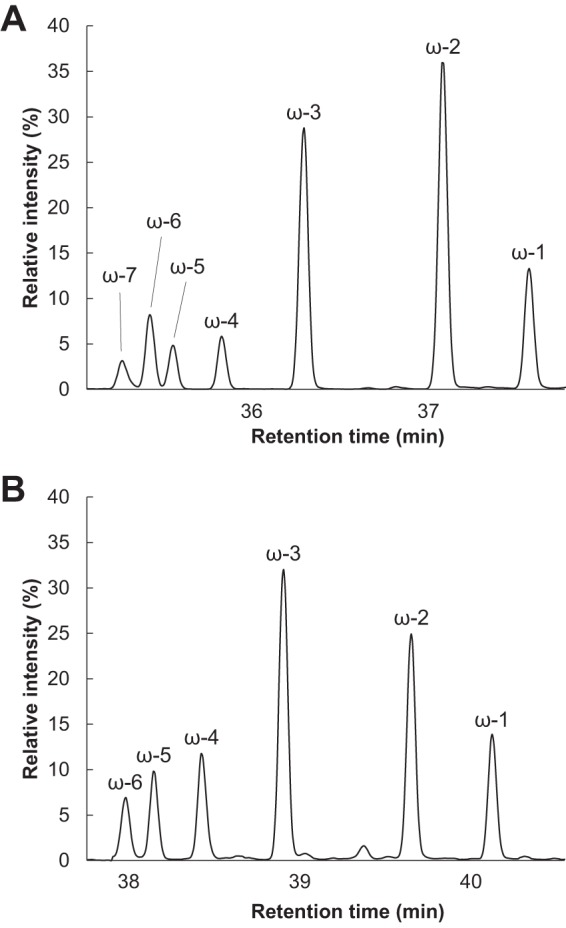
Total ion chromatograms of the products of reactions catalyzed by CYP505D6, with 1-dodecanol and dodecanoic acid as the substrates. The TMS derivation reaction products from reactions with 1-dodecanol (A) and dodecanoic acid (B) as the substrates were analyzed by using GC-MS. The results are representative, and the experiment was performed four times.
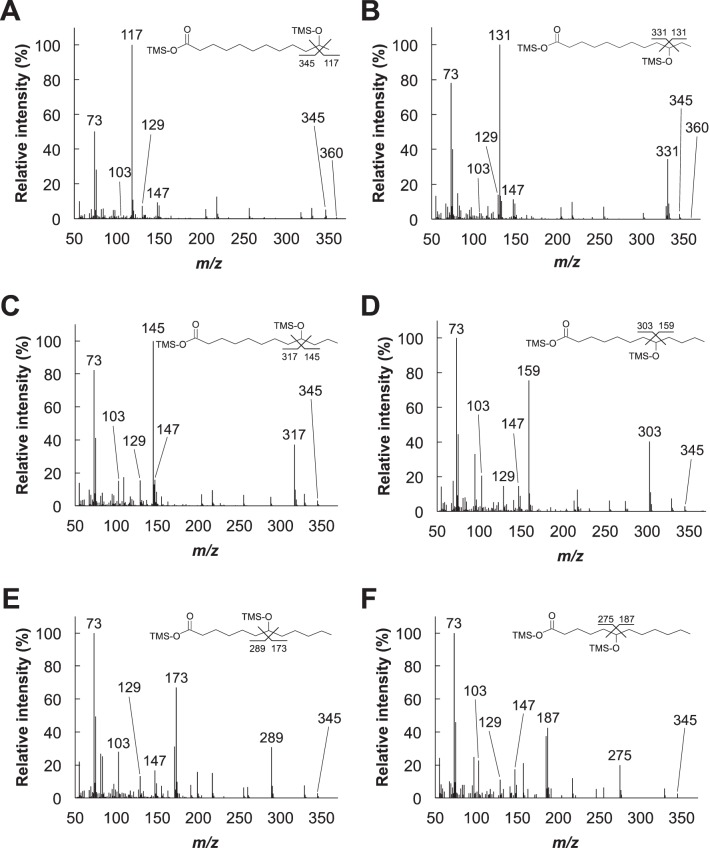
Hydroxylated products of dodecanoic acid conversions were also identified by GC-MS analysis (Fig. 4B and 5). The mass spectra of the TMS derivatives of the reaction products exhibited fragmentation patterns similar to those of dodecanediols hydroxylated at positions ω-1 to ω-6, as previously reported (37). The spectra contained ion fragments common to TMS derivatives of hydroxyl acids at m/z 147, [(CH3)2Si=OSi(CH3)3]+; m/z 129, [(CH3)2SiO=CHC3H6]+; m/z 103, [(CH3)3SiO=CH2]+; and m/z 73, [Si(CH3)3]+. The spectra also contained characteristic ion fragments at m/z 117, [CH3CHOSi(CH3)3]+, for ω-1; m/z 131, [C2H5CHOSi(CH3)3]+, for ω-2; m/z 145, [C3H7CHOSi(CH3)3]+, for ω-3; m/z 159, [C4H9CHOSi(CH3)3]+, for ω-4; m/z 173, [C5H11CHOSi(CH3)3]+, for ω-5; and m/z 187, [C6H13CHOSi(CH3)3]+, for ω-6. This conclusively supported the formation of hydroxydodecanoic acids. Similar to the catalytic conversion of 1-dodecanol, hydroxylation predominantly occurred at the ω-1, ω-2, and ω-3 positions; the yields of ω-4, ω-5, and ω-6 hydroxydodecanoic acids were considerably lower (Fig. 4B). The yields of ω-4- and ω-5-hydroxylated products from dodecanoic acid conversions were higher than those from 1-dodecanol conversions, whereas the yields of products hydroxylated at the ω-6 position were similar.
FIG 5.
Mass spectrometry analysis of the compounds generated by CYP505D6 in a reaction with dodecanoic acid as the substrate. The mass spectra were obtained from the GC peaks at the following retention times (in minutes): 40.08 (11-hydroxydodecanoic acid) (A), 39.62 (10-hydroxydodecanoic acid) (B), 38.88 (9-hydroxydodecanoic acid) (C), 38.41 (8-hydroxydodecanoic acid) (D), 38.13 (7-hydroxydodecanoic acid) (E), and 37.95 (6-hydroxydodecanoic acid) (F) (Fig. 4B).
The kinetic parameters of CYP505D6 were then investigated, using saturated fatty alcohols and acids with various carbon chain lengths (Table 1). The rates of NADPH consumption and substrate oxidation by CYP505D6 were measured spectrophotometrically by monitoring the change in the absorbance at 340 nm and by quantifying the conversion of dodecanoic acid using GC-MS and liquid chromatography-tandem mass spectrometry (LC-MS/MS), respectively (data not shown). The stoichiometry between the consumption of NADPH and the monooxygenation of dodecanoic acid was 1.04:1.00. CYP505D6 showed substantial activity against fatty alcohols and acids with carbon chain lengths of C9 (nonanol and nonanoic acid) to C18 (octadecanol and octadecanoic acid). The highest catalytic efficiency (kcat/Km) was observed with pentadecanol (C15), among fatty alcohols, and pentadecanoic acid (C15), among fatty acids (Table 1 and Fig. S3). When fatty alcohol and fatty acid substrates with carbon chain lengths of C9–15 were used as the substrates, the hydroxylation reaction occurred at the ω-1 to ω-6 positions of fatty alcohols, except for 1-dodecanol, and at the ω-1 to ω-6 positions of fatty acids (Table 1). However, a hydroxylation reaction occurred at the ω-1 to ω-4 but not at the ω-5 to ω-7 positions with substrates with chain lengths of C16–18 (Table 1). The Km values for fatty alcohols were slightly lower than those for fatty acids, when substrates with the same carbon chain length were compared (Table 1). The Kd and Km values for 1-dodecanol and dodecanoic acid were of similar magnitudes (Table 1 and Fig. S1). The NADPH-cytochrome c reductase activity of CYP505D6 was 860 ± 90 nmol min−1 nmol−1, which was similar to those of CYP102A1 and CYP505A1 (18, 20).
