Carbon chain elongation means the conversion of short-chain volatile fatty acids to medium-chain carboxylates, such as n-caproate and n-caprylate with electron donors under anaerobic condition. This bio-reaction can both expand the resource of valuable biochemicals and broaden the utilization of low-grade organic residues in a sustainable biorefinery context. Clostridium kluyveri is conventionally considered model microbe for carbon chain elongation which uses the reverse β-oxidation pathway. However, little is known about the detailed microbial structure and function of other abundant microorganism in a mixed culture (or open culture) of chain elongation. We conducted the comparative metagenomic and metatranscriptomic analysis of a chain elongation microbiome to throw light on the underlying functional microbes and alternative pathways.
KEYWORDS: chain elongation, metagenomic, metatranscriptomic, microbial diversity, metabolic pathways, genome reconstruction
ABSTRACT
Carbon chain elongation (CCE), a reaction within the carboxylate platform that elongates short-chain to medium-chain carboxylates by mixed culture, has attracted worldwide interest. The present study provides insights into the microbial diversity and predictive microbial metabolic pathways of a mixed-culture CCE microbiome on the basis of a comparative analysis of the metagenome and metatranscriptome. We found that the microbial structure of an acclimated chain elongation microbiome was a highly similar to that of the original inoculating biogas reactor culture; however, the metabolic activities were completely different, demonstrating the high stability of the microbial structure and flexibility of its functions. Additionally, the fatty acid biosynthesis (FAB) pathway, rather than the well-known reverse β-oxidation (RBO) pathway for CCE, was more active and pivotal, though the FAB pathway had more steps and consumed more ATP, a phenomenon that has rarely been observed in previous CCE studies. A total of 91 draft genomes were reconstructed from the metagenomic reads, of which three were near completion (completeness, >97%) and were assigned to unknown strains of Methanolinea tarda, Bordetella avium, and Planctomycetaceae. The last two strains are likely new-found active participators of CCE in the mixed culture. Finally, a conceptual framework of CCE, including both pathways and the potential participators, was proposed.
IMPORTANCE Carbon chain elongation means the conversion of short-chain volatile fatty acids to medium-chain carboxylates, such as n-caproate and n-caprylate with electron donors under anaerobic condition. This bio-reaction can both expand the resource of valuable biochemicals and broaden the utilization of low-grade organic residues in a sustainable biorefinery context. Clostridium kluyveri is conventionally considered model microbe for carbon chain elongation which uses the reverse β-oxidation pathway. However, little is known about the detailed microbial structure and function of other abundant microorganism in a mixed culture (or open culture) of chain elongation. We conducted the comparative metagenomic and metatranscriptomic analysis of a chain elongation microbiome to throw light on the underlying functional microbes and alternative pathways.
INTRODUCTION
Carbon chain elongation (CCE) was recently highlighted as a promising biorefinery technology for converting hydrophilic short-chain carboxylates (SCCAs; C2 to C5) derived from biowaste into more valuable and hydrophobic medium-chain carboxylates (MCCAs; mainly C6 or C8), which serve as precursors for a range of promising biorefinery products (1–5).
Until now, Clostridium kluyveri has been the most studied and accepted model microbe for MCCA production from ethanol and acetate via the reverse β-oxidation (RBO) pathway (6–8). Eubacterium pyruvativorans, another model organism isolated from the rumen, can form caproate as well (9). E. pyruvativorans has been reported to harbor metabolic properties analogous to those of C. kluyveri (10). Additionally, n-caproic acid can be produced from lactate by a ruminal Ruminococcaceae bacterium (11) and Megasphaera elsdenii (12).
Unlike pure culture, open culture with mixed microorganisms has been described for the chain elongation of biowaste, because a high microbial diversity improves the process stability and resilience under changing conditions. Additionally, no sterilization is required for open culture, making it more applicable for biowaste utilization (3, 13, 14). Unfortunately, little information is available regarding the microecology involved in an open culture chain elongation system. The dominant player responsible for chain elongation and the secondary consumers or predators are unclear. Furthermore, it is unknown whether additional microorganisms are capable of chain elongation and whether there are other alternative chain elongation metabolic pathways.
The majority of microbial knowledge in the context of mixed culture chain elongation is based on 16S rRNA gene analysis. Steinbusch et al. analyzed the microbial composition of open culture chain elongation microbiomes using denaturing gradient gel electrophoresis combined with 16S rRNA gene amplification (15) and found that the chain elongation bacterium C. kluyveri dominated, comprising up to 57.8% of the composition, while another predominant bacterium, Dechlorosoma oryzae (up to 40%), may only convert ethanol to CO2. Agler et al. observed that C. kluyveri comprised only 4% of a mixed culture, and Clostridium spp. contributed more than 50% of all assigned reads, while the abundance of Ruminococcaceae correlated significantly with increasing n-caproic acid production rates (1). Similar abundances of 47.6% for Clostridium spp. (Clostridium ljungdahlii, Clostridium autoethanogenum, and C. kluyveri) were detected by Zhang et al. (16). Liu et al. (17) observed that C. kluyveri was a minor component of the microbiome (2.1%), but Sporanaerobacter acetigenes (24.7%, a class of Clostridia) was abundant. Additionally, abundant methanogens were present, including acetoclastic Methanosaeta concilii (44.8%), although negligible CH4 was produced in the biosystem due to the addition of the methanogenesis inhibitor 2-bromoethanesulfonate (2-BES). Coma et al. (18) found phylotypes related to Bacteroidaceae and microorganisms or unclassified bacteria directly or indirectly involved in CCE, except for C. kluyveri. Our previous research identified one-step MCCA production from CO, which is likely realized by CO-utilizing chain-elongating microorganisms such as Alcaligenes, Acinetobacter, and Rhodobacteraceae (19). Therefore, the relationship between these unexpected, highly abundant organisms and chain elongation remains ambiguous. A comprehensive and complete characterization of the microbial community structure has also been hindered by the low sequencing depth of 16S rRNA sequencing. Agler et al. (1) conducted a metagenomic analysis of genes with >1,000 assigned reads that were highly correlated with n-caproic acid production. However, the full microbial structure of a mixed culture system must be elucidated. Additionally, key genes with low abundance but high transcription might be ignored.
In terms of chain elongation metabolic pathways, the RBO pathway based on pure culture of C. kluyveri was expounded in two reviews (2, 3) and acknowledged by nearly all studies. However, the actual metabolic pathways of a microbially diverse mixed culture are more complex than the RBO pathway alone based on pure culture. There also may be additional pathways and enzyme alternatives. Therefore, more direct proof of the functional genes related to chain elongation is needed.
To explore the dominant chain elongation players in an open culture system and understand the responsibilities of other active components, we applied a pipeline of combined metagenomic and metatranscriptomic approaches (20). By simultaneously investigating the taxonomic composition, functional genes, and gene transcription activities, the overall microbial structure and functional activities were elucidated to achieve a better understanding of specific metabolic pathways for chain elongation in a mixed culture.
RESULTS AND DISCUSSION
Operational performance of the chain elongation bioreactor.
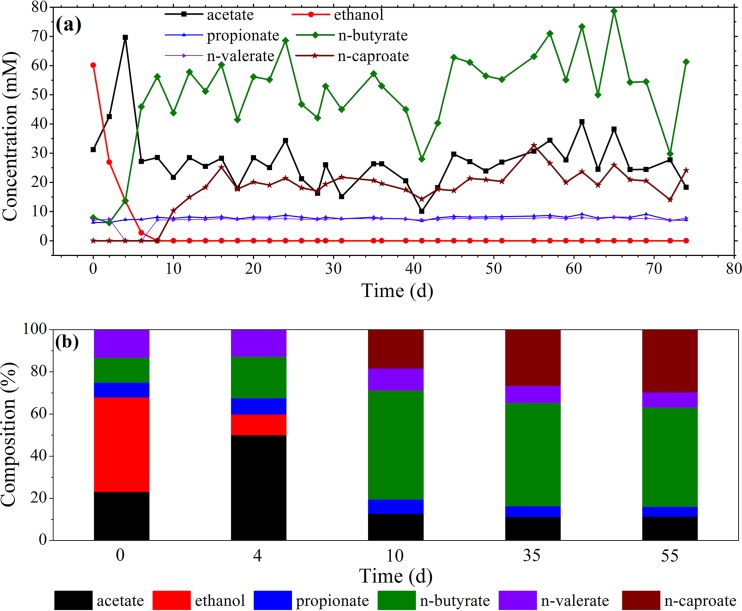
The active CCE microbiome sample for analysis was collected from a laboratory-scale reactor. The carboxylate production profile from the 74-day experimental period is shown in Fig. 1a, and the compositions of the substrates and key products (in mmol/liter carbon) are shown in Fig. 1b. The changes of pH and oxidation-reduction potential (ORP) can be found in Fig. S1 in the supplemental material. Gas production is shown in Table S1. After day 20, there was no gas produced. Overall, negligible CH4 (0.22% of the substrate) was produced as expected. Ethanol oxidation was the major reaction during the first 10 days. Then, n-caproate clearly emerged starting on the 10th day, and the concentration peaked at 26.5 mM on the 55th day, corresponding to a yield of 0.33 mol caproate per mol substrate (ethanol and acetic acid), which is comparable to yields reported in the literature (15, 21, 22). Therefore, the sample collected on the 55th day was considered the active chain elongation microbiome for subsequent metagenomic and transcriptomic analyses.
