Abstract
Royal jelly has received attention because of its necessity for the development of queen honeybees as well as claims of benefits on human health; this product of the hypopharyngeal glands of worker bees contains a large number of proteins, some of which have been claimed to have various biological effects only in their glycosylated state. However, although there have been glycomic and glycoproteomic analyses in the past, none of the glycan structures previously defined would appear to have potential to trigger specific biological functions. In the current study, whole royal jelly as well as single protein bands were subject to off-line LC-MALDI-TOF MS glycomic analyses, complemented by permethylation, Western blotting and arraying data. Similarly to recent in-depth studies on other insect species, previously overlooked glucuronic acid termini, sulfation of mannose residues and core β-mannosylation of the N-glycans were found; additionally, a relatively rare zwitterionic modification with phosphoethanolamine is present, in contrast to the phosphorylcholine occurring in lepidopteran species. Indicative of tissue-specific remodelling of glycans in the Golgi apparatus of hypopharyngeal gland cells, only a low amount of fucosylated or paucimannosidic glycans were detected as compared with other insect samples or even bee venom. The unusual modifications of hybrid and multiantennary structures defined here may not only have a physiological role in honeybee development, but represent epitopes recognized by pentraxins with roles in animal innate immunity.
Keywords: Glycomics, HPLC, Mass Spectrometry, Array analysis, N-Glycosylation, glucuronic acid, phosphoethanolamine, royal jelly, sulphate, T antigen
Dedicated to Dr. Hubert Paschinger (1941–2011), chemist, lover of art and music and avid beekeeper.
Royal jelly is a product of the hypopharyngeal and mandibular glands of worker honeybees (1); its natural role is as the food source for worker and drone larvae for the first 3 days but induces queen development when fed beyond that time and remains, thereafter, the food for a queen's entire life. The observed fertility and longer lifespan of queens, as compared with worker bees, has been correlated with this special diet. However, royal jelly has attracted especial attention because of its putative effects on human health. Indeed, it has been used since ancient times as a food supplement or cosmetic and is considered to have a range of antibiotic, anti-inflammatory and other health-relevant effects (2), although these are controversial and allergic reactions are also known (3). Royal jelly contains a wide range of compounds including carbohydrates, lipids, vitamins, and proteins, many of which are post-translationally modified (4, 5). Because of their multiple and variable monosaccharide units and their size (typically 2000 Da), N-glycans represent the most diverse and complex set of protein modifications known (6) and modulate a wide range of biological processes.
Based on proteomic and glycoproteomic studies of royal jelly, up to 100 different proteins have been identified including the “major royal jelly proteins” (MRJPs)1. Of these, MRJP1 (also known as apisin or royalactin) has a controversial role in queen development (7, 8), but is reported to extend life-span in other invertebrates (9). Furthermore, MRJP1 and MRJP2 (apalbumin2) have been shown to have antihypertensive and antibiotic activity in their glycosylated state (10). Although the sites of glycosylation on MRJPs have been analyzed, there is less information regarding the structures of the glycans on the specific glycoproteins. Although mass spectrometric analyses (11) indicated potentially complex N-glycosylation of MRJP2, HPLC and NMR data showed the presence of oligomannosidic and hybrid glycans (12, 13) on monomeric and oligomeric forms of MRJP1. There have also been a series of glycan analyses on royal jelly, culminating in a report that Galβ1,3GalNAcβ1,4GlcNAc units are detectable on N-glycans and that there is a relevant galactosyltransferase activity (14–17). However, none of the hybrid or biantennary glycans described were especially unusual and so it was concluded that a role for these glycans in the biological activity of royal jelly is unlikely.
Our recent data have proven that insect glycosylation is more complicated than previously thought and that a range of anionic and zwitterionic modifications is present on dipteran and lepidopteran N-glycans (18, 19). Therefore, we have reappraised the N-glycome of royal jelly using an off-line LC-MALDI-TOF MS workflow; we have also examined the glycosylation of individual major royal jelly proteins and have immobilized N-glycan pools in probably the first study to ever test interactions of natural insect glycans in an array format. A significant proportion of structures was thereby found to be of either the hybrid or complex type with up to three antennae. About 4% of the total N-glycans carry novel combinations of glucuronic acid, sulfate, β-mannose and phosphoethanolamine, whereby these epitopes have potential for physiological activity in both insects and humans.
EXPERIMENTAL PROCEDURES
Enzymatic Release and Purification of N-glycans
Royal jelly (Wald und Wiese, Vienna, Austria; ca. 9 g wet weight per glycan preparation) was suspended in water and heat-treated before proteolysis with thermolysin (Promega, Madison, WI) (20) followed by cation exchange (Dowex AG50, Bio-Rad; elution with 0.5 m ammonium acetate, pH 6) and gel filtration (Sephadex G25; GE Healthcare) chromatography of the proteolysate. Thereafter, N-glycans were released from glycopeptides using peptide/N-glycosidase F (PNGase F, 3 U; Roche) at pH 8 as previously described (20), with a subsequent digestion of the remaining glycopeptides using native PNGase A (0.15 mU; Roche) after adjusting to pH 5; the result was a combined PNGase F/A digest. Alternatively, in a second preparation, the PNGase F-released glycans were isolated from the glycopeptides by Dowex AG50 chromatography and the latter were treated with recombinant PNGase Ar (15 U; New England Biolabs, Ipswich); in this case, the pools of PNGase F- and PNGase Ar-released glycans were separately analyzed. After another round of cation-exchange chromatography (Dowex AG50; flow-through), the glycans were subject to solid-phase extraction on nonporous graphitised carbon (SupelClean ENVICarb; Sigma-Aldrich, Merck) as described (20); the “neutral” and “anionic-enriched” fractions were then eluted with 40% (v/v) acetonitrile and 40% (v/v) acetonitrile containing 0.1% trifluoroacetic acid (v/v) respectively. The pools of glycans were subject to MALDI-TOF MS before fluorescent labeling by reductive amination using 2-aminopyridine (PA; Sigma-Aldrich) (20). Similar results were obtained for two different lots of royal jelly from the same supplier. For the workflow in schematic form, refer to the Scheme in the supplement.
Permethylation
The neutral and anionic N-glycans (70% of each pool from one PNGase F-released preparation) were separately permethylated as previously reported (21) with slight modifications. Briefly, the samples were dried in a glass tube before permethylation with 0.2 ml of a slurry of ground NaOH pellets in dimethyl sulfoxide and 0.1 ml of ICH3. The reaction mixture was shaken for 4 h at 4 °C and then quenched on ice with 0.2 ml of cold water, followed by neutralization with 30% aqueous acetic acid, and then applied to a pre-equilibrated C18 solid phase extraction column. Hydrophilic salts and contaminants were stepwise washed off with 3 ml each of water, 2.5 and 10% (v/v) acetonitrile. Subsequently, permethylated N-glycans were eluted serially with 3 ml each of 25 and 50% (v/v) acetonitrile.
HPLC Fractionation
Complete pyridylaminated N-glycomes (10% of the neutral and 90% of the anionic pools) were fractionated by reversed-phase HPLC (Ascentis Express RP-amide; 150 × 4.6 mm, 2.7 μm; Sigma-Aldrich) and a gradient of 30% (v/v) methanol (buffer B) in 100 mm ammonium acetate, pH 4 (buffer A) was applied at a flow rate of 0.8 ml/min as follows: 0–4 min, 0% B; 4–14 min, 0–5% B; 14–24 min, 5–15% B; 24–34 min, 15–35% B; 34–35 min, return to starting conditions (20). The RP-HPLC column was calibrated daily in terms of glucose units using a pyridylaminated dextran hydrolysate and the degree of polymerization of single standards was verified by MALDI-TOF MS. Selected RP-HPLC fractions were subject to HIAX-HPLC as a second dimension as described (20) with an IonPac AS11 column (Dionex, Sunnyvale; 4 × 250 mm, combined with a 4 × 50 mm guard column). A two solvent gradient with buffer A (0.8 m ammonium acetate, pH 3) and buffer B (80% acetonitrile; LC-MS grade) was applied at a flow rate of 1 ml/min: 0–5 min, 99% B; 5–50 min, 90% B; 50–65 min, 80% B; 65–85 min, 75% B. Detection for both columns was by fluorescence (Shimadzu RF-20A XS detector; excitation/emission at 320/400 nm). All manually collected HPLC glycan fractions were lyophilized, redissolved in water and analyzed by MALDI-TOF MS and MS/MS.
Glycan Mass Spectrometry
Monoisotopic MALDI-TOF MS was performed using an Autoflex Speed (Bruker Daltonics, Bremen, Germany) instrument in either positive or negative reflectron modes with 6-aza-2-thiothymine (ATT; Alfa-Aesar, Thermo Scientific) as matrix. MS/MS was in general performed by laser-induced dissociation of the [M+H]+ or [M-H]− pseudomolecular ions (except for permethylated structures analyzed as [M+Na]+ with 2,5-dihydroxybenzoic acid as matrix); typically 2000 shots were summed for MS (reflector voltage, lens voltage and gain respectively 27 kV, 9 kV and 2217 V) and 4000 for MS/MS (reflector voltage, lift voltage and gain respectively 27 kV, 19 kV and 2174 V). Spectra were processed with the manufacturer's software (Bruker Flexanalysis 3.3.80) using the SNAP algorithm with a signal/noise threshold of 6 for MS (unsmoothed) and 3 for MS/MS (four-times smoothed). Glycan spectra were manually interpreted based on the masses of the predicted component monosaccharides, differences of mass in glycan series, fragmentation patterns, comparison with coeluting structures from other insects and nematodes and chemical treatments or exoglycosidase digestions (see also pages S2-S4 of the Supplement). Negative-ion mode LC-MSn of a 2D-HPLC-enriched glycan was performed as previously described, using a 5 μm porous graphitized carbon column (10 cm × 150 μm) coupled to a Thermo Scientific LTQ ion trap mass spectrometer (22); refer also to page S5 of the Supplement for further details. A list of theoretical m/z values for each glycan composition is presented in the supplemental Table S1; mzXML files of raw MS/MS data are available as supplementary information.
Enzymatic and Chemical Treatments
Glycans were treated, before re-analysis by MALDI-TOF MS, with α-fucosidase (bovine kidney from Sigma-Aldrich), α-mannosidases (jack bean from Sigma-Aldrich, Aspergillus α1,2-specific from Prozyme, Hayward, CA, and Xanthomonas α1,2/3-specific from New England Biolabs), β-galactosidase (β1,3-specific from New England Biolabs), β-glucuronidases (E. coli from Megazyme, Bray, Ireland, and Helix pomatia from Sigma-Aldrich; desalted and concentrated before use), β-N-acetylhexosaminidases (jack bean from Sigma-Aldrich, Xanthomonas β1,2-specific N-acetylglucosaminidase from New England Biolabs, Streptomyces β1,3/4-specific N-acetylhexosaminidase (chitinase) from New England Biolabs or in-house produced recombinant forms of Caenorhabditis elegans HEX-4 specific for β1,4-linked GalNAc residues or Apis mellifera FDL specific for the product of GlcNAc-transferase I (23)) in 50 mm ammonium acetate, pH 5, at 37 °C overnight (except for pH 6.5 in the case of HEX-4, pH 7 in the case of E. coli β-glucuronidase or an incubation time of only 3 h in the case of FDL); these incubations were performed in PCR tubes with a final volume of 3 μl (for further details about conditions and specificities, refer to the supplement). Hydrofluoric acid was used for removal of phosphoethanolamine or α1,3-linked fucose (20). As appropriate, treated glycans were re-chromatographed by RP-HPLC to ascertain retention time shifts before MALDI-TOF-MS; otherwise, an aliquot (generally one-fifth) of any digest was analyzed by MALDI-TOF-MS without further purification.