TABLE 1.
Kinetic parameters of saturated fatty alcohol (C9–18) and fatty acid (C9–18) conversions by CYP505D6a
| Substrate | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) |
|---|---|---|---|
| Fatty alcohol | |||
| C9 nonanol | 3,800 | 250 | 6.6 × 10−2 |
| C10 decanol | 1,100 | 220 | 2.0 × 10−3 |
| C11 undecanol | 420 | 310 | 0.74 |
| C12 dodecanol | 350 | 350 | 1.0 |
| C13 tridecanol | 120 | 370 | 3.1 |
| C14 tetradecanol | 20 | 450 | 23 |
| C15 pentadecanol | 10 | 490 | 49 |
| C16 hexadecanol | 18 | 510 | 28 |
| C17 heptadecanol | 15 | 340 | 23 |
| C18 octadecanol | 19 | 330 | 17 |
| Fatty acid | |||
| C9 nonanoic acid | 4,500 | 200 | 4.4 × 10−2 |
| C10 decanoic acid | 1,400 | 230 | 0.16 |
| C11 undecanoic acid | 610 | 230 | 0.38 |
| C12 dodecanoic acid | 450 | 250 | 0.56 |
| C13 tridecanoic acid | 230 | 310 | 1.4 |
| C14 tetradecanoic acid | 65 | 320 | 4.9 |
| C15 pentadecanoic acid | 30 | 390 | 13 |
| C16 hexadecanoic acid | 38 | 330 | 8.7 |
| C17 heptadecanoic acid | 36 | 280 | 7.8 |
| C18 octadecanoic acid | 28 | 260 | 9.3 |
Data are presented as mean values from three independent experiments. The standard errors were <11%.
The thermostabilities of CYP505D6 at 4°C and 30°C were determined using dodecanoic acid as a substrate (Fig. S4). After preincubation at 30°C for 48 h, the activity of CYP505D6 decreased to 29% of its maximal activity without preincubation (Fig. S4). After preincubation at 4°C for 48 h, the activity decreased to 77%, indicating that CYP505D6 is stable at low temperature.
Hydroxylated dodecanoic acid products generated by CYP505D6, CYP102A1, and their variants.
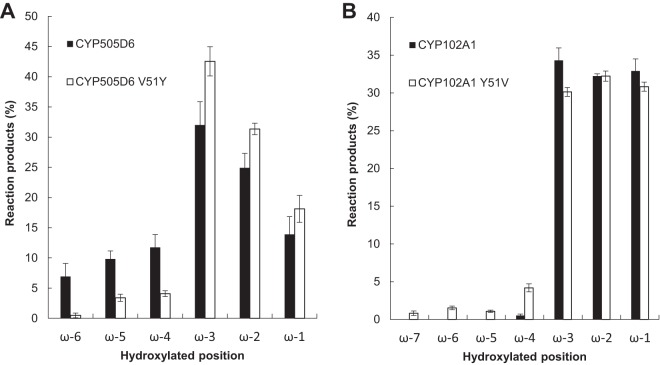
It has been reported that Tyr51 in CYP102A1 plays a crucial role in substrate recognition by interacting with fatty acids (43). In CYP505D6, based on the sequence alignment (Fig. 1), the corresponding amino acid of Tyr51 in CYP102A1 was Val51. Therefore, the V51Y variant of CYP505D6 (CYP505D6 V51Y) was characterized with dodecanoic acid as a substrate. CYP505D6 V51Y produced ω-4, ω-5, and ω-6 hydroxydodecanoic acids; nevertheless, their yields were considerably lower than those obtained using the wild-type enzyme (Fig. 6A; see also Fig. S5A in the supplemental material). Therefore, Val51 is a key residue in CYP505D6 that influences the regiospecificity of hydroxylation with alkyl substrates.
FIG 6.
Ratios of reaction products from reactions catalyzed by CYP505D6, CYP102A1, and protein variants with dodecanoic acid as a substrate. The TMS derivation reaction products from reactions catalyzed by wild-type CYP505D6 and its V51Y variant (A) and by wild-type CYP102A1 and its Y51V variant (B) were analyzed by using GC-MS. Data are presented as mean values ± standard deviations (error bars) from four independent experiments.
CYP102A1 and its Y51V variant (CYP102A1 Y51V) were also generated, and the reaction products from dodecanoic acid conversions were analyzed. Compared with the wild type, the yield of the product hydroxylated at the ω-4 position produced by CYP102A1 Y51V increased (Fig. 6B). The products hydroxylated at ω-5 to ω-7 by CYP102A1 Y51V were not generated by the wild type (Fig. 6B and Fig. S5B and S5C), indicating that Tyr51 at the entrance of the active-site pocket of CYP102A1 represses the hydroxylation of dodecanoic acid at the ω-4 to ω-7 positions. Three-dimensional structures of wild-type CYP102A1 and the Y51V variant were generated by using the available structure of the heme domain of CYP102A1 (Fig. 7). The space around the entrance to the active-site pocket of CYP102A1 Y51V was larger than that in the wild type and, hence, would more flexibly accommodate the aliphatic head of fatty acids, which would result in a diverse hydroxylation at the ω-5 to ω-7 positions of fatty acids (Fig. 7).
FIG 7.
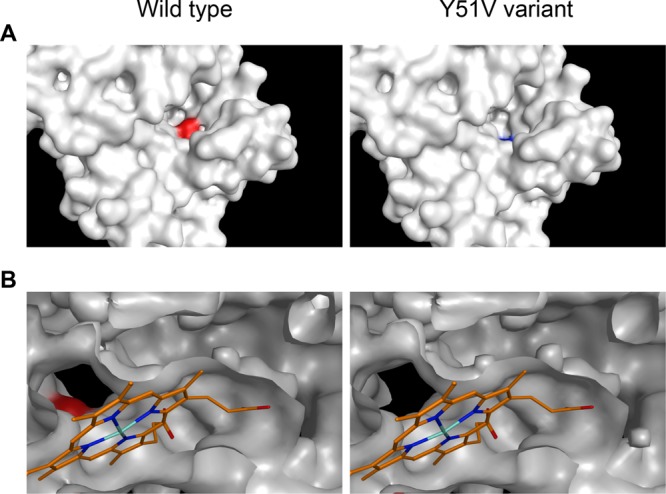
Structural models of wild-type CYP102A1 and its Y51V variant. Shown is a comparison of the overall structure (A) and the entrance of the catalytic pocket (B) of the heme domains. The heme molecule is represented by orange, cyan, and blue lines. The colored residues indicate Tyr51 (red) in CYP102A1 and Val51 (blue) in CYP102A1 Y51V. Tyr51 is located at the entrance of the substrate access channel. An expanded view of the channel entrance in the CYP102A1 Y51V variant is shown.