FIG 1.
(a) Production profiles for carboxylates (acetate, n-butyrate, n-caproate, propionate, and n-valerate) and ethanol. (b) Compositions of the substrates (acetate and ethanol) and key products (n-butyrate, n-caproate, propionate, and n-valerate) based on mM carbon. d, days.
Microbial composition and transcriptionally active taxa of the mixed culture.
Taxonomic annotation was conducted by performing a best hit classification at an E value cutoff of 10−5 with a minimum alignment length of 50 bp on the basis of the entire available source database in MG-RAST (23, 24). The results from the RefSeq database, which provided the most hits compared with those from the other databases (see Table S2), were used for subsequent analyses of the DNA library. For a parallel comparison, the RefSeq database was also used for the cDNA library. The taxonomic assignment of sequences in the DNA and cDNA data sets revealed hundreds of genera and thousands of species in the community (Table 1), demonstrating that the population within the studied mixed culture was extraordinarily microbially diverse. Whole interactive Krona charts of the full taxonomy, showing the panorama of the microbial structure, are shown in Fig. S2 and S3 for the metagenome and metatranscriptome, respectively.
TABLE 1.
Microbial diversity of the studied mixed culture at various phylogenetic levels
| Taxonomy level | No. identified |
|
|---|---|---|
| Metagenome | Metatranscriptome | |
| Domaina | 4 | 4 |
| Phylum | 62 | 57 |
| Class | 145 | 136 |
| Order | 303 | 266 |
| Family | 508 | 479 |
| Genus | 996 | 912 |
| Species | 2,323 | 1,970 |
| Strain | 3,544 | 3,006 |
The four domains refer to bacteria, archaea, eukarya, and viruses. Viruses were excluded at other taxonomic levels because of their low abundance (<0.1%).
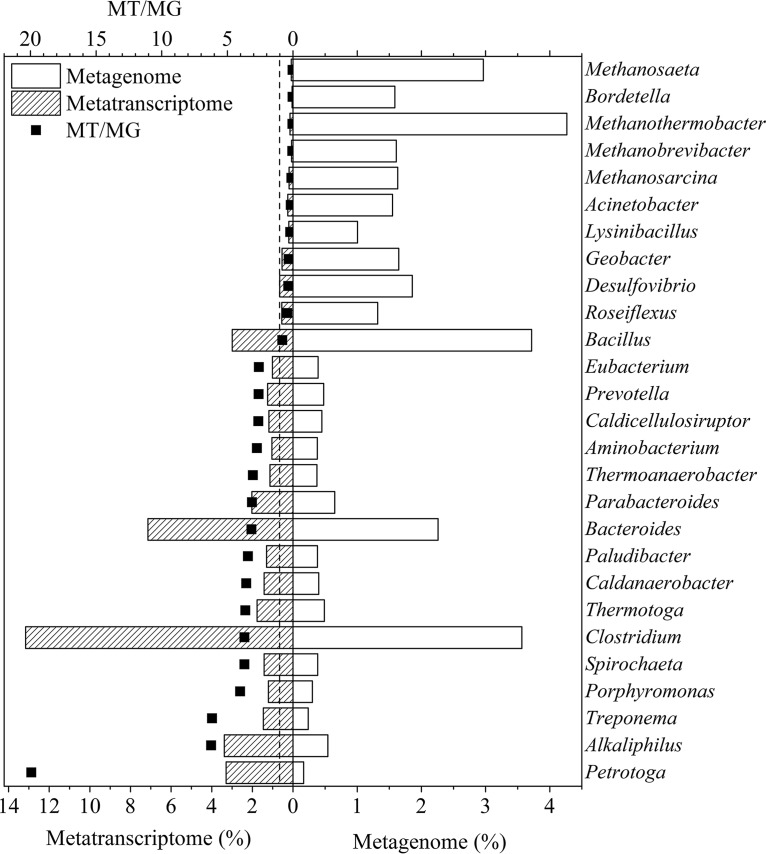
Taxonomic assignment of the DNA gene sequences (see Table S3) revealed that Methanothermobacter (4.3%), Bacillus (3.7%), Clostridium (3.6%), Methanosaeta (3.0%), and Bacteroides (2.3%) were the most abundant populations at the genus level, which is similar to the results from previous metagenomic analyses of biogas-producing cultures (25–27). The dominance of the methanogens Methanothermobacter and Methanosaeta was confusing, since 2-BES was added as methanogenesis inhibitor, negligible CH4 generation was detected (Table S1), and the above methanogens are rarely reported to produce acids. When performing 16S rRNA gene sequencing, Liu et al. also observed abundant methanogens in their CCE reactors used for long-term operation with the addition of 2-BES, which might take part in ethanol oxidation (17).
Comparatively, the taxonomic assignment of the cDNA gene sequences demonstrated that Clostridium (13.2%), Bacteroides (7.1%), Alkaliphilus (3.4%), Petrotoga (3.3%), Bacillus (3.0%), and Parabacteroides (2.0%) (see Table S4) were the most transcriptionally active genera. Suspected methanogens were not assigned. This result was reasonable, since the tested culture was derived from the biogas reactor and operated in batches with the addition of a methanogen inhibitor, which resulted in an abundant number of methanogens but lower metabolic activities of methanogens.
Carboxylate metabolism is tightly associated with the predominance of Clostridium; in particular, we detected an abundance of C. kluyveri, which is a model organism that converts acetate and ethanol to butyrate and caproate (8). Several researchers have also found C. kluyveri to be the dominant microorganism in chain elongation functional cultures on the basis of 16S rRNA technology (1, 15). However, other species of Clostridium were also identified, including Clostridium botulinum, Clostridium thermocellum, Clostridium difficile, Clostridium beijerinckii, Clostridium carboxidivorans, and others, all of which likely participated in the metabolic progression of chain elongation because they had similar abundances to that of C. kluyveri.
Meanwhile, the ratio of the relative abundances of those taxa from the metatranscriptome and metagenome (defined as the MT/MG ratio) varied from 0.0 to 6.2, with the exception of the extraordinarily high MT/MG ratio (19.9) for Petrotoga (Fig. 2). Some other groups with lower genomic abundances, such as Alkaliphilus, Treponema, Porphyromonas, and Spirochaeta, also exhibited much higher relative transcriptomic abundances (Fig. 2), suggesting more active gene transcription per cell than other taxa.
FIG 2.
Relative abundances of the dominant taxa in the metagenome and metatranscriptome at the genus level. Only relative abundances of identified genera higher than 1% in DNA or cDNA library are listed. ■, the ratio of relative abundances from the metatranscriptome and metagenome (MT/MG).
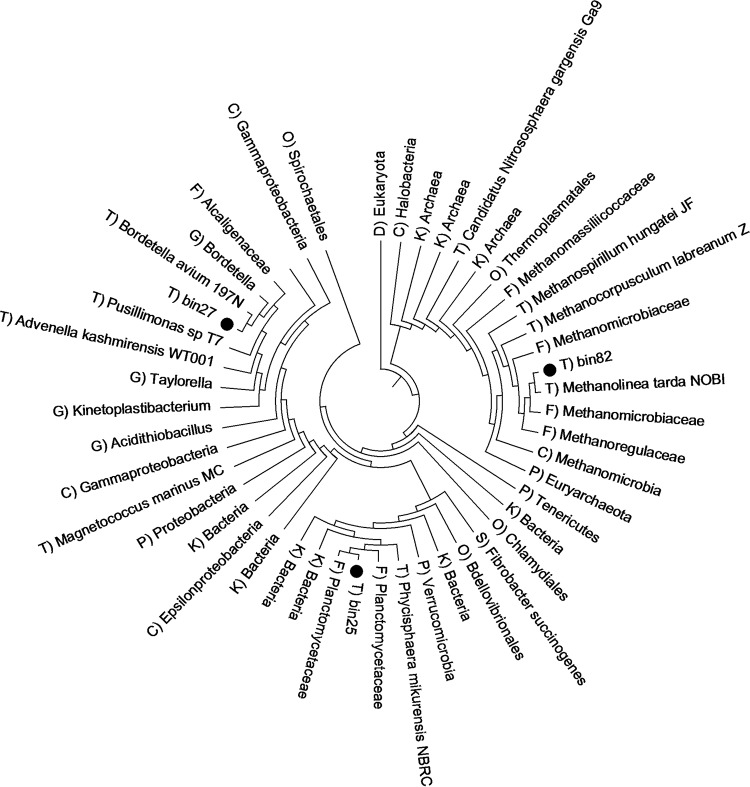
The taxonomic analysis described above, based on metagenomic and metatranscriptomic reads, was limited by the cultured and known genomes in the database. However, there are enormous numbers of uncultured microbes whose genomes are completely unknown and are not included in the database. We successfully reconstructed three near-complete genomes from the metagenomic reads (Table 2). A phylogenetic tree was constructed on the basis of the whole genome (Fig. 3). All three genome bins were assigned to unknown microorganisms. Bin82 demonstrated the highest number of closed relationships with the strain Methanolinea tarda NOBI; thus, we deduced that Bin82 was a methanogen, specifically, a strain of Methanolinea tarda. In the same way, Bin27 was similar to Bordetella avium 197N and was designated Bordetella avium. Bin25 was taxonomically assigned to Planctomycetaceae at the family level.
