Abstract
Glioblastoma multiforme (GBM) is the most aggressive brain tumor with poor prognosis to most patients. Immunotherapy of GBM is a potentially beneficial treatment option, whose optimal implementation may depend on familiarity with tumor specific antigens, presented as HLA peptides by the GBM cells. Furthermore, early detection of GBM, such as by a routine blood test, may improve survival, even with the current treatment modalities. This study includes large-scale analyses of the HLA peptidome (immunopeptidome) of the plasma-soluble HLA molecules (sHLA) of 142 plasma samples, and the membranal HLA of GBM tumors of 10 of these patients' tumor samples. Tumor samples were fresh-frozen immediately after surgery and the plasma samples were collected before, and at multiple visits after surgery. In total, this HLA peptidome analysis involved 52 different HLA allotypes and resulted in the identification of more than 35,000 different HLA peptides. Strong correlations were observed in the signal intensities and in the repertoires of identified peptides between the tumors and plasma-soluble HLA peptidomes of the individual patients, whereas low correlations were observed between these HLA peptidomes and the tumors' proteomes. HLA peptides derived from Cancer/Testis Antigens (CTAs) were selected based on their presence among the HLA peptidomes of the patients and absence of expression of their source genes from any healthy and essential human tissues, except from immune-privileged sites. Additionally, peptides were selected as potential biomarkers if their levels in the plasma-sHLA peptidome were significantly reduced after the removal of tumor mass. The CTAs identified among the analyzed HLA peptidomes provide new opportunities for personalized immunotherapy and for early diagnosis of GBM.
Keywords: Cancer biomarker(s), Cancer therapeutics, Glioblastoma, Peptidomics, Plasma or serum analysis, Immunology, Immunoaffinity, CTA - cancer/testis antigens, HLA - human leukocytes antigen, immunotherapy, MHC - major histocompatibility complex
Glioblastoma multiforme (GBM)1 is the most common and aggressive primary brain tumor, with a median survival of about 15 months (1, 2). The grim prognosis facing most GBM patients calls for improvements in targeted therapeutic treatments, as well as in discovery of proper biomarkers for early detection of the disease. Recently, immunotherapy of different cancers, including GBM, has drawn significant attention (3, 4), reviewed in (5–13). Much of the recent excitement about cancer immunotherapy stems from the success in treating patients with immune checkpoint modulators, which induce anti-cancer T cell immune reactions that can break tolerance and bring about complete responses in increasingly larger percentages of patients (14, 15). Identification of tumor antigens is needed for development of effective cancer immunotherapy, including GBM, and a good source for such antigens are the pools of HLA-bound peptides presented preferentially, or even exclusively, by the tumor cells (16), reviewed in (17–20). Tumor-specific antigens (TSA) and tumor-associated antigens (TAA) that can serve as candidates for cancer immunotherapeutics have been searched for extensively. Indeed, many such antigens were already identified, yet none has induced sufficiently strong anti-cancer immune reaction to eradicate the tumors in large cohorts of patients (18, 20, 21). One subset of preferred TAAs are Cancer/Testis Antigens (CTA), which are abnormally expressed in the malignant cells, and are normally expressed only in fetal tissues and in immune privileged sites, such as the male germ cells, placenta and ovary, but are absent from the normal somatic cells of any healthy tissue (22), reviewed in (23). Another special groups of tumor antigens are neoantigens, which are attracting significant attention as potential targets for cancer immunotherapy (24). These are more likely common in tumors with higher mutational load and therefore less frequently found in GBM.
HLA class I molecules are predominantly expressed as membrane anchored, cell-surface proteins (mHLA). Yet, the HLA molecules are also released from cells (25, 26), reviewed in (27), and can be recovered from the soluble HLA molecules in human plasma (sHLA) for analysis of their bound peptidome (28–30). Confounding factors that limit the use of the sHLA peptidome as a source for biomarkers are the presence of small amounts of sHLA molecules in the plasma of all healthy individuals (27, 31), and in addition, some of the sHLA alleles are released to the circulation in larger amounts than others (32, 33).
It has been suggested that shedding of the sHLA molecules represents a mechanism for evasion of immune recognition of tumor cells, which release larger amounts of sHLA molecules relative to the healthy cells (33). Because the sHLA molecules carry with them their original load of bound peptides, the immunopeptidomes of the sHLA molecules of cancer patients include disproportionally large fractions of peptides originating from the tumor cells, even though the tumors constitute just small fraction of the body mass. It is therefore intuitive to assume that biomarkers of disease can be found among those sHLA peptides. Such plasma-sHLA molecules could then be a useful source of cancer biomarkers for early detection or recurrence during follow-up (28).
Analyses of the HLA peptidomes, based on immunoaffinity purification of the mHLA molecules from cell lines, tumor tissues or patients' plasma, followed by analysis of the bound peptides by chromatography and tandem mass spectrometry (34) currently results in identification of thousands of HLA bound peptides (35–38). HLA peptides were previously isolated from GBM cell lines (39) and fresh tumors, including GBM (4, 16). Similarly, the HLA peptidomes of tumors, such as melanoma (40, 41), renal cell carcinoma (42, 43), leukemia (44) and multiple myeloma (45) have been characterized. Furthermore, several thousands of peptides have successfully been isolated from cancer patients' plasma-sHLA molecules (28, 30).
In this research, more than 35,000 plasma-sHLA and tumor-mHLA peptides from GBM patients were identified. These large HLA peptidomes included several hundred peptides, derived from both known TAAs and newly defined potential CTAs. We propose that these may serve as useful biomarkers and candidates for immunotherapeutics for GBM, as well as for other cancers.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
One hundred six plasma samples were obtained from 52 different GBM patients, collected prior to tumor removal and at different time points after the surgery (supplemental Table S1). Additionally, control plasma of healthy donors (n = 6) and patients diagnosed with ankylosing spondylitis (n = 30), as an exemplar inflammatory process, were used for this study. In addition, tumor samples were obtained from 10 of the 52 GBM patients for mHLA peptidome analysis, as well as for tumor proteome analysis.
Patient Characterization
Peripheral blood (PB) and tumor tissues samples from GBM patients (University Hospital Heidelberg, Leiden University Medical Centre, Vall d'Hebron University Hospital, Université de Genève, Southampton Universities Hospital, Heidelberg University Medical Center, Herlev Hospital and Rigshospitalet) and non-cancerous controls (Universitario Central de Asturias and Bnei-Zion Hospital). The HLA tissue typing of the GBM patients and noncancerous controls, healthy, and Ankylosing Spondylitis patients are listed only with gender, age, and stage of disease in supplemental Table S1. All human bio-specimens were obtained with informed consent, and after approval of the relevant ethics committees.
Plasma and Tumor Cells Collection
Peripheral blood samples were collected into EDTA tubes and cleared of the cells by centrifugation for 10 min at 1200 × g, at room temperature. The cleared plasma was stored frozen at −80 °C until use for sHLA purification. Tumor samples were frozen in liquid nitrogen immediately after dissection and stored at −80 °C until use, for both mHLA peptidome and proteome analyses of the same tissue extracts.
Affinity Purification of HLA Molecules
Membranal HLA class I molecules were purified from GBM tumor tissues by immunoaffinity from the cells after lysis with 0.25% sodium deoxycholate, 0.2 mm iodoacetamide, 1 mm EDTA, 1:200 Protease Inhibitors Mixture (Sigma Aldrich, St. Louis, MO), 1 mm PMSF, and 1% octyl-d-glucopyranoside (Sigma Aldrich) in PBS at 4 °C for 1 h. The lysate was cleared by centrifugation for 45 min at 18,000 rpm (40,000g, SS-34 rotor, Sorval, Thermo Fisher Scientific, Waltham, MA), at 4 °C. HLA class I molecules from the cleared lysate or from the fresh human plasma were immunoaffinity purified using the W6/32 mAb bound to Amino-Link beads (Thermo-Fisher Scientific) as in (28, 46). The HLA molecules with their bound peptides were eluted from the affinity column with five column volumes of 1% TFA. The eluted HLA class I proteins, and the released peptides were loaded on disposable C18 micro-columns (Harvard Apparatus, Holliston, MA), and the peptides fraction was recovered with 30% acetonitrile in 0.1% TFA, whereas the protein fraction was recovered with 80% acetonitrile in 0.1% TFA, as in (46). The peptide fractions were dried using vacuum centrifugation, reconstituted in 100 μl of 0.1% TFA, reloaded on C18 Stage-Tips (47), eluted with 80% acetonitrile, dried, and reconstituted with 0.1% formic acid for LC-MS/MS analysis.
Proteomics analysis was performed after trypsin digestion of the proteins in gel slices. Briefly, 40 μg protein sample from each tissue was ran in 10% acrylamide gel, stained with Coomassie, and each lane of the gel was sliced into five slices. The proteins in the gel slices were reduced with 2.8 mm DTT at 60 °C for 30 min and carbamidomethylated with 8.8 mm iodoacetamide in 100 mm ammonium bicarbonate at room temperature for 30 min and digested overnight at 37 °C in 10% acetonitrile and 10 mm ammonium bicarbonate with modified trypsin (Promega, Madison, WI) at a 1:10 (wt/wt) enzyme-to-substrate ratio. This was repeated for another 4 h with a similar amount of trypsin, followed by 150 min gradients of LC-MS/MS of the tryptic peptides from each gel slice.