Hydroxylation of naphthalene by CYP505D6.
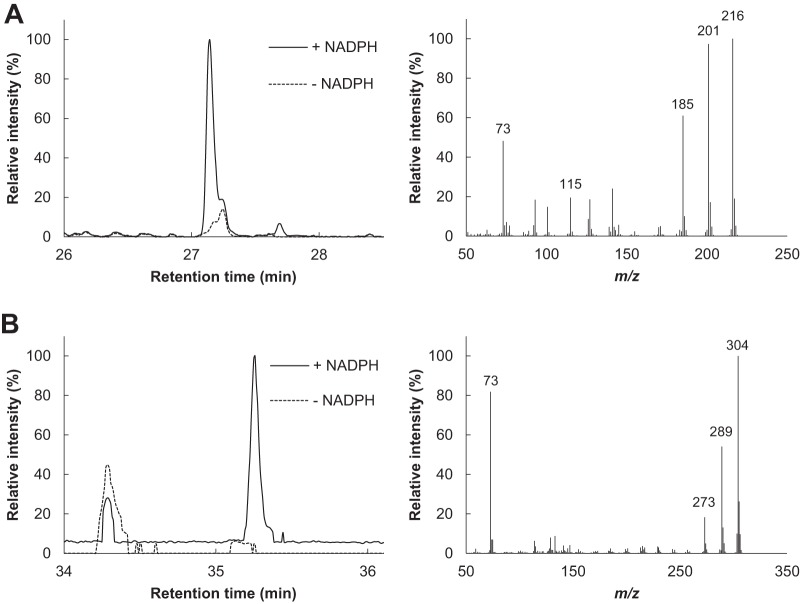
The hydroxylation of aromatic compounds by CYP505D6 was further examined by using naphthalene and 1-naphthol as the substrates. Naphthalene was converted to 1-naphthol and 1,3-dihydroxynaphthalene by CYP505D6 (Fig. 8). 1-Naphthol was also converted to 1,3-dihydroxynaphthalene (data not shown). The specific activity of CYP505D6 toward 1-naphthol (107 ± 7 mol min−1 mol−1) was 4.7-fold higher than that toward naphthalene (23 ± 2 mol min−1 mol−1).
FIG 8.
Total ion chromatograms and mass spectra of products generated by CYP505D6 from naphthalene as a substrate. The TMS derivation reaction products from a reaction with naphthalene as a substrate were analyzed by using GC-MS. The mass spectra (1-naphthol [A] and 1,3-dihydroxynaphthol [B]) were obtained from the GC peaks appearing at retention times of 27.14 min and 35.26 min. The results are representative, and the experiment was performed three times.
DISCUSSION
In the present study, the biochemical characterization of CYP505D6 from P. chrysosporium was reported. Among the seven analyzed fused protein-encoding genes, CYP505D6 was the most strongly induced gene after a 2-h fungal incubation with 1-dodecanol and dodecanoic acid. CYP505D6 was capable of hydroxylating fatty alcohols (C9–15) as well as fatty acids (C9–15) at the ω-1 to ω-7 or ω-1 to ω-6 positions, respectively. Naphthalene and 1-naphthol were also hydroxylated, indicating that CYP505D6 was more versatile than CYP102A1 and CYP505A1.
Quantitative RT-PCR was used to evaluate the expression of class III P450-encoding genes from P. chrysosporium. All the analyzed genes were expressed, and differently regulated, by exogenous 1-dodecanol and dodecanoic acid (Fig. 2). CYP505D1, CYP505D2, CYP505D3, CYP505D4, and CYP505D5 comprise a gene cluster in the P. chrysosporium genome (10, 11). However, these tandem P450 genes showed different expression profiles (Fig. 2). Noncooperative regulation of tandem P450 genes from P. chrysosporium has been previously reported (44). The differential gene expression of P450 underpins the metabolic diversity of basidiomycetes.
P450-dependent hydroxylation of 1-dodecanol by several class III P450 proteins was previously reported albeit demonstrating ω-1 to ω-3 hydroxylation (45–48). In the present study, the data clearly indicated that CYP505D6 catalyzed the hydroxylation of 1-dodecanol to form ω-1- to ω-7-hydroxylated 1-dodecanol. The 1,9-, 1,10-, and 1,11-dodecanediols were produced as the major products. The level of production of 1,5-, 1,6-, 1,7-, and 1,8-dodecanediols was considerably lower (Fig. 4A). These observations strongly supported the notion that P. chrysosporium metabolizes 1-dodecanol to 1,5-, 1,6-, 1,7-, 1,8-, 1,9-, 1,10-, and 1,11-dodecanediols using CYP505D6 and that CYP505D6 is responsible for the fungal metabolic conversions of 1-dodecanol previously reported (37). However, the product ratios for fungal cells and purified CYP505D6 appeared to be different (37). These observations suggest that other CYPs, including CYP505D isozymes, might be involved in hydroxylation reactions of 1-dodecanol in P. chrysosporium.
White-rot fungi are capable of degrading a wide variety of recalcitrant aromatic compounds, including naphthalene (7, 37, 49). An engineered CYP102A1 with three amino acid substitutions efficiently catalyzes the oxidation of polycyclic aromatic hydrocarbons (50). The present study revealed that even wild-type CYP505D6 catalyzed the hydroxylation of naphthalene to form 1-naphthol and that of 1-naphthol to form 1,3-dihydroxynaphthalene (Fig. 8). This indicated that CYP505D6 was involved in aromatic hydrocarbon degradation in P. chrysosporium.
The catalytic properties of CYP505D6 were considerably different from those of other class III P450 proteins, such as CYP102A1 and CYP505A1. Sequence alignment suggested that the amino acid residues at the entrance to the active-site pocket in CYP505D6 may be responsible for the unique catalytic performance of CYP505D6. According to crystallographic and mutational studies, several amino acid residues of CYP102A1 are critical for substrate binding (43, 51–55). Arg47 and Tyr51 residues located at the entrance to the substrate access channel play important roles in fatty acid substrate binding by interacting with the carboxyl group of the substrate (43, 51–55). Inside the entrance, the hydrophobic residues Leu75, Phe87, Leu181, Ile263, and Leu437 form a hydrophobic stretch in CYP102A1 that allows access to the aliphatic chains of fatty acids (43, 51–55). All of the residues around the entrance to the catalytic channel are conserved in CYP505D6 except for Try51.