Western Blotting
Before SDS-PAGE, resuspended royal jelly was precipitated (mixed with a 5-fold volume excess of methanol), incubated at −80 °C for one hour, centrifuged at 4 °C, 21,000 × g and dissolved in a reducing sample buffer. After electrophoresis (10 μg/lane) and blotting to a nitrocellulose membrane, the following reagents for detection of glycan epitopes were employed: anti-horseradish peroxidase (Sigma-Aldrich; 1:10,000 diluted in Tris buffered saline with 0.05% Tween and 0.5% BSA, to detect core α1,3-fucose (24)) and serum amyloid P protein (Fitzgerald, Acton, MA; 1:200, to detect phosphoethanolamine (25)) as well as C-reactive protein (MP Biochemicals, Santa Ana; 1:200, which binds preferentially to phosphorylcholine (25)) followed by the relevant peroxidase-conjugated secondary antibodies and development with SigmaFAST 3,3′-diaminobenzidine tetrahydrochloride (19). Other glycan determinants were detected with biotinylated forms of Aleuria aurantia, wheat germ and peanut agglutinins (Vector Labs, Burlingame, CA; 1:1000) followed by phosphatase-conjugated anti-biotin (Sigma-Aldrich; 1:10,000) and development with SigmaFAST BCIP/NBT (26).
Protein-specific Glycan Analysis
After performing SDS-PAGE (10 μg/lane) and staining with Coomassie Blue, protein bands were excised and washed/destained (twice with 50% acetonitrile in water and successively once with 1:1 0.1 m ammonium bicarbonate/acetonitrile and 100% acetonitrile only) before drying (26). The proteins in the gel pieces were reduced for one hour at 56 °C with 10 mm dithiothreitol and alkylated for 45 min with 55 mm iodoacetamide in the dark. After a second round of washing (as above), the gel pieces were dried, covered with a 1:2 mixture of ammonium bicarbonate/trypsin (100 ng/μl) and incubated at 37 °C overnight. The (glyco)peptides were extracted with a mixture of acetonitrile/water/trifluoroacetic acid (660/330/1 (v/v/v)), dried and dissolved in water before MALDI-TOF MS using either α-cyanocinnamic acid (ACH) or 6-aza-thiothymine (ATT) as matrices, using similar mass spectrometer settings as above. Tryptic peptide fingerprint data were analyzed using the Mascot webserver (version 2.6.0) and the Swissprot database (release 2017_06) as described on page S5 of the Supplement.
For protein-specific glycan analysis, the (glyco)peptides were first heat-treated to inactivate trypsin and 90% of the samples were subject to PNGase Ar treatment (2.5 U; New England Biolabs) in 20 mm ammonium acetate, pH 5 overnight at 37 °C. Whereas the deglycosylated peptides were analyzed by MALDI-TOF MS, the released N-glycans were purified using two different columns packed respectively with Lichroprep C18/Dowex AG50 and nonporous graphitized carbon/Lichroprep C18. The C18/AG50 column was washed with 2% acetic acid and 60% isopropanol; after equilibration of the column with 2% acetic acid, the N-glycans were acidified with 10% acetic acid before application. The N-glycans were eluted (three column volumes of 2% acetic acid) before use of the carbon/C18 column, which was pre-washed with 100% acetonitrile and equilibrated with water. The N-glycans (combined neutral and anionic pools) were then eluted with 40% acetonitrile containing 0.1% trifluoro-acetic acid. After drying, the glycans were fluorescently labeled by reductive amination using 2-aminopyridine and then analyzed with MALDI-TOF MS and HPLC as above.
Immobilization of N-glycans
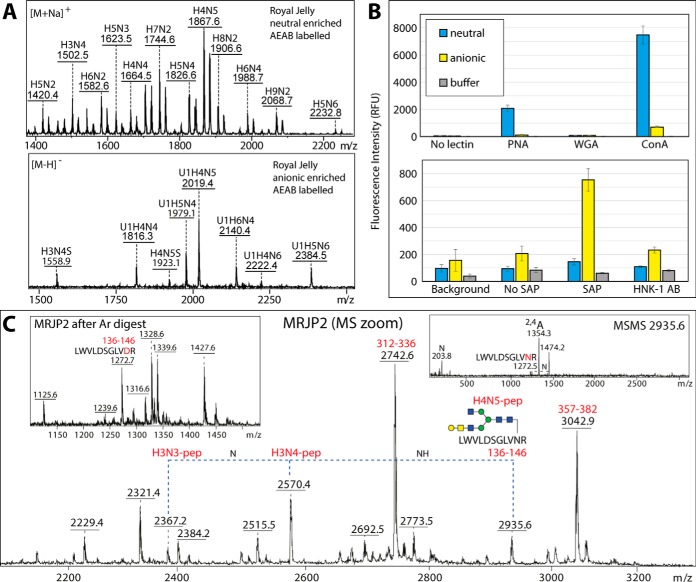
Free N-glycans of the neutral pool were modified reductively with 2-amino-N-(2-amino-ethyl)-benzamide (AEAB; excitation/emission of 330/420 nm) as described by Song (27). Briefly, the dried N-glycan pool was dissolved in 0.35 m AEAB and 1 m NaCNBH3 and incubated for 2 h at 65 °C. To remove the excess AEAB, the labeled glycans were precipitated three times with 100% acetonitrile followed by solid phase extraction on LC-NH2 (Supelco, Sigma-Aldrich) normal phase material. Successful derivatization of the N-glycans was determined by MALDI-TOF MS and HPLC (also to normalize the concentration of glycans based on fluorescence intensity). The glycan pool was mixed 1:1 with spotting buffer (300 mm sodium phosphate pH 7.5, 0.005% Tween-20) then spotted (n = 10) by noncontact printing (Flexarrayer S1; Scienion, Berlin, Germany) onto NHS-derivatised Nexterion H glass slides (Schott, Jena, Germany). After 16 h of hybridization, slides were blocked (50 mm ethanolamine in 50 mm sodium borate, pH 9.0) for 1 h at RT, washed (TBS + 0.05% Tween-20, TBS, and H2O) and dried (28). The slides were incubated with (1) biotinylated forms of peanut agglutinin, wheat germ agglutinin or concanavalin A (VectorLabs; 10 μg/ml or 5 μg/ml in TBS + 0.05% Tween-20 + 1% BSA, i.e. TTBSA) followed by incubation with anti-biotin FITC conjugate (Sigma-Aldrich) (28), (2) serum amyloid protein (amyloid P component from human serum, SAP; Fitzgerald, diluted 1:200 in TTBSA) followed by incubation with anti-amyloid P IgG from rabbit (Calbiochem, Merck; in TTBSA) and finally anti-rabbit IgG AlexaFluor-647 conjugate (Invitrogen, Carlsbad, CA; in TTBSA), or (3) anti-L2/HNK-1 (clone 412; diluted 1:1000 in TTBSA) followed by incubation with anti-mouse IgG AlexaFluor-647 conjugate (Invitrogen; in TTBSA). Slides were scanned with an Agilent G2565AA Microarray Scanner (multiple photomultiplier tube (PMT) gain values from 10–100%) and raw fluorescence values (green for FITC and red for AlexaFluor-647) were used to calculate (in Excel) the mean and standard deviation from all ten spots. The negative controls (either only TBS-Tween, no primary lectin or pentraxin for respectively “background” and “no lectin/pentraxin” controls) show fluorescence because of either the AEAB label itself or nonspecific binding of the fluorescent secondary antibodies. Galactosidase (recombinant Aspergillus niger β1,3/4-specific) and glucuronidase (Helix pomatia β-specific) treatments of AEAB-labeled glycans were respectively performed in solution before printing or directly on the printed slides before probing with lectins or antibodies. For further details, refer to pages S6 and S7 of the Supplement.
RESULTS
Strategy for the N-glycan Analysis
Analysis of glycan-based post-translational modifications of proteins remains a challenge and experience has shown that a thorough analysis of unknown invertebrate glycomes requires multiple fractionation steps in order to prevent suppression effects and to enable separation of isomeric/isobaric structures of the same or similar mass (29). Thus, we first separated the free N-glycans of honeybee royal jelly into “neutral” and “anionic” pools, before fluorescent labeling, fractionation on an RP-amide HPLC column (calibrated in terms of glucose units) and MALDI-TOF-MS/MS in combination with chemical and enzymatic treatments (20). Based on the fluorescence intensities of the two glycan pools, it is estimated that at least 3% of the N-glycans are “anionically” modified.
The initial screen, before HPLC, of the pyridylaminated pools by MALDI-TOF MS indicated that the neutral pool contained, in accordance with previous studies (14), a range of oligomannosidic and potentially hybrid or complex structures as judged by mass (supplemental Fig. S1). On the other hand, MALDI-TOF MS of the anionic pool suggested the presence of glycans potentially modified with sulfate or glucuronic acid, neither of which have been reported for royal jelly glycans, but of a type familiar from our recent data on dipteran and lepidopteran N-glycomes (18, 19). However, unusually for N-glycans from an insect source, fucosylation was not obvious from the complete spectra despite using both PNGase F and PNGase A in series to release the glycans. Relatively low amounts of core mono- and difucosylation were found only upon HPLC analysis of the glycans; also, paucimannosidic glycans (Man2–4GlcNAc2), generally dominant in insect glycomes (18, 30), were present at only a low level.
The subsequent RP-amide fractionation was necessary to resolve the fine detail of the N-glycome of royal jelly; nearly 70 structures were detected in the neutral pools (see Fig. 1) with a partial overlap with the over 80 structures in the various anionic-enriched pools; in total, over 100 different compositions could be verified (see the supplemental Table S1 for the theoretical m/z values). Considering a widely held belief that MALDI-TOF-MS of nonpermethylated glycans leads to “in source” artifacts, the major neutral RP-HPLC peaks were subject to both NP-HPLC (i.e. HIAX; Fig. 2) and permethylation; also, aliquots of the whole N-glycome were permethylated (supplemental Figs. S2 and S3). The results of these additional procedures confirmed the validity of our general approach. Furthermore, the analysis was extended to the individual glycoproteins by using Western blotting and tryptic peptide mapping as well as release of the N-glycans from the single protein bands followed by fluorescent labeling and HPLC fractionation.