TABLE 2.
Characteristics of high-quality genome bins
| Characteristic | Bin25 | Bin27 | Bin82 |
|---|---|---|---|
| No. of markers | 142 | 427 | 234 |
| Completeness (%) | 98.85 | 97.66 | 97.71 |
| Contamination (%) | 0.45 | 0.86 | 0 |
| Genome size (bp) | 6,924,786 | 4,321,078 | 1,805,897 |
| No. of contigs | 678 | 421 | 178 |
| Mean contig length | 10,213 | 10,263 | 10,145 |
| No. of predicted genes | 5,362 | 4,242 | 1,956 |
| GC content (%) | 65.00 | 65.70 | 56.18 |
| Coding density | 85.35 | 91.19 | 87.19 |
FIG 3.
Phylogenetic tree of the reconstructed genome bins: Bin25, Bin27, and Bin82. D, domain; K, kingdom; P, phylum; C, class; O, order; F, family; G, genus; S, species; T, strain.
Annotated functions of genes from the DNA and cDNA data sets.
Metagenomic and metatranscriptomic sequencing also provided information to evaluate potential microbial functions and enzymatic activities. To perform functional profiling and then predict possible microbial metabolic pathways of the mixed culture, the total reads were annotated on the basis of the categories of all accessible relevant databases in MG-RAST, including Subsystems, COG, and KO. The results from various databases (Table S2) were coincident in general; however, there were differences due to the unique characteristics of the given databases. The results from the Subsystems database were used for the subsequent discussion, because this database had the highest number of annotated hits and it was convenient to compare the Subsystem results of many other researchers.
Subsystems.
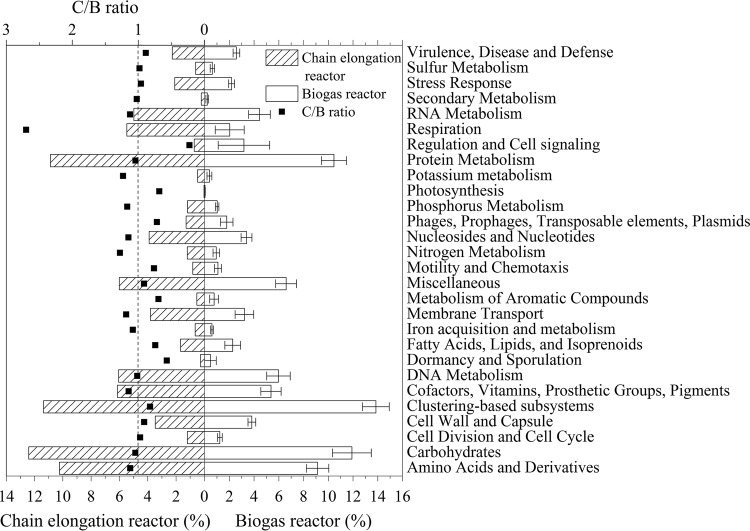
The functional genes in the metagenome of the present studied culture were compared with those from biogas reactor cultures in the literature. The value of the biogas reactor was the average and standard deviation of four metagenomic studies with 17 total biogas reactors: 14 full-scale biogas reactors treating manure and sludge (27), a full-scale anaerobic reactor digesting activated sludge from wastewater treatment (25) (MG-RAST identifier [ID], mgm4536159.3), an anaerobic granules upflow reactor in Delft (MG-RAST ID, mgm4559623.3), and a biogas reactor treating digestate (MG-RAST ID, mgm4446507.3). As shown in Fig. 4, the comparison demonstrated that the major functional categories of the chain elongation culture and of the biogas reactor cultures were similar in general. The relative abundance ratios of the categories of annotated genes from the chain elongation culture to those of the biogas reactor cultures (C/B ratio) varied from 0.7 to 1.2, except for the categories of respiration (2.7), regulation and cell signaling (0.2), and dormancy and sporulation (0.5), indicating that the metabolic activities of chain elongation encompass a significantly enhanced respiration function compared with the activities of biogas production. In contrast, the functions of regulation and cell signaling and of dormancy and sporulation were weakened. There were no significant differences for other functional categories. This was consistent with the similar microbial compositions of the chain elongation and biogas reactor cultures.
FIG 4.
Distribution of the functional genes obtained in the present study and in biogas reactor microbiome as determined with Subsystems. ■, the ratio of relative abundances of functional genes between chain elongation microbiome and biogas reactor microbiome based on metagenomic data (C/B ratio). The data for the biogas reactor are averages from 17 biogas reactors, and the error bars indicate the standard deviations.
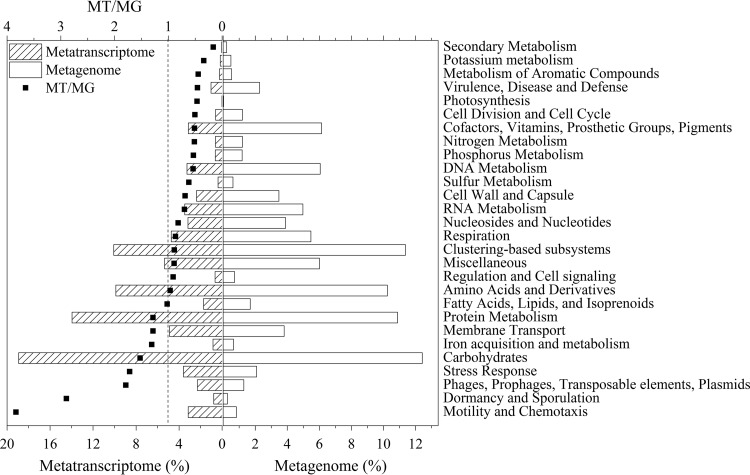
To identify the transcriptionally active and inactive functional genes, the distributions of the functional genes from DNA and cDNA sequences were compared (Fig. 5). Overall, the categories of functional genes had similar distributions between the DNA and cDNA libraries. The carbohydrates category was the most abundant, followed by protein metabolism, amino acids and derivatives, and clustering-based subsystems (encompassing functions such as proteasomes, ribosomes, and recombination-related clusters [28]) for both the DNA and cDNA libraries, implying that these genes were not only abundant but also highly transcribed. Thus, the involved genes in these categories play a vital role in functional metabolic pathways, with the exception of basic metabolic activities.
FIG 5.
Distribution of the functional genes in the metagenome and metatranscriptome as determined with Subsystems. ■, the ratio of relative abundances between metatranscriptome and metagenome (MT/MG).
The numerical ratios of the relative abundances of the different categories from the DNA and cDNA reads (MT/MG), shown in Fig. 5, were calculated to indicate the transcriptional activity of the annotated genes. Genes from the categories of motility and chemotaxis, dormancy and sporulation, phages, prophages, transposable elements, plasmids, stress response, and carbohydrates were actively transcribed, because the ratios of the relative abundances from the DNA and cDNA reads within these categories were all above 1.5. Since the carbohydrates category was not only actively transcribed but also had the highest abundance (Fig. 5), it was further analyzed. As shown in Table S5 and Table S6, central carbohydrate metabolism was the major function in the level 2 subsystems for both the DNA and cDNA profiles. Central carbohydrate metabolism is used to describe the integration of the transport and oxidation pathways for the main carbon sources inside the cell (29). This involves a complex series of enzymatic steps to convert external substrates into significant metabolic precursors, such as acetyl coenzyme A (acetyl-CoA) and pyruvate (30). These precursors, especially acetyl-CoA, which is the core intermediate in this metabolic process according to a previous study (30), are tightly associated with the process of chain elongation (3, 14).
