Abstract
Pseudomyxoma peritonei (PMP) is a subtype of mucinous adenocarcinoma that most often originates from the appendix, and grows in the peritoneal cavity filling it with mucinous ascites. KRAS and GNAS mutations are frequently found in PMP, but other common driver mutations are infrequent. As altered glycosylation can promote carcinogenesis, we compared N-linked glycan profiles of PMP tissues to those of normal appendix. Glycan profiles of eight normal appendix samples and eight low-grade and eight high-grade PMP specimens were analyzed by mass spectrometry. Our results show differences in glycan profiles between PMP and the controls, especially in those of neutral glycans, and the most prominent alteration was increased fucosylation. We further demonstrate up-regulated mRNA expression of four fucosylation-related enzymes, the core fucosylation performing fucosyltransferase 8 and three GDP-fucose biosynthetic enzymes in PMP tissues when compared with the controls. Up-regulated protein expression of the latter three enzymes was further observed in PMP cells by immunohistochemistry. We also demonstrate that restoration of fucosylation either by salvage pathway or by introduction of an expression of intact GDP-mannose 4,6-dehydratase enhance expression of MUC2, which is the predominant mucin molecule secreted by the PMP cells, in an intestinal-derived adenocarcinoma cell line with defective fucosylation because of deletion in the GDP-mannose 4,6-dehydratase gene. Thus, altered glycosylation especially in the form of fucosylation is linked to the characteristic mucin production of PMP. Glycomic data are available via ProteomeXchange with identifier PXD010086.
Keywords: Glycomics, Mass Spectrometry, Gastrointestinal disease, Glycosylation, Gene Expression, Immunohistochemistry, fucosylation, GNAS, mucin, pseudomyxoma peritonei
Pseudomyxoma peritonei (PMP)1 is a rare mucinous adenocarcinoma with an incidence of one to two cases per million per year (1). PMP originates most commonly from a low-grade appendiceal mucinous neoplasm (LAMN), and through its rupture neoplastic cells spread into the peritoneal cavity. In the peritoneal cavity PMP cells grow relatively slowly, but they secrete abundant amounts of extracellular mucin, which in the terminal phase of the disease causes progressive obstruction of the bowel that finally leads to death of the patient. PMP is classified into low-grade (LG) and high-grade (HG) subtypes (2, 3), the five-year survival being 63% for the LG and only 23% for the HG disease (4). The current standard treatment for PMP is aggressive cytoreductive surgery followed by hyperthermic intraperitoneal chemotherapy (5). However, this treatment is amenable only for 60–70% of the patients (6, 7) and a significant proportion of treated PMP patients get relapse (8). Thus, biomarkers for estimation of the aggressiveness of the disease and novel targeted treatments are needed. Further, therapeutic interventions that would reduce secretion of mucinous ascites could potentially be beneficial for the patients.
We and others have previously shown that PMP contains activating mutations of KRAS (70–100%) and GNAS (38–69%) genes (9–16). We have also reported that several components of the cyclic AMP-protein kinase A pathway are mutated in PMP disease (16), which stimulates mucin production (17, 18). Glycosylation is an important post-translational modification of proteins, and all human cells have a glycan-rich coating composed of glycoproteins, glycolipids, and proteoglycans, which affect cell differentiation, adhesion, and proliferation (19). In line with that, altered glycosylation has been observed in many types of malignant diseases (19), and recently gastrointestinal cancers have been shown to express changes in asparagine-linked glycan (N-glycan) branching and increased fucosylation and sialylation (20). Thus, glycomic profiling of neoplastic diseases has potential to reveal novel, effective molecular targets for diagnostics and as therapeutics.
In this study we characterize, for the first time, the N-linked glycan profiles of PMP by mass spectrometry and show differences in glycan profiles between the PMP tissue and normal appendix. Further, we found differentially expressed glycosylation-related gene products, and analyzed expression of fucosylation-related enzymes and fucosylated glycans by immunohistochemistry and lectin histochemistry in the PMP tissue specimens. Finally, we confirmed the relationship between fucosylation and mucin expression in an intestinal-derived adenocarcinoma cell line.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
We used previously developed workflow (21–24) to analyze the N-glycan profiles from formalin-fixed, paraffin-embedded (FFPE) tissue specimens of PMP patients and normal control appendices. For statistical comparisons we used eight control appendices and 16 PMP specimens, of which eight were LG PMPs and eight were HG PMPs. For analysis of glycosylation-related gene expression, we used previously published microarray data of 26 fresh frozen PMP specimens and three appendix controls (25). The in vitro experiments were repeated at least three times, and in qPCR assays, three biological replicates and three to four technical replicates were used. The statistical analyses performed are detailed below in the corresponding chapters.
Tissue Specimens
For glycomic profiling, we selected FFPE tissue blocks from eight normal appendices, and eight LG and eight HG PMP specimens originating from the appendix. Detailed information of each specimen is listed in supplemental Table S1. The patients received no preoperative oncological treatments. To avoid fucosyltransferase 3 (FUT3) mutated cases, occurring in 5–10% of the population, only CA19–9 immunopositive cases were included based on our previous study (26). All tissue specimens were routinely fixed and embedded in paraffin at the Department of Pathology, HUSLAB, Helsinki University Hospital, between 2007 and 2016, and the study was approved by the Ethics Committee of the Helsinki University Hospital. Areas containing the highest percentages of epithelial cells were first marked on HE slides and macrodissection was used to cut 30 μm thick flakes from the most representative areas, after which a new HE slide was stained to confirm the representativeness of the flakes. In addition, FFPE tissue specimens of appendix-derived PMPs (n = 100; 60 LG and 40 HG) and control appendices (n = 26) were used for immunohistochemistry and lectin histochemistry. For appendix the mean age is 57 and the median 60, range 31–80, and for PMP the mean age is 58 and the median 60, range 29–88; in respect of gender appendix group consisted of 58% females, the value for PMP being 60%. The appendiceal origin of the tumors was judged from the appendix tissue block and/or the surgery report, and grading of the tumors was re-evaluated according to the WHO 2010 classification and the content of neoplastic cells in the tissue specimens was estimated by AR (2). LG morphology was defined by strips of cells or small islands of relatively low cellularity with typically only low-grade atypia, whereas HG morphology contained more complex structures (invasive small cell clusters, cribriform structures and/or signet ring cell morphology), stromal reaction, and typically high-grade atypia with high cellularity. Any amount of HG morphology lead to classification of the tumor as HG PMP.
Glycan Isolation and Mass Spectrometry
FFPE tissue flakes were deparaffinized and disrupted in microcentrifuge tubes using a fitting plastic micro pestle, and N-linked glycans were detached by N-glycosidase F (PNGase F) digestion (Glyko; ProZyme Inc., Hayward, CA) and purified as previously reported (23). Briefly, the detached glycans were passed in water through C18 silica and absorbed to graphitized carbon material, both in 96-well format. The carbon wells were washed with water, and neutral glycans were eluted with 25% acetonitrile and acidic glycans with 0.05% trifluoroacetic acid in 25% acetonitrile in water. The acidic glycans were further purified by hydrophilic interaction solid-phase extraction, also in 96-well format, after which both glycan fractions were additionally passed in water through strong cation-exchange resin. Matrix-assisted laser desorption-ionization time-of-flight (MALDI-TOF) mass spectrometry was then performed with a Bruker Ultraflex TOF/TOF instrument (Bruker Daltonics Inc, Bremen, Germany) with neutral N-glycans detected in positive ion reflector mode as (M+Na)+ ions and acidic N-glycans in negative ion reflector mode as (M-H)− ions. Examples of unprocessed MALDI-TOF mass spectra of both neutral and acidic N-glycans are shown in supplemental Fig. S1, and annotated, mass-labeled spectra of all samples are provided in supplemental Data S1 (neutral glycans) and supplemental Data S2 (acidic glycans). Relative molar abundances of neutral and acidic glycan components were assigned based on their relative signal intensities in the mass spectra when analyzed separately as neutral and acidic N-glycan fractions, and the raw data were processed into the present glycan profiles as previously (21, 22). The resulting glycan signals in the presented glycan profiles were normalized to 100% to allow comparison between samples and assigned to biosynthetic groups based on their proposed monosaccharide compositions (supplemental Tables S2 and S3) (21, 22).