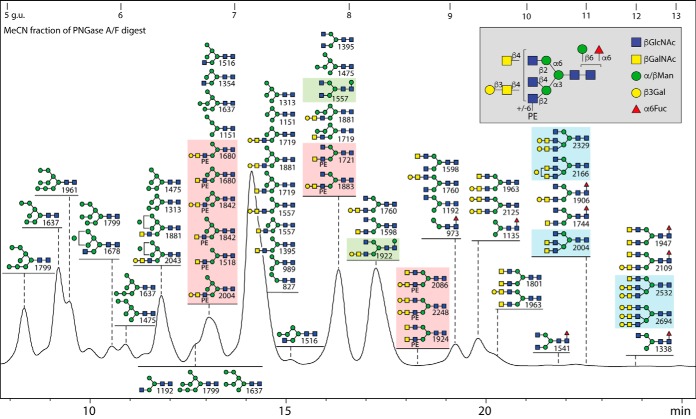
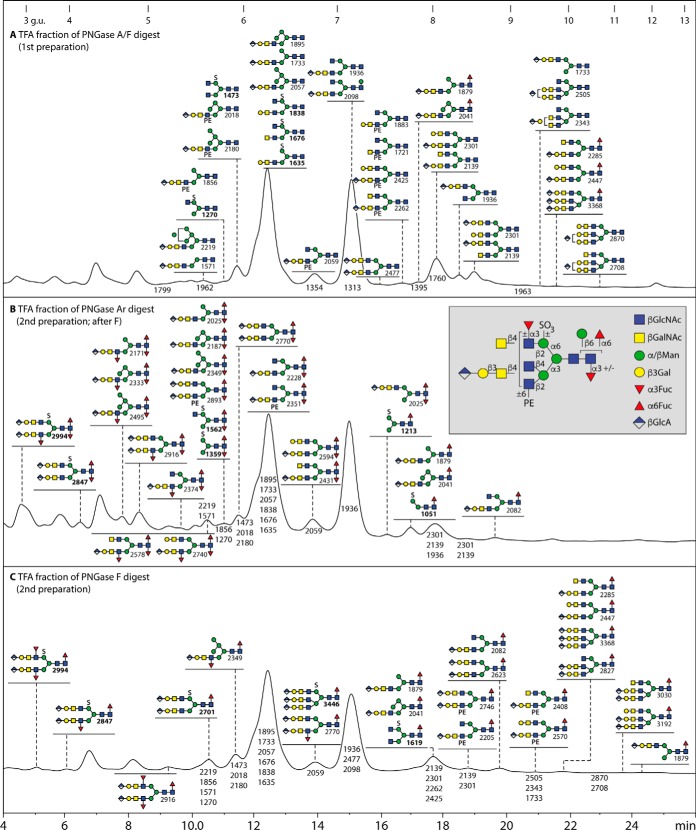
Fig. 1.
RP-HPLC fractionation of the neutral N-glycans from royal jelly. Glycans were released by PNGase F and A before solid phase extraction (to separate the neutral and anionic glycans), pyridylamination and finally fractionation on an RP-amide column (refer to supplemental Fig. S1 for overall MALDI-TOF MS profiles). The chromatogram is annotated with glycan structures (each with the positive mode m/z value for the protonated ion in order of abundance for each fraction) according to the Symbolic Nomenclature for Glycans (circles, galactose or mannose; squares, GalNAc or GlcNAc; triangles, fucose; PE, phosphoethanolamine; see also inset for symbols and linkage information). The HPLC column was calibrated in terms of glucose units (g.u.). Oligomannosidic glycans are annotated by comparison to previous studies on insect glycomes using the same column (unusual isomers were also subject to specific α-mannosidase digestion), whereas other glycans are defined based on MS/MS and digest data. The PNGase F alone and PNGase Ar (after F) digests of the second preparation yielded qualitatively similar chromatograms, but Man9GlcNAc2 was more dominant than Man5GlcNAc2 (see also supplemental Fig. S1A and S1B). The hymenoptera-specific zwitterionic, β-mannosylated and triantennary glycans are highlighted in red, green or blue boxes; the presence of phosphoethanolamine (rather than phosphorylcholine) and of β-mannose, but the relative lack of fucose in this specific glycome, contrasts to the larval N-glycomes of lepidopteran species (19).
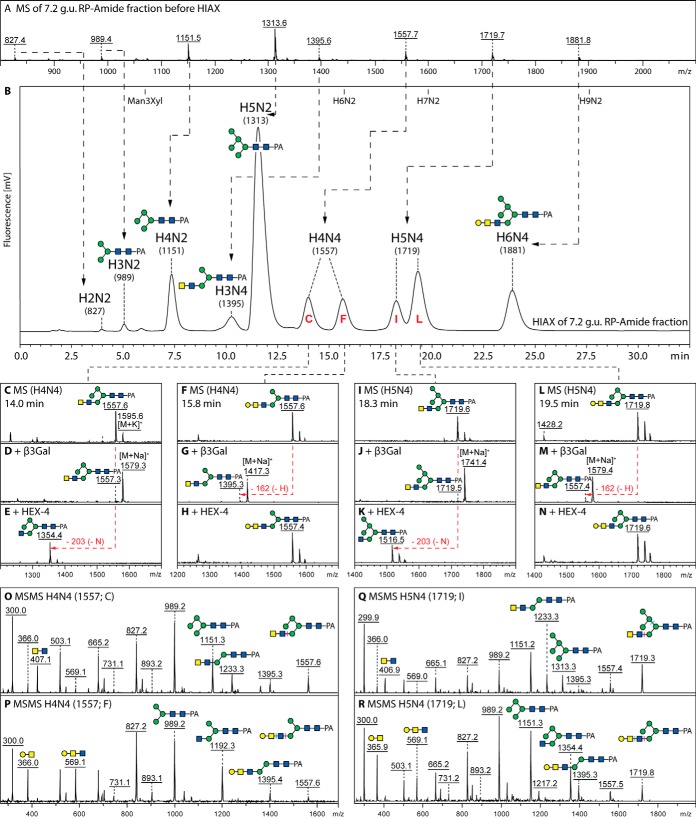
Fig. 2.
Isomeric separation of hybrid N-glycans by two-dimensional HPLC. The major RP-HPLC fraction (7.2 g.u.) was analyzed by MALDI-TOF MS (A) and subfractionated by HIAX-HPLC (B); thereby, the HIAX column resolves glycans not just based on mass, but even with a different isomeric composition of mannose and galactose residues. This fractionation results in peaks containing single glycans with no evidence of in source MS fragmentation with even the fluorescent intensities correlating to those for the MALDI-TOF MS data. The pairs of isomeric glycans (Hex4–5HexNAc4 with m/z 1557 and 1719; C/F and I/L) display contrasting susceptibilities to β1,3-specific galactosidase (D, G, J, and M) and β1,4-N-acetylgalactosamine-specific HEX-4 treatments (E, H, K, and N) as well as apparently different MS/MS spectra (O–R) for the undigested glycans. The m/z 407 B-fragments correlate with terminal LacdiNAc (O and Q), but those at m/z 569 with the galactosylated version (P and R), whereas the pairs of isomers also differ in the pattern of m/z 989/1151/1313 Man3–5-based Y-fragments. Based on these data the presence of LacdiNAc motifs with or without a terminal β1,3-galactose substitution can be demonstrated. Refer to supplemental Fig. S4 for further data on digestion of hybrid glycans, including α-mannosidase or combined galactosidase/HEX-4 treatment of the 7.2 g.u. fraction.
Oligomannosidic N-glycans
In two independent preparations, the most abundant N-glycans in royal jelly were found to be, according to mass and retention time data (Fig. 1 and supplemental Figs. S1 and S2), either the typical “Golgi-processed” isomer of Man5GlcNAc2 or Man9GlcNAc2 (respectively eluting at 7.2 or 5.5 g.u. with m/z 1313 or 1961). The MALDI-TOF MS analysis of individual HPLC fractions revealed the significant presence of several other oligomannosidic structures (Man6–8GlcNAc2) as well as one other isomer of Man5GlcNAc2 (6.4 g.u.) and low amounts of paucimannosidic glycans (Man2–4GlcNAc2). In total, around 20 distinct pauci- and oligomannosidic isomers can be proposed based on MS/MS data, comparisons to retention times of such glycans from other species (19, 31) and selected α-mannosidase digests (see, e.g. supplemental Fig. S4). Comparison of the MALDI-TOF MS data on the major RP-amide HPLC fraction with HIAX size fractionation (Fig. 2) and permethylation (data not shown) prove that no “in source” fragmentation of oligomannosidic N-glycans occurs in our MALDI-TOF MS analyses.
Neutral Hybrid and Complex N-glycans
About 50 of the proposed royal jelly neutral glycans were predicted on the basis of mass to contain three or more N-acetylhexosamine residues; the initial mass spectrometric screen of the neutral pool revealed a rich set of hybrid and complex structures ranging from Hex3HexNAc3 (m/z 1192) through to Hex5HexNAc6 (m/z 2125; supplemental Fig. S1). The more in-depth analysis of the neutral HPLC fractions (Fig. 1) showed the presence of further masses including minor amounts of triantennary Hex3–6HexNAc7–8 (m/z 2004, 2166, 2329, 2532, and 2694) and fucosylated complex Hex3–4HexNAc5–6Fuc1 (m/z 1744, 1906, 1947, and 2109) structures, the latter being some of the few fucosylated glycans found upon PNGase F release. MS/MS of glycans of m/z ≥1395 generally revealed B-fragments at m/z 407 and/or 569 (Hex0–1HexNAc2), which was taken as an indication for the presence of LacdiNAc units (Fig. 2 and supplemental Figs. S5–S6).
To resolve the individual neutral glycans in terms of the modifications and isomeric status, selected HPLC fractions were treated in a targeted manner with a number of exoglycosidases. For glycans containing four or five hexose residues, a major question was as to whether these hybrid structures were terminally galactosylated as previously suggested (14). However, isomers of the same observed masses (e.g. two isomers each of Hex4–5HexNAc4 with m/z 1557 and 1719) were identified by their different 2D-HPLC elution and MS/MS properties (Fig. 2). Man4–5-based hybrid glycans lost maximally three residues with jack bean α-mannosidase (dependent on the efficiency of removal of the “core” α1,6-mannose), whereas the pattern of β1,3-specific galactosidase and HEX-4 β1,4-specific N-acetylgalactosaminidase (23) sensitivity depended on the isomer (Fig. 2 and supplemental Fig. S4). Successful galactosidase digestion correlated with the distinct presence of an MS/MS fragment at m/z 569, whereas HEX-4 sensitivity was observed for glycans with fragments at m/z 407 (Fig. 2C–2R). The presence of either a fourth/fifth mannose or a galactose substitution of antennal GalNAc was corroborated by MS/MS of permethylated glycans showing Hex4–5HexNAc2 Y-fragments at m/z 1361 and 1565 or Hex0–1HexNAc1–2 B-fragments at m/z 486, 527 and 731 (supplemental Fig. S3A and S3B). For those hybrid glycans containing a sixth mannose (one isomer each of m/z 1881 and 2043; 6.4 g.u.), four mannose residues were lost upon jack bean mannosidase digestion, whereas a single one was removed by an α1,2-specific mannosidase (data not shown).
In the case of biantennary glycans, the definition of which antenna was elongated was based on the retention time (isomers with longer lower arms, i.e. on the α1,3-mannose, elute earlier (32)), fragmentation pattern and enzyme sensitivity (supplemental Fig. S5), whereby the FDL N-acetylglucosaminidase is specific for removal of unsubstituted β1,2-linked GlcNAc from the lower arm (23). As for the hybrid glycans, the longer antennae (with m/z 569 B-fragments for, e.g. isomers of m/z 1760) were digested by a combination of β1,3-galactosidase and HEX-4 as for the hybrid forms, whereas glycans with a terminal GalNAc were sensitive to HEX-4 alone (with m/z 407 B-fragments as for isomers of m/z 1598). The cumulative data show that LacdiNAc (GalNAcβ1, 4GlcNAc), with or without a β1,3-linked galactose cap, is a frequent motif of royal jelly N-glycans and that galactose was present on HexdiNAc units.