Identification of the HLA and Tryptic Peptides
Both the HLA and tryptic peptides were resolved by capillary chromatography and electrospray tandem mass spectrometry using a Q-Exactive-Plus mass spectrometer fitted with a capillary Ultimate 3000 RSLC nano-capillary UHPLC (Thermo-Fisher Scientific). The reversed phase chromatographs were with about 30 cm long, 75 micron inner diameter capillary columns, home-packed with 3.5 m silica ReproSil-Pur C18-AQ resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) as in (48). HLA and tryptic peptides were eluted with a linear gradient of 5–28% of acetonitrile in 0.1% formic acid. For both the HLA peptides and for the in-gel digested fractions, the capillary HPLC gradients were run at flow rates of 0.15 μl/minutes during 120 min. Data was acquired using a data-dependent “top 10” method, fragmenting the peptides by higher-energy collisional dissociation (HCD). Full scan MS spectra were acquired at a resolution of 70,000 at 200 m/z with a target value of 3 × 106 ions. MS/MS ions were accumulated to an AGC target value of 105 with a maximum injection time of 100 msec. For the HLA peptides with unassigned precursor ion charge states, or charge states of four and above, no fragmentation was performed. For tryptic peptides, the fragmentation was performed on charge states between 2 to 7. The peptide match option was set to Preferred. Normalized collision energy was set to 25% and MS/MS resolution was 17,500 at 200 m/z. Fragmented m/z values were dynamically excluded from further selection for 20 s.
Data Analysis
The MS data was analyzed by the MaxQuant computational proteomics platform (49) version 1.5.3.8 and searched with the Andromeda search engine (50). Peptide identifications were based on the human section of the Uniprot database (http://www.uniprot.org/) of July 2015 containing 69,693 entries. Proteins were declared positive identification with at least two identified tryptic peptides per protein. Mass tolerance of 4.5 ppm for the precursor masses and 20 ppm for the fragments were allowed. Methionine oxidation was accepted as variable modification for both tryptic and HLA peptides. Carbamidomethyl cysteine was accepted as a fixed modification for the proteomics data and as a variable modification for the HLA peptidome data. Methionine sulfoxide and n-acetylation were set as variable modifications for both the proteomics and HLA peptidomics analyses. Minimal peptide length was set to seven amino acids and a maximum of two miscleavages was allowed for tryptic peptides. The false discovery rate (FDR) was set to 0.01 for protein identifications, and 0.05 for the HLA peptides, because it resulted in identification of about twice as many true HLA ligands (see Discussion). The resulting identified protein tables were filtered to eliminate the identifications derived from the reverse database, as well as common contaminants.
Normalization of the HLA Peptidome and Proteome Data
The HLA peptidome of each of the LC-MS/MS runs was normalized according to its median. The medians were zeroed, and downregulated HLA peptides were defined as those changing by at least two folds in the plasma samples collected immediately before surgery, relative to samples collected at later clinic visits. The proteomics LC-MS/MS data was normalized using the LFQ and iBAQ tools of the MaxQuant software (49).
HLA Typing
DNA was extracted from blood of GBM patients using the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) as per the kit instructions. The HLA typing was conducted with this DNA at the DKMS Life Science Lab (Dresden, Germany). The samples were processed within high-throughput workflow using next generation sequencing. HLA typing was based on the regions covering the peptide-binding domains. Regions sequenced were Exons 2 and 3 for HLA-A, -B, -C, -DQB1, -DRB1, and -DPB1 (51).
Definition of the Tumor Associated Antigen (TAA) Group
Selection of TAAs was based on the cancer tumor database (CT gene database) (http://www.cta.lncc.br/) (52) comprised of 277 different TAAs (data accumulated between the years 2005–2009) and the Tumor T cell Antigen database (TANTIGEN) (http://cvc.dfci.harvard.edu/tadb), comprised of 259 different TAAs (53).
RESULTS
Large HLA Peptidomes are Identified From Tumors and Plasma of GBM Patients
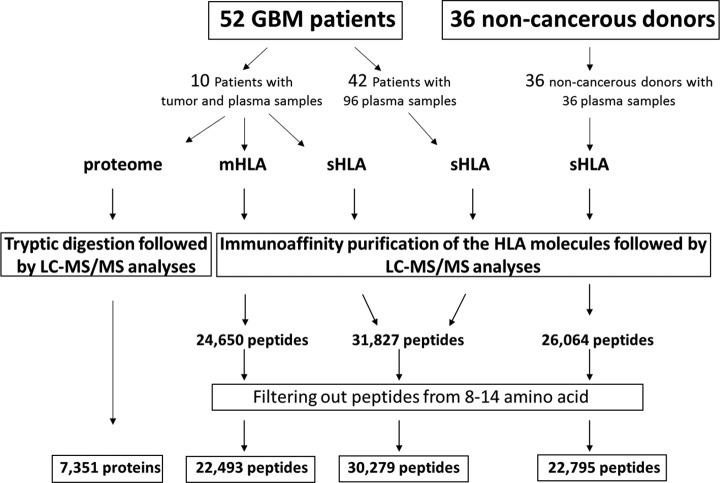
Tumor samples were obtained from 10 different patients undergoing surgery. These freshly frozen tumor tissues were used for analysis of their membranal HLA peptidomes and their tumor proteomes. In addition, 106 plasma samples from these 10 patients, and other 42 GBM patients (52 patients in total) were used for the analysis of their plasma-sHLA bound peptidomes. Additionally, control plasma of healthy donors (n = 6) and patients diagnosed with ankylosing spondylitis (n = 30), as an exemplar inflammatory process, were used for this study (Fig. 1, supplemental Table S1). The study was performed as part of the GAPVAC project (The European Glioma Actively Personalized Vaccine Consortium), which aimed to develop a personal immunotherapy treatment based on administration of synthetic copies of selected HLA peptides derived from tumor antigens to GBM patients. Here we describe the results of the plasma-soluble HLA (sHLA) peptidome analyses, the tumor membranal HLA (mHLA) peptidome and proteome analyses, performed in parallel with the GBM tumors. Overall, the tumor-mHLA peptidome analyses resulted in the identification of 24,650 different HLA peptides, derived from 7217 different source proteins, and the plasma-sHLA peptidome analyses resulted in the identification of 32,089 different HLA peptides, derived from 8281 different source proteins (supplemental Table S2). The proteomics analysis of these 10 GBM tumor samples resulted in identification of 7351 different proteins (supplemental Table S3).
Fig. 1.
Overview of the sHLA and mHLA peptidome and proteome analyses schemes, and selection of likely HLA ligands.
The GBM HLA Peptidomes Include Many Peptides from Multiple Tumor Antigens
Among the patients' plasma-sHLA and the tumor-mHLA peptidomes, 904 different HLA peptides derived from 143 known TAAs were identified. The reference set of TAAs was based on the cancer tumor database (CT gene database) (http://www.cta.lncc.br/) comprising 277 different TAAs (data accumulated between 2005 and 2009) (52) and the Tumor T cell Antigen database (TANTIGEN) (http://cvc.dfci.harvard.edu/tadb) (53) with 259 different TAAs, for a total set of 496 TAAs.
Derived from these 496 TAAs, a total of 763 plasma-sHLA and 611 tumor-membranal HLA peptides were identified. Importantly, up to 78.8% of the plasma-sHLA peptides, derived from this TAA group, were also detected among the tumor-mHLA peptidomes of the different patients. These results indicate that the plasma-sHLA peptidome may indeed provide a useful source of tumor antigens for diagnosis and immunotherapeutics (selected examples in Table I, and the entire list in supplemental Table S4). Four hundred five out of the 904 identified HLA peptides derived from TAAs were detected only in GBM plasma and tissue samples but not in any noncancerous donors' plasma samples, supporting their potential significance as authentic tumor antigens.
Table I. List of HLA peptides derived from the TAA group, identified among the GBM sHLA and mHLA peptidomes.