Analysis of N-glycan Profiles
N-glycan data was analyzed in four different study group settings: control appendices, LG PMPs, HG PMPs, and all PMP samples. For statistical analyses, two-sided Wilcoxon Rank Sum test for the mean value of relative intensities of N-glycan signals was used to compare the differences between the groups. To visualize the differences between the groups, we used Principal component analysis (PCA). It was first performed by using all glycan structures, but in this analysis the sample groups were not statistically significantly separated (result not shown). Next, we performed the PCA using neutral and acidic compositions whose relative intensities differed significantly (p < 0.05) between the control appendices and all PMP samples, as well as between the LG PMPs and the HG PMPs. Two components were extracted, and Bartlett's and Kaiser-Meyer-Olkin tests were used to test the adequacy of PCA for the data. Both the normalized signal intensity data and the assigned biosynthetic groupings were used for the analyses. Analyses were performed by Anduril2 workflow framework (manuscript under preparation) and R (27).
Validation of N-glycan Structures
To validate the proposed molecular structures, non-reducing terminal glycan epitopes of neutral N-glycan fraction from a HG PMP sample were analyzed by digestion with specific exoglycosidase enzymes: α-mannosidase from Jack beans (Canavalia ensiformis; Sigma-Aldrich, St. Louis, MO), β-N-acetylglucosaminidase from Streptococcus pneumoniae (recombinant in Escherichia coli; Calbiochem, San Diego, CA), β-N-acetylhexosaminidase from Jack beans (Seikagaku, Tokyo, Japan),α1,2-fucosidase from Xanthomonas manihotis (New England Biolabs, Ipswich, MA), and α1,3/4-fucosidase from Prunus dulcis (expressed in Pichia pastoris; New England Biolabs). Enzyme specificity toward isomeric structures was controlled with defined purified oligosaccharides. Reactions were carried out by overnight digestion at 37 °C in 50 mm sodium acetate buffer (pH 5.0). Digested glycan fractions were purified for analysis by solid-phase extraction with graphitized carbon, analyzed by MALDI-TOF mass spectrometry as described above, and interpreted as previously (21). For further verification of glycan structures the N-glycans were permethylated (28) for MS/MS fragmentation analysis.
Analysis of Microarray Data
The microarray data of 26 fresh frozen PMP specimens (24 LG, 2 HG) and three appendix controls (25) were kindly provided by Prof. Edward A. Levine (Wake Forest University, NC), and the analysis was performed with the Anduril workflow framework (29) and R. The raw expression data were normalized using the RMA (30) and quantile methods. Differentially expressed genes between the PMP samples and the appendix controls were calculated by comparing the log2 median values of normalized expressions. A gene was considered differentially expressed when at least 1.2-fold increase or decrease in the expression was observed. Statistical significance for the differentially expressed genes was calculated by two-sided t test, and False Discovery Rate (FDR) (31) was used for the multiple test correction. All the glycosylation-related differentially expressed genes with the FDR corrected p value < 0.01 were further analyzed by using The Consortium for Functional Glycomics (CFG) list of over 1200 glycosylation-relevant genes (Human Gene Lists GLYCOv4; http://www.functionalglycomics.org/static/consortium/resources/resourcecoree.shtml). From the 30 differentially expressed glycosylation-related genes we then selected those fitting with the observed N-glycan profiles.
Immunohistochemistry
Three-μm tissue sections were immunostained with antibodies specific to fucosylation-related enzymes fucosyltransferase 8 (FUT8) (66118–1-Ig; mouse monoclonal; Proteintech Group, Rosemont, IL; 1:500 dilution), GDP-mannose 4,6-dehydratase (GMDS) (clone 2A1; mouse monoclonal; LSBio, Seattle, WA; 1:1000 dilution), GDP-mannose pyrophosphorylase A (GMPPA) (NBP1–85904; rabbit polyclonal; Novus Biologicals, Littleton, CO; 1:100 dilution), and GDP-l-fucose synthase (TSTA3) (LS-B9837; rabbit polyclonal; LSBio; 1:100 dilution). Further, complex fucosylation containing glycan structure sialyl-Lewis x (sLex) was stained using α-CD15s monoclonal antibody (clone CSLEX1; BD Biosciences, San Jose, CA; 1:500 dilution). In addition to 95–100 PMP specimens, all the proteins and sLex glycan were further immunostained from 25–26 normal appendix specimens. The stainings were performed with Ventana BenchMark XT immunostainer (Ventana Medical Systems, Tucson, AZ) utilizing OptiView DAB IHCv3 kit (Ventana) (FUT8, GMDS, and sLex) or UltraView DAB kit (Ventana) with (GMPPA) or without (TSTA3) Amplification kit (Ventana). The staining intensities were scored as negative, weak (+), moderate (++), or strong (+++). In case two staining intensities were detected, the one covering the largest area was selected as the overall score. For statistical analysis the scoring was divided to low (negative and weak) and high (moderate and strong) immunopositivity. The scoring was performed by HL and PN, and discrepant scores were evaluated separately, after which consensus score was used for the statistical analysis with Fisher's exact test.
Lectin Histochemistry
Three-μm tissue sections of 28 PMP tumors (14 LG, 14 HG) and ten control appendices were stained with Aleuria aurantia lectin (AAL), that binds to fucosylated glycans with a preference to α1,6-linked core fucose residues attached by FUT8 enzyme (32). The sections were first deparaffinized and rehydrated, after which they were heated in Tris-HCl buffer (pH 8.5) in a microwave oven for 4 × 5 min to unmask the glycans. Endogenous peroxidase activity was blocked using 0.3% (v/v) hydrogen peroxide in methanol for 30 min and nonspecific binding was reduced by blocking with 3% (w/v) BSA in PBS for 1 h. Horseradish peroxidase (HRP)-conjugated AAL (Alpha Diagnostic International, San Antonio, TX) was then incubated at a dilution of 1:1000 for 1 h in a moist chamber. After washing with PBS, the signal was developed with 3,3′-diaminobenzidine (DAB) as the chromogen. Finally, the slides were counterstained with hematoxylin, dehydrated in ascending ethanol series, and embedded in Mountex (Histolab Products AB, Gothenburg, Sweden).
Cell Culture
Parental intestinal-derived carcinoma cell line HCT116 and a HCT116 cell line bearing activating GNAS mutation (R201C/+; both from Horizon Discovery Ltd., Cambridge, UK) were cultured in Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12 Ham medium (Sigma-Aldrich) supplemented with Penicillin-Streptomycin, GlutaMAXTM Supplement, and 10% Fetal Bovine Serum (Thermo Fisher Scientific, Waltham, MA). The cell lines were routinely tested for mycoplasma infection using the Venor®GeM Classic Mycoplasma PCR detection Kit (Minerva Biolabs GmbH, Berlin, Germany). HCT116 cell line has been demonstrated to show deficient fucosylation because of large deletion in the gene coding for the GDP-fucose biosynthetic enzyme GMDS (33), and to restore fucosylation in these cells, we used two approaches. First, to restore fucosylation via salvage pathway, 20 mm l-fucose (Carbosynth Ltd., Newbury, UK) was added into the growth medium and the cells were grown for 8–11 days (subculture twice per week) before analyses. To restore fucosylation via de novo pathway, pcDNA3.1/Hyg vector carrying human wildtype GMDS (33) (kindly provided by Prof. E. Miyoshi, Osaka University Graduate School of Medicine, Japan), or an empty vector serving as a control, were transfected into both parental and GNAS mutant HCT116 cells with LipofectamineTM 3000 Reagent (Thermo Fisher Scientific) according to the manufacturer's protocol. Selection was performed with Hygromycin B (Thermo Fisher Scientific) at 200 μg/ml and the resistant cells were analyzed.
Western Blotting and Lectin Blot Analysis
Cells grown to near confluency were collected and lysed with RIPA buffer (150 mm NaCl, 1% NP-40, 1% DOC, 0.1% SDS, 50 mm Tris-HCl (pH 8.0), 1 mm EDTA (pH 8.0)) containing protease inhibitor mixture (Roche, Basel, Switzerland) on ice for 30 min. After 10 s sonication the samples were centrifuged at 14,000 rpm for 15 min at 4 °C, and the protein concentrations of the supernatants were quantitated using BCA Protein Assay Kit (Pierce/Thermo Scientific, Rockford, IL). Samples containing 15 or 20 μg (lectin blot) or 60 μg (Western blotting) total proteins were then resolved by 12% SDS-PAGE gels and transferred onto nitrocellulose membranes (Amersham Biosciences, GE Healthcare, Little Chalfont, UK). The membranes were blocked with 5% milk and immunoblotted using rabbit anti-GMDS antibody (Proteintech Group) and HRP-conjugated anti-rabbit secondary antibody (DAKO/Agilent, Santa Clara, CA), or blocked with 3% BSA and lectin blotted using the above mentioned HRP-conjugated AAL. To verify equal loading of each lane, the membranes were blotted with mouse anti-α-Tubulin antibody (Abcam, Cambridge, UK) and HRP-conjugated anti-mouse secondary antibody (DAKO).