In the case of triantennary glycans eluting at 11 g.u. (see Fig. 1), a combination of galactosidase and specific hexosaminidase digests together with reinjection onto the HPLC column revealed that there could be a “free” GlcNAc on the upper α1,6-mannose and two branches on the lower α1,3-mannose (supplemental Fig. S6A–S6E). On treatment with Xanthomonas β1,2-specific N-acetylglucosaminidase, removal of the GlcNAc on the α1,6-arm resulted in conversion of the m/z 1030 Y-fragment to one at m/z 827; the third antenna is based on a β1,4-linked GlcNAc as shown by co-elution (9.5 g.u.) of the m/z 1598 galactosidase/HEX-4 digestion product of the 11 g.u. fraction with a degalactosylated bovine fetuin N-glycan. Later eluting glycans (12 g.u.) having two or three fully galactosylated antennae displayed intense MS/MS fragment ions at m/z 1233 or 1395 resulting from loss of the “heavy” α1,3-arm as indicated (supplemental Fig. S6F and S6G). All defined hybrid, bi- and triantennary structures were the basis for various anionic/zwitterionic-modified glycans subsequently found in the anionic pool.
Core Mannosylated N-glycans
A hallmark of many invertebrate N-glycans, when pyridylaminated, is the presence of an m/z 446 (Fuc1GlcNAc1-PA) fragment indicative of core fucosylation (see below); recently, in an oyster and a marine snail (22, 33) we have found glycans with an m/z 462 fragment (Hex1HexNAc1-PA). In the snail this fragment was abolished on β-mannosidase digestion of relevant glycans; this rules out that this is a Y-fragment of an endoglycosidase digestion product, which would elute before 5 g.u. rather than at ∼8 g.u. In the present study, the m/z 462 fragment was also found for the first time in an insect species and could also be verified by the MS/MS pattern of 2D-HPLC-purified glycans followed by glycosidase digestion of selected structures including one carrying antennal glucuronic acid (Fig. 3 and supplemental Fig. S7). Initially, we presumed that the β-mannose in royal jelly would also be 1,3-linked to the proximal GlcNAc as in molluscs (22), but parallel re-injection experiments with two RP-HPLC columns (supplemental Fig. S7A and S7B) as well as the LC-MSn fragmentation pattern (Fig. 3C and 3D) indicated that the royal jelly m/z 1557 mannosylated structure was not identical to that from the snail; thus, we conclude that the β-mannose in royal jelly glycans is 1,6-linked.
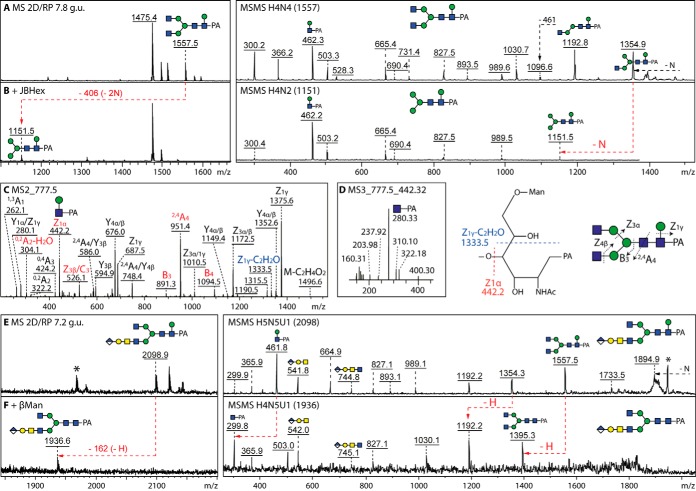
Fig. 3.
Core β1,6-mannose as a modification of 2D-HPLC-fractionated royal jelly glycans. A and B, The m/z 1557 glycan (Hex4HexNAc4-PA, 7.8 g.u.; see Fig. 1) was subject to MALDI-TOF MS/MS before and after removal of the terminal GlcNAc residues with jack bean β-hexosaminidase (JBHex); the m/z 462 Y-fragment is indicative of the β-mannose modification as verified by β-mannosidase digestion. C and D, LC-MS2 and LC-MS3 analyses of Hex4HexNAc4-PA (m/z 777.5 as [M-2H]2−) confirm hexosylation of the reducing terminal GlcNAc residue, but the presence of an m/z 1333.5 MS2 fragment and the absence of a m/z 221 MS3 fragment contrast with the previously-published data on an isomeric glycan from a marine snail (22); on the basis of the LC-MS data as well as the RP-HPLC comparisons (supplemental Fig. S7), we conclude that the royal jelly core mannose is β1,6-linked to the proximal GlcNAc. E–F, The m/z 2098 anionic glycan (7.2 g.u.; see Fig. 5) was analyzed by MALDI-TOF MS/MS before and after β-mannosidase treatment which resulted in loss of the m/z 462 fragment, thereby confirming the core modification, whereas the glucuronylated m/z 542 and 745 fragments indicate the presence of a terminal glucuronic acid in this structure; asterisks indicate ions of unknown origin. It is to be noted that β-mannosylation has only a minor effect on N-glycan RP-amide HPLC elution properties as compared with the non-β-mannosylated structure, thus 2D-HPLC was necessary before the presented digests.
Zwitterionic N-glycans
On analysis of the individual “neutral” fractions, it was observed that some glycans could be detected in both positive and negative ion modes and had compositions suggestive of a modification with a moiety of 123 Da, which is reminiscent of the presence of phosphoethanolamine on N-glycans from Trichomonas vaginalis or Penicillium species (34, 35). The most distinct fragment ions (Fig. 4 and supplemental Fig. S8 and S9) were previously unknown ones at m/z 1153, 1315, 1477, 1518 and 1639 in positive mode (Hex2–5HexNAc3–4PE1-PA Y-fragments) or at m/z 528/530 and 690/692 in negative/positive modes (Hex0–1HexNAc2PE1 B-fragments, which reflect the antennal capping by LacdiNAc with or without galactose). To resolve isomers, HIAX separation of the RP-HPLC 6.6–6.8 g.u. was performed (supplemental Fig. S8). Thereafter, it was found that the Hex4–5HexNAc4PE1 isomers (m/z 1678/1680 and 1840/1842) display different MS/MS spectra because of variations in the positions of galactose and mannose residues, similarly to the “parental” later-eluting nonzwitterionic glycans (compare with Fig. 2). The position of the phosphoethanolamine on “internal” GlcNAc, rather than on GalNAc, became obvious on removal of antennal galactose and GalNAc residues as judged by the alterations in the B-fragments, i.e. conversion to m/z 325/327 or 528/530 (Fig. 4A–4F and supplemental Fig. S9).
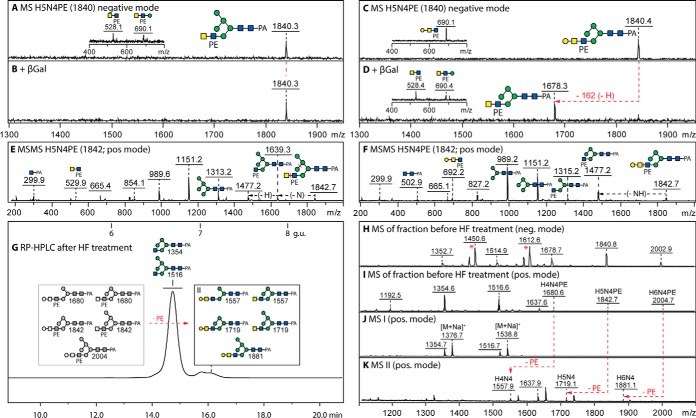
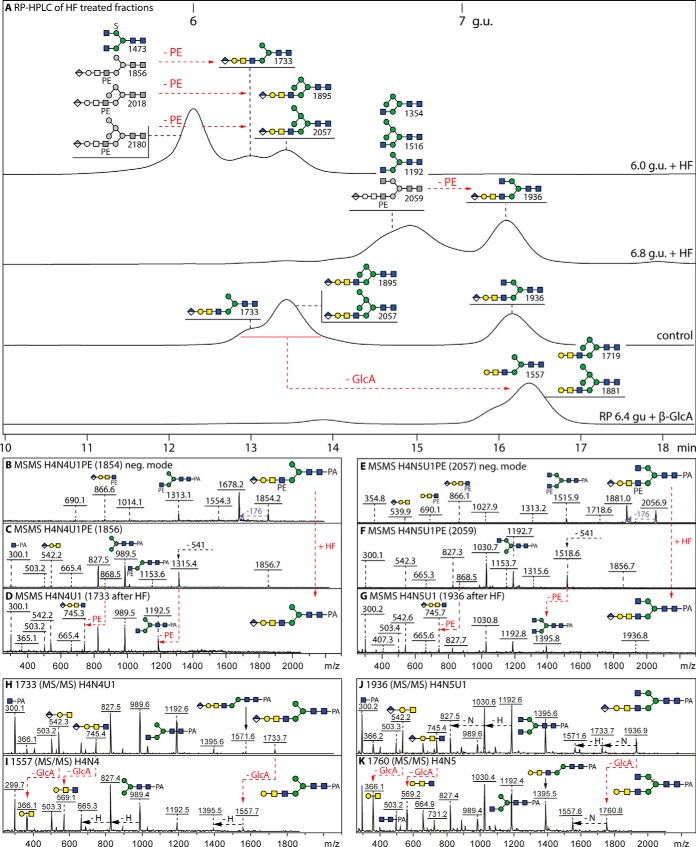
Fig. 4.
MALDI-TOF MS analysis of phosphoethanolamine-modified N-glycans. A–F, Analysis of two 2D-HPLC separated glycans of m/z 1840/1842 (Hex5HexNAc4PE1; for the HIAX chromatogram, see supplemental Fig. S8) showing negative mode spectra before and after treatment with β1,3-specific galactosidase (β3Gal), which (together with the effects of HEX-4 β-N-acetylgalactosaminidase and jack bean α-mannosidase; see supplemental Fig. S9A–S9N) aid definition of the isomeric positions for mannose or galactose; insets show a region of the negative mode MS/MS spectrum highlighting changes in the phosphoethanolamine-containing B-fragments. G–K, RP-amide HPLC and MALDI-TOF MS of the 6.6–6.8 g.u. fraction after hydrofluoric acid treatment showing the shift to later elution times on loss of phosphoethanolamine of a small proportion (10%) of glycans in this particular fraction (peak II with smaller m/z), whereas the elution position of the major m/z 1354/1516 glycans (peak I) remains unaffected; the phosphoethanolamine-modified glycans are observable in negative and positive modes, whereas the simple hybrid structures are also detected as adducts in negative mode (indicated as *). Alterations in MS because of digestion are indicated with red arrows; greyscale annotations in panel G indicate original structures before removal of phosphodiesters. For further MS/MS data, refer to supplemental Figs. S8 and S9.
Further proof for the presence of the phosphodiester was provided by sensitivity toward hydrofluoric acid (Fig. 4H–4K) and a subsequent shift to higher RP-HPLC retention time correlating with the loss of the hydrophilic moiety (Fig. 4G). Thereby, rather than the phosphorylcholine substitution found on N-glycans from two lepidopteran species (19), we conclude that several hybrid and complex glycans in royal jelly are modified with phosphoethanolamine, a moiety which also decorates in 6-linkage internal GlcNAc residues of dipteran glycolipids (36–38). Despite an in-depth examination and the ability to find phosphoethanolamine-modified glycans when permethylating Penicillium glycomes (unpublished data) which display a 30% degree of such structures, no zwitterionic glycans were found on permethylation of the royal jelly glycome, which may be because of losses of these low-abundance structures (<0.5%) during the chemical or separation procedures; thus, the off-line LC-MALDI approach was necessary to detect this class of glycans, which were not detected in any spectrum of an entire glycome (supplemental Figs. S1 and S2).