| Gene name | Sequence | Count # |
||
|---|---|---|---|---|
| sHLA non-cancerous | sHLA GBM | mHLA GBM tissue | ||
| ETV5 | FPDNQRPFL | 0 | 0 | 1 |
| FSSSHEGFSY | 0 | 1 | 0 | |
| GPAPAPHSL | 0 | 1 | 1 | |
| KVAGERYVY | 0 | 3 | 3 | |
| LPYAEGFAY | 0 | 6 | 2 | |
| MPGPPAHGF | 0 | 0 | 2 | |
| RPAMNYDKL | 0 | 2 | 1 | |
| VPRPPHQPLQM | 0 | 0 | 0 | |
| SOX4 | KIMEQSPDM | 0 | 10 | 4 |
| SAASASAAL | 0 | 19 | 0 | |
| SISNLVFTY | 0 | 4 | 1 | |
| MDM2 | GEISEKAKL | 0 | 9 | 2 |
| YTMKEVLFY | 1 | 0 | 1 | |
| GEISEKAKL | 0 | 9 | 2 | |
| SEQETLVRP | 0 | 3 | 0 | |
| DDR1 | AVGDGPPRV | 0 | 8 | 6 |
| DSPQDLVSL | 1 | 0 | 0 | |
| FLAEDALNTV | 0 | 2 | 6 | |
| LPPPPQNSV | 0 | 0 | 0 | |
| YLQVDLQRL | 0 | 0 | 3 | |
| CSF1 | HSSGSVLPLGELE | 2 | 1 | 0 |
| HTVDPGSAKQR | 1 | 7 | 3 | |
| TMEM108 | TVFSTSSLK | 0 | 17 | 2 |
| UBE2A | REYEKRVSA | 2 | 2 | 0 |
| RLQEDPPAGV | 0 | 14 | 6 | |
| BST2 | DASAEVERL | 2 | 5 | 0 |
| AAAPQLLIV | 1 | 5 | 0 | |
| VPLIIFTI | 0 | 14 | 1 | |
| NPM1 | DENEHQLSL | 0 | 3 | 1 |
| EAIQDLWQW | 0 | 0 | 1 | |
| EGSPIKVTL | 3 | 3 | 0 | |
| EITPPVVLR | 0 | 4 | 0 | |
| FEITPPVVL | 0 | 0 | 2 | |
| HQLSLRTV | 1 | 0 | 0 | |
| MSPLRPQNYLFG | 1 | 0 | 0 | |
| NYEGSPIKVTL | 0 | 0 | 0 | |
| QLSLRTVSL | 0 | 3 | 1 | |
| SPIKVTLATL | 1 | 13 | 1 | |
| VEAEAMNY | 0 | 6 | 1 | |
| VEAKFINY | 1 | 8 | 1 | |
| YEGSPIKVTL | 0 | 2 | 2 | |
| BCAP31 | AESASEAAKKY | 1 | 2 | 1 |
| KLDVGNAEV | 0 | 7 | 3 | |
| KYMEENDQL | 1 | 3 | 0 | |
| LEKAENQVL | 0 | 1 | 2 | |
| THVLEGAGNKL | 0 | 5 | 1 | |
| TLISQQATL | 0 | 0 | 0 | |
| CDKN2A | AAPGAPAAV | 0 | 0 | 2 |
| LPVDLAEEL | 1 | 7 | 2 | |
| SART3 | AAFTRALEY | 0 | 9 | 1 |
| AEAPRLAEY | 2 | 2 | 1 | |
| ARLEKVHSL | 0 | 1 | 4 | |
| AYIDFEMKI | 2 | 3 | 0 | |
| DHQVISVTF | 0 | 5 | 1 | |
| TVFVSNLPYSM | 1 | 0 | 0 | |
| PA2G4 | AEDLVVTKY | 0 | 1 | 0 |
| DVAQGTQVTGR | 1 | 1 | 0 | |
| IAFPTSISV | 2 | 21 | 5 | |
| KEGEFVAQF | 1 | 4 | 1 | |
| TPIEGMLSH | 1 | 9 | 1 | |
| COTL1 | FVISDRKEL | 1 | 8 | 3 |
| VYVLESWHL | 0 | 0 | 0 | |
| ATIC | KTLTPISAAY | 1 | 2 | 0 |
| NLYPFVKTV | 0 | 8 | 6 | |
| GPNMB | AARLPLDAA | 0 | 3 | 0 |
| ALTSTLISV | 0 | 7 | 3 | |
| GPNMB | AYMREHNQL | 2 | 0 | 1 |
| IENSPGNVV | 0 | 3 | 0 | |
| IPTEVCTII | 0 | 0 | 1 | |
| KGLSVFLNR | 3 | 6 | 1 | |
| VFLNRAKAVFF | 1 | 1 | 0 | |
| VLTSDSPAL | 0 | 1 | 1 | |
| CDK4 | ALTPVVVTL | 0 | 6 | 5 |
| FGLARIYSY | 1 | 4 | 1 | |
| ISTVREVAL | 2 | 3 | 1 | |
| SOX11 | AHSASEQQL | 1 | 0 | 1 |
| NFSDLVFTY | 0 | 1 | 1 | |
Among the TAAs expressed in the tumor cells, a subset of genes, normally expressed only in germline, embryonic and placenta cells, can be defined as CTAs, which are expressed at levels below a threshold of nine gcrma units (expression units using background adjustment: GC content adjusted with Robust Multiarray Average, described in the BioGPS website) in all normal and essential tissues. Such CTAs were further defined as those derived from genes whose transcripts are expressed at significantly higher levels in the tumors tissues (according to BioGPS (55, 56), see Discussion). Using this filtration, a list of 145 CTAs was established (supplemental Table S5). In the HLA peptidome analysis described here, 19 different HLA peptides, belonging to 11 CTAs, were identified. Of these, 15 HLA peptides were identified only in the GBM plasma and/or tumor samples (for example ODF1, supplemental Fig. S1A and the entire list in supplemental Table S2). In addition, HLA peptides derived from proteins expressed only in fetal tissues were observed in the GBM samples, and not at all, or in less than 5% of the control plasma samples. An example for such antigen is the transcription factor SOX11 gene (SOX11) that is expressed only in fetal brain, according to BioGPS (supplemental Fig. S1B). In this analysis, two of its derived HLA peptides, AHSASEQQL and NFSDLVFTY, were observed in the plasma-sHLA and tumor-mHLA of the GBM patients and were detected in less than 5% of the noncancerous blood donors' plasma-sHLA peptidomes (Table I). Moreover, HLA peptides derived from male tissues such as testis or prostate, and identified in women's plasma or GBM tissue, or HLA peptides derived from female tissues, such as placenta and ovaries, and identified in the men's plasma or GBM tissue, can serve as potential immunotherapeutics candidates (Table II). For example, eight different HLA peptides originating from the ETV5 gene were identified in this analysis. This gene is expressed only in placenta but not at any other tissue in the body. Moreover, the eight ETV5 HLA peptides were identified mostly among the male GBM sHLA and mHLA samples but not at any of the non-cancerous donors, suggesting their relevance to serve as proper biomarkers for the disease.
Table II. HLA peptides derived from typical male tissues and identified in women's plasma or GBM tumor tissue, and HLA peptides derived from typical female tissues and identified in men's plasma or GBM tumor tissue.
| Gene name | Sequence | CTA description | # of identifications |
|||||
|---|---|---|---|---|---|---|---|---|
| mHLA male | mHLA female | sHLA GBM male | sHLA GBM female | sHLA non-cancerous male | sHLA non-cancerous female | |||
| ETV5 | FPDNQRPFL | Expressed only in placenta | 0 | 1 | 0 | 0 | 0 | 0 |
| FSSSHEGFSY | 0 | 0 | 1 | 0 | 0 | 0 | ||
| GPAPAPHSL | 0 | 1 | 0 | 1 | 0 | 0 | ||
| KVAGERYVY | 3 | 0 | 3 | 0 | 0 | 0 | ||
| LPYAEGFAY | 1 | 1 | 5 | 1 | 0 | 0 | ||
| MPGPPAHGF | 1 | 1 | 0 | 0 | 0 | 0 | ||
| RPAMNYDKL | 0 | 1 | 2 | 0 | 0 | 0 | ||
| VPRPPHQPLQM | 0 | 1 | 0 | 0 | 0 | 0 | ||
| ODF1 | AHIKGVETI | Expressed only in testis | 1 | 0 | 0 | 2 | 0 | 0 |
| MESPCSPCSPC | 2 | 0 | 0 | 0 | 0 | 0 | ||
| RIIDTPISEMGF | 1 | 0 | 0 | 0 | 0 | 0 | ||
| ACPP | SAHDTTVSGLQ | Expressed only in prostate | 0 | 0 | 0 | 0 | 1 | 0 |
| AHDTTVSGL | 1 | 0 | 0 | 0 | 0 | 0 | ||
| TTK | SPNSILKAA | Expressed only in testis and fetal organs | 0 | 0 | 0 | 2 | 0 | 0 |
| SPAG17 | DQEKEKEKEK | Expressed only in testis | 0 | 0 | 1 | 0 | 0 | 0 |
The Levels of Potential Biomarker Plasma-sHLA Peptides Are Reduced Following Surgical Removal of the Tumors
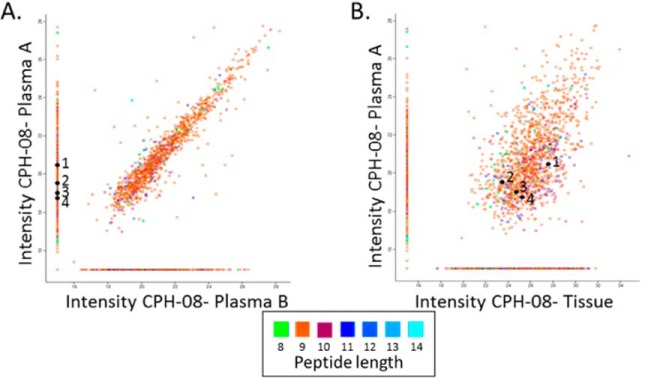
The presence of the plasma-sHLA peptides derived from tumor antigens may serve as surrogate biomarkers for different cancers, including GBM. Peptides detected among the plasma-sHLA peptidome, derived from tumor antigens that are not normally expressed at elevated levels in normal tissues, should be reduced in their levels after removal or reduction of the tumor load by treatments (57) such as the surgery performed for the GBM patients studied here. Most of the plasma-sHLA peptides of the individual patients remain relatively stable before and after surgery, and even a few months later, because these are self-peptides of the healthy tissues (example in Fig. 2A). This facilitated focusing on the minority of HLA peptides that were reduced in their levels relative to the rest of the sHLA peptidome, to search among them for potential biomarkers. In this study, 94 different plasma samples were collected before, and following surgery of the same 34 GBM patients as part of the GAPVAC project. The sHLA molecules of the different plasma samples were affinity purified and their bound peptidomes were analyzed separately by LC-MS/MS. As many as 250 sHLA peptides, derived from 236 proteins (out of the 8532 sHLA peptides identified only in the GBM plasma samples), were downregulated by at least two folds in the plasma samples collected before and a few weeks after surgery, and remained low in the plasma samples collected at subsequent visits to the clinic. These HLA peptides were derived from genes/proteins from which no sHLA peptides were detected in any of the plasma samples of the noncancerous donors (supplemental Table S2, column B). Examples for such downregulated peptides are displayed in Fig. 2A. Some of the downregulated peptides were derived from known cancer related genes, such as the HLA-A*32:01 peptide RVNPLVKSF of FRMD3, which is not expressed in any of the normal tissues of the body, according to BioGPS. The disappearance of sHLA peptides from the plasma samples after surgery can be because of real reduction in their amounts, or because of chance misidentifications caused by the shotgun LC-MS/MS approach used here (examples in Fig. 2A).