Quantitative Polymerase Chain Reaction (qPCR) of MUC2
Total RNA was extracted from the cell pellets using NucleoSpin® RNA kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany), and the RNA quality and quantity were measured with NanoDrop spectrophotometer (Thermo Fischer Scientific). For further quality control, the RNA samples were analyzed with Agilent 2200 TapeStation using RNA ScreenTapes (Agilent) and the RIN values were detected to vary between 9.5 and 10.0. For cDNA synthesis, 1 μg of total RNA was taken and the synthesis was accomplished with SuperScript® IV First-Strand Synthesis System (Thermo Fisher Scientific) according to the kit manual, using the Oligo(dT)20 primers included and incubating the reactions at 50 °C. For qPCR reaction, 1.5 μl of the 20 μl cDNA synthesis reaction was combined with TaqMan® Universal Master Mix II (with UNG), and TaqMan® Gene Expression Assays for MUC2 (Hs00894041_g1 with FAM-MGB) and RPL13A (Hs04194366_g1 with VIC-MGB, used as an endogenous reference gene) in a volume of 10 μl (Thermo Fisher Scientific). The qPCR run was performed with Applied Biosystems® 7500 Fast Real-Time PCR System (Thermo Fischer Scientific) using following conditions: 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles of 95 °C denaturation for 15 s and annealing/extension for 1 min and analyzing each sample in triplicates. The raw data were analyzed using ΔΔCt analysis. All the experiments were repeated more than three times with three biological replicates and three to four technical replicates each time. Statistical significance for the differential expression (p < 0.05) was calculated by paired-samples t test, after first confirming the normality with Shapiro-Wilk test (IBM SPSS Statistics 24; IBM Corporation, Armonk, NY).
RESULTS
Neutral N-linked Glycan Profiles
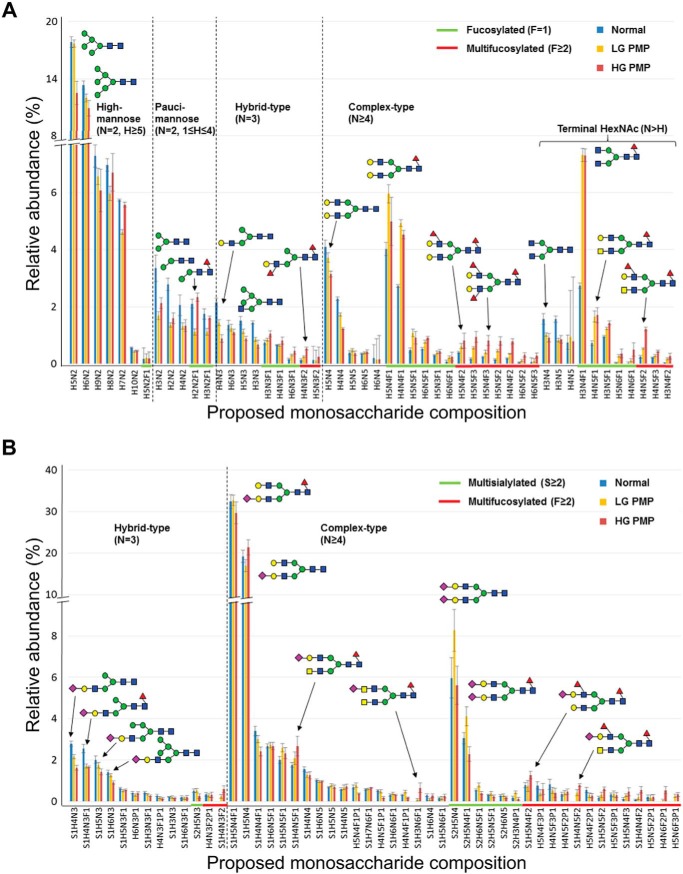
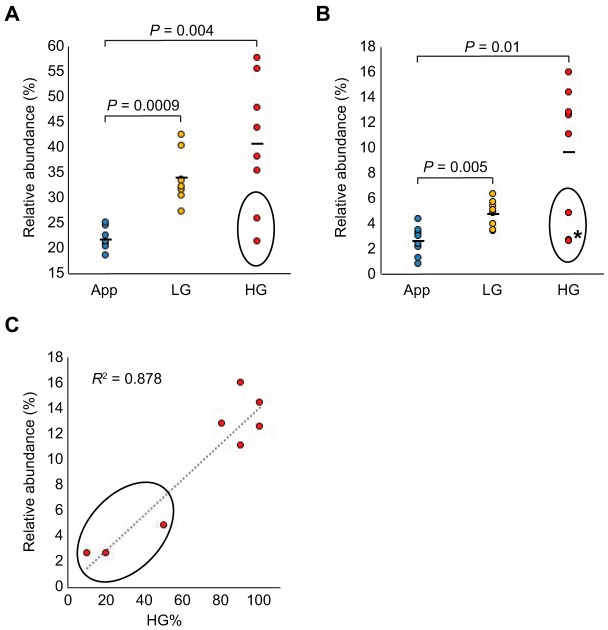
Neutral N-linked glycan profiles of PMP patient specimens were analyzed using mass spectrometry. Glycomic profiling of normal appendix controls and LG and HG PMP samples (n = 8 in each group) are shown as relative abundancy of the fifty most prominent proposed monosaccharide compositions in Fig. 1. When neutral and acidic N-linked glycan profiles were stratified by biosynthetic classification rules based on the amounts of hexose (H), N-acetylhexosamine (N), and deoxyhexose (F) residues in the proposed monosaccharide compositions, and shown as proportion of major glycan structural classes between the control tissues and the PMP samples, we found several fucosylation-related glycan classes to be elevated in the PMP samples when compared with the controls (Table I). Overall fucosylation (all structures containing F≥1) was increased in the PMP (p = 0.0003) along with several fucosylated subgroups (fucosylation in hybrid-type (p < 0.0001), fucosylation in complex-type (p = 0.0001), complex fucosylation (multifucosylation; p = 0.002), and complex fucosylation in hybrid-type (p = 0.023)). Furthermore, complex fucosylation in hybrid-type showed higher levels in the HG PMP when compared with the LG PMP specimens (supplemental Table S4). In addition to fucosylation, complex-type structures (p = 0.0005), terminal HexNAc (p = 0.001), and large N-glycans (p = 0.04) were elevated in PMPs when compared with the controls, whereas biantennary-size (p = 0.009), and high- (p = 0.023) and low-mannose (p = 0.032) type structures were found in lower abundance in PMPs than in the controls. Relative abundance of two subgroups (overall fucosylation (F≥1) and complex fucosylation (F≥2)) are shown in Fig. 2A and 2B. As HG samples contained different proportions of HG and LG morphology (any amount of HG morphology being criteria for grading HG type of PMP), we tested correlation between the abundance of complex fucosylation and the percentage of HG morphology, and found a positive correlation (R2 = 0.878; Fig. 2C). Our data suggest that complex fucosylation is elevated in PMP and that it may be especially high in PMP cells of HG morphology.
Fig. 1.
Relative abundance of monosaccharide compositions in normal appendix controls (blue bars) and in pseudomyxoma peritonei samples of low-grade (LG, yellow bars) and of high-grade (HG, brown bars) as analyzed by mass spectrometry (n = 8 in each sample group). A, On X-axis are shown the proposed neutral N-glycan structures of which fucosylated and multifucosylated are highlighted by green and by red lines, respectively. B, On X-axis are shown the proposed acidic N-glycan structures of which multisialylated and multifucosylated ones are highlighted by green and by red lines, respectively. Each three sample groups consisted of eight samples and the results are shown as means ±S.E. Glycan structures are depicted by green circle (d-mannose), blue square (N-acetyl-d-glucosamine), red triangle (l-fucose), yellow circle (d-galactose), and purple diamond (N-acetylneuraminic acid), and major structural subgroups are separated by dotted line. H = Hexose; N = N-Acetylhexosamine; F = Deoxyhexose; S = Sialic acid; P = Acid ester.