Glucuronylated N-glycans
One of the key steps in our analyses to reveal the fine details of royal jelly glycans was the separation on graphitized carbon of the neutral from the anionic species. Thus, after pyridylamination, the anionic pools (∼3% of the total) obtained from different preparations were separately applied to the RP-amide HPLC column (Fig. 5); obvious was the presence of glycans modified with glucuronic acid, which (as in our previous studies of insect N-glycans (18, 19)) could be detected in both positive and negative ion modes, but fragment best in the positive mode to yield MS/MS fragments at, e.g. m/z 542 and 745 (HexA1Hex1HexNAc1–2; see Figs. 6–8 as well as supplemental Figs. S6H–S6K and S10–S12). On this basis, mono-, di- and triantennary glycans with up to three glucuronic acid residues were identified in the PNGase F-released anionic pool (Hex3–6HexNAc4–8Fuc0–1GlcA1–3; Fig. 5A). Glucuronylated glycans were also found on permethylation of the anionic pool and displayed relevant HexA1Hex1HexNAc0–2 fragments at m/z 476, 704 and 949 indicative of linear antennae (supplemental Figs. S2C and S3F).
Fig. 5.
RP-HPLC fractionation of the anionic N-glycans from royal jelly. Glycans were released by (A) PNGase F and A, (B) PNGase Ar after PNGase F or (C) PNGase F alone before solid phase extraction to separate the neutral and anionic glycans, pyridylamination and finally separation on an RP-amide column. The chromatograms are annotated with glycan structures (each with the positive mode m/z value for glucuronylated glycans or the negative mode m/z value, in bold, for sulfated structures) according to the Symbolic Nomenclature for Glycans (circles, galactose or mannose; diamonds, glucuronic acid; squares, GalNAc or GlcNAc; triangles, fucose; PE, phosphoethanolamine; S, sulfate; see inset for symbol and linkage information). The HPLC column was calibrated in terms of glucose units (g.u.). The chromatogram A (of the PNGase F/A-released anionic glycans) is annotated with all structures determined in that pool, whereas chromatograms B and C (i.e. the PNGase Ar and PNGase F released anionic glycans from a second preparation) highlight the fucosylated glycans (the elution positions of other detected glycans being indicated by the m/z values alone); in the case of PNGase Ar release, the low abundance fucosylated structures can be core difucosylated and even carry one Lewis-like antennal fucose, whereas PNGase F can only release glycans with core α1,6- and up to two antennal (α1,3-) fucose residues.
Fig. 6.
RP-HPLC and MS/MS analysis of glycans modified with both glucuronic acid and phosphoethanolamine. A, RP-amide chromatograms of the two hydrofluoric acid (HF) treated 6.0 and 6.8 g.u. fractions are shown compared with a control co-injection of aliquots of 6.4 and 7.2 g.u. glucuronylated glycans. The effect of E. coli β-glucuronidase on the retention time of the 6.4 g.u. control glycans, corresponding to the HF products of the 6.0 g.u. fraction, is also indicated; see supplemental Figs. 10 and 11 for further MS and HPLC data on glucuronidase and HF digests. B–G, The MALDI-TOF MS/MS of untreated glycans in negative (B and E) and positive modes (C and F) and of the HF-treated products (positive mode only; D and G) highlight changes in the fragmentation patterns because of removal of phosphoethanolamine residues (loss of m/z 868 fragments correlating to the later retention times; indicated with red arrows). (H–K) Positive mode MALDI-TOF MS/MS of the 6.4 and 7.2 g.u. glucuronylated glycans before and after β-glucuronidase digestion indicating loss of the m/z 542 and 745 fragments and appearance of ones at m/z 366 and 569. For space reasons, the phosphoethanolamine group is depicted underneath the antenna, although a 6-linkage is assumed based on analogy to insect glycolipids (36–38); the greyscale structures indicate the original elution times, those in color the products'. Based on these analyses, glycans carrying both phosphoethanolamine and glucuronic acid are estimated to represent 30% of these particular anionic fractions. It is to be noted that sulfated glycans (e.g. m/z 1473) are resistant to hydrofluoric acid treatment, unlike those modified with phosphomono- or diesters.
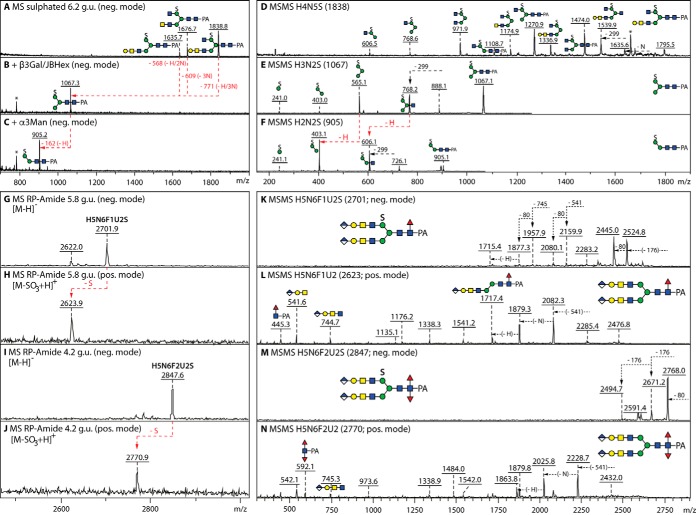
Fig. 7.
Glycosidase digestion and MALDI-TOF MS/MS analysis of sulfated N-glycans. A–C, Negative mode MALDI-TOF MS of the 6.2 g.u. anionic fraction before and after combined β1,3-galactosidase (β3Gal) and jack bean hexosaminidase (JBHex) digestion and subsequent α1,3-mannosidase (α3Man) treatment; the removal of galactose and HexNAc residues results in a single m/z 1067 product from which the α1,3-mannose can be digested, thereby localizing the sulfate to the α1,6-mannose as in other monosulphated glycans from insects. D–F, Negative mode MALDI-TOF MS/MS of a sulfated glycan and of its digestion products. G–N, MS and MS/MS in negative and positive modes of sulfated glycans modified with also fucose and glucuronic acid; in contrast to the ability of phosphorylated glycans to ionize in both positive and negative modes (54), sulfated structures are detectable in the negative mode as [M-H]− and the indicated in-source fragmentation in positive ion mode results in the presence of an ion of 78 mass units lower than the negative ion detected in the same fraction. MS/MS of the [M-SO3+H]+ ion aided definition of the core modifications (m/z 446 and 592 mono- and difucosylated Y-fragments), whereas the sulfation of the 6-linked mannose is assumed by analogy to the smaller structures and to the permethylation data in Supplemental Fig. 3.
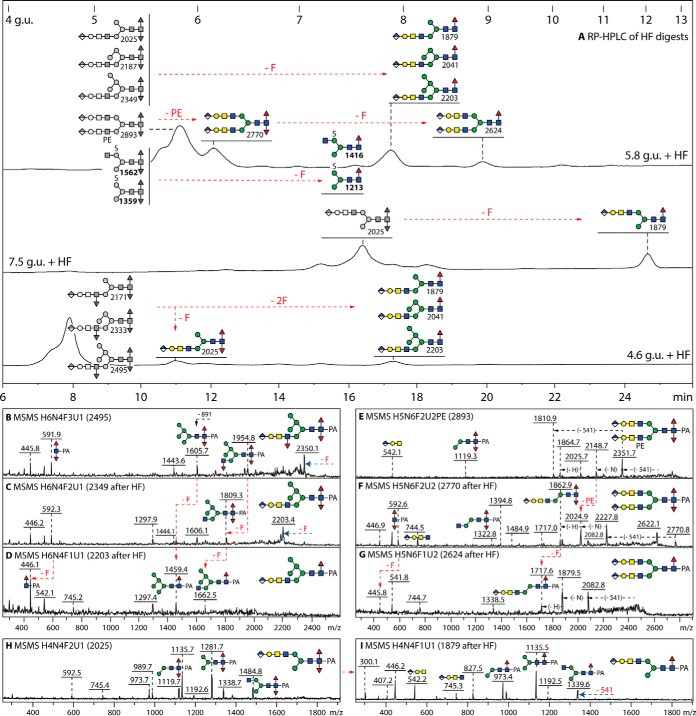
Fig. 8.
HPLC and MS/MS analysis of N-glycans modified with both glucuronic acid and fucose residues. A, RP-HPLC of three hydrofluoric acid treated anionic fractions (originally eluting at 4.6, 5.8 and 7.5 g.u.) highlighting the shifts to higher retention times because of the loss of core and antennal α1,3-fucose and of phosphoethanolamine (either -F, -2F or -PE; respective shifts of 2, 1 and 0.5 g.u. for the removal of core α1,3-fucose, antennal α1,3-fucose and phosphoethanolamine); the loss of antennal α1,3-fucose or phosphoethanolamine is more efficient, whereas the loss of the core α1,3-fucose is partial for the employed hydrolysis time. The greyscale structures indicate the original elution times, those in color the products'; note that sulfated difucosylated glycans merely lose a fucose, but not the sulfate, as previously seen for the same structures isolated from High Five cells (19). B–G, Positive mode MALDI-TOF MS/MS of a trifucosylated glycan and of a difucosylated phosphoethanolamine-modified glycan showing the spectra for the original glycan as well as the intermediate and final HF-hydrolysis products. H and I, Positive mode MALDI-TOF MS/MS of a difucosylated glycan with an “upper” arm before and after hydrofluoric acid treatment. The loss of core α1,3-fucose correlates with the absence of the difucose-containing m/z 592 Y-fragment on hydrofluoric acid treatment; see also further MS/MS in supplemental Fig. S11.
Verification that the 176 Da modification is glucuronic acid rather than methylhexose was also offered by sensitivity to β-glucuronidases of three HPLC fractions containing monoglucuronylated hybrid and biantennary glycans; this treatment resulted in a shift to later RP-amide retention times (typically an increase of about 1 g.u.; see chromatograms in Fig. 6 and supplemental Fig. S10), loss of the negative ion mode signal and alterations in the MS/MS spectra (i.e. loss of the m/z 542 and 745 fragments and increased intensities at m/z 366 and 569; Fig. 6H–6K and supplemental Fig. S10A–S10L). As deglucuronylation resulted in retention times and MS/MS spectra akin to those for glycans found in the neutral pools, it is assumed that the structures with antennal Galβ1, 3GalNAcβ1,4GlcNAc underlie the glucuronylated forms.
Glucuronylated N-glycans Modified With Phosphoethanolamine
A further set of modifications is represented by glucuronylated structures also carrying phosphoethanolamine present in the anionic pool. Such glycans elute one HPLC fraction earlier than the “parent” structure lacking the zwitterion; hydrofluoric acid treatment to release the phosphoethanolamine results in a shift backwards in terms of retention time (Fig. 6A). This treatment also results in alterations in the MS/MS spectra (i.e. losses of negative or positive mode HexA1Hex1HexNAc2PE1 B-fragments at m/z 866/868 and appearance of positive ones at m/z 745; Fig. 6B–6G and supplemental Fig. S11A–S11D) indicative of removal of phosphoethanolamine (123 Da) from glucuronylated antennae. As judged by re-application onto HPLC, phosphoethanolamine-containing structures account for some 5% of the anionic pool.