COG.
The joint reads were also annotated according to the COG database to perform a more detailed analysis of the functional genes. Overall, the results from the DNA and cDNA sequences were coincident at level 1 of the COG categories. In detail, large numbers of reads (48.21% for DNA, 50.53% for cDNA) were associated with metabolism, followed by information storage and processing (21.06% for DNA, 21.55% for cDNA) and cellular processes and signaling (18.05% for DNA, 17.88% for cDNA). The specific functions annotated by COG are listed in Table S7 for DNA reads and Table S8 for cDNA reads. In the metabolism category, the dominating metabolism type was energy production and conversion, followed by amino acid transport and metabolism and carbohydrate transport and metabolism. These metabolic activities are closely associated with the conversion of short-chain volatile fatty acids to medium-chain fatty acids during the fermentation process. These results were also consistent with the results obtained from Subsystems.
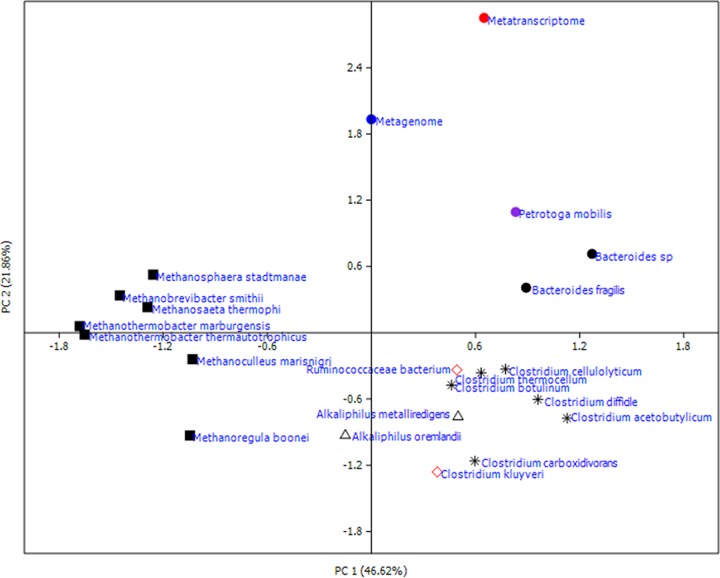
To identify potential metabolic patterns on the basis of the annotated genes from COG, the functional categories of the annotated DNA and cDNA reads were compared with those from specific strains whose metabolic features have been well studied using principal-component analysis (PCA) based on Bray-Curtis distances. Twenty species were chosen, and their annotated genes according to COG were downloaded from UniProt (http://www.uniprot.org/). Afterwards, the COG categories of these species were determined (see Table S9). C. kluyveri (8, 31) and Ruminococcaceae bacterium (11) were selected because they both elongate short-chain acids via the RBO pathway. According to the metagenomic taxonomy at the species level (Table S3), there were 7 methanogens among the top 10 most abundant species, and these were chosen as well. From the metatranscriptomic taxonomy (Table S4), the dominant species Petrotoga mobilis, Alkaliphilus metalliredigens, and Alkaliphilus oremlandii from Alkaliphilus, Bacteroides spp. and Bacteroides fragilis from Bacteroides, and Clostridium botulinum, Clostridium thermocellum, Clostridium difficile, Clostridium carboxidivorans, Clostridium cellulolyticum, and Clostridium acetobutylicum from Clostridium were also taken into consideration.
As shown in Fig. 6, there are clear differences between Clostridium and the methanogens; this finding was expected because these microbes utilize different metabolic pathways. P. mobilis had a metabolic pattern similar to that of Clostridium, which is reasonable because P. mobilis was the most abundant species among the metatranscriptomic taxa. P. mobilis ferments a wide variety of carbohydrates, which are isolated from hot oil field water of a North Sea oil reservoir (32). The DNA-based data set unexpectedly clustered apart from the methanogens. This again suggests that the quantitatively dominant microbes do not contribute according to the functional metabolic activities, which is consistent with the microbial composition analysis described above. However, the cDNA-based data set was distant from both methanogens, Clostridium, and the two chain elongation model bacteria. On the basis of annotated functional genes, the above results indicated that the functional culture derived from the biogas reactor has the potential for chain elongation. However, the actual metabolic pattern was surprisingly complex and differed from well-studied patterns. Therefore, more efforts should be made to reveal the complex and interactive metabolic pathways.
FIG 6.
PCA of the DNA and cDNA data sets and core species of the taxonomic profile based on COG functional gene categories. CE model bacteria (red open diamond): Clostridium kluyveri and Ruminococcaceae bacterium; Clostridium ( ): C. botulinum, C. thermocellum, C. difficile, C. carboxidivorans, C. cellulolyticum, and C. acetobutylicum; Alkaliphilus (△): A. metalliredigens and A. oremlandii; Bacteroides (●): Bacteroides sp. and B. fragilis; methanogens (■): Methanosaeta thermophila, Methanobrevibacter smithii, Methanoculleus marisnigri, Methanoregula boonei, Methanosphaera stadtmanae, Methanothermobacter marburgensis, and Methanothermobacter thermautotrophicus; Petrotoga: Petrotoga mobilis.
): C. botulinum, C. thermocellum, C. difficile, C. carboxidivorans, C. cellulolyticum, and C. acetobutylicum; Alkaliphilus (△): A. metalliredigens and A. oremlandii; Bacteroides (●): Bacteroides sp. and B. fragilis; methanogens (■): Methanosaeta thermophila, Methanobrevibacter smithii, Methanoculleus marisnigri, Methanoregula boonei, Methanosphaera stadtmanae, Methanothermobacter marburgensis, and Methanothermobacter thermautotrophicus; Petrotoga: Petrotoga mobilis.
Key enzymes and potential metabolic pathways for chain elongation.
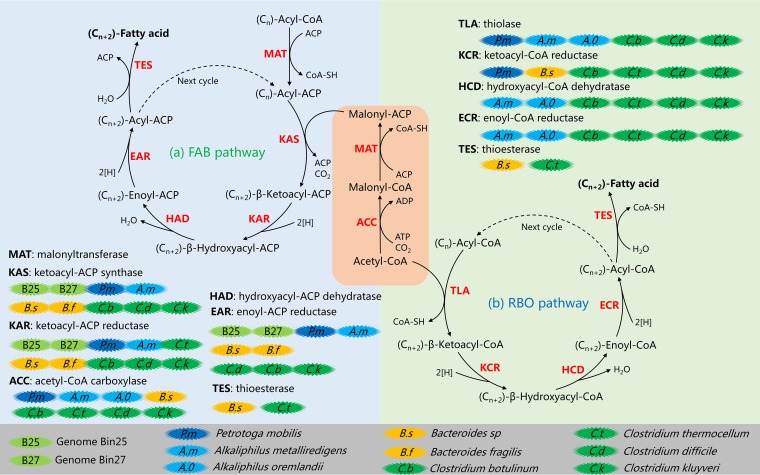
All the functional genes of the metagenome or metatranscriptome were annotated against the KO database to carry out enzyme assays to further investigate the chain elongation pathways. Functional enzymes involved in MCCA formation were specifically studied and are listed in Table 3. Two cyclic pathways were constructed (Fig. 7). The RBO pathway was identified on the basis of information in the literature (2, 8, 33, 34), and the fatty acid biosynthesis (FAB) pathway (below) was generated on the basis of the information from previous studies (35–39) and the KEEG Pathway database (http://www.genome.jp/kegg/pathway.html). The enzymes involved and annotated in this study were labeled during the process.
TABLE 3.