Table 1. Relative abundance of neutral and acidic N-linked glycan structure groups in pseudomyxoma peritonei specimens of low- and high-grade and in the appendix control samples. Only statistically different structures are shown (p < 0.05).
| Mean value % |
p value for Control vs. PMP (LG and HG) | |||
|---|---|---|---|---|
| Control (n = 8) | LG (n = 8) | HG (n = 8) | ||
| Neutral N-glycan structure | ||||
| Fucosylation in hybrid-type | 27.1 | 36.9 | 50.9 | 0.00003 |
| Fucosylation in complex-type | 56.9 | 74.7 | 78.4 | 0.0001 |
| Overall fucosylation | 22.0 | 33.8 | 40.7 | 0.0003 |
| Complex-type | 26.1 | 37.7 | 40.2 | 0.0005 |
| Terminal HexNAc | 9.1 | 14.8 | 16.9 | 0.001 |
| Complex fucosylation | 2.6 | 4.8 | 9.6 | 0.002 |
| Terminal HexNAc (bisecting-size) | 5.5 | 8.0 | 9.1 | 0.004 |
| Biantennary-size | 33.6 | 28.6 | 24.9 | 0.009 |
| High-mannose type | 52.2 | 47.6 | 42.6 | 0.023 |
| Complex fucosylation in hybrid-type | 4.1 | 5.9 | 13.2 | 0.023 |
| Low-mannose type | 12.5 | 7.0 | 9.4 | 0.032 |
| Large-N-glycans | 10.4 | 12.7 | 16.1 | 0.040 |
| Acidic N-glycan structure | ||||
| Hybrid-type | 12.1 | 9.7 | 8.8 | 0.006 |
| Complex-type | 87.6 | 89.8 | 90.9 | 0.007 |
| Terminal HexNAc (bisecting-size) | 5.4 | 6.3 | 6.7 | 0.029 |
| Complex fucosylation in hybrid-type | 3.2 | 5.5 | 11.2 | 0.046 |
LG: low-grade; HG: high-grade; PMP: pseudomyxoma peritonei.
Fucosylation here is defined as structures containing ≥1 deoxyhexoses (F).
Complex fucosylation is defined as structures containing ≥2 deoxyhexoses (multifucosylation).
Fig. 2.
Relative abundances of neutral glycans containing fucosylation in pseudomyxoma peritonei. Relative abundance of (A) overall fucosylation and (B) complex fucosylation in pseudomyxoma peritonei samples of low-grade (LG) and of high-grade (HG) as compared with appendix controls (App). In (C) relative abundance of complex fucosylation is plotted against the percentage of HG morphology. Two HG samples not separated because of very close values (2.6 and 2.8) are marked by an asterisk. The circled values represent samples with ≤ 50% HG component.
To validate the proposed monosaccharide compositions, a PMP sample was further analyzed with a combination of MS/MS fragmentation analysis directed to specific glycan signals (supplemental Fig. S2) and by exoglycosidase digestions of the glycan mixture (supplemental Fig. S3 and supplemental Table S5). As an example, glycan signal H3N4F1 was putatively classified as a complex-type N-glycan with terminal N-acetylhexosamine and fucosylation, because it contains at least four N-acetylhexosamine residues (N≥4), more N-acetylhexosamine than hexose residues (N≥H), and at least one deoxyhexose residue (F≥1), respectively. In MS/MS fragmentation analysis of a permethylated glycan sample (supplemental Fig. S2A), H3N4F1 produced characteristic fragments of two non-reducing terminal N-acetylhexosamine residues (m/z 1317 and 866) as well as N-glycan core fucosylation (m/z 474 for core fragment and 1384 for antenna fragment resulting from cleavage of the N-glycan core GlcNAc-GlcNAc linkage). The core fucose residue was shown to be α1,6-linked by the well-established specificity of the N-glycosidase F enzyme that was used to liberate the N-glycans from tissue (34). A set of exoglycosidase digestions was used to further characterize the glycan structures in the N-glycan mixture (supplemental Table S5). H3N4F1 was sensitive to both β-N-acetylglucosaminidase and β-N-acetylhexosaminidase enzyme digestions, which cleaved two terminal β-N-acetylglucosamine (β-GlcNAc) residues from the glycan converting most of the signal intensity to glycan signal H3N2F1 (supplemental Fig. S3A and S3B). In contrast, H3N4F1 was not sensitive to α-mannosidase, α1,2-fucosidase or α1,3/4-fucosidase digestions (supplemental Table S5). Taken together, these analyses confirmed the molecular structure of H3N4F1 as an α1,6-core-fucosylated biantennary N-glycan with two non-reducing terminal β-GlcNAc residues. Similarly, glycan signal H5N4F3 was initially classified for complex fucosylation (multifucosylation), because it contained three deoxyhexose residues (F≥2). In MS/MS fragmentation analysis (supplemental Fig. S2B), H5N4F3 produced fragments that were characteristic to both core fucosylation (m/z 717 and 1315) and antenna fucosylation (m/z 658 and 864). H5N4F3 was completely digested by α1,3/4-fucosidase digestion (supplemental Fig. S3C), but not by the other exoglycosidases, and it was only partially sensitive to α1,2-fucosidase (supplemental Fig. S3D and supplemental Table S5), establishing the majority of the antenna fucose residues as either α1,3-linked or α1,4-linked. Human galactosyltransferase specificities toward biantennary N-glycan structures suggest that the underlying antenna structures are most likely type II N-acetyllactosamine and the fucosylated epitopes thus α1,3-fucosylated Lewis x blood group epitopes i.e. Galβ1–4(Fucα1–3)GlcNAc. Similar analyses demonstrated the presence of N-acetyllactosediamine (LacdiNAc; supplemental Fig. S3B) and bisecting GlcNAc glycan structures in major N-glycan signals associated with PMP tumors (supplemental Table S5 and data not shown). These data indicate that the proposed monosaccharide compositions could be validated as expected.
Acidic N-linked Glycan Profiles
Acidic N-linked glycan profiles, composed of glycans containing acid esters (sulfate or phosphate) or sialic acid residues, of the PMP patient specimens and the normal appendix controls are shown as relative abundancy of the fifty most prominent proposed monosaccharide compositions in Fig. 1B. Similarly to the neutral glycans, complex fucosylation in hybrid-type (p = 0.046), terminal HexNAc (bisecting-size; p = 0.029), and complex-type structures (p = 0.007) were elevated in the acidic glycan profiles of PMP samples (Table I). The only structure which abundance was decreased in PMP in the acidic glycan group was the hybrid-type (p = 0.006). However, altered sialylation was not a prominent feature in PMP profiles (supplemental Table S6).
Principal Component Analysis of Differentially Expressed N-Linked Glycans
To further visualize the difference between PMP patient specimens and the controls, we selected from both the neutral and acidic N-glycan profiles those glycan signals that were significantly different (p < 0.05) between the appendix controls and PMP samples, as well as between LG and HG PMPs (supplemental Table S7). Relative intensity of these structures for each sample were then subjected to PCA analysis with two components extracted (Bartlett's test: p < 0.0001, Kaiser-Meyer-Olkin test: 0.744, PC1 (57.1%) versus PC2 (12.4%)), resulting in clustering of samples into separate groups (Fig. 3). Two HG samples containing only 10–20% proportion of HG morphology (the rest of the HG samples containing 50–100% of HG morphology) clustered into the group of LG samples.
Fig. 3.

Principal component analysis (PCA) of control appendices (triangle) and low-grade (open square) and high-grade (filled square) pseudomyxoma peritonei samples. PCA was used for visualization of the differences between sample groups and for that, neutral and acidic glycan signals whose relative intensities differed significantly (p < 0.05) between the control appendices and all PMP samples, as well as between the LG PMPs and the HG PMPs were used. Bartlett's test: p < 0.001, Kaiser-Meyer-Olkin test: 0.744, Principal component 1 is 57.1% and component 2 is 12.4%.