Sulfated N-glycans
In previous studies on dipteran and lepidopteran glycomes, the sulfated N-glycome within the anionic pool was dominated by fucosylated structures such as Hex3HexNAc2–3Fuc1S1 (m/z 1213 and 1416); these were not present in royal jelly. Rather nonfucosylated Hex3HexNAc3–4S1 (m/z 1270 and 1473; see Fig. 5) were the major sulfated glycans detected, reflecting the general lack of fucose in royal jelly. Serial digestions of larger structures (Hex3–4HexNAc4–5S1) with β1,3-galactosidase and jack bean β-hexosaminidase removed the antennae; as only one underlying mannose was sensitive to both the unspecific jack bean α-mannosidase and α1,3-specific mannosidase, it was possible to define the α1,6-mannose as being sulfated (Fig. 7A–7F). Confirming our assignments were the typical negative mode fragments at m/z 606, 768 and 971 (Hex2–3HexNAc1–2S1), as seen for other cases of sulfation of mannose residues, including glycans of the same elution time previously identified in lepidopteran species (18, 19). Finally, on using solid phase extraction following permethylation of the anionic pool, it was also possible to detect certain sulfated glycans; MS/MS thereof supported the conclusion that the α1,6-linked mannose is sulfated (supplemental Figs. S2B and S3C). Otherwise, sulfated forms of the fucosylated and glucuronylated glycans were only observed in the off-line LC-MS analysis (see below).
Core Mono- and Di-fucosylated Anionic N-glycans
In the combined PNGase A and F-released neutral pools or when PNGase F alone was employed, only traces of a variety of singly fucosylated glycans were detected (Fig. 1); however, most fucosylated glycans were glucuronylated and therefore present in the anionic pool (Fig. 5), including the largest structures observed in this study, i.e. triantennary monofucosylated forms without and with sulfate (m/z 3366 and 3446 as [M-H]−). The presence of a core α1,6-fucose on these can be inferred from their release by PNGase F and late elution time as well as by the m/z 446 Y-fragment absent after bovine α-fucosidase treatment (supplemental Fig. S12F–S12J and S12K–S12N). However, when the glycans were released from the second sample of royal jelly using PNGase Ar, we noted traces of glycans with two or three fucose residues in the anionic pool (Fig. 5B). A key Y-fragment of m/z 592 (Fuc2GlcNAc1-PA; Fig. 8 and supplemental Figs. S11E and S11H and S12A–S12E), as well as a degree of anti-horseradish peroxidase staining (supplemental Fig. S13A), indicates that there are core α1,3/α1,6-difucosylated N-glycans; these glycans elute earlier than those carrying a single core α1,6-fucose. The third fucose residue is present on the antennae as seen, e.g. on bee venom glycoproteins (39), and is defined by Y-fragments such as m/z 1630, 1792 and 1954 (Hex3–5HexNAc3Fuc-PA) or losses of 891 mass units from the parent; hydrofluoric acid treatments and relevant shifts to later elution time or alterations in MS/MS spectra confirmed the presence of core and/or antennal α1,3-fucose (Fig. 8 and Supplemental Fig. S11E–S11J). Another isomeric trifucosylated variation, because of a single fucose on the core and two antennal fucoses, was found in the PNGase F-released pool (compare the two glycans of m/z 2916; supplemental Fig. S12E and S12I). Some of the fucosylated/glucuronylated glycans even contained phosphoethanolamine (Fig. 8E and supplemental Fig. S12B and S12G).
Rare, but detectable, were fucosylated glycans containing both glucuronic acid and sulfate observed as [M-H]− ions in negative mode and [M-SO3+H]+ in positive mode (Fig. 7G–7J); here also the fragmentation patterns indicate the sulfation of an α-mannose, as serial losses of an glucuronic acid antenna and then of sulfate are observed in negative mode MS/MS spectra, whereas the positive mode MS/MS show the nature of the “backbone” core fucosylated structure (Fig. 7K–7N). Smaller sulfated difucosylated glycans akin to those in High Five cells (19) were also detected (m/z 1359 and 1562; Fig. 8A) and one fucose could be removed from these with hydrofluoric acid; as this treatment is also known to remove phosphoesters (29) but not sulfate (40), the resistance of the 80 Da moiety is another confirmation that these glycans are sulfated.
Analysis of Protein-specific Glycosylation and Binding to Glycan Arrays
To test whether there are variations in the glycosylation of individual glycoproteins in royal jelly, SDS-PAGE was performed before either Western blotting or release of glycans from trypsin-treated gel bands. In terms of blotting, we used reagents capable of binding core α1,3-fucose, zwitterionic or antennal modifications. Thereby, the presence of core α1,3-fucose and of phosphoethanolamine as judged by the glycomic data correlates with the staining with anti-horseradish peroxidase and serum amyloid P; correspondingly the binding to C-reactive protein, which recognizes preferentially phosphorylcholine over phosphoethanolamine (25), is low (supplemental Fig. S13A). Also, reactivity toward fucose-specific Aleuria aurantia lectin, HexNAc-specific wheat germ agglutinin and Galβ1,3GalNAc-specific peanut agglutinin (41) was detected. The AEAB-labeled neutral and anionic pools of royal jelly N-glycans (Fig. 9A) were immobilized and specifically recognized in microarray format by peanut agglutinin (42), serum amyloid P (25) and an anti-L2/HNK-1 antibody (43, 44), whereby the reactivity to peanut agglutinin and anti-L2/HNK-1 was reduced on respective β-galactosidase or β-glucuronidase treatment (Fig. 9B and supplemental Fig. S14) and is compatible to the MS-based glycomic analyses.
Fig. 9.
A, MALDI-TOF MS of AEAB-labeled royal jelly neutral and anionic N-glycans; refer to supplemental Fig. S14 for further selected MS and MS/MS data. B, The binding of peanut aggulutin (PNA) to neutral and anionic N-glycans and of the human pentraxin serum amyloid P (SAP) and the anti-L2/HNK-1 antibody clone 412 (HNK-1) to anionic N-glycans is proven by probing the AEAB-labeled N-glycan pools on NHS-modified glass slides (see corresponding MALDI-TOF MS); the chart indicates the uncorrected fluorescence values with the standard deviations (green or red channels; mean of 10 spots) as compared with concanavalin A (as a nonspecific lectin) and to negative controls (spotting buffer or no primary lectin, antibody or pentraxin). The recognition by PNA, SAP and anti-L2/HNK-1 correlates, respectively, with the presence of “pseudo” T antigen epitopes in both pools and of phosphoethanolamine and terminal glucuronic acid in the anionic pool as proven by the analyses of the PA-labeled glycans, but the binding of the glycans to wheat germ agglutinin (WGA) is minimal. As shown in supplemental Fig. S14, binding to PNA decreases and to WGA increases after β-galactosidase treatment, whereas binding to anti-L2/HNK-1 is β-glucuronidase sensitive. Refer to supplemental Fig. S13A for Western blotting data with PNA, WGA and SAP. C, A zoomed-in region of the MRJP2 spectrum (see supplemental Fig. S13D) containing ions corresponding to a peptide modified with different complex N-glycans, including one with a T antigen epitope of the type recognized by PNA; the insets show respectively a region of the spectrum after PNGase Ar-treatment and one example MS/MS showing the 2,4A cross-ring GlcNAc-containing fragment for the m/z 2935 glycopeptide. Raw mzXML files corresponding to the full MRJP2 peptide spectra before and after PNGase Ar treatment as well as two MS/MS glycopeptide spectra are included in the supplementary zip archive.
For analysis of protein-specific glycosylation, the five most dominant Coomassie Blue-stained protein bands were excised as indicated and subject to trypsin peptide mass fingerprinting to verify their identity (coverage of 29–46%; supplemental Fig. S13B–S13D and Supplemental Table 2); MRJP1 is the major band, MRJP2 the second most dominant component and MRJP3 present in three protein bands, consistent with its heterogeneity as reported in the literature (45). We could detect forms of the arginine-containing peptide 136–146 from MRJP2 modified by complex N-glycans (Fig. 9C). The tryptic peptides were treated with PNGase Ar to release the N-glycans and these protein-specific glycomes were separately pyridylaminated, HPLC fractionated and analyzed by MALDI-TOF MS and MS/MS (supplemental Fig. S15). Considering the difference in the amount of each protein in the sample, it is not possible to conclude that there is much variation in the types of glycans on the individual glycoproteins. Each protein (MRJP1, MRJP 2 and MRJP 3) is modified with a range of glycans despite having each a limited number (n = 1–3) of glycosylation sites with the most dominant glycan being Man9GlcNAc2 (in terms of MALDI-TOF MS as well as fluorescence intensity at 5.7 g.u.); on the other hand, the heterogeneous sets of hybrid and complex glycans probably account for 50% of the structures as is the case in the overall royal jelly glycome. The ability to detect phosphoethanolamine (correlating with the binding to serum amyloid P), glucuronic acid and core β-mannose in the MRJP1 glycome (supplemental Fig. S15D–S15K) is possibly a result of this protein being the most abundant.
DISCUSSION
N-glycan Analysis of Insects
The Insecta is one of the most diverse classes of invertebrates with about thirty different orders, four of which contain the vast majority of insect species: Coleoptera (beetles), Diptera (true flies), Hymenoptera (wasps, bees and ants) and Lepidoptera (moths and butterflies). Among these, scientifically, economically and medically significant species include mosquitoes (e.g. Anopheles and Aedes spp.) which transmit pathogens, the model fruit fly Drosophila melanogaster and the honeybee Apis mellifera as well as Trichoplusia ni and Spodoptera frugiperda whose cell lines are used for baculovirus-based production of recombinant proteins. N-glycan structures from all these species have been described, but until recently only “simple” paucimannosidic and oligomannosidic glycans were reported (30, 46, 47) except for the more complex fucosylated antennae of bee venom glycoproteins (39). Of other insects, there are singular studies on glycomes or glycoproteins isolated from the silkworm Bombyx mori (48, 49), the flour beetle Tribolium castaneum (50) and the locust Locusta migratoria (51), of which the latter was most noteworthy as aminoethylphosphonate was detected as an antennal modification.
The most recent data indicate that the N-glycomic potential of insects is indeed underestimated. For many years, a major question (now resolved (52)) was whether insects sialylate their glycoconjugates, but perhaps the search for sialylated structures distracted attention from considering other anionic modifications. Our workflow based on separating the neutral and anionic fractions by solid phase extraction on graphitized carbon followed by off-line MALDI-TOF MS (in both modes) is relatively unbiased, circumvents suppression-dependent underestimation of anionic modifications and allows for further chemical or enzymatic treatments in order to confirm fine details of glycan structures (20, 29). Not only does our approach result in detection of typically one hundred different glycans but has vastly increased the range of structures now known in different species including molluscs, nematodes, slime molds and insects (18, 19, 22, 33, 53, 54). We have also examined binding of three lectins (Con A, PNA and WGA), one antibody (anti-L2/HNK-1) and one pentraxin (serum amyloid P) to the neutral and anionic N-glycomes of royal jelly in a microarray format.