The first interaction of CYP102A1 with fatty acid substrates would be the one between the carboxylate anion of the substrate and Arg47 and Tyr51 residues of the protein (43, 51–55). Arg47 is essential for fixing the carboxylate of fatty acids and is conserved in CYP505D6. However, the cavity around the substrate access entrance of CYP505D6 would be larger than that of CYP102A1 because of the Tyr51-to-Val51 substitution, since the Y51V substitution in CYP102A1 enlarged the entrance (Fig. 7). The difference in the catalytic properties of these P450 proteins may be explained by this substitution. The aliphatic head of fatty acids would turn and then penetrate the access channel. In CYP102A1, Tyr51 replaces Val51 in CYP505D6, making it impossible for the hydrogen bonding that supports the electrostatic interaction with the carboxylate to occur. Because CYP505D6 harbors Val51 at the corresponding position, the interaction of CYP505D6 with the carboxylate anion of fatty acids at the entrance to the substrate access channel would be relatively weaker than that of CYP102A1. Furthermore, a large entrance in CYP505D6 would be flexible enough to accommodate the aliphatic heads of fatty acids, resulting in a diverse hydroxylation reaction at the ω-1 to ω-6 positions of fatty acids (C9–15). CYP505D6 V51Y produced fewer ω-4, ω-5, and ω-6 hydroxylation products of dodecanoic acid than wild-type CYP505D6 (Fig. 6A). CYP102A1 catalyzed the hydroxylation of dodecanoic acid at the ω-1 to ω-4 positions, whereas CYP102A1 Y51V catalyzed hydroxylation at the ω-1 to ω-7 positions (Fig. 6B). These observations supported the notion that Val51 at the entrance of the active-site pocket of CYP505D6 plays an important role in determining the regiospecificity of fatty acid hydroxylation. Given that these enzymes perform a wide range of biochemical reactions on physiological and industrial scales (51), the results of the present study suggest that CYP505D6 and CYP102A1 Y51V might be promising starting points for a further understanding and innovative protein engineering of the class III cytochrome P450 family.
CYP505A1 catalyzes the subterminal (ω-1 to ω-3) hydroxylation of fatty acids (16–18), whereas Krac9955 hydroxylates tetradecanoic acid at the ω-1 to ω-7 positions (21). In CYP102A1, Tyr51 replaces Phe51 and Val51 in CYP505A1 and Krac9955, respectively. The diverse hydroxylation reaction at the ω-4 to ω-6 positions of fatty acids might require a nonaromatic amino acid at the entrance to the active-site pocket in class III P450. Although Krac9955 was unable to oxidize hexadecanoic acid, 1-dodecanol, or 1-tetradecanol, fatty alcohols and acids with carbon chain lengths of C9 to C18 were hydroxylated by CYP505D6. These are interesting observations, but it is still unclear why CYP505D6 and Krac9955 exhibit diverse hydroxylation of fatty compounds and differences in substrate specificity. Unraveling the details underlying the unique catalytic features of CYP505D6 and Krac9955 will require further clarification by site-directed mutagenesis and three-dimensional structural studies. Compared with CYP102A1 and CYP505A1, CYP505D6 and Krac9955 show low activity toward saturated fatty acids with the same carbon chain length (56). The poor regioselectivity of the fatty acid hydroxylation resulting from a large entrance might cause low catalytic activity. The kcat/Km values of CYP505D6 for fatty alcohols were higher than those for fatty acids when substrates with the same carbon chain length were compared (Table 1). Because the steric hindrance of the aliphatic heads of fatty alcohols is smaller than that of fatty acids, fatty alcohols should be more accessible to the substrate-accessing channel of CYP505D6 than fatty acids.
Hydroxy fatty acids and their derivatives are of particular interest for food science and biomedical applications (21–25) and also play physiological and pharmacological roles (26–33). Although hydroxyl fatty acids can be chemically synthesized, harsh reaction conditions and by-product formation render their production and purification processes costly (34, 35). To overcome these limitations, biological processes have been developed (35). P450-mediated enzymatic oxidation reactions can be used for the efficient generation of alcohols from alkanes, while these conversions are not feasible using current organic synthetic methods (57–60). The catalytic efficiencies of class III P450 proteins, which are fused proteins of P450 and its reductase, are higher than those of other P450 classes (16–20, 58). In the present study, CYP505D6 was shown to be versatile, generating fatty acids hydroxylated at more positions than those produced by CYP102A1 and CYP505A1. Although poor regioselectivity of hydroxylation means that more resources (financial and material) have to be devoted to purifying individual metabolites, CYP505D6 would be useful for the synthesis of hydroxyl fatty alcohols and hydroxyl fatty acids that are difficult to synthesize on an industrial scale. Furthermore, the long-term stability of CYP505D6 at 30°C is clearly higher than those of CYP102A1 and CYP102A3 (see Fig. S4 in the supplemental material), which were completely inactivated after 48 h at 30°C (61). These properties suggest that CYP505D6 should be considered a potential candidate biocatalyst with additional bioengineering possibilities (62).
In this report, the biochemical characterization of a class III P450 enzyme from a basidiomycetous fungus was presented, providing a better understanding of the fungal biology associated with the functional diversity of P450 enzymes. The broad substrate specificity of CYP505D6 not only is attractive for biotechnological applications but also provides insights into the biochemistry of this enzyme class.
MATERIALS AND METHODS
Chemicals.
Fatty acids and alcohols were purchased from Wako Pure Chemicals Co. Ltd. (Osaka, Japan). Naphthalene, 1-naphthol, and 1,3-dihydroxynaphthalene were obtained from Sigma-Aldrich (St. Louis, MO, USA). All chemicals were of analytical grade. Deionized water was obtained by using the MilliQ system (Merck-Millipore, Billerica, MA, USA).
Culture conditions.
P. chrysosporium (ATCC 34541) was typically maintained in HCLN medium (pH 4.5) (37, 63) at 37°C in a stationary culture. For the experiments, fungal conidia were inoculated in 20 ml of HCLN medium (pH 4.5) (37, 63) in a 200-ml Erlenmeyer flask and incubated at 37°C in a stationary culture. The medium was supplemented with 28 mM d-glucose and 1.2 mM ammonium tartrate as carbon and nitrogen sources, respectively, as previously described (37, 63). After a 2-day incubation, the mycelia were harvested by filtration and then immediately frozen in liquid nitrogen and stored at −80°C.
Quantitative RT-PCR analysis of class III P450 genes.
After a 2-day preincubation in HCLN medium (pH 4.5) at 37°C in a stationary culture, the appropriate substrates (2 mM 1-dodecanol or dodecanoic acid) were added to the cultures. The mycelia were incubated for an additional 2 h. Total RNA was extracted from the mycelia grown in the absence or presence of substrates by using the RNeasy minikit (Qiagen, Venlo, The Netherlands). Single-stranded cDNA was then synthesized from the total RNA. Quantitative RT-PCR was performed using gene-specific primer sets (Table 2) designed using the genomic sequence of P. chrysosporium (http://genome.jgi.doe.gov/Phchr2/Phchr2.home.html). The CYP505D1-specific primer set was designed based on an earlier P. chrysosporium genome sequence (JGI genome version 2.0) because of nonavailability in the updated genome database. Gene expression was normalized to the expression of the actin gene (64). The expression of each gene in the presence of a substrate is shown relative to its expression in the absence of the substrate.
TABLE 2.