Expression of Glycosylation-related Enzymes
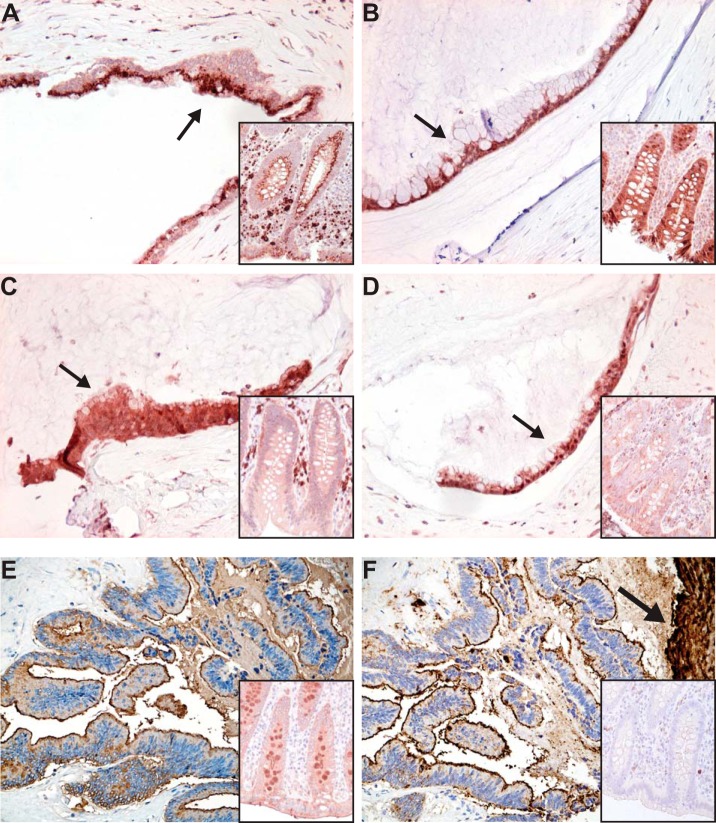
Expression of glycosylation-related genes was analyzed from appendiceal PMP tissue samples (n = 26) and compared with appendix control samples (n = 3) using DNA microarray (25). From over 1200 glycosylation-related genes studied, 30 genes were found to be differentially expressed in PMP samples, and from these we identified four fucosylation-related enzymes, i.e. fucosyltransferase 8 (FUT8) that performs α1,6-linked core fucosylation and three enzymes of the GDP-fucose de novo biosynthetic pathway (GMDS, GMPPA, and TSTA3), to be up-regulated in the PMP samples. A schematic representation of the pathway for these enzymes is shown in the Fig. 4. Expression of FUT8 was increased 2.3-fold and for the three GDP-fucose biosynthetic enzymes 1.8-fold in the PMP sample group (p < 0.0001 for all). When these fucosylation-related enzymes FUT8 (n = 100), GMDS (n = 99), GMPPA (n = 95), and TSTA3 (n = 100) were studied at the protein level by immunohistochemistry, the expression was shown to localize to the PMP cells (Fig. 5A–5D). FUT8 immunopositivity was high in 75% of LG and 78% of HG PMPs, and low in rest of the samples, but the staining was not significantly differing from normal appendiceal epithelium (p = 0.795). GMDS was high in 85% of LG and 90% of HG PMPs, and low in rest of the samples. GMPPA was high in 52% of LG and 66% of HG PMPs, and low in rest of the samples. TSTA3 was high in 47% of LG and 53% of HG PMPs, and low in rest of the samples. All these three proteins showed higher expression in PMP samples than in control appendix specimens (p = 0.018 for GMDS, p < 0.0001 for GMPPA, and p = 0.0002 for TSTA3). Thus, the majority of the PMP samples overexpressed these enzymes, but no significant differences were found between frequency of expression of these markers between LG and HG samples.
Fig. 4.
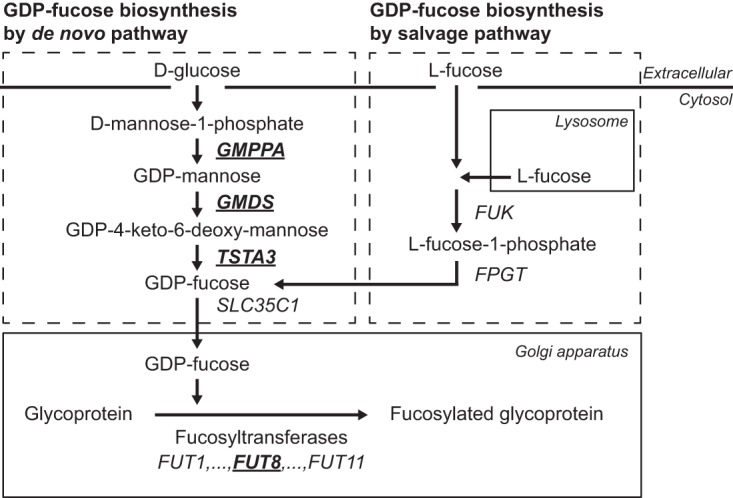
Schematic representation of the biosynthetic pathways of GDP-fucose, the substrate of fucosyltransferase enzymes. Pseudomyxoma peritonei samples (n = 26) show increased mRNA expression of four fucosylation-related enzymes, FUT8, GMPPA, GMDS, and TSTA3 (underlined and in bold) as compared with normal appendixes (n = 3) as analyzed by Affymetrix HG U133A 2.0 arrays.
Fig. 5.
Expression of fucosylation-related enzymes and fucosylated glycans in pseudomyxoma peritonei as analyzed by immunohistochemistry and lectin histochemistry. Cytoplasmic expression in tumor cells is shown in Fig. (A) for FUT8, (B) for GMDS, (C) for GMPPA, and (D) for TSTA3 protein. Expression of core fucosylated glycans (E) detected by lectin histochemistry utilizing AAL that preferentially binds to α1,6-linked core fucose residues attached by FUT8 enzyme, and complex fucosylation containing glycan sialyl Lewis x (F) detected by immunohistochemistry. Staining of a control appendix is shown in the insert of each figure. Note the intense staining of mucus in (F) marked by an arrow. Original magnifications, 200X, DAB was used as a chromogen.
Expression of Fucosylated Glycans
Expression of core fucosylated glycans was found in PMP cells and in their secreted mucus as detected by AAL lectin histochemistry (n = 28 for PMP and n = 10 for appendix) (Fig. 5E). To further investigate complex fucosylation of glycans, we immunostained sLex from the PMP sample set (n = 100), and, similarly, the immunopositivity localized in both PMP cells and mucus (Fig. 5F). sLex staining was high in 92% of LG and 97% of HG PMPs, and low in rest of the samples, and it showed significant overexpression as compared with normal appendiceal epithelium (n = 25) (p < 0.0001). In addition, mucus lakes stained very strongly (Fig. 5F). In contrast to the fucosylation-related enzymes, which showed expression in normal appendiceal epithelial cells, sLex staining was mainly negative in these cells with only some positive areas detected in 20% of the specimens. Also, PMP samples (n = 8) previously found to demonstrate negative or weak immunostaining of sialyl-Lewis a (sLea, also known as CA19–9) because of mutation in the biosynthetic enzyme FUT3 (26), showed intense staining of sLex (data not shown).
Effect of Fucosylation on MUC2 Expression
To study the possible relationship between fucosylation and mucin production, we used the intestine-derived cancer cell line HCT116, which has defective fucosylation because of deletion in the GDP-fucose biosynthetic enzyme GMDS coding gene (33). To restore fucosylation in these cells through salvage pathway (Fig. 4), we added l-fucose into the growth medium or to restore fucosylation through de novo pathway, in a separate experiment, we transfected the cells with a wildtype copy of the GMDS cDNA. Both interventions increased core fucosylation as demonstrated by AAL lectin blotting (transfectants shown in Fig. 6A). In a previous study, activating GNAS mutation was shown to up-regulate expression of MUC2, the predominant mucin molecule secreted by PMP tumor cells, when introduced into HT29 colon cancer cells (18). In HCT116 cell, we found GNAS mutation to cause a moderate up-regulation of MUC2 expression, which was significantly enhanced after restoring fucosylation (Fig. 6B).
Fig. 6.