Fig. 2.
Relative LC-MS signal intensities of the sHLA peptides identified in plasma A (before surgery) and plasma B (at least 4 weeks after surgery) of patient CPH-08. The group of HLA peptides detected only in plasma A are indicated on the vertical line and those observed only in plasma B are on the horizontal line with arbitrary numbers of 15 (A). Comparison between the plasma A sHLA peptidome and the tumor mHLA peptidome of patient CPH-08 (B). The LC-MS signals of the peptides are indicated on a log2 scale. The numbered HLA peptides (1–4) were identified in plasma A and tissue but not in plasma B: 1) YERDEDNNL; 2) EHTEVLQLL; 3) NHALPLPGF; 4) EIIEKDTKY.
The Plasma-sHLA and Tumor-mHLA Peptidomes of the Same Patients Are Highly Correlated
The large HLA peptidomes of the patients' tumors-mHLA and plasma-sHLA enabled comparisons between these patients' peptidomes. The peptidome analyses of the ten patients, from which both types of samples were available, indicated overlaps of up to 50% between the identified peptides of the individual patients (Fig. 2B). Multiple plasma samples of each patient were also considered as biological replica, and their LC-MS intensities were averaged. Many of the mHLA and the sHLA peptides of the individual patients were shared between both types of samples (supplemental Fig. S2A). In addition, the LC-MS signal intensities of these shared sHLA and mHLA peptides, of the individual patients, were significantly correlated with Pearson correlations between 0 and 0.53 (supplemental Fig. S2B). Most of these shared peptides were true HLA ligands and received NetMHC scores below rank of 2%, according to the patient's HLA alleles. For example, in patient “11–002”, as many as 1083, out of the 1204 (about 90%) shared sHLA peptides of plasma A (taken before surgery) and tissue mHLA peptidome, fitting this definition as HLA ligands (supplemental Table S2). Importantly, many of the shared HLA peptides were derived from the TAA group (supplemental Table S4).
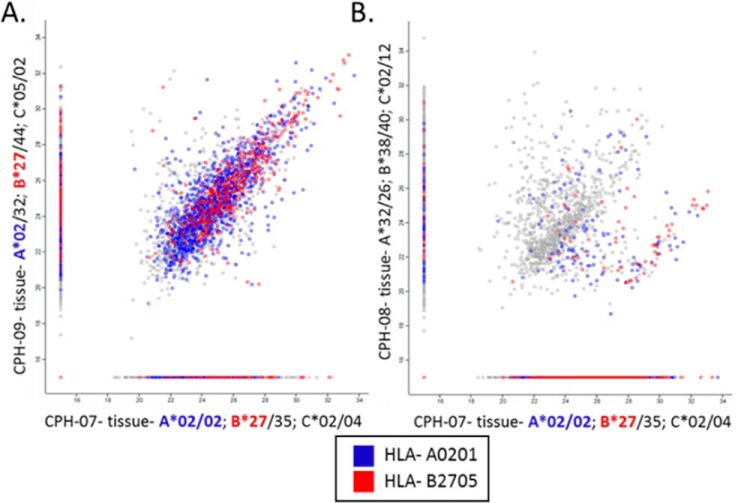
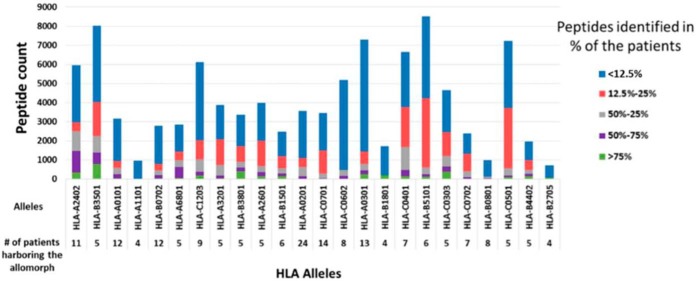
Similarity Between HLA Peptidomes of Individuals Sharing HLA Allotypes
As expected, both the tumor-mHLA and plasma-sHLA peptidomes of different blood donors were more similar when they shared HLA allotypes (examples in Fig. 3) and these patients shared more TAAa and CTAs peptides (supplemental Table S6) even though only small fractions of the peptidomes were shared among most patients (Fig. 4, supplemental Table S7). The shared peptides identified in the plasma-sHLA peptidomes of people harboring very different HLA allotypes that belong to dissimilar HLA supertypes, are likely contaminating peptides, copurifying with the HLA molecules during the affinity purification. Furthermore, peptides that fit the sequence motifs of the HLA allotypes of the patients are more likely authentic HLA ligands of these allotypes (Fig. 3).
Fig. 3.
Comparisons of the mHLA peptidomes of three patients' tumors. The HLA alleles of the patients are as following: CPH-07- A*02:01/02:01; B*35:01/27:05; C*02:02/04:01, CPH-09- A*02:01/32:01; B*27:05/44:02; C*05:01/02:02, CPH-08- A*32:01/26:01; B*38:01/40:02; C*02:02/12:03. Patients CPH-07 and CPH-09 share the same A*02:01 and B*27:05 HLA alleles (A). Patients CPH-08 and CPH-07 have completely different HLA alleles (B). The NAN values were replaced with arbitrary value of 15, the axes are on a Log2 scale and the color represents the HLA allomorphs.
Fig. 4.
Numbers of HLA peptides shared between different patients harboring the same HLA allomorphs. The bars represent the peptides counts that were detected in multiple mHLA and sHLA peptidomes of different GBM patients and the colors indicate the percentage of patients in which such peptides were detected.
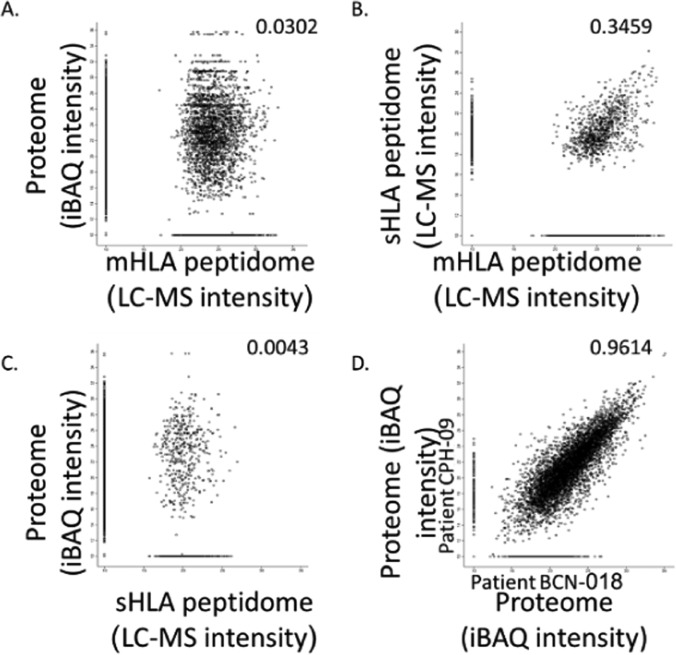
The HLA Peptidomes Do Not Correlate with the Proteomes of the Tumors
The protein repertoires and their levels (measured in iBAQ values) of the different tumors, were much better correlated than the HLA peptidomes of the different tumors (Fig. 5, supplemental Fig. S3). As many as 4855 different proteins were identified in all ten tumors analyzed, out of the total number of 7351 proteins identified (supplemental Table S3). Importantly, 1302 different HLA peptides, derived from 706 of the proteins identified in all of the tumor samples, were also detected in the tumor tissues mHLA and plasma sHLA peptidomes. None of these peptides was derived from the source genes of the sHLA peptides that were detected in the non-cancerous donors' plasma (supplemental Table S2, column E). Among these 1302 different peptides, a subset of 32 HLA peptides belong to the TAA group (Table III). An example for such peptide is AHIKGVETI of Outer dense fiber protein 1 (ODF1). The mRNA of ODF1 is not expressed in any of the essential healthy tissues, except the testis in healthy individuals, according to the BioGPS (supplemental Fig. S1A) and its protein was observed by the proteome analyses in all the ten analyzed tumors. Furthermore, no peptides derived from this gene were observed in any of the plasma-sHLA peptidome of the noncancerous donors. HLA peptides derived from this gene were detected in the plasma-sHLA of two female GBM patients (Table II). Therefore, this gene and similar genes like it, whose proteins products are expressed in the tumor tissues and their derived sHLA peptides appear in the plasma-sHLA peptidomes, may serve as both, potential biomarkers and candidates for immunotherapeutics.
Fig. 5.
Comparison between the tumor proteomes and HLA peptidomes: Comparison between the tumor proteome and mHLA peptidome (A), plasma sHLA and tumor mHLA peptidome (B), between the tumor proteome and plasma sHLA peptidome (C) of patient CPH-09, and comparison between the tumor proteomes of two different patients, CPH-09 and BCN-018 (D). Only gene products observed as both proteins and HLA peptides are displayed. Pearson correlations are indicated in each panel and the protein levels are calculated as iBAQ intensities on a Log2 scales. Missing values were replaced with arbitrary value of 10.