Oligonucleotide primersa used in this study
| Primer | Gene | Nucleotide sequence |
|---|---|---|
| Quantitative RT-PCR | ||
| CYP505D1-f | CYP505D1 | 5′-CAAGTCCATTTGGGACGACT-3′ |
| CYP505D1-r | 5′-ACGAGACCAGGAGATGGAGA-3′ | |
| CYP505D2-f | CYP505D2 | 5′-ATGGAGAAGGCCAAGAACCT-3′ |
| CYP505D2-r | 5′-AGGAGGATGGACATGACGAC-3′ | |
| CYP505D3-f | CYP505D3 | 5′-ATGGAGAAGGCCAAGAACCT-3′ |
| CYP505D3-r | 5′-AGGAGGATGGACATGACGAC-3′ | |
| CYP505D4-f | CYP505D4 | 5′-ATGGAGAAGGCCAAGAACCT-3′ |
| CYP505D4-r | 5′-AGGAGGATGGACATGACGAC-3′ | |
| CYP505D5-f | CYP505D5 | 5′-ATGGAGAAGGCCAAGAACCT-3′ |
| CYP505D5-r | 5′-AGGAGGATGGACATGACGAC-3′ | |
| CYP505D6-f | CYP505D6 | 5′-CAGCTCCTCAAGTCCTACGG-3′ |
| CYP505D6-r | 5′-TCCTTCACAAGCTCCTCGAT-3′ | |
| CYP505D7-f | CYP505D7 | 5′-CATGGTCTGTGGTGTTGAGC-3′ |
| CYP505D7-r | 5′-CGAAGTTGTCGTCCTCCTTC-3′ | |
| ACT1-f | ACT1 | 5′-CCAAGGCTAACCGTGAGAAG-3′ |
| ACT1-r | 5′-CACCAGAGTCGAGCACGATA-3′ | |
| Cloning for recombinant protein production | ||
| CYP505D6-f | CYP505D6 | 5′-ATGACGTCTACCATTCCGACGCCGCCGTCCAT-3′ |
| CYP505D6-r | 5′-CTATTCGAAGATATCGGTAGCGAA-3′ | |
| CYP102A1-f | CYP102A1 | 5′-ATGACAATTAAAGAAATGCCTCAGCCAAAAAC-3′ |
| CYP102A1-r | 5′-TTACCCAGCCCACACGTCTTTTGC-3′ | |
| Site-directed mutagenesis | ||
| CYP505D6 V51Y-f | CYP505D6 | 5′-TACGTCAGCTCGGCGAAGCTCAT-3′ |
| CYP505D6 V51Y-r | 5′-GAGCAGCTTCCTCCCGAGAA-3′ | |
| CYP102A1 Y51V-f | CYP102A1 | 5′-GTCTTATCAAGTCAGCGTCTAAT-3′ |
| CYP102A1 Y51V-r | 5′-GCGCGTTACACGACCAGGCG-3′ |
Gene-specific primers were designed based on genomic sequence data (http://genome.jgi.doe.gov/Phchr2/Phchr2.home.html).
Construction of the gene expression system.
A full-length P. chrysosporium CYP505D6 gene was PCR amplified by using the primer combination specified in Table 2, using a 2400 DNA thermal cycler (TaKaRa Bio, Otsu, Japan), as follows: an initial denaturation step at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 62°C for 30 s, and extension at 72°C for 3 min. The PCR products were separated by agarose gel (0.8%) electrophoresis. The gels were stained with ethidium bromide and visualized by using Molecular Imager FX (Bio-Rad, CA, USA). The amplified CYP505D6 gene was inserted into the pBAD/TOPO vector (Invitrogen, Carlsbad, CA, USA) by TA cloning, using the pBAD/TOPO TA cloning kit (Invitrogen). The recombinant plasmid was used to transform E. coli strain TOP10 (Invitrogen) using the heat shock method, and the transformants were selected based on ampicillin resistance on LB medium. Plasmid DNA was extracted from 4 colonies. The identity of the plasmid containing the CYP505D6 gene (pBAD-CYP505D6) was verified by sequencing. The plasmid for the production of recombinant CYP102A1 protein was constructed using essentially the same strategy.
Based on amino acid sequence analyses, sequences encoding the V51Y variant of CYP505D6 (CYP505D6 V51Y) and the Y51V variant of CYP102A1 (CYP102A1 Y51V) were generated by overlap extension PCR using the QuikChange site-directed mutagenesis kit (Stratagene, San Diego, CA, USA) and primers shown in Table 2.
Heterologous expression and purification of CYP505D6 and CYP102A1.
E. coli TOP10 cells harboring each of the P450 expression plasmids were grown at 37°C with constant shaking in the Terrific broth (TB) supplemented with 100 μg/ml ampicillin, until the optical density of the cultures at 600 nm reached 0.5. The expression of P450 was induced by the addition of arabinose (final concentration of 0.2%, wt/vol) to the medium. The cultures were allowed to continue for up to 48 h at 30°C. Cells were harvested by centrifugation (4,000 × g at 4°C for 10 min), and the cell pellet was resuspended in buffer A [50 mM 3-(N-morpholino)propanesulfonic acid (pH 7.4), 1 mM EDTA, 1 mM dithiothreitol, 10% (vol/vol) glycerol, and 0.25 mM phenylmethylsulfonyl fluoride]. The cells were then broken by sonication (5 times for 30 s) with a Q700 sonicator (Qsonica, Melville, NY, USA). The insoluble debris was removed by centrifugation (15,000 × g at 4°C for 15 min), and the supernatant was stored at 4°C.
The crude extract was loaded onto a DEAE-Sepharose column (Sigma-Aldrich) equilibrated with buffer A. The column was washed with buffer A, and proteins were then eluted with a 0 to 0.3 M KCl gradient in buffer A. The fractions containing heme were collected, dialyzed against buffer A, and loaded onto a 2′,5′-ADP-Sepharose column (GE Healthcare, Waukesha, WI, USA) equilibrated with the same buffer. A dark brown fraction that was eluted in buffer A supplemented with 2 mM NADPH was loaded directly onto a Superdex 200 HR 10/30 column (GE Healthcare) equilibrated with buffer A. The resulting eluate was considered to contain purified recombinant proteins.
Spectral characterization.
The P450 content of purified CYP505D6 was determined by carbon monoxide difference spectral analysis using an extinction coefficient of 91 mM−1 cm−1 (65). The contents of FAD and FMN in CYP505D6 were determined by a fluorometric method (41, 42). Type I difference spectral determination of the Kd values for the binding of 1-dodecanol or dodecanoic acid to CYP505D6 was performed as described previously (41). The absorption and fluorometric spectra were analyzed by using a SpectraMax instrument (Molecular Devices, San Jose, CA, USA).
P450 enzyme assay.
Hydroxylase activities were determined as previously described (19, 59), with slight modifications. Briefly, the activity and substrate specificity of CYP505D6 were determined in reaction mixtures (1.0 ml) containing 50 nM CYP505D6, 125 μM NADPH, 1 μM FMN, 1 μM FAD, 10% (vol/vol) glycerol, and 5 μl of a substrate solution (0 to 600 mM in ethanol or dimethyl sulfoxide) in 50 mM sodium phosphate buffer (pH 7.5). The reaction was initiated by the addition of the substrate, and the decrease in the absorbance at 340 nm was monitored by using a spectrophotometer. The reactions proceeded at 30°C for 15 min, and 10 μl of 1 M HCl was then added. The background rate of NADPH consumption in reaction mixture without the substrate was subtracted from the observed rates of those with various substrates. The kinetic parameters Km and kcat were calculated by fitting the obtained initial rates using the Michaelis-Menten equation in Origin version 6.0 software (OriginLab, Northampton, MA, USA). The residual substrate and reaction products were analyzed by GC-MS after extraction with 1 ml of chloroform (three times), evaporation, and TMS derivation using N,O-bis(TMS)trifluoroacetamide–pyridine (2:1, vol/vol) (37).