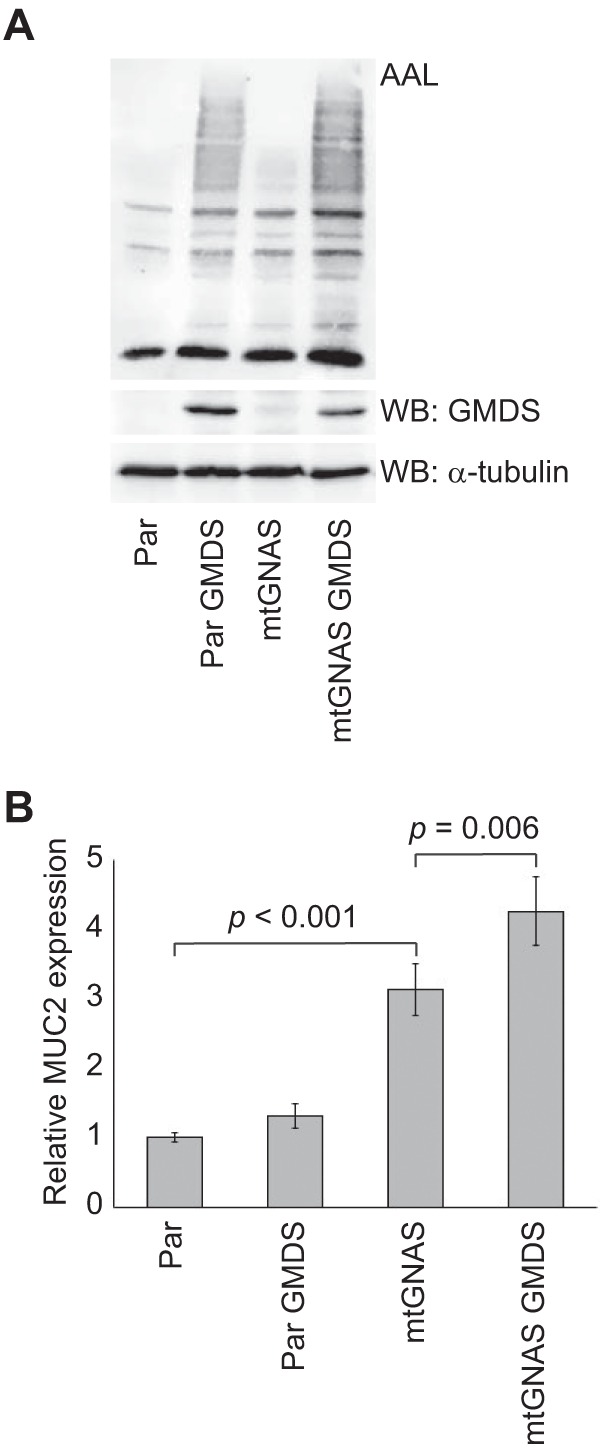
Restoration of fucosylation and expression of MUC2 in HCT116 cells. Lectin blotting with AAL (A) and MUC2 qPCR analysis (B) of parental and GNAS mutant HCT116 cells, which have defective fucosylation because of deletion in GMDS gene, before and after restoration of fucosylation by transfection of wildtype GMDS containing vector. The values in B are means ±S.E. from three transfection experiments (n = 9). α-tubulin serves as a loading control in A.
DISCUSSION
We found altered N-linked glycan profiles in PMP tissue specimens when compared with appendix controls, and increased fucosylation was an especially prominent feature. In addition to overall fucosylation, complex fucosylation (multifucosylation) was increased in PMP samples. Previously, altered fucosylation has been associated with aggressiveness of colorectal cancer (35, 36), which may depend on increased EGFR-mediated signaling (37) and modulation of adhesion molecule function (38, 39). In addition, core fucosylation was recently shown to drive melanoma metastasis by preventing the proteolytic cleavage of adhesion molecule L1CAM (40).
Next we studied fucosylation-related gene expression in PMP tissue specimens compared with control appendices using DNA microarray (25). In line with glycan profiles, we found increased expression of the core fucosylation performing enzyme FUT8, as well as three enzymes needed in the biosynthesis of its donor substrate GDP-fucose. With immunohistochemistry we could demonstrate up-regulated protein expression of the GDP-fucose biosynthetic enzymes (GMDS, GMPPA, and TSTA3) in PMP cells as compared with normal appendiceal epithelium. Fucosylated glycans, in turn, were detected in both PMP cells and the mucus secreted by them as shown by AAL lectin and sLex immunohistochemical staining. Of these glycans, sLex was significantly overexpressed in PMP cells, being mostly negative in normal appendiceal epithelial cells. These data support fucosylation to be a prominent feature of the PMP disease. Supporting our data, FUT8 has previously been shown to be up-regulated in hepatocellular carcinoma (41), and knock-down of this enzyme inhibits malignant behavior of lung cancer cells (42). In addition, pancreatic intraductal papillary mucinous neoplasm (IPMN) that is closely related to PMP in respect of mutation profiling (43, 44) show elevated FUT8 and GMDS expression, and fucosylation (45, 46).
The complex fucosylation containing glycan CA19–9 (sLea) is a widely used serum tumor marker. However, CA19–9 cannot be used in patients whose FUT3 is mutated (47, 48), because other FUT enzymes do not compensate for the lack of α1,4-fucosylation activity of this enzyme. In our previous study, we demonstrated that 9% (8/92) of PMP cases showed impaired CA19–9 expression because of FUT3 mutation (26). In our present study, we found that all these cases showed intense staining of sLex, which can be synthesized by FUT 3–7 enzymes. Thus, overexpression of sLex and lack of sLea is most likely accounted by the fact that other FUT enzymes (FUT4–7) can compensate the deficient α1,3-fucosylation activity of mutated FUT3, whereas they do not compensate the deficient α1,4-fucosylation activity of this α1,3/4-fucosyltransferase (49). Importantly, sLex measurement might help in monitoring of patients with CA19–9 negative tumors, as has been already proposed for pancreatic cancer (50), and could be a useful tumor marker also in PMP.
Over 60% of PMPs contain an activating GNAS mutation (9–15, 18), which has been demonstrated to result in constitutive activation of the membrane protein adenylyl cyclase and increased expression of extracellular mucins MUC2 and MUC5AC in colon cancer HT29 cell line (18). In contrast to HT29 cells, we found only modest induction of MUC2 expression induced by mutated GNAS in HCT116 cells, which has been reported to have defective fucosylation because of deletion in the GMDS gene (33). To test the possible relationship between GNAS mutation-induced MUC2 expression and fucosylation, we restored fucosylation through GDP-fucose de novo or salvage pathway. According to these results, GNAS mutation induced significantly higher MUC2 expression in HCT116 cell after restoration of fucosylation. Because elevated fucosylation and GNAS mutation are frequently found in PMP, this could partially explain the high mucin production in PMP disease.
In addition to fucosylation, we detected an increase in individual structural features previously associated with carcinogenesis, such as terminal GlcNAc (e.g. H3N4F1) (21), bisecting GlcNAc (e.g. H5N5F1) (51), and potentially also LacdiNAc (e.g. H3N6F1) (51). The proposed monosaccharide compositions were further validated with both MS/MS fragmentation analysis and exoglycosidase digestions. Increased sialylation is common in several cancer types (19, 20). Although we detected increased multisialylated N-glycans in LG PMP, they were not a feature of HG PMP. It is possible that highly increased fucosylation can compete for the substrate and thus lead to limited sialylation output of PMP cells. On the other hand, unaltered sialylation might explain the slow progression and rarely metastazising nature of the PMP disease. Further, a possibility exists that there might be changes in the proportions of different linkages of sialic acids, or in the overall sialylation level, which should be explored by future studies.
Taken together, we show for the first time that the N-glycan profiles differ between control appendices and appendix-originating PMPs. Fucosylation was found to be significantly increased in PMP samples, which could depend on elevated expression of fucosylation-related enzymes. Our data also show that GNAS-induced MUC2 expression is at least partially fucosylation dependent. Because inhibition of fucosylation by l-fucose analogs has been suggested as a method to suppress neoplastic properties of liver cancer cells (52), this approach might also slow PMP disease progression.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (www.proteomexchange.org) via the PRIDE (53) partner repository with the dataset identifier PXD010086.
Supplementary Material
Acknowledgments
We thank Prof. Edward A. Levine (Wake Forest University, NC) for providing the appendiceal PMP gene expression data, Prof. Eiji Miyoshi (Osaka University Graduate School of Medicine, Japan) for providing wildtype GMDS plasmid, and Carita Liikanen, Merja Haukka, and Matilda Holm for excellent technical assistance.
Footnotes
* This study was supported by University of Helsinki, the Sigrid Jusélius Foundation, the Finnish Cancer Foundation, Helsinki University Central Hospital Research Funds, the Academy of Finland (Center of Excellence in Cancer Genetics Research), Finska Läkaresällskapet, Finnish Cultural Foundation (PN), Ida Montin Foundation (LS), the Finnish Cancer Foundation (LS), Orion Research Foundation (LS), K. Albin Johansson Foundation (LS), Paulo Foundation (LS), and Biomedicum Helsinki Foundation (LS).
 This article contains supplemental Figures and Tables. TS is shareholder of Glykos Finland Ltd, which performed the mass spectrometric analysis services of N-glycans. The other authors declare that they have no competing interests.
This article contains supplemental Figures and Tables. TS is shareholder of Glykos Finland Ltd, which performed the mass spectrometric analysis services of N-glycans. The other authors declare that they have no competing interests.