Detailed information on the functional enzymes involved in MCCA formation
| Abbreviation | Name | Gene | Definition | KO entry | EC no. | COG | MT |
MG |
MT/MG | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of hits | % | No. of hits | % | ||||||||
| Reverse β oxidation | |||||||||||
| TLA | Thiolase | fadA | Acetyl-CoA acyltransferase | K00632 | 2.3.1.16 | COG0183 | 78 | 0.0157 | 1,429 | 0.0487 | 0.32 |
| HADHB | Acetyl-CoA acyltransferase | K07509 | 2.3.1.16 | 1 | 0.0002 | 3 | 0.0001 | 1.96 | |||
| KCR | Ketoacyl-CoA reductase | phbB | Acetoacetyl-CoA reductase | K00023 | 1.1.1.36 | COG1028 | 36 | 0.0072 | 241 | 0.0082 | 0.88 |
| HCD | Hydroxyacyl-CoA dehydratase | fadB | 3-Hydroxybutyryl-CoA dehydrogenase | K00074 | 1.1.1.157 | COG1250 | 1,404 | 0.2820 | 2,089 | 0.0713 | 3.96 |
| fadJ | 3-Hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase/3-hydroxybutyryl-CoA epimerase | K01782 | 1.1.1.35, 4.2.1.17, 5.1.2.3 | COG1250, COG1024 | 8 | 0.0016 | 201 | 0.0069 | 0.23 | ||
| fadN | 3-Hydroxyacyl-CoA dehydrogenase | K07516 | 1.1.1.35 | COG1250, COG1024 | 29 | 0.0058 | 1,338 | 0.0456 | 0.13 | ||
| HADH | 3-Hydroxyacyl-CoA dehydrogenase | K00022 | 2.3.1.16 | 4 | 0.0008 | 29 | 0.0010 | 0.81 | |||
| ECR | Enoyl-CoA reductase | ysiB, fadB | Enoyl-CoA hydratase | K13767 | 4.2.1.17 | COG1024 | 14 | 0.0028 | 109 | 0.0037 | 0.76 |
| fadJ | 3-Hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase/3-hydroxybutyryl-CoA epimerase | K01782 | 1.1.1.35, 4.2.1.17, 5.1.2.3 | COG1250, COG1024 | 8 | 0.0016 | 201 | 0.0069 | 0.23 | ||
| paaF, echA | Enoyl-CoA hydratase | K01692 | 4.2.1.17 | COG1024 | 85 | 0.0171 | 2,867 | 0.0978 | 0.17 | ||
| echA | Ech hydrogenase subunit A | K14086 | 6 | 0.0012 | 55 | 0.0019 | 0.64 | ||||
| crt | 3-Hydroxybutyryl-CoA dehydratase | K01715 | 4.2.1.17 | 716 | 0.1438 | 563 | 0.0192 | 7.49 | |||
| TES | Thioesterase | yciA | Acyl-CoA thioesterase YciA | K10806 | 3.1.2.− | COG1607 | 7 | 0.0014 | 71 | 0.0024 | 0.58 |
| Fatty acid biosynthesis | |||||||||||
| ACC | Acetyl-CoA carboxylase | accA | Acetyl-CoA carboxylase carboxyl transferase subunit alpha | K01962 | 6.4.1.2 | COG0825 | 128 | 0.0257 | 1,284 | 0.0438 | 0.59 |
| accB | Acetyl-CoA carboxylase biotin carboxyl carrier protein | K02160 | COG0511 | 52 | 0.0104 | 331 | 0.0113 | 0.93 | |||
| accC | Acetyl-CoA carboxylase, biotin carboxylase subunit | K01961 | 6.4.1.2, 6.3.4.14 | COG0439 | 256 | 0.0514 | 3,059 | 0.1043 | 0.49 | ||
| accD | Acetyl-CoA carboxylase carboxyl transferase subunit beta | K01963 | 6.4.1.2 | COG0777 | 94 | 0.0189 | 1,067 | 0.0364 | 0.52 | ||
| MAT | Malonyltransferase | fas | Fatty acid synthase, bacteria type | K11533 | 2.3.1.− | 3 | 0.0006 | 3 | 0.0001 | 5.89 | |
| fabD | [Acyl-carrier-protein] S-malonyltransferase | K00645 | 2.3.1.39 | COG0331 | 283 | 0.0568 | 1,436 | 0.0490 | 1.16 | ||
| KAS | Ketoacyl-ACP synthase | fas | Fatty acid synthase, bacteria type | K11533 | 2.3.1.− | 3 | 0.0006 | 3 | 0.0001 | 5.89 | |
| fabB | 3-Oxoacyl-[acyl carrier protein] synthase I | K00647 | 2.3.1.41 | COG0304 | 20 | 0.0040 | 227 | 0.0077 | 0.52 | ||
| fabF | 3-Oxoacyl-[acyl carrier protein] synthase II | K09458 | 2.3.1.179 | COG0304 | 1,224 | 0.2458 | 3,931 | 0.1341 | 1.83 | ||
| fabH | 3-Oxoacyl-[acyl carrier protein] synthase III | K00648 | 2.3.1.180 | COG0332 | 617 | 0.1239 | 3,341 | 0.1140 | 1.09 | ||
| KAR | Ketoacyl-ACP reductase | fas | Fatty acid synthase, bacteria type | K11533 | 2.3.1.− | 3 | 0.0006 | 3 | 0.0001 | 5.89 | |
| fabG | 3-Oxoacyl-[acyl carrier protein] reductase | K00059 | 1.1.1.100 | COG1028 | 1,003 | 0.2014 | 5,275 | 0.1799 | 1.12 | ||
| HAD | Hydroxyacyl-ACP dehydratase | fas | Fatty acid synthase, bacteria type | K11533 | 2.3.1.− | 3 | 0.0006 | 3 | 0.0001 | 5.89 | |
| fadZ | 3-Hydroxyacyl-[acyl carrier protein] dehydratase | K02372 | 4.2.1.59 | COG0764 | 241 | 0.0484 | 1,007 | 0.0343 | 1.41 | ||
| EAR | Enoyl-ACP reductase | fas | Fatty acid synthase, bacteria type | K11533 | 2.3.1.− | 3 | 0.0006 | 3 | 0.0001 | 5.89 | |
| fabI | Enoyl-[acyl carrier protein] reductase I | K00208 | 1.3.1.9, 1.3.1.10 | COG0623 | 150 | 0.0301 | 777 | 0.0265 | 1.14 | ||
| fabK | Enoyl-[acyl carrier protein] reductase II | K02371 | 1.3.1.− | COG2070 | 163 | 0.0327 | 884 | 0.0302 | 1.09 | ||
| fabL | Enoyl-[acyl carrier protein] reductase III | K10780 | 1.3.1.− | COG1028 | 6 | 0.0012 | 216 | 0.0074 | 0.16 | ||
FIG 7.
Metabolic pathways and possible participants in chain elongation. The FAB pathway (top left) and RBO pathway (top right) are presented. The two pathways were constructed with information from enzymes annotated against the KO database. Abbreviations: ACP, acyl carrier protein; CoA, coenzyme A; 2[H], NADH/NADPH/ferredoxin. Dashed lines indicate the next cycle of elongation, of which (Cn+2)-acyl-CoA/ACP plays the role of starter molecule. Key enzymes are highlighted in red, and involved enzymes are listed in Table 3. (Cn), the number of carbon atoms of the molecule [for example, in (Cn)-acyl-CoA, an n of 1 means formyl-CoA, an n of 2 means acetyl-CoA, an n of 3 means propionyl-CoA, etc.]. Each turn of the cycle generates an acyl-CoA/ACP (indicated as Cn+2) which is two carbons longer than the initial acyl-CoA/ACP (indicated as Cn).
Chain elongation research (2, 8, 33, 34) has shown that the process of chain elongation in mixed culture is achieved via the RBO pathway, which centers on acetyl-CoA. Acetyl-CoA serves as a 2-C donor, adding two carbon atoms to the initial acyl-CoA molecule every cycle to achieve CCE. In the present study, acetyl-CoA was primarily derived from ethanol oxidation. The substrate acetate is also an alternative acetyl-CoA source. Meanwhile, the 2-C donor acetyl-CoA is generated from various molecules, such as lactate (40), methanol (41), and even hydrogen and carbon dioxide (16, 42), in open culture fermentation, according to previous work.
Although the RBO pathway was treated as a model pathway for chain elongation in mixed culture, in the present study, the FAB pathway also attracted our attention. All the required functional enzymes for the FAB pathway were identified in both the metagenomic and metatranscriptomic profiles. Thus, the FAB pathway likely contributed to MCCA production in this study. Unlike acetyl-CoA in the RBO pathway, malonyl-CoA plays the role of a 2-C donor in the FAB process. However, both the RBO and FAB pathways are cyclic processes, and two carbon atoms are added to the starter molecule in each cycle. The detailed processes are shown in Fig. 7. MCCA products host both even-carbon chains and odd-carbon chains, which are determined by the substrate. For example, if acetate (C2) acts as a substrate, its possible products are n-butyrate (C4), n-caproate (C6), and n-caprylate (C8) (1, 2, 17, 43). If the substrate is propionate (C3), n-valerate (C5) or n-heptylate (C7) is produced (44).