The NADPH cytochrome c reductase activity was determined as an NADPH-dependent cytochrome c-reducing activity in reaction mixtures (1.0 ml) containing 10 nM CYP505D6, 50 μM cytochrome c, 0.5 mM EDTA, 125 μM NADPH, 1 μM FMN, 1 μM FAD, and 10% (vol/vol) glycerol in 50 mM sodium phosphate buffer (pH 7.5). The reaction was initiated by the addition of NADPH. One unit of activity was defined as the amount of enzyme that catalyzes the reduction of cytochrome c at an initial rate of 1 pmol min−1. The rate of cytochrome c reduction was determined spectrophotometrically at 30°C using 21.1 mM−1 cm−1 as the extinction coefficient for reduced cytochrome c minus that for oxidized cytochrome c at 550 nm (16–19, 59).
The thermostability of CYP505D6 was determined using dodecanoic acid as a substrate. Purified CYP505D6 was preincubated at 4°C and 30°C for 3, 6, 12, 24, 36, and 48 h. Residual CYP505D6 activity was assayed in reaction mixtures (0.5 ml) containing 125 μM dodecanoic acid, 125 μM NADPH, 1 μM FMN, 1 μM FAD, 10% (vol/vol) glycerol, and purified enzyme in 50 mM sodium phosphate (pH 7.5). The amount of generating NADP+ produced by CYP505D6 was measured spectrophotometrically by monitoring absorbance changes at 340 nm.
Instrumentation.
GC-MS was performed at 70 eV using a GCMS-QP2010 apparatus (Shimadzu, Kyoto, Japan) equipped with a 30-m fused silica column (DB-5; J&W Scientific). The oven temperature was programmed to ramp from 80°C to 320°C at 8°C/min, with an injection temperature of 280°C. Naphthalene conversion products were identified by comparing their respective GC retention times and mass fragmentation patterns with those of authentic standards (37). Compounds for which authentic standards are not available were identified by comparing the mass data with those available from the National Institute of Standards and Technology (NIST) mass spectral library.
Homology modeling of CYP102A1.
Homology modeling of wild-type CYP102A1 and CYP102A1 Y51V was performed by using the Molecular Operating Environment (MOE) homology program (Chemical Computing Group Inc., Montreal, Canada) based on the template of the heme domain of CYP102A1 (PDB accession number 2IJ2).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants-in-aid for scientific research (17K07734 to M.S. and 16K07679 to M.K.) and partially supported by a grant from the Takahashi Industrial and Economic Research Foundation. There are no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01091-18.
REFERENCES
- 1.Gold MH, Wariishi H, Valli K. 1989. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium, p 127–140. In Whitaker JR, Sonnet PE (ed), ACS symposium series: biocatalysis in agricultural biotechnology, vol 389 American Chemical Society, Washington, DC. [Google Scholar]
- 2.Kirk TK, Farrell RL. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol 41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 3.Bezalel L, Hadar Y, Fu PP, Freeman JP, Cerniglia C. 1996. Initial oxidation products in the metabolism of pyrene, anthracene, fluorene and dibenzothiophene by the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol 62:2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiratsuka N, Wariishi H, Tanaka H. 2001. Degradation of diphenyl ether herbicides by the lignin-degrading basidiomycete Coriolus versicolor. Appl Microbiol Biotechnol 57:563–571. doi: 10.1007/s002530100789. [DOI] [PubMed] [Google Scholar]
- 5.Hiratsuka N, Oyadomari M, Shinohara H, Tanaka H, Wariishi H. 2005. Metabolic mechanisms involved in hydroxylation reactions of diphenyl compounds by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Biochem Eng J 23:241–246. doi: 10.1016/j.bej.2005.01.008. [DOI] [Google Scholar]
- 6.Ichinose H, Wariishi H, Tanaka H. 1999. Biotransformation of recalcitrant 4-methyldibenzothiophene to water-extractable products using lignin-degrading basidiomycete Coriolus versicolor. Biotechnol Prog 15:706–714. doi: 10.1021/bp990082z. [DOI] [PubMed] [Google Scholar]
- 7.Masaphy S, Levanon D, Henis Y, Venkateswarlu K, Kelly SL. 1996. Evidence for cytochrome P-450 and P-450-mediated benzo(a)pyrene hydroxylation in the white rot fungus Phanerochaete chrysosporium. FEMS Microbiol Lett 135:51–55. doi: 10.1111/j.1574-6968.1996.tb07965.x. [DOI] [PubMed] [Google Scholar]
- 8.Teramoto H, Tanaka H, Wariishi H. 2004. Fungal cytochrome P450s catalyzing hydroxylation of substituted toluenes to form their hydroxymethyl derivatives. FEMS Microbiol Lett 234:255–260. doi: 10.1111/j.1574-6968.2004.tb09541.x. [DOI] [PubMed] [Google Scholar]
- 9.Teramoto H, Tanaka H, Wariishi H. 2004. Degradation of 4-nitrophenol by the white-rot basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol 66:312–317. doi: 10.1007/s00253-004-1637-z. [DOI] [PubMed] [Google Scholar]
- 10.Doddapaneni H, Chakraborty R, Yadav JS. 2005. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genomics 14:92. doi: 10.1186/1471-2164-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Syed K, Yadav JS. 2012. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit Rev Microbiol 38:339–363. doi: 10.3109/1040841X.2012.682050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinose H, Wariishi H. 2013. High-level heterologous expression of fungal cytochrome P450s in Escherichia coli. Biochem Biophys Res Commun 23:289–294. doi: 10.1016/j.bbrc.2013.07.057. [DOI] [PubMed] [Google Scholar]
- 13.Mgbeahuruike AC, Kovalchuk A, Ubhayasekera W, Nelson DR, Yadav JS. 2017. CYPome of the conifer pathogen Heterobasidion irregulare: inventory, phylogeny and transcriptional analysis of the response to biocontrol. Fungal Biol 121:158–171. doi: 10.1016/j.funbio.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Syed K, Porollo A, Lam YW, Grimmett PE, Yadav JS. 2013. CYP63A2, a catalytically versatile fungal P450 monooxygenase capable of oxidizing higher-molecular-weight polycyclic aromatic hydrocarbons, alkylphenols, and alkanes. Appl Environ Microbiol 79:2692–2702. doi: 10.1128/AEM.03767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol 22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- 16.Narhi LO, Fulco AJ. 1986. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem 261:7160–7169. [PubMed] [Google Scholar]