1 The abbreviations used are:
- PMP
- pseudomyxoma peritonei
- LG
- low-grade
- HG
- high-grade
- FFPE
- formalin-fixed, paraffin-embedded
- FUT3
- fucosyltransferase 3
- MALDI-TOF
- matrix-assisted laser desorption-ionization time-of-flight
- PCA
- principal component analysis
- FUT8
- fucosyltransferase 8
- GMDS
- GDP-mannose 4,6-dehydratase
- GMPPA
- GDP-mannose pyrophosphorylase A
- TSTA3
- tissue specific transplantation antigen P35B/GDP-L-fucose synthase
- sLex
- sialyl Lewis x antigen
- AAL
- Aleuria aurantia lectin
- DAB
- 3,3′-diaminobenzidine
- GlcNAc
- N-acetylglucosamine.
REFERENCES
- 1. Smeenk R. M., van Velthuysen M. L., Verwaal V. J., and Zoetmulder F. A. (2008) Appendiceal neoplasms and pseudomyxoma peritonei: a population based study. Eur. J. Surg. Oncol. 34, 196–201 [DOI] [PubMed] [Google Scholar]
- 2. Carr N. J., and Sobin L. H. (2010) Adenocarcinoma of the appendix. In Bosman F. T., Carneiro F., Hruban R. H., and Theise N. D. editors. WHO classification of tumours of the digestive system, pp. 122–125. IACR, Lyon, France [Google Scholar]
- 3. Carr N. J., Cecil T. D., Mohamed F., Sobin L. H., Sugarbaker P. H., Gonzalez-Moreno S., Taflampas P., Chapman S., and Moran B. J. (2016) A consensus for classification and pathologic reporting of pseudomyxoma peritonei and associated appendiceal neoplasia: the results of the Peritoneal Surface Oncology Group International (PSOGI) modified Delphi process. Am. J. Surg. Pathol. 40, 14–26 [DOI] [PubMed] [Google Scholar]
- 4. Carr N. J., Finch J., Ilesley I. C., Chandrakumaran K., Mohamed F., Mirnezami A., Cecil T., and Moran B. (2012) Pathology and prognosis in pseudomyxoma peritonei: a review of 274 cases. J. Clin. Pathol. 65, 919–923 [DOI] [PubMed] [Google Scholar]
- 5. Sugarbaker P. H., Kern K., and Lack E. (1987) Malignant pseudomyxoma peritonei of colonic origin. Natural history and presentation of a curative approach to treatment. Dis. Colon Rectum 30, 772–779 [DOI] [PubMed] [Google Scholar]
- 6. Järvinen P., Ristimäki A., Kantonen J., and Lepistö A. (2013) Feasibility of radical cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for pseudomyxoma peritonei of appendiceal origin. Scand. J. Surg. 102, 145–151 [DOI] [PubMed] [Google Scholar]
- 7. Taflampas P., Dayal S., Chandrakumaran K., Mohamed F., Cecil T. D., and Moran B. J. (2014) Pre-operative tumour marker status predicts recurrence and survival after complete cytoreduction and hyperthermic intraperitoneal chemotherapy for appendiceal pseudomyxoma peritonei: analysis of 519 patients. Eur. J. Surg. Oncol. 40, 515–520 [DOI] [PubMed] [Google Scholar]
- 8. Järvinen P., Ristimäki A., Kantonen J., Aronen M., Huuhtanen R., Järvinen H., and Lepistö A. (2014) Comparison of serial debulking and cytoreductive surgery with hyperthermic intraperitoneal chemotherapy in pseudomyxoma peritonei of appendiceal origin. Int. J. Colorectal Dis. 29, 999–1007 [DOI] [PubMed] [Google Scholar]
- 9. Sio T. T., Mansfield A. S., Grotz T. E., Graham R. P., Molina J. R., Que F. G., and Miller R. C. (2014) Concurrent MCL1 and JUN amplification in pseudomyxoma peritonei: a comprehensive genetic profiling and survival analysis. J. Hum. Genet. 59, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alakus H., Babicky M. L., Ghosh P., Yost S., Jepsen K., Dai Y., Arias A., Samuels M. L., Mose E. S., Schwab R. B., Peterson M. R., Lowy A. M., Frazer K. A., and Harismendy O. (2014) Genome-wide mutational landscape of mucinous carcinomatosis peritonei of appendiceal origin. Genome Med. 6, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu X., Mody K., de Abreu F. B., Pipas J. M., Peterson J. D., Gallagher T. L., Suriawinata A. A., Ripple G. H., Hourdequin K. C., Smith K. D., Barth R. J. Jr, Colacchio T. A., Tsapakos M. J., Zaki B. I., Gardner T. B., Gordon S. R., Amos C. I., Wells W. A., and Tsongalis G. J. (2014) Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin. Chem. 60, 1004–1011 [DOI] [PubMed] [Google Scholar]
- 12. Nummela P., Saarinen L., Thiel A., Järvinen P., Lehtonen R., Lepistö A., Järvinen H., Aaltonen L. A., Hautaniemi S., and Ristimäki A. (2015) Genomic profile of pseudomyxoma peritonei analyzed using next-generation sequencing and immunohistochemistry. Int. J. Cancer 136, E282–E289 [DOI] [PubMed] [Google Scholar]
- 13. Noguchi R., Yano H., Gohda Y., Suda R., Igari T., Ohta Y., Yamashita N., Yamaguchi K., Terakado Y., Ikenoue T., and Furukawa Y. (2015) Molecular profiles of high-grade and low-grade pseudomyxoma peritonei. Cancer. Med. 4, 1809–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pietrantonio F., Perrone F., Mennitto A., Gleeson E. M., Milione M., Tamborini E., Busico A., Settanni G., Berenato R., Caporale M., Morano F., Bossi I., Pellegrinelli A., Di Bartolomeo M., de Braud F., Baratti D., Bowne W. B., Kusamura S., and Deraco M. (2016) Toward the molecular dissection of peritoneal pseudomyxoma. Ann. Oncol. 27, 2097–2103 [DOI] [PubMed] [Google Scholar]
- 15. Gleeson E. M., Feldman R., Mapow B. L., Mackovick L. T., Ward K. M., Morano W. F., Rubin R. R., and Bowne W. B. (2017) Appendix-derived pseudomyxoma peritonei (PMP): molecular profiling toward treatment of a rare malignancy. Am. J. Clin. Oncol. 41, 777–783 [DOI] [PubMed] [Google Scholar]
- 16. Saarinen L., Nummela P., Thiel A., Lehtonen R., Järvinen P., Järvinen H., Aaltonen L. A., Lepistö A., Hautaniemi S., and Ristimäki A. (2017) Multiple components of PKA and TGF-beta pathways are mutated in pseudomyxoma peritonei. PLoS ONE 12, e0174898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bradbury N. A. (2000) Protein kinase-A-mediated secretion of mucin from human colonic epithelial cells. J. Cell. Physiol. 185, 408–415 [DOI] [PubMed] [Google Scholar]
- 18. Nishikawa G., Sekine S., Ogawa R., Matsubara A., Mori T., Taniguchi H., Kushima R., Hiraoka N., Tsuta K., Tsuda H., and Kanai Y. (2013) Frequent GNAS mutations in low-grade appendiceal mucinous neoplasms. Br. J. Cancer 108, 951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinho S. S., and Reis C. A. (2015) Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer. 15, 540–555 [DOI] [PubMed] [Google Scholar]
- 20. Mereiter S., Balmana M., Gomes J., Magalhaes A., and Reis C. A. (2016) Glycomic approaches for the discovery of targets in gastrointestinal cancer. Front. Oncol. 6, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satomaa T., Heiskanen A., Leonardsson I., Ångström, Olonen J. A., Blomqvist M., Salovuori N., Haglund C., Teneberg S., Natunen J., Carpen O., and Saarinen J. (2009) Analysis of the human cancer glycome identifies a novel group of tumor-associated N-acetylglucosamine glycan antigens. Cancer Res. 69, 5811–5819 [DOI] [PubMed] [Google Scholar]
- 22. Satomaa T., Heiskanen A., Mikkola M., Olsson C., Blomqvist M., Tiittanen M., Jaatinen T., Aitio O., Olonen A., Helin J., Hiltunen J., Natunen J., Tuuri T., Otonkoski T., Saarinen J., and Laine J. (2009) The N-glycome of human embryonic stem cells. BMC Cell Biol. 10, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaprio T., Satomaa T., Heiskanen A., Hokke C. H., Deelder A. M., Mustonen H., Hagström J., Carpen O., Saarinen J., and Haglund C. (2015) N-glycomic profiling as a tool to separate rectal adenomas from carcinomas. Mol. Cell. Proteomics 14, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leijon H., Kaprio T., Heiskanen A., Satomaa T., Hiltunen J. O., Miettinen M. M., Arola J., and Haglund C. (2017) N-glycomic profiling of pheochromocytomas and paragangliomas separates metastatic and non-metastatic disease. J. Clin. Endocrinol. Metab. 102, 3990–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levine E. A., Blazer D. G. 3rd, Kim M. K., Shen P., Stewart J. H. 4th, Guy C., and Hsu D. S. (2012) Gene expression profiling of peritoneal metastases from appendiceal and colon cancer demonstrates unique biologic signatures and predicts patient outcomes. J. Am. Coll. Surg. 214, 599–606; discussion 606–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nummela P., Leinonen H., Järvinen P., Thiel A., Järvinen H., Lepistö A., and Ristimäki A. (2016) Expression of CEA, CA19–9, CA125, and EpCAM in pseudomyxoma peritonei. Hum. Pathol. 54, 47–54 [DOI] [PubMed] [Google Scholar]