The Variety of N-glycans in Royal Jelly
Some of the basic backbones of the neutral glycans were shown to be biantennary, others hybrid: isomeric variation could be detected because of the 1D- or 2D-HPLC separation approach. In terms of the neutral N-glycome, our data refines previous interpretations of those structures beyond the oligomannosidic, specifically showing that terminal galactosylation in royal jelly is in the context of LacdiNAc units, resulting in the previously-reported “pseudo” T-antigen Galβ1,3GalNAcβ1,4GlcNAc (16), but not as Galβ1,3GlcNAc as proposed in two other studies (15, 55); our data is also compatible with the presence of terminal LacdiNAc as well as of β1,4-linked branching GlcNAc (15, 56). The galactosylation of LacdiNAc, rather than also of GlcNAc, is a hallmark of royal jelly as opposed to honeybee larvae (unpublished data), whereas the venom is well known for both core and antennal α1,3-fucose (39). Interestingly, we detect core β-mannosylation in an insect for the first time, whereas fucosylation is at a low level in royal jelly as compared with other insect glycomes.
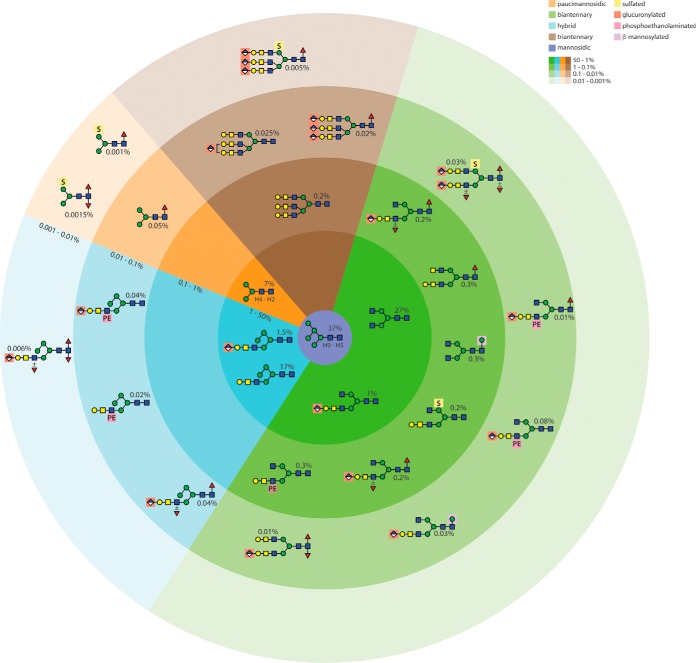
In the present study on royal jelly, the variety of N-glycan modifications is again larger than previously appreciated and this “expansion” is primarily because of the detection of some new and unusual anionic and zwitterionic structures (summarized in Fig. 10). We have detected anionic hybrid and bi/tri-antennary glycans with sulfate, phosphoethanolamine and glucuronic acid singly or in combination, some of which also have multiple fucose residues; indeed, unlike previously-defined di- and trifucosylated glycans from insects (19, 39), all such structures in the royal jelly glycome were glucuronylated and/or sulfated. Although phosphoethanolamine has been found on wasp and mosquito O-glycans (18, 57) and insect glycolipids (38), this is the first demonstration of this modification on any N-glycan from an animal species. Otherwise, phosphoethanolamine is a component of trichomonad, fungal and bacterial N-glycans (34, 35, 58), protist and bacterial lipid-linked oligosaccharides (59, 60) and glycosylphosphatidylinositol membrane anchors from various species (61); sulfation of mannose is known in, e.g. slime molds and insects (18, 54, 62), but sulfates are linked to other monosaccharides in mammalian glycoconjugates (e.g. Gal on thyroglobulin (63), GalNAc on pituitary glycoproteins (64), GlcNAc in the context of sulfo-Lewis X (65) and GlcA of the HNK-1 epitope (66)). In all animals, glucuronic acid is a component of chondroitin and heparan sulfates (67), whereas it is also present in mammals within the extended matriglycan chains on dystroglycan (68) as well as on various neural N-glycans (66); the binding of some anti-L2/HNK-1 monoclonals to blowfly glycoproteins and glycolipids, presumed to be because of nonsulfated GlcA (69), would correlate with our structural and array data for royal jelly N-glycans.
Fig. 10.
Summary of the N-glycome of honeybee royal jelly. The N-glycans are categorized based on structural type with color code indicative of either oligomannosidic, paucimannosidic, hybrid, biantennary and triantennary structures; a further highlighting is used for the β-mannose, sulfate, glucuronic acid or phosphoethanolamine motifs. Approximate abundances were calculated based on HPLC and MS intensities. The order of abundance of the glycans decreases from the center (with less intense background colors for the least abundant), whereby the oligomannosidic Man5–9GlcNAc2 are estimated as accounting for 37% of the total N-glycome. For simplicity not all glycans of a certain type are shown, rather a representative major example, whereby, e.g. the biantennary class (27%) includes structures with Gal and GalNAc residues or the hybrid class (17%) pertains also to Man3–5-based isomers without Gal/GalNAc modification.
Previously, a minor MRJP2 homologue was concluded to have Hex9HexNAc2 on one glycosylation site and Hex4–5HexNAc3–4 on the other (11) and here we obtained a similar result for the second site, whereas the N-terminal glycosylation site of MRJP1 is apparently modified by Hex3–5HexNAc4–6 and the other two sites by oligomannosidic structures (16), which may be because of differential exposure to Golgi processing enzymes. Because of the off-line LC-MS approach, we have revealed a heterogeneity in the protein-specific glycosylation pattern of MRJP1 and MRJP2 previously not observed by MS or other analyses. The protein-specific glycans identified reflect the higher amount of Man9GlcNAc2 in the second preparation as compared with the first (compare supplemental Fig. S1A and S1B); this could be because of variations in hypopharyngeal gland glycan processing in individual bees or the presence of α-mannosidase, a proteomically determined royal jelly component (5). Furthermore, the 3D-structures of the major glycoproteins present in royal jelly may play a role in determining the accessibility of glycans to Golgi enzymes and so effect the overall status of the secreted glycome. It is also of interest that some of these royal jelly proteins are also present in honeybee venom and, in recombinant form, are recognized by the IgE of allergic patients (70), whereas MJRP1 in honey possesses glycan-dependent IgE epitopes (71). On the other hand, in contrast to our data showing a low degree of core α1,3-fucosylation, Western blotting with an anti-plant-glycoprotein antiserum was considered negative in a previous study (14), whereas wheat germ and peanut agglutinin binding (respectively indicative for terminal HexNAc and Galβ1,3GalNAc modifications) to royal jelly proteins was positive (the latter correlating with the glycan array data). Certainly, modifications with fucose, glucuronic acid, core mannose or phosphoethanolamine were previously not defined on single royal jelly glycoproteins.
In contrast to mosquitoes and lepidopterans (18, 19), sulfation and glucuronylation were almost mutually exclusive in royal jelly, with the latter again being the dominant “charged” modification; parallel (unpublished) analyses of honeybee venom and larvae indicate that these glycomes are different in terms of the degree and type of antennal modifications, which suggests that royal jelly glycans are the product of a tissue-specific glycosylation pattern. Thereby, whereas fucosyltransferases (FucTA, FucTC and FucT6) are predicted to be expressed at low levels in hypopharyngeal glands, other gland-specific transcription events may be responsible for the increase in anionic/zwitterionic glycans. Indeed, FucTA core α1,3-fucosyltransferase mRNA levels are seemingly far lower in hypopharyngeal as compared with venom glands (72).
Potential Biological Relevance of Royal Jelly N-glycans
As anionic and zwitterionic moieties have different biophysical properties and so be differently recognized as compared with neutral saccharides, it is to be expected that they have distinct bioactivities. For instance, sulfation of mannose of the N-glycans of a lepidopteran protein has been claimed to have a pro-inflammatory role in leukocyte activation in dermatitis caused by setae of a tropical moth (62), whereas in various species, sulfation of glycosaminoglycans, Lewis X or terminal GalNAc is associated with processes such as blood clotting, morphogenesis, cell adhesion or hormone clearance (73–76). Furthermore, the glucuronic acid-containing L2/HNK-1-type epitope (present in nonsulfated form on royal jelly glycans as verified by the array data in Fig. 9B) may be recognized by the human cytokine IL-6, so having relevance to interleukin signaling (77), as well as by HMGB1 or amphoterin (78) which is also known to interact with Toll-like receptors (79). Phosphoethanolamine is one of the ligands for serum amyloid P protein, a pentraxin known to bind, e.g. bacteria, but also to activate complement (80) and here we show by blotting and array experiments that glycoproteins and the anionic N-glycans thereof in particular in royal jelly are indeed recognized by serum amyloid P (Fig. 9B and supplemental Fig. S13A); it would certainly be of interest to know which ligands are recognized by insect members of the pentraxin family (81), including a homologue in the honeybee (gene LOC102656294), as such proteins from another arthropod (the horseshoe crab) do bind zwitterions (82). Phosphoethanolamine is the nonmethylated form of phosphorylcholine, which is present on N- and O-glycans of insect cell lines used for recombinant protein production (19, 83), but which is also known to possess immunomodulatory function as a modification of nematode glycoproteins (84). Thus, although there are no systematic studies as to the protein ligands or receptors for such moieties, for each “charged” modification of royal jelly N-glycans there is potential for bioactivity, immunoreactivity or immunoactivity.
DATA AVAILABILITY
Mass spectrometric data in mzXML format are appended in a supplementary zip file.
Supplementary Material
Acknowledgments
We thank Dr. Hans Bakker (Medizinische Hochschule Hannover) for the aliquot of the anti-L2/HNK-1 antibody (clone 412).
Footnotes
* This work was supported by the Austrian Science Fund (FWF) and by the European Union; A.H. and K.P. are FWF fellows (grants P26662 and P25058) and B.E. a student within the FWF-funded BioTOP doctoral program W1224; J.V. was an experienced researcher within the Glycopar Initial Training Network (PITN-GA-2013-608295). The LTQ mass spectrometer was obtained by a grant from the Swedish Research Council (342–2004-4434).
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- MRJP
- major royal jelly proteins.