Table III. List of the HLA peptides and their source proteins, belonging to the TAA group. Peptides and proteins were selected only if they were detected in all ten GBM tumors, and their derived HLA peptides were not detected in any of the non-cancerous donors' plasma-sHLA peptidomes.
| Gene name | Protein name | Sequence |
|---|---|---|
| ADAM17 | Disintegrin and metalloproteinase domain-containing protein 17 | THVETLLTF |
| CTNNA2 | Catenin alpha-2 | SEFKAMDSF |
| SPVQALSEF | ||
| CTSH | Pro-cathepsin H | ESAIAIATGK |
| TQDFMMYRT | ||
| EGFR | Epidermal growth factor receptor | AENAEYLRV |
| ALAVLSNYDA | ||
| DEYLIPQQGF | ||
| GEAPNQALL | ||
| HNQPLNPAPSRDPH | ||
| IPLENLQII | ||
| KEILDEAYVM | ||
| NPTTYQMDVNPEGK | ||
| QEILHGAVRF | ||
| STFDSPAHW | ||
| TEFKKIKVL | ||
| TPLLSSLSATSNN | ||
| VERIPLENL | ||
| YENSYALAVL | ||
| YQDPHSTAV | ||
| KRAS | GTPase KRas | FVDEYDPTI |
| KSFEDIHHY | ||
| ODF1 | Outer dense fiber protein 1 | AHIKGVETI |
| MESPCSPCSPC | ||
| RIIDTPISEMGF | ||
| PAK2 | Serine/threonine-protein kinase PAK 2 | GQGASGTVF |
| TVKQKYLSF | ||
| PPIB | Peptidyl-prolyl cis-trans isomerase B | LLLPGPSAA |
| SCRN1 | Secernin-1 | AIIESDQEQGR |
| AIIESDQEQGRKLR | ||
| SNRPD1 | Small nuclear ribonucleoprotein Sm D1 | HETVTIEL |
| WNK2 | Serologically defined colon cancer antigen 43 | QEHVPTSSA |
Some HLA Allomorphs Present Larger Diversity of Peptides In Both Tumors and Plasma
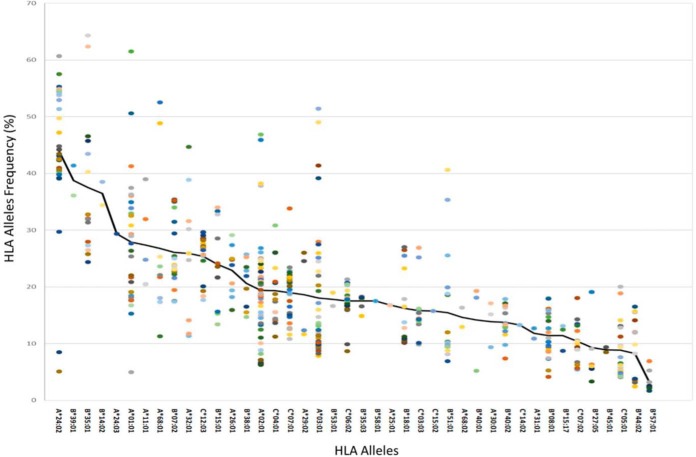
The 52 different GBM samples and 36 different noncancerous samples have undergone a complete HLA typing analysis based on DNA sequencing (51). This analysis facilitated the use of the NetMHC platform to fit the HLA peptides sequences to their likely presenting HLA allomorphs by selecting peptides with NetMHC rank equal or better than 2 (http://www.cbs.dtu.dk/services/NetMHC/). HLA allomorphs with a consensus sequence motif such as HLA-B*35:01 and HLA-A*24:02 presented larger numbers of peptides relative to allomorphs such as HLA-B*27:05 or HLA-B*40:01 (Fig. 4 and supplemental Table S8). This phenomenon was observed in both GBM and in the non-cancerous donors' plasma samples, as well as the GBM tissues (Fig. 6, supplemental Fig. S4). The HLA-A*24:02 allele is common among glioma patients (58) and therefore HLA peptides presented by these alleles may become useful for treatment of larger groups of patients.
Fig. 6.
Fitness of HLA peptides to different HLA alleles presenting them. Percentages of peptides discovered in each of the plasma sHLA peptidomes of the GBM patients that fit the sequence motifs of the HLA alleles of the individual patients, with scores below or equal 2 according to NetMHC. The colored dots represent different patients.
DISCUSSION
This study is an extensive immunopeptidome analysis of 106 GBM plasma samples, 10 GBM tumor tissues, and 36 control plasma samples. The analysis led to high confidence identification of 35,545 unique HLA peptides. It was suggested before that the plasma-sHLA peptidome represents the tumor-mHLA peptidome, because the tumors' cells shed larger amounts of sHLA molecules relative to the uninvolved tissues (28). Similarly, in this study, a significant overlap was observed between mHLA peptidomes of the tumor tissues and matched plasma-sHLA peptidomes of each of the patients, demonstrating that the plasma-sHLA peptidome contains many peptides derived from the tumors. These correlations, between the repertoires and LC-MS intensities of the mHLA and sHLA peptidomes of the individual patients, were much larger than the correlations between the HLA peptidomes and the proteomes of the patients' tumors (supplemental Fig. S3). Low correlations in both repertoires and expression levels, between HLA peptidomes, proteomes, and transcriptomes, of the same cells, were suggested in previous publications (59, 60) but were found to be higher in others (61–64). Additionally, we demonstrate that the plasma-sHLA peptidomes are relatively stable, can be analyzed reproducibly, and represent the tumor-mHLA peptidomes. Comparative analysis of both mHLA and sHLA peptidomes may provide candidate peptides, potentially useful for immunotherapy and as biomarkers. Precision medicine based on large-scale body-fluid biomarkers may help to identify patients that are most likely to benefit from specific treatments, including, but not limited to immunotherapy (65–67). It is expected that different HLA peptides derived from the TAA and CTA genes will be presented among the patients' sHLA allomorphs' peptidomes. Thus, the search among the sHLA peptidomes of different people for disease biomarkers is not limited to specific peptides, but to the presence of different peptides derived from the selected TAA/CTA genes. A few of the 250 downregulated sHLA peptides observed in this study may serve as biomarkers for early detection of the disease or relapse. Indeed, potentially useful TAA can be selected based on different criteria. In our opinion, first and outmost are peptides that are derived from genes that not expressed in any healthy adult tissue, other than the immune privileged sites, such as the germline cells. Such HLA peptides are potentially useful for both immunotherapy and diagnosis. Plasma sHLA peptides that disappear after treatment or remission are potentially also useful for diagnosis, for early detection and for relapse. sHLA and mHLA peptides that appear in tumor tissues and not in the sHLA peptidome of the healthy donors are clearly more significant targets for further research because such peptides are more likely (but not necessarily) tumor specific. However, only a few of the downregulated peptides observed here are TAAs. Such selected biomarker peptides do not need to be derived from bona fide CTAs, but need to be affected similarly in a large cohort of patients in order to become useful for clinical exploitation. One should consider that many peptides were not detected in this study in the plasma-sHLA peptidomes after surgery, also because of the nature of shotgun peptidomics methodology used here. Performing targeted LC-MS/MS analysis with the same or with different sets of samples is still required to exclude this possibility. The alternative use of data-independent analysis of HLA peptidome was already used for analyses of HLA peptidomes at higher reproducibility (68–71), and SRM were used to obtain more accurate presentation levels for the selected peptides in multiple analyses (72, 73). Smaller numbers of HLA peptides were identified with the LC-MS/MS data collected in this study when the FDR was set to 0.01 instead of 0.05 in the Andromeda search (50), performed within the MaxQuant analysis tool (49). This is expected, because the MS/MS fragmentations of many peptides are suboptimal in the data-dependent (shotgun) analysis performed in this study. The use of data-independent analysis for the HLA peptidome analysis may help to solve some of the data loss incurred (69, 71, 74). Alternatively, one can increase the FDR to 0.05 and facilitate this way the discovery of candidates for vaccine or biomarkers HLA peptides that would have been lost with 0.01 FDR (75). Here as well, a significant fraction of the peptides that were lost by use of 0.01 FDR, instead of 0.05, are true ligands of the HLA allomorphs of the patient (supplemental Table S9), as defined by the similarity of sequence motifs to the peptides included in the 0.01 FDR (supplemental Fig. S5).
The blood-brain barrier (BBB) prevents entrance and exit of some cells and molecules (5, 6) to the brain and it is unknown if it allows passage of the circulating sHLA-peptide complexes. However, the BBB in GBM patients is partially broken by the local inflammatory conditions and differs in its tumor vessels morphology and in the hyper-permeability of its endothelial cells (5). The loss of functional integrity of the BBB allows passage of low molecular weight compounds, including different chemotherapeutics into the brain (76). Such changes in the BBB may also facilitate the release of sHLA molecules with their bound peptides into the circulation, thus allowing their detection in the plasma for further evaluations as tumor markers.