The FAB pathway is less efficient because it is longer than the RBO pathway. Additionally, the FAB pathway consumes more ATP because the use of acyl-acyl carrier protein (ACP) intermediates and malonyl-ACP as the 2-C donor limits its ATP efficiency. The net consumption of 1 ATP per molecule is required for the synthesis of malonyl-ACP (33, 35, 37). However, in the present study, the FAB pathway was likely even more active than the RBO pathway, because the enzymes of the FAB pathway had higher numbers of hits (as well as a high proportion in the annotated enzyme profile) against the KO database than the enzymes of the RBO pathway, in both the DNA and cDNA profiles (Table 3). This indicated that the transcription of these genes was also active, concomitant with the high abundance of the genes responsible for the FAB pathway. Additionally, the MT/MG ratios in Table 3 indicated the transcriptional activity of the relative genes. The overall value of the MT/MG ratios for the FAB enzymes exceeded 1.0, while the same value for the RBO enzymes was less than 1.0, indicating higher levels of transcription for the FAB pathway enzymes than for the RBO pathway enzymes. This, in turn, also explained the distance between the cDNA-based data set and the two RBO model bacteria in the PCA analysis (Fig. 6).
Corresponding microbes for chain elongation.
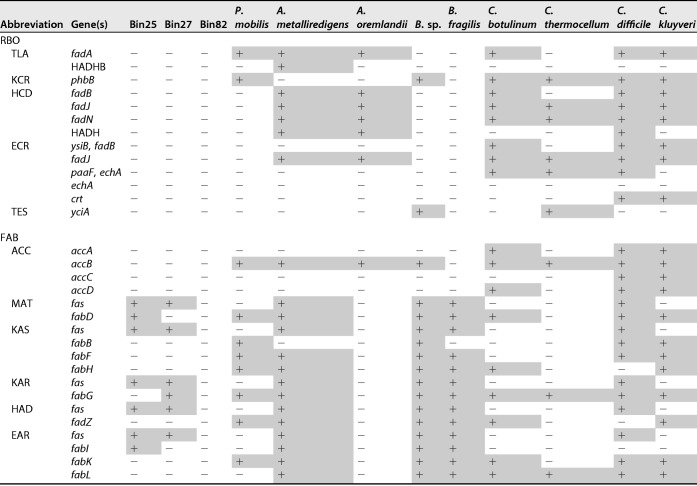
Although the integrated metagenomic and metatranscriptomic analysis provided us with more direct proof of the pathway involved in chain elongation in this study, the enzymes mentioned above are hosted by numerous microbes in mixed culture. Furthermore, some microbes have all the necessary enzymes for these pathways (i.e., a pure culture of C. kluyveri achieves MCCA production via the RBO pathway [8]). It is difficult to determine all the actual hosts for each specific enzyme and bioreaction. However, the dominant species in the metatranscriptomic taxonomy and with a high value of MT/MG, for example, P. mobilis, likely contributed to MCCA production. The enzyme composition of these suspected microbes, including the reconstructed bins, was investigated, and MCCA production-related enzymes are listed in Table 4. Some of the microbes possess all the necessary enzymes for the FAB pathway, and they are probably the contributors for CCE, such as P. mobilis and newly reconstructed Bin27 and Bin25. Bin82 was assigned to a strain of Methanolinea tarda, which is a methanogen. But Bin82 lacks all the necessary enzymes for both RBO and FAB pathways and thus has no relationship with CCE. The probable contributions of these microbes are shown in Fig. 7. However, the actual contributions of various microbes to MCCA production are still unclear and warrant further investigation through systematic multi-omic analysis (including metaproteomics and metabolomics), isotope-labeled flux analysis, or genetic engineering manipulation to obtain direct proof.
TABLE 4.
Presence of MCCA production-related enzymes in the microbes suspected of contributing to MCCA productiona
Shading indicates the enzyme is included in the microbe. B. sp., Bacteroides sp.
Conclusion.
In the present study, the obtained results showed there is a high abundance of microorganisms as deduced from a metagenomic analysis, which does not necessarily indicate high transcriptional or metabolic activity, and vice versa. Biogas reactor-derived and chain elongation-acclimated microbiomes shared similar microbial structures in comparison to those of a biogas reactor culture; however, the metabolic activities were completely different.
Using a metagenomic assembly approach, we retrieved 91 draft genomes in total, 3 of which were nearly complete and were assigned to unknown strains of Methanolinea tarda, Bordetella avium, and Planctomycetaceae, which, except for the methanogen Methanolinea tarda, were probably the active participants of medium-chain carboxylate production. By investigating the abundances and transcriptional activities of the genes associated with chain elongation, we reconstructed the potential pathway for MCCA production. The well-known RBO pathway for chain elongation was unexpectedly less active, and the FAB pathway instead played a more significant role in MCCA production. Finally, a conceptual framework of chain elongation including two pathways and potential participators was proposed.
Further studies utilizing more powerful methods such as a systematic multi-omic analysis (including metaproteomics and metabolomics), isotope-labeled flux analysis, or genetic engineering manipulation are required to elucidate the large number of currently unknown species and the insufficiently characterized metabolic pathways involved in chain elongation.
MATERIALS AND METHODS
Reactor setup and incubation.
The chain elongation experiment was carried out in a 21-liter upflow blanket filter reactor, which had an effective volume of 18 liters. A 2-liter gas bag was attached to collect any gas produced. The inoculum was anaerobic sludge obtained from a mesophilic reactor treating paper mill wastewater. The substrates were 60 mM ethanol and 20 mM acetic acid, which were added to the basic culture medium, while 50 mM 2-BES was added as a methanogenesis inhibitor. The concentration of the inoculum was 8 g · liter−1 based on volatile solids (VS). The pH of the medium was only initially adjusted to 7.00 by adding 4 M KOH and was not controlled afterwards. Figure S1 in the supplemental material shows that the pH values were in the range of 6.37 to 7.15 during the operation period. The reactor was purged with high-purity (99.999%) nitrogen gas to guarantee anaerobic conditions after it was sealed. The basic culture medium per liter contained NaHCO3, 5,040 mg; NH4H2PO4, 3,600 mg; MgCl2·6H2O, 330 mg; MgSO4·7H2O, 200 mg; KCl, 150 mg; CaCl2·2H2O, 200 mg; MnCl2·4H2O, 0.5 mg; H3BO3, 0.05 mg; ZnCl2, 0.05 mg; CuCl2, 0.03 mg; Na2MoO4·2H2O, 0.01 mg; CoCl2·6H2O, 1 mg; NiCl2·6H2O, 0.1 mg; Na2SeO3, 0.05 mg; biotin, 0.02 mg; pyridoxine-HCl, 0.1 mg; thiamine-HCl·2H2O, 0.05 mg; d-capantothenate, 0.05 mg; folic acid, 0.02 mg; riboflavin, 0.05 mg; nicotinic acid, 0.05 mg; p-aminobenzoic acid, 0.05 mg; vitamin B12, 0.001 mg; and lipoic acid, 0.05 mg. All the chemicals used in the study were of analytical grade or better.
The reactor was operated in batch mode at 35 ± 1°C, and no additional substrates were added throughout the 74-day incubation. Periodic recirculation in upflow mode at a frequency of 15 min · h−1 and a flow rate of 300 ml · min−1 was applied through a peristaltic pump. The gas bag was emptied when it was necessary, and the components and volume of the gas in the gas bag were tested. Ten milliliters of the solid-liquid mixture was collected from the reactor every 2 days and stored at −80°C for physiochemical analysis after pH and ORP were determined with a pH meter (PXSJ-216F; Shanghai Precision and Scientific Instrument Co., Ltd., Shanghai, China) and an automatic potentiometric titrator (ZD-2; Shanghai Precision and Scientific Instrument Co., Ltd., Shanghai, China), respectively. The samples collected for DNA and RNA extraction were immediately frozen in liquid nitrogen and stored at −80°C for future use, while RNase-free tubes were used to store samples for RNA extraction.
Measurement of liquid and gaseous compounds.
The concentrations of carboxylates (C2 to C8, i.e., acetate, n-butyrate, n-caproate, propionate, i-butyrate, n-valerate, i-valerate, and n-heptanoate) and ethanol were measured using a gas chromatograph (Focus GC; Thermo Scientific Co., Waltham, MA, USA) equipped with a flame ionization detector and a 30 m by 0.25 mm DB-WAX UI polyethylene glycol capillary column (122-7032UI; Agilent, USA). The gas chromatograph column operating temperature was maintained at 60°C for 1 min, increased by 30°C/min to 130°C, increased again by 15°C/min to 220°C, and maintained at 220°C for 2 min. The injector temperature was set at 250°C, and the detector was set at 280°C. An automatic sampler injected 1 μl of each sample, while the split ratio was 10. Prior to injection, the liquid samples were centrifuged at 16,000 rpm for 10 min at 4°C in a high-speed refrigerated centrifuge (TL-18 M; Shanghai, China) and then diluted and acidified with 3% (vol/vol) phosphoric acid. The conversion efficiency was evaluated by the production of targeted and nontargeted fermentation products.