- 17.Nakayama N, Takemae A, Shoun H. 1996. Cytochrome P450foxy, a catalytically self-sufficient fatty acid hydroxylase of the fungus Fusarium oxysporum. J Biochem 119:435–440. doi: 10.1093/oxfordjournals.jbchem.a021260. [DOI] [PubMed] [Google Scholar]
- 18.Kitazume T, Takaya N, Nakayama N, Shoun H. 2000. Fusarium oxysporum fatty-acid subterminal hydroxylase (CYP505) is a membrane-bound eukaryotic counterpart of Bacillus megaterium cytochrome P450BM3. J Biol Chem 275:39734–39740. doi: 10.1074/jbc.M005617200. [DOI] [PubMed] [Google Scholar]
- 19.Ho PP, Fulco AJ. 1976. Involvement of a single hydroxylase species in the hydroxylation of palmitate at the omega-1, omega-2 and omega-3 positions by a preparation from Bacillus megaterium. Biochim Biophys Acta 27:249–256. doi: 10.1016/0005-2760(76)90145-4. [DOI] [PubMed] [Google Scholar]
- 20.Matson RS, Hare RS, Fulco AJ. 1977. Characteristics of a cytochrome P-450-dependent fatty acid omega-2 hydroxylase from Bacillus megaterium. Biochim Biophys Acta 22:487–494. doi: 10.1016/0005-2760(77)90218-1. [DOI] [PubMed] [Google Scholar]
- 21.Munday SD, Maddigan NK, Young RJ, Bell SG. 2016. Characterisation of two self-sufficient CYP102 family monooxygenases from Ktedonobacter racemifer DSM44963 which have new fatty acid alcohol product profiles. Biochim Biophys Acta 1860:1149–1162. doi: 10.1016/j.bbagen.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Vandamme EJ, Soetaert W. 2002. Bioflavours and fragrances via fermentation and biocatalysis. J Chem Technol Biotechnol 77:1323–1332. doi: 10.1002/jctb.722. [DOI] [Google Scholar]
- 23.Metzger JO, Bornscheuer U. 2006. Lipids as renewable resources: current state of chemical and biotechnological conversion and diversification. Appl Microbiol Biotechnol 71:13–22. doi: 10.1007/s00253-006-0335-4. [DOI] [PubMed] [Google Scholar]
- 24.Kirilov P, Gauffre F, Franceschi-Messant S, Perez E, Rico-Lattes I. 2009. Rheological characterization of a new type of colloidal dispersion based on nanoparticles of gelled oil. J Phys Chem B 113:11101–11108. doi: 10.1021/jp905260s. [DOI] [PubMed] [Google Scholar]
- 25.Kirilov P, Lukyanova L, Franceschi-Messant S, Perier V, Perez E, Rico-Lattes I. 2008. A new type of colloidal dispersions based on nanoparticles of gelled oil. Colloids Surf A Physicochem Eng Asp 328:1–7. doi: 10.1016/j.colsurfa.2008.06.011. [DOI] [Google Scholar]
- 26.Martin B, Brouillet F, Franceschi S, Perez E. 2017. Evaluation of organogel nanoparticles as drug delivery system for lipophilic compounds. AAPS PharmSciTech 18:1261–1269. doi: 10.1208/s12249-016-0587-y. [DOI] [PubMed] [Google Scholar]
- 27.Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, Dhaneshwar AS, Hammarstedt A, Smith U, McGraw TE, Saghatelian A, Kahn BB. 2014. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159:318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YI, Hirai S, Goto T, Ohyane C, Takahashi H, Tsugane T, Konishi C, Fujii T, Inai S, Iijima Y, Aoki K, Shibata D, Takahashi N, Kawada T. 2012. Potent PPARα activator derived from tomato juice, 13-oxo-9,11-octadecadienoic acid, decreases plasma and hepatic triglyceride in obese diabetic mice. PLoS One 7:e31317. doi: 10.1371/journal.pone.0031317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuda O, Brezinova M, Rombaldova M, Slavikova B, Posta M, Beier P, Janovska P, Veleba J, Kopecky J Jr, Kudova E, Pelikanova T, Kopecky J. 2016. Docosahexaenoic acid-derived fatty acid esters of hydroxy fatty acids (FAHFAs) with anti-inflammatory properties. Diabetes 65:2580–2590. doi: 10.2337/db16-0385. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Moraes-Vieira PM, Castoldi A, Aryal P, Yee EU, Vickers C, Parnas O, Donaldson CJ, Saghatelian A, Kahn BB. 2016. Branched fatty acid esters of hydroxy fatty acids (FAHFAs) protect against colitis by regulating gut innate and adaptive immune responses. J Biol Chem 291:22207–22217. doi: 10.1074/jbc.M115.703835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan YW, Hong WJ, Chu JJ. 2016. Inhibition of enterovirus VP4 myristoylation is a potential antiviral strategy for hand, foot and mouth disease. Antiviral Res 133:191–195. doi: 10.1016/j.antiviral.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Guo L, Zhang X, Zhou D, Okunade AL, Su X. 2012. Stereospecificity of fatty acid 2-hydroxylase and differential functions of 2-hydroxy fatty acid enantiomers. J Lipid Res 53:1327–1335. doi: 10.1194/jlr.M025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo C, Yoon J, Rhee Y, Kim JJ, Nam SJ, Lee W, Lee G, Yee ST, Paik MJ. 2015. Simultaneous analysis of seven 2-hydroxy fatty acids as tert-butyldimethylsilyl derivatives in plasma by gas chromatography-mass spectrometry. Biomed Chromatogr 29:156–160. doi: 10.1002/bmc.3251. [DOI] [PubMed] [Google Scholar]
- 34.Nagano M, Takahara K, Fujimoto M, Tsutsumi N, Uchimiya H, Kawai-Yamada M. 2012. Arabidopsis sphingolipid fatty acid 2-hydroxylases (AtFAH1 and AtFAH2) are functionally differentiated in fatty acid 2-hydroxylation and stress responses. Plant Physiol 159:1138–1148. doi: 10.1104/pp.112.199547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaper DGM, Mitchell DL. 1960. An improved preparation of omega-hydroxy aliphatic acids and their esters. Can J Chem 38:1976–1982. doi: 10.1139/v60-266. [DOI] [Google Scholar]
- 36.Jung E, Park BG, Ahsan MM, Kim J, Yun H, Choi KY, Kim BG. 2016. Production of ω-hydroxy palmitic acid using CYP153A35 and comparison of cytochrome P450 electron transfer system in vivo. Appl Microbiol Biotechnol 100:10375–10384. doi: 10.1007/s00253-016-7675-5. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzaki F, Wariishi H. 2004. Functional diversity of cytochrome P450s of the white-rot fungus Phanerochaete chrysosporium. Biochem Biophys Res Commun 324:387–393. doi: 10.1016/j.bbrc.2004.09.062. [DOI] [PubMed] [Google Scholar]
- 38.Sevrioukova IF, Hazzard JT, Tollin G, Poulos TL. 1999. The FMN to heme electron transfer in cytochrome P450BM-3. J Biol Chem 274:36097–36106. doi: 10.1074/jbc.274.51.36097. [DOI] [PubMed] [Google Scholar]
- 39.Gustafsson MC, Roitel O, Marshall KR, Noble MA, Chapman SK, Pessegueiro A, Fulco AJ, Cheesman MR, von Wachenfeldt C, Munro AW. 2004. Expression, purification, and characterization of Bacillus subtilis cytochromes P450 CYP102A2 and CYP102A3: flavocytochrome homologues of P450 BM3 from Bacillus megaterium. Biochemistry 43:5474–5487. doi: 10.1021/bi035904m. [DOI] [PubMed] [Google Scholar]