Ideally, HLA peptides useful to serve as immunotherapeutics or biomarkers should be derived from genes expressed at sufficient levels in the malignant tissues, but not at all in any of the other essential healthy tissues (18, 23). Fresh-frozen tumors can be used for exome, transcriptome, proteome and HLA peptidome analyses, allowing identification of neoepitopes and tumor antigens (4, 16, 40–45, 63, 77). Selection of CTA candidates for immunotherapy can be based on high expression levels of their HLA peptides, mRNA and proteins in the tumors, and no expression in healthy tissues. Even though tumors and plasma are good sources of HLA peptides, healthy tissues are not normally available for analysis of their HLA peptidomes, whereas data about the gene expression levels is available in public databases. Very large and relatively accurate databases are publicly available, including data on gene and protein expression in many tissues, of numerous people. Examples include BioGPS (55, 56), TANTIGEN (53), and HPR (https://www.proteinatlas.org/) (78, 79). Although these databases are based on numerous studies, some discrepancies were observed between them in regard to different selected CTAs studied here. Using additional gene expression databases may alleviate some of these concerns and provide candidate immunotherapeutics with lower risk of inducing adverse effects.
The preferred CTAs, providing potentially useful HLA peptides discovered in this study, were defined as those derived from genes whose transcripts are expressed at levels below nine gcrma units in all normal, essential tissues (according to BioGPS) and are expressed at significantly higher levels in the tumors tissues. Nine gcrma units of mRNA levels of expression was selected here as sufficiently low, because this is the highest measured mRNA expression level of many well-characterized CTAs in healthy (nontestis) adult tissues. Some of these CTAs, including CTAG1A (NY-ESO-1) and MAGE-A1 are well known TAAs, whose expression levels in healthy tissues are below 9 gcrma units. These CTAs were already used in multiple clinical studies without observable autoimmune reactions (19, 80–82) suggesting that genes expressed below these levels are possibly safe for clinical use.
The tumor-mHLA and plasma-sHLA peptidomes of different people can be compared while looking for HLA peptides shared between larger cohorts of individuals. HLAs with completely distinct binding motifs are not expected to bind and present shared peptide ligands. Importantly, significant similarities were observed in this study between the HLA peptidomes of different patients that share some of their HLA alleles (Fig. 3). In contrast, shared peptides, detected in the HLA peptidomes of different people who do not have any common HLA alleles or have HLA alleles belonging to different HLA supertypes (83) are more likely to be defined as contaminants, rather than true HLA ligands. For example, it is very unlikely that HLAs such as HLA-A*2, B*7 and B*27 will share any peptide ligands (83). On the other hand, shared tumor antigens that are detected in multiple patients that have similar HLA allotypes, are extremely important, because of their potential to become useful for treatment of multiple patients.
The HLA molecules of some HLA allomorphs, such as HLA A*24:02, are more abundant in the plasma of carriers of these alleles (32). In addition, some HLA allomorphs present more numerous peptides than others do (84). Indeed, here we observe similar phenomena at the HLA peptidome levels, some HLA allomorphs, such as HLA-A24, HLA-B35, and HLA-B51 present more diverse repertoires of peptides than others do, in both the tumors and the plasma. This implies that the use of plasma-sHLA peptidome analysis is probably more efficient for carriers of these alleles, because larger sHLA peptidomes can be recovered and identified from their plasma samples (Fig. 4 and Fig. 6). It may also mean that there is larger immune tolerance in the HLA-A*24 patients and, therefore, it may be more difficult to break the immune tolerance induced by circulating sHLA molecules, when attempting immunotherapy. Importantly, HLA-A*24 was claimed to be associated genetically with glioma (58). Therefore, the use of the sHLA peptidome analysis to search for disease biomarkers will need to be adjusted accordingly, to consider the HLA allotypes of the individual patients.
In conclusion, the data described here suggests a useful method for selection of biomarkers and cancer immune-therapeutics for GBM and provides large lists of such candidates. Such methodologies are likely useful for discovery of biomarkers and immune-therapeutics candidates for other cancers. Most importantly, the identification of numerous sHLA peptides derived from CTAs can represent a promising noninvasive strategy for the monitoring of patients' response and progression during standard treatment modalities.
Data Availability
The mass spectrometry proteomics data of the tissue proteomes, the mHLA and the sHLA peptidomes have been deposited to the ProteomeXchange Consortium (54) (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD008127.
Supplementary Material
Acknowledgments
We thank Ilana Navon from the Smoler Proteomics Center at the Technion for performing the LC-MS/MS experiments. We acknowledge the advice and discussion with the members of the GAPVAC consortium, Stefan Stevanovic and Cécile Gouttefangeas.
Footnotes
* The GAPVAC project was funded by the European Union Framework 7 Program. Funding to Carlos López-Larrea were from the European Union Fondos Feder and Instituto de Salud Carlos III (Spain, grant number PI16/01318). http://gapvac.eu/funfing/.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GBM
- glioblastoma
- MHC
- major histocompatibility complex
- HLA
- human leukocytes antigen
- CTA
- cancer/testis antigen
- TSA
- tumor specific antigens
- TAA
- tumor associated antigens
- sHLA
- soluble human leukocytes antigen
- mHLA
- membranal human leukocytes antigen
- FDR
- false discovery rate
- BBB
- blood brain barrier.
REFERENCES
- 1. Alifieris C., and Trafalis D. T. (2015) Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol. Ther. 152, 63–82 [DOI] [PubMed] [Google Scholar]
- 2. Thakkar J. P., Dolecek T. A., Horbinski C., Ostrom Q. T., Lightner D. D., Barnholtz-Sloan J. S., and Villano J. L. (2014) Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol. Biomarkers Prev. 23, 1985–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Terasaki M., Shibui S., Narita Y., Fujimaki T., Aoki T., Kajiwara K., Sawamura Y., Kurisu K., Mineta T., Yamada A., and Itoh K. (2011) Phase I trial of a personalized peptide vaccine for patients positive for human leukocyte antigen-A24 with recurrent or progressive glioblastoma multiforme. J. Clin. Oncol. 29, 337–344 [DOI] [PubMed] [Google Scholar]
- 4. Neidert M. C., Schoor O., Trautwein C., Trautwein N., Christ L., Melms A., Honegger J., Rammensee H. G., Herold-Mende C., Dietrich P. Y., and Stevanović S. (2013) Natural HLA class i ligands from glioblastoma: Extending the options for immunotherapy. J. Neurooncol. 111, 285–294 [DOI] [PubMed] [Google Scholar]
- 5. Patel M. A., and Pardoll D. M. (2015) Concepts of immunotherapy for glioma. J. Neurooncol. 123, 323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohen-Inbar O., and Zaaroor M. (2016) Immunological aspects of malignant gliomas. Can. J. Neurol. Sci. / J. Can. des Sci. Neurol. 43, 494–502 [DOI] [PubMed] [Google Scholar]
- 7. Polivka J., Holubec L., Kubikova T., Priban V., Hes O., Pivovarcikova K., and Treskova I. (2017) Advances in experimental targeted therapy and immunotherapy for patients with glioblastoma multiforme. Anticancer Res. 37, 21–33 [DOI] [PubMed] [Google Scholar]
- 8. Swartz A. M., Batich K. A., Fecci P. E., and Sampson J. H. (2015) Peptide vaccines for the treatment of glioblastoma. J. Neurooncol. 123, 433–440 [DOI] [PubMed] [Google Scholar]
- 9. Ampie L., Woolf E. C., and Dardis C. (2015) Immunotherapeutic advancements for glioblastoma. Front. Oncol. 5, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oh T., Sayegh E. T., Fakurnejad S., Oyon D., Lamano J. B., DiDomenico J. D., Bloch O., and Parsa A. T. (2015) Vaccine therapies in malignant glioma. Curr. Neurol. Neurosci. Rep. 15, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Srinivasan V. M., Ferguson S. D., Lee S., Weathers S.-P., Kerrigan B. C. P., and Heimberger A. B. (2017) Tumor Vaccines for Malignant Gliomas. Neurotherapeutics 14, 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kamran N., Calinescu A., Candolfi M., Chandran M., Mineharu Y., Asad A. S., Koschmann C., Nunez F. J., Lowenstein P. R., and Castro M. G. (2016) Recent advances and future of immunotherapy for glioblastoma. Expert Opin. Biol. Ther. 16, 1245–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ott P. A., Hu Z., Keskin D. B., Shukla S. A., Sun J., Bozym D. J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., Chen C., Olive O., Carter T. A., Li S., Lieb D. J., Eisenhaure T., Gjini E., Stevens J., Lane W. J., Javeri I., Nellaiappan K., Salazar A. M., Daley H., Seaman M., Buchbinder E. I., Yoon C. H., Harden M., Lennon N., Gabriel S., Rodig S. J., Barouch D. H., Aster J. C., Getz G., Wucherpfennig K., Neuberg D., Ritz J., Lander E. S., Fritsch E. F., Hacohen N., and Wu C. J. (2017) An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharma P., and Allison J. P. P. (2015) Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161, 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Palucka A. K., and Coussens L. M. (2016) The basis of oncoimmunology. Cell 164, 1233–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dutoit V., Herold-Mende C., Hilf N., Schoor O., Beckhove P., Bucher J., Dorsch K., Flohr S., Fritsche J., Lewandrowski P., Lohr J., Rammensee H.-G., Stevanovic S., Trautwein C., Vass V., Walter S., Walker P. R., Weinschenk T., Singh-Jasuja H., and Dietrich P.-Y. (2012) Exploiting the glioblastoma peptidome to discover novel tumour-associated antigens for immunotherapy. Brain 135, 1042–1054 [DOI] [PubMed] [Google Scholar]