The components of the gas (O2, H2, CH4, and CO2) were measured using a gas chromatograph (Trace1300; Thermo Fisher Scientific, Waltham, MA, USA) equipped with a flame ionization detector and thermal conductivity detector. The volume of the gas was determined by a gas meter (TG05/6; Ritter, Bochum, Germany).
DNA and RNA extraction and sequencing.
Total DNA extraction was conducted using an E.Z.N.A. Soil DNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA), according to the manufacturer's instructions. DNA quality was assessed by gel electrophoresis (1% agarose), and DNA concentrations were determined by a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA).
Total RNA was extracted using a RNeasy minikit (Qiagen, Germany) according to the manufacturer's protocol with a minor modification: RNA-free sterile zirconia beads were added to the samples to improve the extraction yield (45). Any residual DNA was then removed from the RNA sample using an RNase-free DNase set (Qiagen, Germany) according to the manufacturer's instructions. The quality and quantity of total RNA produced were also estimated with a NanoDrop 2000 spectrophotometer and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Thereafter, rRNA was depleted using a Ribo-Zero rRNA removal kits (meta-bacteria and plant leaf; Epicenter Biotechnologies, Madison, WI). Poly(A) RNA was removed using an Oligotex mRNA kit (Qiagen, Valencia, CA). A TruSeq RNA Sample Prep kit (Illumina, San Diego, CA) was used for sample preparation according to the TruSeq sample preparation guide. Thereafter, reverse-transcribed cDNA was obtained and prepared for sequencing.
Metagenomic and metatranscriptomic sequencing was performed using an Illumina HiSeq 2500 platform by Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China.
Total DNA and cDNA were first sheared into fragments of approximately 300 bp with a M220 Focused-ultrasonicator (Covaris Inc., Woburn, MA, USA). The fragments were used for paired-end library construction. DNA templates were prepared by using a TruSeq DNA Sample Pre-kit according to the standard protocol (Illumina, San Diego, CA). Dual-index adapters containing the full complement of sequencing primer hybridization sites were ligated to blunt-end fragments. Following amplification (10 cycles) and denaturation with sodium hydroxide, the libraries were pooled and loaded onto an Illumina cBot (46). Paired-end sequencing (2 × 101 bp) was performed on an Illumina genome analyzer (HiSeq 2500; Illumina Inc., San Diego, CA, USA) using TruSeq PE cluster kit v3-cBot-HS and TruSeq SBS kit v3-HS, according to the manufacturer's instructions (Illumina, San Diego, CA).
After sequencing, metagenome and metatranscriptome libraries consisting of fragments of approximately 150 bp in size were constructed. Sequencing depths of 8.6 Gb and 10.2 Gb were applied for the metagenomic and metatranscriptomic data sets, respectively.
Bioinformatic analyses using the MG-RAST pipeline.
Unassembled DNA and cDNA sequences were loaded onto the Metagenomics Rapid Annotation (MG-RAST) server (v4.0) (http://metagenomics.anl.gov) to enable phylogenetic and metabolic reconstructions and to perform a protein similarity analysis, including both functional annotation and function classification (23, 24). Quality control and assembly information are available in Fig. S4 and MG-RAST.
Taxonomic profiles were calculated according to the best hit classification at an E value cutoff of 10−5 with a minimum alignment length of 50 bp on the basis of all annotation source databases available in MG-RAST (including RefSeq, IMG, TrEMBL, SEED, KEEG, and GenBank). The distributions of the taxonomic domains, namely, the phyla, orders, families, genera, species, and strains, for the annotations were analyzed in detail. For taxonomic profiles, the percentages shown in the study refer to classifications at a specific taxonomic level.
Functional profiling was conducted by performing gene annotation with the Subsystems, Clusters of Orthologous Group (COG), and KO databases using hierarchical classification at an E value cutoff of 10−5 and a minimum alignment length of 17 amino acids (47, 48). Detailed statistical information regarding hits against different databases is summarized in Table S2.
Binning and draft genome reconstruction.
Metagenomic shotgun sequencing reads were trimmed and screened to remove adapters and to filter sequences with a minimum length of 50 bp and no ambiguous bases using SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle), respectively. Trimmed and screened paired-end reads were individually assembled into contigs using SOAPdenovo (http://soap.genomics.org.cn/, version 1.06). To give more weight to larger contigs and mitigate the effects of assembly errors, we cut contigs into chunks of 10 kb. Then, Bowtie (http://bowtie-bio.sourceforge.net, version 2.3.3) was used to map the contigs back to generate a coverage file. The coverage file and contigs were used to bin and refine genomes using CONCOCT (49). Ninety-one genome bins were gained after binning, and 39 bins were selected after screening bins whose sizes were less than 1 Mbp. The selected genome bins were then checked for completeness and contamination using CheckM (50). Three genome bins were further analyzed for their high quality (Table 2). The detailed genomic characteristics of all the 39 bins are found in Data Set S1.
Selected genome bins (Bin25, Bin27, and Bin82) were used for further phylogenetic analysis with CVtree3 (http://tlife.fudan.edu.cn/cvtree/cvtree/). CVtree3 constructs prokaryotic phylogeny trees by using a composition vector and whole genomes, facilitating the analysis of many large-scale genomes. Thus, this method is better than the traditional phylogenetic analysis based on a single gene or a few genes (51). Then, a phylogenetic tree was constructed using MEGA7 (52). Genome assembly and annotation were also performed by applying the MG-RAST pipeline for Bin25, Bin27, and Bin82.
Accession number(s).
In the present study, raw DNA and cDNA nucleotide sequences were deposited onto MG-RAST (http://metagenomics.anl.gov) under the accession numbers 4719096.3 and 4725988.3, respectively. The reconstructed draft gnome of Bin25, Bin27, and Bin82 can also be found on MG-RAST under accession numbers 4779432.3, 4779427.3, and 4779428.3, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundation of China (51622809).
We thank Ke Yu (Shenzhen Graduate School, Peking University) for discussions on binning strategies.
W. Han conducted the experiment, analyzed the data, and drafted the manuscript. P. He and F. Lü developed the general research question, discussed the results, and revised the manuscript. L. Shao contributed to data analysis. All authors reviewed and edited the manuscript.
The authors declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01614-18.