- 40.Ichinose H, Wariishi H. 2013. High-level heterologous expression of fungal cytochrome P450s in Escherichia coli. Biochem Biophys Res Commun 438:289–294. doi: 10.1016/j.bbrc.2013.07.057. [DOI] [PubMed] [Google Scholar]
- 41.Kitazume T, Tanaka A, Takaya N, Nakamura A, Matsuyama S, Suzuki T, Shoun H. 2002. Kinetic analysis of hydroxylation of saturated fatty acids by recombinant P450foxy produced by an Escherichia coli expression system. Eur J Biochem 269:2075–2082. doi: 10.1046/j.1432-1033.2002.02855.x. [DOI] [PubMed] [Google Scholar]
- 42.Aliverti A, Curti B, Vanoni MA. 1999. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol Biol 131:9–23. [DOI] [PubMed] [Google Scholar]
- 43.Noble MA, Miles CS, Chapman SK, Lysek DA, MacKay AC, Reid GA, Hanzlik RP, Munro AW. 1999. Roles of key active-site residues in flavocytochrome P450BM3. Biochem J 329:371–379. doi: 10.1042/bj3390371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doddapaneni H, Yadav JS. 2005. Microarray-based global differential expression profiling of P450 monooxygenases and regulatory proteins for signal transduction pathway in the white rot fungus Phanerochaete chrysosporium. Mol Genet Genomics 274:454–466. doi: 10.1007/s00438-005-0051-2. [DOI] [PubMed] [Google Scholar]
- 45.Scheller U, Zimmer T, Becher D, Schauer F, Schunck W. 1998. Oxygenation cascade in conversion of n-alkanes to alpha,omega-dioic acids catalyzed by cytochrome P450 52A3. J Biol Chem 273:32528–32534. doi: 10.1074/jbc.273.49.32528. [DOI] [PubMed] [Google Scholar]
- 46.Miura Y. 1981. Omega- and (omega-1)-hydroxylation of 1-dodecanol by frog liver microsomes. Lipids 16:721–725. doi: 10.1007/BF02535338. [DOI] [PubMed] [Google Scholar]
- 47.Miura Y, Fulco AJ. 1974. (Omega-2) hydroxylation of fatty acids by a soluble system from Bacillus megaterium. J Biol Chem 249:1880–1888. [PubMed] [Google Scholar]
- 48.Miura Y, Fulco AJ. 1975. Omega-1, omega-2 and omega-3 hydroxylation of long-chain fatty acids, amides and alcohols by a soluble enzyme system from Bacillus megaterium. Biochim Biophys Acta 388:305–317. doi: 10.1016/0005-2760(75)90089-2. [DOI] [PubMed] [Google Scholar]
- 49.D'Annibale A, Ricci M, Leonardi V, Quaratino D, Mincione E, Petruccioli M. 2005. Degradation of aromatic hydrocarbons by white-rot fungi in a historically contaminated soil. Biotechnol Bioeng 90:723–731. doi: 10.1002/bit.20461. [DOI] [PubMed] [Google Scholar]
- 50.Li QS, Ogawa J, Schmid RD, Shimizu S. 2001. Engineering cytochrome P450 BM-3 for oxidation of polycyclic aromatic hydrocarbons. Appl Environ Microbiol 67:5735–5739. doi: 10.1128/AEM.67.12.5735-5739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravichandran KG, Boddupalli SS, Hasermann CA, Peterson JA, Deisenhofer J. 1993. Crystal structure of hemoprotein domain of P450BM-3, a prototype for microsomal P450's. Science 261:731–736. doi: 10.1126/science.8342039. [DOI] [PubMed] [Google Scholar]
- 52.Li HY, Poulos TL. 1997. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat Struct Biol 4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 53.Oliver CF, Modi S, Primrose WU, Lian LY, Roberts GC. 1997. Engineering the substrate specificity of Bacillus megaterium cytochrome P-450 BM3: hydroxylation of alkyl trimethylammonium compounds. Biochem J 327:537–544. doi: 10.1042/bj3270537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graham-Lorence S, Truan G, Peterson JA, Falck JR, Wei S, Helvig C, Capdevila JH. 1997. An active site substitution, F87V, converts cytochrome P450 BM-3 into a regio- and stereoselective (14S,15R)-arachidonic acid epoxygenase. J Biol Chem 272:1127–1135. doi: 10.1074/jbc.272.2.1127. [DOI] [PubMed] [Google Scholar]
- 55.Oliver CF, Modi S, Sutcliffe MJ, Primrose WU, Lian LY, Roberts GC. 1997. A single mutation in cytochrome P450 BM3 changes substrate orientation in a catalytic intermediate and the regiospecificity of hydroxylation. Biochemistry 36:1567–1572. doi: 10.1021/bi962826c. [DOI] [PubMed] [Google Scholar]
- 56.Maddigan NK, Bell SG. 2017. The self-sufficient CYP102 family enzyme, Krac9955, from Ktedonobacter racemifer DSM44963 acts as an alkyl- and alkyloxy-benzoic acid hydroxylase. Arch Biochem Biophys 615:15–21. doi: 10.1016/j.abb.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 57.Bell SG, Hoskins N, Whitehouse CJC, Wong LL. 2007. Design and engineering of cytochrome P450 systems, p 437–476. In Sigel A, Sigel H, Sigel RKO (ed), Metal ions in life sciences, vol 3 John Wiley & Sons Ltd, Chichester, England. [Google Scholar]
- 58.Whitehouse CJ, Bell SG, Wong LL. 2012. P450(BM3) (CYP102A1): connecting the dots. Chem Soc Rev 41:1218–1260. doi: 10.1039/C1CS15192D. [DOI] [PubMed] [Google Scholar]
- 59.Roiban GD, Reetz MT. 2015. Expanding the toolbox of organic chemists: directed evolution of P450 monooxygenases as catalysts in regio- and stereoselective oxidative hydroxylation. Chem Commun (Camb) 51:2208–2224. doi: 10.1039/C4CC09218J. [DOI] [PubMed] [Google Scholar]
- 60.Fasan R. 2012. Tuning P450 enzymes as oxidation catalysts. ACS Catal 2:647–666. doi: 10.1021/cs300001x. [DOI] [Google Scholar]
- 61.Eiben S, Bartelmäs H, Urlacher VB. 2007. Construction of a thermostable cytochrome P450 chimera derived from self-sufficient mesophilic parents. Appl Microbiol Biotechnol 75:1055–1061. doi: 10.1007/s00253-007-0922-z. [DOI] [PubMed] [Google Scholar]
- 62.Yun CH, Kim KH, Kim DH, Jung HC, Pan JG. 2007. The bacterial P450 BM3: a prototype for a biocatalyst with human P450 activities. Trends Biotechnol 25:289–298. doi: 10.1016/j.tibtech.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 63.Kirk TK, Schultz E, Connors WJ, Lorenz LF, Zeikus JG. 1978. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch Microbiol 117:277–285. doi: 10.1007/BF00738547. [DOI] [Google Scholar]
- 64.Shimizu M, Yuda N, Nakamura T, Tanaka H, Wariishi H. 2005. Metabolic regulation at the tricarboxylic acid and glyoxylate cycles of the lignin-degrading basidiomycete Phanerochaete chrysosporium against exogenous addition of vanillin. Proteomics 5:3919–3931. doi: 10.1002/pmic.200401251. [DOI] [PubMed] [Google Scholar]
- 65.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.