- 17. Bassani-Sternberg M., and Coukos G. (2016) Mass spectrometry-based antigen discovery for cancer immunotherapy. Curr. Opin. Immunol. 41, 9–17 [DOI] [PubMed] [Google Scholar]
- 18. Rammensee H.-G., and Singh-Jasuja H. (2013) HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev. Vaccines 12, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pol J., Bloy N., Buqué A., Eggermont A., Sautès-fridman, Galon C. J., Tartour E., Kroemer G., Galluzzi L., Pol J., Bloy N., Buqué A., Eggermont A., Cremer I., Sautès-fridman, Galon C. J., Tartour E., Zitvogel L., Kroemer G., Galluzzi L., Watch T., Pol J., Bloy N., Buqué A., Eggermont A., Cremer I., Zitvogel L., Pol J., Bloy N., Buqu A., Buque A., Eggermont A., Cremer I., Sautes-Fridman C., Galon J., Tartour E., Zitvogel L., Kroemer G., and Galluzzi L. (2015) Trial Watch : Peptide-based anticancer vaccines Trial Watch : Peptide-based anticancer vaccines. Oncoimmunology 4, e974411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Comber J. D., and Philip R. (2014) MHC class I antigen presentation and implications for developing a new generation of therapeutic vaccines. Ther Adv Vaccines 2, 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heemskerk B., Kvistborg P., and Schumacher T. N. M. (2012) The cancer antigenome. EMBO J. 32, 194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y. T., Scanlan M. J., Sahin U., Türeci O., Gure A. O., Tsang S., Williamson B., Stockert E., Pfreundschuh M., and Old L. J. (1997) A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. U.S.A. 94, 1914–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whitehurst A. W. (2014) Cause and consequence of cancer/testis antigen activation in cancer. Annu. Rev. Pharmacol. Toxicol. 54, 251–272 [DOI] [PubMed] [Google Scholar]
- 24. Schumacher T. N., and Schreiber R. D. (2015) Neoantigens in cancer immunotherapy. Science 348, 69–74 [DOI] [PubMed] [Google Scholar]
- 25. Charlton R. K., and Zmijewski C. M. (1970) Soluble HL-A7 antigen: localization in the beta-lipoprotein fraction of human serum. Science 170, 636–637 [DOI] [PubMed] [Google Scholar]
- 26. van Rood J. J., van Leeuwen A., and van Santen M. C. (1970) Anti HL-A2 inhibitor in normal human serum. Nature 226, 366–367 [DOI] [PubMed] [Google Scholar]
- 27. Tabayoyong W. B., and Zavazava N. (2007) Soluble HLA revisited. Leuk. Res. 31, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bassani-Sternberg M., Barnea E., Beer I., Avivi I., Katz T., and Admon A. (2010) Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc. Natl. Acad. Sci. U.S.A. 107, 18769–18776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ritz D., Gloger A., Weide B., Garbe C., Neri D., and Fugmann T. (2016) High-sensitivity HLA class I peptidome analysis enables a precise definition of peptide motifs and the identification of peptides from cell lines and patients' sera. Proteomics 16, 1570–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ritz D., Gloger A., Neri D., and Fugmann T. (2017) Purification of soluble HLA class I complexes from human serum or plasma deliver high quality immuno peptidomes required for biomarker discovery. Proteomics 17, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puppo F., Scudeletti M., Indiveri F., and Ferrone S. (1995) Serum HLA class I antigens: markers and modulators of an immune response? Immunol. Today 16, 124–127 [DOI] [PubMed] [Google Scholar]
- 32. Adamashvili I. M., Fraser P. A., and McDonald J. C. (1996) Association of serum concentration of soluble class I HLA with HLA allotypes. Transplantation 61, 984–987 [DOI] [PubMed] [Google Scholar]
- 33. Campoli M., and Ferrone S. (2008) Tumor escape mechanisms: potential role of soluble HLA antigens and NK cells activating ligands. Tissue Antigens 72, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hunt D. F., Henderson R. A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox a Appella L. E., and Engelhard V. H. (1992) Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 [DOI] [PubMed] [Google Scholar]
- 35. Granados D. P., Laumont C. M., Thibault P., and Perreault C. (2015) The nature of self for T cells-a systems-level perspective. Curr. Opin. Immunol. 34, 1–8 [DOI] [PubMed] [Google Scholar]
- 36. Schumacher F.-R., Delamarre L., Jhunjhunwala S., Modrusan Z., Phung Q. T., Elias J. E., and Lill J. R. (2017) Building proteomic tool boxes to monitor MHC class I and class II peptides. Proteomics 17, 1600061. [DOI] [PubMed] [Google Scholar]
- 37. de Verteuil D., Granados D. P., Thibault P., and Perreault C. (2012) Origin and plasticity of MHC I-associated self peptides. Autoimmun. Rev. 11, 627–635 [DOI] [PubMed] [Google Scholar]
- 38. Fritsche J., Rakitsch B., Hoffgaard F., Römer M., Schuster H., Kowalewski D. J., Priemer M., Stos-Zweifel V., Hoerzer H., Satelli A., Sonntag A., Goldfinger V., Song C., Mahr A., Ott M., Schoor O., Weinschenk T., Hörzer H., Satelli A., Sonntag A., Goldfinger V., Song C., Mahr A., Ott M., Schoor O., and Weinschenk T. (2018) Translating Immunopeptidomics to Immunotherapy-decision-making for patient and personalized target selection. Proteomics 18, e1700284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shraibman B., Kadosh D. M., Barnea E., and Admon A. (2016) Human leukocyte antigen (HLA) peptides derived from tumor antigens induced by inhibition of DNA methylation for development of drug-facilitated immunotherapy. Mol. Cell. Proteomics 15, 3058–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weinschenk T., Gouttefangeas C., Schirle M., Obermayr F., Walter S., Schoor O., Kurek R., Loeser W., Bichler K., Wernet D., Stevanović S., and Rammensee H.-G. (2002) Integrated functional genomics approach for the design of patient-individual antitumor vaccines. Cancer Res. 62, 5818–5827 [PubMed] [Google Scholar]
- 41. Bassani-Sternberg M., Bräunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., Specht K., Martignoni M. E., Werner A., Hein R., Busch H. D., Peschel C., Rad R., Cox J., Mann M., Krackhardt A. M., Busch H D, Peschel C., Rad R., Cox J., Mann M., and Krackhardt A. M. (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seliger B., Dressler S. P., Massa C., Recktenwald C. V., Altenberend F., Bukur J., Marincola F. M., Wang E., Stevanovic S., and Lichtenfels R. (2011) Identification and characterization of human leukocyte antigen class I ligands in renal cell carcinoma cells. Proteomics 11, 2528–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klatt M. G., Kowalewski D. J., Schuster H., Di Marco M., Hennenlotter J., Stenzl A., Rammensee H.-G., and Stevanović S. (2016) Carcinogenesis of renal cell carcinoma reflected in HLA ligands: A novel approach for synergistic peptide vaccination design. Oncoimmunology 5, e1204504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kowalewski D. J., Schuster H., Backert L., Berlin C., Kahn S., Kanz L., Salih H. R., Rammensee H.-G., Stevanovic S., and Stickel J. S. (2015) HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL). Proc. Natl. Acad. Sci. U.S.A. 112, E166–E175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walz S., Stickel J. S., Kowalewski D. J., Schuster H., Weisel K., Backert L., Kahn S., Nelde A., Stroh T., Handel M., Kohlbacher O., Kanz L., Salih H. R., Rammensee H., and Stevanović S. (2015) The antigenic landscape of multiple myeloma: mass spectrometry (re)defines targets for T-cell-based immunotherapy. Blood 126, 1203–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Milner E., Gutter-Kapon L., Bassani-Strenberg M., Barnea E., Beer I., and Admon A. (2013) The effect of proteasome inhibition on the generation of the human leukocyte antigen (HLA) peptidome. Mol. Cell. Proteomics 12, 1853–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ishihama Y., Rappsilber J., and Mann M. (2006) Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J. Proteome Res. 5, 988–994 [DOI] [PubMed] [Google Scholar]
- 48. Ishihama Y., Rappsilber J., Andersen J. S., and Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 49. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 50. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 51. Lange V., Böhme I., Hofmann J., Lang K., Sauter J., Schöne B., Paul P., Albrecht V., Andreas J. M., Baier D. M., Nething J., Ehninger U., Schwarzelt C., Pingel J., Ehninger G., and Schmidt A. H. (2014) Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics 15, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Almeida L. G., Sakabe N. J., deOliveira A. R., Silva M. C., Mundstein A. S., Cohen T., Chen Y. T., Chua R., Gurung S., Gnjatic S., Jungbluth A. A., Caballero O. L., Bairoch A., Kiesler E., White S. L., Simpson A. J., Old L. J., Camargo A. A., and Vasconcelos A. T. (2009) CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 37, D816–D819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang G. L., Sun J., Chitkushev L., and Brusic V. (2014) Big data analytics in immunology: a knowledge-based approach. Biomed Res. Int., 10.1155/2014/437987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Serrano A., Tanzarella S., Lionello I., Mendez R., Traversari C., Ruiz-Cabello F., and Garrido F. (2001) Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2′-deoxycytidine treatment. Int. J. Cancer 94, 243–251 [DOI] [PubMed] [Google Scholar]