REFERENCES
- 1.Agler MT, Spirito CM, Usack JG, Werner JJ, Angenent LT. 2012. Chain elongation with reactor microbiomes: upgrading dilute ethanol to medium-chain carboxylates. Energy Environ Sci 5:8189–8192. doi: 10.1039/c2ee22101b. [DOI] [Google Scholar]
- 2.Spirito CM, Richter H, Rabaey K, Stams AJM, Angenent LT. 2014. Chain elongation in anaerobic reactor microbiomes to recover resources from waste. Curr Opin Biotechnol 27:115–122. doi: 10.1016/j.copbio.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Angenent LT, Richter H, Buckel W, Spirito CM, Steinbusch KJJ, Plugge CM, Strik DPBT, Grootscholten TIM, Buisman CJN, Hamelers HVM. 2016. Chain elongation with reactor microbiomes: open-culture biotechnology to produce biochemicals. Environ Sci Technol 50:2796–2810. doi: 10.1021/acs.est.5b04847. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Strik DPBT, Buisman CJN, Kroeze C. 2017. Production of caproic acid from mixed organic waste: an environmental life cycle perspective. Environ Sci Technol 51:7159–7168. doi: 10.1021/acs.est.6b06220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin Y, Zhang Y, Karakashev DB, Wang J, Angelidaki I. 2017. Biological caproate production by Clostridium kluyveri from ethanol and acetate as carbon sources. Bioresour Technol 241:638–644. doi: 10.1016/j.biortech.2017.05.184. [DOI] [PubMed] [Google Scholar]
- 6.Barker HA, Kamen MD, Bornstein BT. 1945. The synthesis of butyric and caproic acids from ethanol and acetic acid by Clostridium kluyveri. Proc Natl Acad Sci U S A 31:373–381. doi: 10.1073/pnas.31.12.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornstein BT, Barker HA. 1948. The nutrition of Clostridium kluyveri. J Bacteriol 55:223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seedorf H, Fricke WF, Veith B, Brueggemann H, Liesegang H, Strittimatter A, Miethke M, Buckel W, Hinderberger J, Li F, Hagemeier C, Thauer RK, Gottschalk G. 2008. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A 105:2128–2133. doi: 10.1073/pnas.0711093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace RJ, McKain N, McEwan NR, Miyagawa E, Chaudhary LC, King TP, Walker ND, Apajalahti J, Newbold CJ. 2003. Eubacterium pyruvativorans sp. nov., a novel non-saccharolytic anaerobe from the rumen that ferments pyruvate and amino acids, forms caproate and utilizes acetate and propionate. Int J Syst Evol Microbiol 53:965–970. doi: 10.1099/ijs.0.02110-0. [DOI] [PubMed] [Google Scholar]
- 10.Wallace RJ, Chaudhary LC, Miyagawa E, McKain N, Walker ND. 2004. Metabolic properties of Eubacterium pyruvativorans, a ruminal “hyper-ammonia-producing” anaerobe with metabolic properties analogous to those of Clostridium kluyveri. Microbiology 150:2921–2930. doi: 10.1099/mic.0.27190-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhu X, Zhou Y, Wang Y, Wu T, Li X, Li D, Tao Y. 2017. Production of high-concentration n-caproic acid from lactate through fermentation using a newly isolated Ruminococcaceae bacterium CPB6. Biotechnol Biofuels 10:102. doi: 10.1186/s13068-017-0788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsden SR, Gilchrist FM, Lewis D, Volcani BE. 1956. Properties of a fatty acid forming organism isolated from the rumen of sheep. J Bacteriol 72:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleerebezem R, van Loosdrecht MCM. 2007. Mixed culture biotechnology for bioenergy production. Curr Opin Biotechnol 18:207–212. doi: 10.1016/j.copbio.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Agler MT, Wrenn BA, Zinder SH, Angenent LT. 2011. Waste to bioproduct conversion with undefined mixed cultures: the carboxylate platform. Trends Biotechnol 29:70–78. doi: 10.1016/j.tibtech.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Steinbusch KJJ, Hamelers HVM, Plugge CM, Buisman CJN. 2011. Biological formation of caproate and caprylate from acetate: fuel and chemical production from low grade biomass. Energy Environ Sci 4:216–224. doi: 10.1039/C0EE00282H. [DOI] [Google Scholar]
- 16.Zhang F, Ding J, Zhang Y, Chen M, Ding Z, van Loosdrecht MCM, Zeng RJ. 2013. Fatty acids production from hydrogen and carbon dioxide by mixed culture in the membrane biofilm reactor. Water Res 47:6122–6129. doi: 10.1016/j.watres.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, He P, Shao L, Zhang H, Lu F. 2017. Significant enhancement by biochar of caproate production via chain elongation. Water Res 119:150–159. doi: 10.1016/j.watres.2017.04.050. [DOI] [PubMed] [Google Scholar]
- 18.Coma M, Vilchez-Vargas R, Roume H, Jauregui R, Pieper DH, Rabaey K. 2016. Product diversity linked to substrate usage in chain elongation by mixed-culture fermentation. Environ Sci Technol 50:6467–6476. doi: 10.1021/acs.est.5b06021. [DOI] [PubMed] [Google Scholar]
- 19.He P, Han W, Shao L, Lu F. 2018. One-step production of C6-C8 carboxylates by mixed culture solely grown on CO. Biotechnol Biofuels 11:4. doi: 10.1186/s13068-017-1005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. 2015. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. mBio 6:e02288-14. doi: 10.1128/mBio.02288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weimer PJ, Stevenson DM. 2012. Isolation, characterization, and quantification of Clostridium kluyveri from the bovine rumen. Appl Microbiol Biotechnol 94:461–466. doi: 10.1007/s00253-011-3751-z. [DOI] [PubMed] [Google Scholar]
- 22.Roghair M, Liu Y, Strik DPBT, Weusthuis RA, Bruins ME, Buisman CJN. 2018. Development of an effective chain elongation process from acidified food waste and ethanol into n-caproate. Front Bioeng Biotechnol 6:50. doi: 10.3389/fbioe.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. 2010. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc 2010:pdb.prot5368. doi: 10.1101/pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 25.Guo J, Peng Y, Ni B, Han X, Fan L, Yuan Z. 2015. Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb Cell Fact 14:33. doi: 10.1186/s12934-015-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stolze Y, Zakrzewski M, Maus I, Eikmeyer F, Jaenicke S, Rottmann N, Siebner C, Pühler A, Schlüter A. 2015. Comparative metagenomics of biogas-producing microbial communities from production-scale biogas plants operating under wet or dry fermentation conditions. Biotechnol Biofuels 8:14. doi: 10.1186/s13068-014-0193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo G, Fotidis IA, Angelidaki I. 2016. Comparative analysis of taxonomic, functional, and metabolic patterns of microbiomes from 14 full-scale biogas reactors by metagenomic sequencing and radioisotopic analysis. Biotechnol Biofuels 9:51. doi: 10.1186/s13068-016-0465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delmont TO, Prestat E, Keegan KP, Faubladier M, Robe P, Clark IM, Pelletier E, Hirsch PR, Meyer F, Gilbert JA, Le Paslier D, Simonet P, Vogel TM. 2012. Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J 6:1677–1687. doi: 10.1038/ismej.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papagianni M. 2012. Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb Cell Fact 11:50. doi: 10.1186/1475-2859-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noor E, Eden E, Milo R, Alon U. 2010. Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell 39:809–820. doi: 10.1016/j.molcel.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 31.Kenealy WR, Cao Y, Weimer PJ. 1995. Production of caproic acid by cocultures of ruminal cellulolytic bacteria and Clostridium kluyveri grown on cellulose and ethanol. Appl Microbiol Biotechnol 44:507–513. doi: 10.1007/BF00169952. [DOI] [PubMed] [Google Scholar]
- 32.Lien T, Madsen M, Rainey FA, Birkeland NK. 1998. Petrotoga mobilis sp. nov., from a North Sea oil-production well. Int J Syst Bacteriol 48:1007–1013. doi: 10.1099/00207713-48-3-1007. [DOI] [PubMed] [Google Scholar]
- 33.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. 2011. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476:355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 34.Lian J, Zhao H. 2015. Reversal of the β-oxidation cycle in Saccharomyces cerevisiae for production of fuels and chemicals. ACS Synth Biol 4:332–341. doi: 10.1021/sb500243c. [DOI] [PubMed] [Google Scholar]
- 35.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 36.Lennen RM, Pfleger BF. 2013. Microbial production of fatty acid-derived fuels and chemicals. Curr Opin Biotechnol 24:1044–1053. doi: 10.1016/j.copbio.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratledge C. 2004. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie 86:807–815. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Chan DI, Vogel HJ. 2010. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J 430:1–19. doi: 10.1042/BJ20100462. [DOI] [PubMed] [Google Scholar]
- 39.Magnuson K, Jackowski S, Rock CO, Cronan JE. 1993. Regulation of fatty-acid biosynthesis in Escherichia coli. Microbiol Res 57:522–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kucek LA, Nguyen M, Angenent LT. 2016. Conversion of l-lactate into n-caproate by a continuously fed reactor microbiome. Water Res 93:163–171. doi: 10.1016/j.watres.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Chen WS, Ye Y, Steinbusch KJJ, Strik DPBT, Buisman CJN. 2016. Methanol as an alternative electron donor in chain elongation for butyrate and caproate formation. Biomass Bioenergy 93:201–208. doi: 10.1016/j.biombioe.2016.07.008. [DOI] [Google Scholar]
- 42.Zhang F, Ding J, Shen N, Zhang Y, Ding Z, Dai K, Zeng RJ. 2013. In situ hydrogen utilization for high fraction acetate production in mixed culture hollow-fiber membrane biofilm reactor. Appl Microbiol Biotechnol 97:10233–10240. doi: 10.1007/s00253-013-5281-3. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Lu F, Shao L, He P. 2016. Alcohol-to-acid ratio and substrate concentration affect product structure in chain elongation reactions initiated by unacclimatized inoculum. Bioresour Technol 218:1140–1150. doi: 10.1016/j.biortech.2016.07.067. [DOI] [PubMed] [Google Scholar]
- 44.Grootscholten TIM, Steinbusch KJJ, Hamelers HVM, Buisman CJN. 2013. High rate heptanoate production from propionate and ethanol using chain elongation. Bioresour Technol 136:715–718. doi: 10.1016/j.biortech.2013.02.085. [DOI] [PubMed] [Google Scholar]
- 45.Yu ZT, Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 46.Clark MJ, Chen R, Lam HYK, Karczewski KJ, Chen R, Euskirchen G, Butte AJ, Snyder M. 2011. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 29:206–908. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Yu K, Xia Y, Lau FTK, Tang DTW, Fung WC, Fang HHP, Zhang T. 2014. Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biotechnol 98:5709–5718. doi: 10.1007/s00253-014-5648-0. [DOI] [PubMed] [Google Scholar]
- 48.Jung JY, Lee SH, Kim JM, Park MS, Bae J, Hahn Y, Madsen EL, Jeon CO. 2011. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl Environ Microbiol 77:2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 50.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Z, Hao B. 2009. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res 37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S, Stecher G, Tamura K. 2016. Mega7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.