- 27. Core Team R. (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 28. Kang P., Mechref Y., Klouckova I., and Novotny M. V. (2005) Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Commun. Mass Spectrom. 19, 3421–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ovaska K., Laakso M., Haapa-Paananen S., Louhimo R., Chen P., Aittomäki V., Valo E., Nunez-Fontarnau J., Rantanen V., Karinen S., Nousiainen K., Lahesmaa-Korpinen A. M., Miettinen M., Saarinen L., Kohonen P., Wu J., Westermarck J., and Hautaniemi S. (2010) Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme. Genome Med. 2, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., and Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 31. Bejamini Y., and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57, 289–300 [Google Scholar]
- 32. Matsumura K., Higashida K., Hata Y., Kominami J., Nakamura-Tsuruta S., and Hirabayashi J. (2009) Comparative analysis of oligosaccharide specificities of fucose-specific lectins from aspergillus oryzae and aleuria aurantia using frontal affinity chromatography. Anal. Biochem. 386, 217–221 [DOI] [PubMed] [Google Scholar]
- 33. Moriwaki K., Noda K., Furukawa Y., Ohshima K., Uchiyama A., Nakagawa T., Taniguchi N., Daigo Y., Nakamura Y., Hayashi N., and Miyoshi E. (2009) Deficiency of GMDS leads to escape from NK cell-mediated tumor surveillance through modulation of TRAIL signaling. Gastroenterology 137, 188–198, 198.e1-2 [DOI] [PubMed] [Google Scholar]
- 34. Tretter V., Altmann F., and Marz L. (1991) Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1–3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 199, 647–652 [DOI] [PubMed] [Google Scholar]
- 35. Muinelo-Romay L., Villar-Portela S., Cuevas Alvarez E., Gil-Martin E., and Fernandez-Briera A. (2011) Alpha(1,6)fucosyltransferase expression is an independent prognostic factor for disease-free survival in colorectal carcinoma. Hum. Pathol. 42, 1740–1750 [DOI] [PubMed] [Google Scholar]
- 36. Osuga T., Takimoto R., Ono M., Hirakawa M., Yoshida M., Okagawa Y., Uemura N., Arihara Y., Sato Y., Tamura F., Sato T., Iyama S., Miyanishi K., Takada K., Hayashi T., Kobune M., and Kato J. (2016) Relationship between increased fucosylation and metastatic potential in colorectal cancer. J. Natl. Cancer Inst. 108, doi: 10.1093/jnci/djw210 [DOI] [PubMed] [Google Scholar]
- 37. Wang X., Gu J., Ihara H., Miyoshi E., Honke K., and Taniguchi N. (2006) Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J. Biol. Chem. 281, 2572–2577 [DOI] [PubMed] [Google Scholar]
- 38. Zhao Y., Itoh S., Wang X., Isaji T., Miyoshi E., Kariya Y., Miyazaki K., Kawasaki N., Taniguchi N., and Gu J. (2006) Deletion of core fucosylation on alpha3beta1 integrin down-regulates its functions. J. Biol. Chem. 281, 38343–38350 [DOI] [PubMed] [Google Scholar]
- 39. Osumi D., Takahashi M., Miyoshi E., Yokoe S., Lee S. H., Noda K., Nakamori S., Gu J., Ikeda Y., Kuroki Y., Sengoku K., Ishikawa M., and Taniguchi N. (2009) Core fucosylation of E-cadherin enhances cell-cell adhesion in human colon carcinoma WiDr cells. Cancer. Sci. 100, 888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Agrawal P., Fontanals-Cirera B., Sokolova E., Jacob S., Vaiana C. A., Argibay D., Davalos V., McDermott M., Nayak S., Darvishian F., Castillo M., Ueberheide B., Osman I., Fenyo D., Mahal L. K., and Hernando E. (2017) A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer. Cell. 31, 804–819.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cheng L., Gao S., Song X., Dong W., Zhou H., Zhao L., and Jia L. (2016) Comprehensive N-glycan profiles of hepatocellular carcinoma reveal association of fucosylation with tumor progression and regulation of FUT8 by microRNAs. Oncotarget 7, 61199–61214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen C. Y., Jan Y. H., Juan Y. H., Yang C. J., Huang M. S., Yu C. J., Yang P. C., Hsiao M., Hsu T. L., and Wong C. H. (2013) Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amato E., Molin M. D., Mafficini A., Yu J., Malleo G., Rusev B., Fassan M., Antonello D., Sadakari Y., Castelli P., Zamboni G., Maitra A., Salvia R., Hruban R. H., Bassi C., Capelli P., Lawlor R. T., Goggins M., and Scarpa A. (2014) Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J. Pathol. 233, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tan M. C., Basturk O., Brannon A. R., Bhanot U., Scott S. N., Bouvier N., LaFemina J., Jarnagin W. R., Berger M. F., Klimstra D., and Allen P. J. (2015) GNAS and KRAS mutations define separate progression pathways in intraductal papillary mucinous neoplasm-associated carcinoma. J. Am. Coll. Surg. 220, 845–854.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sato N., Fukushima N., Maitra A., Iacobuzio-Donahue C. A., van Heek N. T., Cameron J. L., Yeo C. J., Hruban R. H., and Goggins M. (2004) Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am. J. Pathol. 164, 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Watanabe K., Ohta M., Yada K., Komori Y., Iwashita Y., Kashima K., and Inomata M. (2016) Fucosylation is associated with the malignant transformation of intraductal papillary mucinous neoplasms: a lectin microarray-based study. Surg. Today 46, 1217–1223 [DOI] [PubMed] [Google Scholar]
- 47. Tempero M. A., Uchida E., Takasaki H., Burnett D. A., Steplewski Z., and Pour P. M. (1987) Relationship of carbohydrate antigen 19–9 and lewis antigens in pancreatic cancer. Cancer Res. 47, 5501–5503 [PubMed] [Google Scholar]
- 48. Elmgren A., Mollicone R., Costache M., Borjeson C., Oriol R., Harrington J., and Larson G. (1997) Significance of individual point mutations, T202C and C314T, in the human lewis (FUT3) gene for expression of lewis antigens by the human alpha(1,3/1,4)-fucosyltransferase, fuc-TIII. J. Biol. Chem. 272, 21994–21998 [DOI] [PubMed] [Google Scholar]
- 49. Moriwaki K., and Miyoshi E. (2010) Fucosylation and gastrointestinal cancer. World J. Hepatol. 2, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang H., Singh S., Partyka K., Kletter D., Hsueh P., Yadav J., Ensink E., Bern M., Hostetter G., Hartman D., Huang Y., Brand R. E., and Haab B. B. (2015) Glycan motif profiling reveals plasma sialyl-lewis x elevations in pancreatic cancers that are negative for sialyl-lewis A. Mol. Cell. Proteomics 14, 1323–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anugraham M., Jacob F., Everest-Dass A. V., Schoetzau A., Nixdorf S., Hacker N. F., Fink D., Heinzelmann-Schwarz V., and Packer N. H. (2017) Tissue glycomics distinguish tumour sites in women with advanced serous adenocarcinoma. Mol. Oncol. 11, 1595–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou Y., Fukuda T., Hang Q., Hou S., Isaji T., Kameyama A., and Gu J. (2017) Inhibition of fucosylation by 2-fluorofucose suppresses human liver cancer HepG2 cell proliferation and migration as well as tumor formation. Sci. Rep. 7, 11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (www.proteomexchange.org) via the PRIDE (53) partner repository with the dataset identifier PXD010086.