- 55. Wu C., Jin X., Tsueng G., Afrasiabi C., and Su A. I. (2016) BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 44, D313–D316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., Su A. I., Huss JW 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Admon A., and Bassani-Sternberg M. (2011) The Human Immunopeptidome Project, a suggestion for yet another Postgenome Next Big Thing. Mol. Cell. Proteomics 10, O111.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nitta T., Ebato M., and Sato K. (1994) Association of malignant glioma with the human leukocyte antigen, HLA-A24(9). Neurosurg. Rev. 17, 211–215 [DOI] [PubMed] [Google Scholar]
- 59. Bourdetsky D., Schmelzer C. E. H., and Admon A. (2014) The nature and extent of contributions by defective ribosome products to the HLA peptidome. Proc. Natl. Acad. Sci. U.S.A. 111, E1591–E1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shraibman B., Kadosh D. M., Barnea E., and Admon A. (2016) Human Leukocyte Antigen (HLA) Peptides Derived from Tumor Antigens Induced by Inhibition of DNA Methylation for Development of Drug-facilitated Immunotherapy. Mol. Cell. Proteomics MCP 15, 3058–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bassani-Sternberg M., Pletscher-Frankild S., Jensen L. J., and Mann M. (2015) Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell. Proteomics 14, 658–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pearson H., Daouda T., Granados D. P., Durette C., Bonneil E., Courcelles M., Rodenbrock A., Laverdure J.-P., Côté C., Mader S., Lemieux S., Thibault P., and Perreault C. (2016) MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Invest. 126, 4690–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Khodadoust M. S., Olsson N., Wagar L. E., Haabeth O. A. W., Chen B., Swaminathan K., Rawson K., Liu C. L., Steiner D., Lund P., Rao S., Zhang L., Marceau C., Stehr H., Newman A. M., Czerwinski D. K., Carlton V. E. H., Moorhead M., Faham M., Kohrt H. E., Carette J., Green M. R., Davis M. M., Levy R., Elias J. E., and Alizadeh A. A. (2017) Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 543, 723–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schuster H., Peper J. K., Bösmüller H.-C., Röhle K., Backert L., Bilich T., Ney B., Löffler M. W., Kowalewski D. J., Trautwein N., Rabsteyn A., Engler T., Braun S., Haen S. P., Walz J. S., Schmid-Horch B., Brucker S. Y., Wallwiener D., Kohlbacher O., Fend F., Rammensee H.-G., Stevanović S., Staebler A., and Wagner P. (2017) The immunopeptidomic landscape of ovarian carcinomas. Proc. Natl. Acad. Sci. U.S.A. 114, E9942–E9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sawyers C. L. (2008) The cancer biomarker problem. Nature 452, 548–552 [DOI] [PubMed] [Google Scholar]
- 66. Perez-Gracia J. L., Sanmamed M. F., Bosch A., Patiño-Garcia A., Schalper K. A., Segura V., Bellmunt J., Tabernero J., Sweeney C. J., Choueiri T. K., Martín M., Fusco J. P., Rodriguez-Ruiz M. E., Calvo A., Prior C., Paz-Ares L., Pio R., Gonzalez-Billalabeitia E., Gonzalez Hernandez A., Páez D., Piulats J. M., Gurpide A., Andueza M., de Velasco G., Pazo R., Grande E., Nicolas P., Abad-Santos F., Garcia-Donas J., Castellano D., Pajares M. J., Suarez C., Colomer R., Montuenga L. M., and Melero I. (2016) Strategies to design clinical studies to identify predictive biomarkers in cancer research. Cancer Treat. Rev. 53, 79–97 [DOI] [PubMed] [Google Scholar]
- 67. Tsiatas M., Mountzios G., and Curigliano G. (2016) Future perspectives in cancer immunotherapy. Ann. Transl. Med. 4, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Croft N. P., de Verteuil D. A., Smith S. A., Wong Y. C., Schittenhelm R. B., Tscharke D. C., and Purcell A. W. (2015) Simultaneous quantification of viral antigen expression kinetics using data-independent (DIA) mass spectrometry. Mol. Cell. Proteomics 14, 1361–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Caron E., Espona L., Kowalewski D. J., Schuster H., Ternette N., Alpízar A., Schittenhelm R. B., Ramarathinam S. H., Lindestam Arlehamn C. S., Chiek Koh C., Gillet L. C., Rabsteyn A., Navarro P., Kim S., Lam H., Sturm T., Marcilla M., Sette A., Campbell D. S., Deutsch E. W., Moritz R. L., Purcell A. W., Rammensee H.-G., Stevanovic S., and Aebersold R. (2015) An open-source computational and data resource to analyze digital maps of immunopeptidomes. Elife 4, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Caron E., Kowalewski D. J., Chiek Koh C., Sturm T., Schuster H., and Aebersold R. (2015) Analysis of major histocompatibility complex (MHC) immunopeptidomes using mass spectrometry. Mol. Cell. Proteomics 14, 3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ritz D., Kinzi J., Neri D., and Fugmann T. (2017) Data-independent acquisition of HLA class I peptidomes on the Q Exactive mass spectrometer platform. Proteomics 17, 1700177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tan C. T., Croft N. P., Dudek N. L., Williamson N. A., and Purcell A. W. (2011) Direct quantitation of MHC-bound peptide epitopes by selected reaction monitoring. Proteomics 11, 2336–2340 [DOI] [PubMed] [Google Scholar]
- 73. Croft N. P., Purcell A. W., and Tscharke D. C. (2015) Quantifying epitope presentation using mass spectrometry. Mol. Immunol. 68, 77–80 [DOI] [PubMed] [Google Scholar]
- 74. Schittenhelm R. B., Sivaneswaran S., Lim Kam Sian T. C., Croft N. P., and Purcell A. W. (2016) Human Leukocyte Antigen (HLA) B27 Allotype-Specific Binding and Candidate Arthritogenic Peptides Revealed through Heuristic Clustering of Data-independent Acquisition Mass Spectrometry (DIA-MS) Data. Mol. Cell. Proteomics 15, 1867–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barnea E., Melamed Kadosh D., Haimovich Y., Satumtira N., Dorris M. L., Nguyen M. T., Hammer R. E., Tran T. M., Colbert R. A., Taurog J. D., and Admon A. (2017) The Human Leukocyte Antigen (HLA)-B27 Peptidome in Vivo, in Spondyloarthritis-susceptible HLA-B27 Transgenic Rats and the Effect of Erap1 Deletion. Mol. Cell. Proteomics 16, 642–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Agarwal S., Sane R., Oberoi R., Ohlfest J. R., and Elmquist W. F. (2011) Delivery of molecularly targeted therapy to malignant glioma, a disease of the whole brain. Expert Rev. Mol. Med. 13, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kalaora S., Barnea E., Merhavi-Shoham E., Qutob N., Teer J. K., Shimony N., Schachter J., Rosenberg S. A., Besser M. J., Admon A., and Samuels Y. (2016) Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget 7, 5110–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Uhlén M., Björling E., Agaton C., Szigyarto C. A.-K., Amini B., Andersen E., Andersson A.-C., Angelidou P., Asplund A., Asplund C., Berglund L., Bergström K., Brumer H., Cerjan D., Ekström M., Elobeid A., Eriksson C., Fagerberg L., Falk R., Fall J., Forsberg M., Björklund M. G., Gumbel K., Halimi A., Hallin I., Hamsten C., Hansson M., Hedhammar M., Hercules G., Kampf C., Larsson K., Lindskog M., Lodewyckx W., Lund J., Lundeberg J., Magnusson K., Malm E., Nilsson P., Odling J., Oksvold P., Olsson I., Oster E., Ottosson J., Paavilainen L., Persson A., Rimini R., Rockberg J., Runeson M., Sivertsson A., Sköllermo A., Steen J., Stenvall M., Sterky F., Strömberg S., Sundberg M., Tegel H., Tourle S., Wahlund E., Waldén A., Wan J., Wernérus H., Westberg J., Wester K., Wrethagen U., Xu L. L., Hober S., and Pontén F. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics 4, 1920–1932 [DOI] [PubMed] [Google Scholar]
- 79. Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å Kampf C, Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A.-K., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Pontén F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 80. Dutoit V., Taub R. N., Papadopoulos K. P., Talbot S., Keohan M. L., Brehm M., Gnjatic S., Harris P. E., Bisikirska B., Guillaume P., Cerottini J. C., Hesdorffer C. S., Old L. J., and Valmori D. (2002) Multiepitope CD8+ T cell response to an NY-ESO-1 peptide vaccine results in imprecise tumor targeting. J. Clin. Invest. 110, 1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Marchand M., Van Baren N., Weynants P., Brichard V., Dréno B., Tessier M. H., Rankin E., Parmiani G., Arienti F., Humblet Y., Bourlond A., Vanwijck R., Liénard D., Beauduin M., Dietrich P. Y., Russo V., Kerger J., Masucci G., Jäger E., De Greve J., Atzpodien J., Brasseur F., Coulie P. G., Van Der Bruggen P., and Boon T. (1999) Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Int. J. Cancer 80, 219–230 [DOI] [PubMed] [Google Scholar]
- 82. Rosenberg S. A., Yang J. C., Schwartzentruber D. J., Hwu P., Marincola F. M., Topalian S. L., Restifo N. P., Dudley M. E., Schwarz S. L., Spiess P. J., Wunderlich J. R., Parkhurst M. R., Kawakami Y., Seipp C. A., Einhorn J. H., and White D. E. (1998) Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat. Med. 4, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sidney J., Peters B., Frahm N., Brander C., and Sette A. (2008) HLA class I supertypes: A revised and updated classification. BMC Immunol. 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Paul S., Weiskopf D., Angelo M. A., Peters B., Sette A., Paul S., Weiskopf D., Angelo M. A., Sidney J., Peters B., and Sette A. (2013) HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J. Immunol. 191, 5831–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data of the tissue proteomes, the mHLA and the sHLA peptidomes have been deposited to the ProteomeXchange Consortium (54) (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD008127.