REFERENCES
- 1. Fratini F., Cilia G., Mancini S., and Felicioli A. (2016) Royal Jelly: An ancient remedy with remarkable antibacterial properties. Microbiol. Res. 192, 130–141 [DOI] [PubMed] [Google Scholar]
- 2. McCleskey C. S., and Melampy R. M. (1939) Bactericidal Properties of Royal Jelly of the Honeybee. J. Economic Entomol. 32, 581–587 [Google Scholar]
- 3. Paola F., Pantalea D. D., Gianfranco C., Antonio F., Angelo V., Eustachio N., and Elisabetta D. L. (2014) Oral allergy syndrome in a child provoked by royal jelly. Case Rep. Med. 941248, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang L., Fang Y., Li R., Feng M., Han B., Zhou T., and Li J. (2012) Towards posttranslational modification proteome of royal jelly. J. Proteomics 75, 5327–5341 [DOI] [PubMed] [Google Scholar]
- 5. Zhang L., Han B., Li R., Lu X., Nie A., Guo L., Fang Y., Feng M., and Li J. (2014) Comprehensive identification of novel proteins and N-glycosylation sites in royal jelly. BMC Genomics 15, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paschinger K., and Wilson I. B. H. (2015) Comparative Glycobiology. in Glycoscience: Biology and Medicine (Taniguchi N. ed.), Springer; Japan: pp 795–805 [Google Scholar]
- 7. Kamakura M. (2011) Royalactin induces queen differentiation in honeybees. Nature 473, 478–483 [DOI] [PubMed] [Google Scholar]
- 8. Buttstedt A., Ihling C. H., Pietzsch M., and Moritz R. F. (2016) Royalactin is not a royal making of a queen. Nature 537, E10–E12 [DOI] [PubMed] [Google Scholar]
- 9. Detienne G., De Haes W., Ernst U. R., Schoofs L., and Temmerman L. (2014) Royalactin extends lifespan of Caenorhabditis elegans through epidermal growth factor signaling. Exp. Gerontol. 60, 129–135 [DOI] [PubMed] [Google Scholar]
- 10. Feng M., Fang Y., Han B., Xu X., Fan P., Hao Y., Qi Y., Hu H., Huo X., Meng L., Wu B., and Li J. (2015) In-depth N-glycosylation reveals species-specific modifications and functions of the royal jelly protein from western (Apis mellifera) and eastern honeybees (Apis cerana). J. Proteome Res. 14, 5327–5340 [DOI] [PubMed] [Google Scholar]
- 11. Bíliková K., Mirgorodskaya E., Bukovská G., Gobom J., Lehrach H., and Šimúth J. (2009) Towards functional proteomics of minority component of honeybee royal jelly: the effect of post-translational modifications on the antimicrobial activity of apalbumin2. Proteomics 9, 2131–2138 [DOI] [PubMed] [Google Scholar]
- 12. Kimura Y., Washino N., and Yonekura M. (1995) N-linked sugar chains of 350-kDa royal jelly glycoprotein. Biosci. Biotechnol. Biochem. 59, 507–509 [DOI] [PubMed] [Google Scholar]
- 13. Kimura Y., Kajiyama S., Kanaeda J., Izukawa T., and Yonekura M. (1996) N-linked sugar chain of 55-kDa royal jelly glycoprotein. Biosci. Biotechnol. Biochem. 60, 2099–2102 [DOI] [PubMed] [Google Scholar]
- 14. Kimura Y., Miyagi C., Kimura M., Nitoda T., Kawai N., and Sugimoto H. (2000) Structural features of N-glycans linked to royal jelly glycoproteins: structures of high-mannose type, hybrid type, and biantennary type glycans. Biosci. Biotechnol. Biochem. 64, 2109–2120 [DOI] [PubMed] [Google Scholar]
- 15. Kimura Y., Tsumura K., Kimura M., Okihara K., Sugimoto H., and Yamada H. (2003) First evidence for occurrence of Galβ1-3GlcNAcβ1–4Man unit in N-glycans of insect glycoprotein: β1–3Gal and β1–4GlcNAc transferases are involved in N-glycan processing of royal jelly glycoproteins. Biosci Biotechnol Biochem 67, 1852–1856 [DOI] [PubMed] [Google Scholar]
- 16. Kimura Y., Nagai H., Miyamoto M., Kimura M., and Yonekura M. (2010) Identification of a Royal Jelly Glycoprotein That Carries Unique Complex-Type N-Glycans Harboring the T-Antigen (Galβ1–3GalNAc) Unit. Biosci. Biotechnol. Biochem. 74, 2148–2150 [DOI] [PubMed] [Google Scholar]
- 17. Ichimiya T., Maeda M., Sakamura S., Kanazawa M., Nishihara S., and Kimura Y. (2015) Identification of β1,3-galactosyltransferases responsible for biosynthesis of insect complex-type N-glycans containing a T-antigen unit in the honeybee. Glycoconj. J. 32, 141–151 [DOI] [PubMed] [Google Scholar]
- 18. Kurz S., Aoki K., Jin C., Karlsson N. G., Tiemeyer M., Wilson I. B. H., and Paschinger K. (2015) Targetted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J. Proteomics 126, 172–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stanton R., Hykollari A., Eckmair B., Malzl D., Dragosits M., Palmberger D., Wang P., Wilson I. B. H., and Paschinger K. (2017) The underestimated N-glycomes of lepidopteran species. Biochim. Biophys. Acta 1861, 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hykollari A., Paschinger K., Eckmair B., and Wilson I. B. H. (2017) Analysis of Invertebrate and Protist N-Glycans. Methods Mol. Biol. 1503, 167–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khoo K.-H., and Yu S.-Y. (2010) Mass Spectrometric Analysis of Sulfated N- and O-Glycans. Methods Enzymol. 478, 3–26 [DOI] [PubMed] [Google Scholar]
- 22. Eckmair B., Jin C., Abed-Navandi D., and Paschinger K. (2016) Multi-step fractionation and mass spectrometry reveals zwitterionic and anionic modifications of the N- and O-glycans of a marine snail. Mol. Cell. Proteomics 15, 573–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dragosits M., Yan S., Razzazi-Fazeli E., Wilson I. B. H., and Rendić D. (2015) Enzymatic properties and subtle differences in the substrate specificity of phylogenetically distinct invertebrate N-glycan processing hexosaminidases. Glycobiology 25, 448–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paschinger K., Rendić D., and Wilson I. B. H. (2009) Revealing the anti-HRP epitope in Drosophila and Caenorhabditis. Glycoconj. J. 26, 385–395 [DOI] [PubMed] [Google Scholar]
- 25. Mikolajek H., Kolstoe S. E., Pye V. E., Mangione P., Pepys M. B., and Wood S. P. (2011) Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component. J. Mol. Recognit. 24, 371–377 [DOI] [PubMed] [Google Scholar]
- 26. Paschinger K., Gonzalez-Sapienza G. G., and Wilson I. B. H. (2012) Mass spectrometric analysis of the immunodominant glycan epitope of Echinococcus granulosus antigen Ag5. Int. J. Parasitol. 42, 279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song X., Xia B., Stowell S. R., Lasanajak Y., Smith D. F., and Cummings R. D. (2009) Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem. Biol. 16, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiménez-Castells C., Stanton R., Yan S., Kosma P., and Wilson I. B. H. (2016) Development of a multifunctional aminoxy-based fluorescent linker for glycan immobilization and analysis. Glycobiology 26, 1297–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paschinger K., and Wilson I. B. H. (2016) Analysis of zwitterionic and anionic N-linked glycans from invertebrates and protists by mass spectrometry. Glycoconj. J. 33, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fabini G., Freilinger A., Altmann F., and Wilson I. B. H. (2001) Identification of core α1,3-fucosylated glycans and the requisite fucosyltransferase in Drosophila melanogaster. Potential basis of the neural anti-horseradish peroxidase epitope. J. Biol. Chem. 276, 28058–28067 [DOI] [PubMed] [Google Scholar]
- 31. Yan S., Wilson I. B. H., and Paschinger K. (2015) Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode Pristionchus pacificus. Electrophoresis 36, 1314–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomiya N., Awaya J., Kurono M., Endo S., Arata Y., and Takahashi N. (1988) Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Anal. Biochem. 171, 73–90 [DOI] [PubMed] [Google Scholar]
- 33. Kurz S., Jin C., Hykollari A., Gregorich D., Giomarelli B., Vasta G. R., Wilson I. B. H., and Paschinger K. (2013) Haemocytes and plasma of the eastern oyster (Crassostrea virginica) display a diverse repertoire of sulphated and blood group A-modified N-glycans. J. Biol. Chem. 288, 24410–24428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paschinger K., Hykollari A., Razzazi-Fazeli E., Greenwell P., Leitsch D., Walochnik J., and Wilson I. B. H. (2012) The N-glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology 22, 300–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hykollari A., Eckmair B., Voglmeir J., Jin C., Yan S., Vanbeselaere J., Razzazi-Fazeli E., Wilson I. B. H., and Paschinger K. (2016) More than just oligomannose: An N-glycomic comparison of Penicillium species. Mol. Cell. Proteomics 15, 73–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weske B., Dennis R. D., Helling F., Keller M., Nores G. A., Peter-Katalinic J., Egge H., Dabrowski U., and Wiegandt H. (1990) Glycosphingolipids in insects. Chemical structures of two variants of a glucuronic-acid-containing ceramide hexasaccharide from a pupae of Calliphora vicina (Insecta: Diptera), distinguished by a N-acetylglucosamine-bound phosphoethanolamine sidechain. Eur. J. Biochem. 191, 379–388 [DOI] [PubMed] [Google Scholar]
- 37. Helling F., Dennis R. D., Weske B., Nores G., Peter-Katalinic J., Dabrowski U., Egge H., and Wiegandt H. (1991) Glycosphingolipids in insects. The amphoteric moiety, N-acetylglucosamine-linked phosphoethanolamine, distinguishes a group of ceramide oligosaccharides from the pupae of Calliphora vicina (Insecta: Diptera). Eur. J. Biochem. 200, 409–421 [DOI] [PubMed] [Google Scholar]
- 38. Seppo A., Moreland M., Schweingruber H., and Tiemeyer M. (2000) Zwitterionic and acidic glycosphingolipids of the Drosophila melanogaster embryo. Eur. J. Biochem. 267, 3549–3558 [DOI] [PubMed] [Google Scholar]
- 39. Kubelka V., Altmann F., Staudacher E., Tretter V., März L., Hård K., Kamerling J. P., and Vliegenthart J. F. G. (1993) Primary structures of the N-linked carbohydrate chains from honeybee venom phospholipase A2. Eur. J. Biochem. 213, 1193–1204 [DOI] [PubMed] [Google Scholar]
- 40. Wing D. R., Rademacher T. W., Field M. C., Dwek R. A., Schmitz B., Thor G., and Schachner M. (1992) Use of large-scale hydrazinolysis in the preparation of N-linked oligosaccharide libraries: application to brain tissue. Glycoconjugate J. 9, 293–301 [DOI] [PubMed] [Google Scholar]
- 41. Iskratsch T., Braun A., Paschinger K., and Wilson I. B. H. (2009) Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal. Biochem. 386, 133–146 [DOI] [PubMed] [Google Scholar]
- 42. Lotan R., Skutelsky E., Danon D., and Sharon N. (1975) The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea). J. Biol. Chem. 250, 8518–8523 [PubMed] [Google Scholar]
- 43. Kruse J., Mailhammer R., Wernecke H., Faissner A., Sommer I., Goridis C., and Schachner M. (1984) Neural cell adhesion molecules and myelin-associated glycoprotein share a common carbohydrate moiety recognized by monoclonal antibodies L2 and HNK-1. Nature 311, 153–155 [DOI] [PubMed] [Google Scholar]
- 44. Schmitz B., Schachner M., Ito Y., Nakano T., and Ogawa T. (1994) Determination of structural elements of the L2/HNK-1 carbohydrate epitope required for its function. Glycoconjugate J. 11, 345–352 [DOI] [PubMed] [Google Scholar]
- 45. Albert S., Klaudiny J., and Simuth J. (1999) Molecular characterization of MRJP3, highly polymorphic protein of honeybee (Apis mellifera) royal jelly. Insect Biochem. Mol. Biol. 29, 427–434 [DOI] [PubMed] [Google Scholar]
- 46. Williams P. J., Wormald M. R., Dwek R. A., Rademacher T. W., Parker G. F., and Roberts D. R. (1991) Characterisation of oligosaccharides from Drosophila melanogaster glycoproteins. Biochim. Biophys. Acta Gen. Subj. 1075, 146–153 [DOI] [PubMed] [Google Scholar]
- 47. Shi X., and Jarvis D. L. (2007) Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets. 8, 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kajiura H., Hamaguchi Y., Mizushima H., Misaki R., and Fujiyama K. (2015) Sialylation potentials of the silkworm, Bombyx mori; B. mori possesses an active α2,6-sialyltransferase. Glycobiology 25, 1441–1453 [DOI] [PubMed] [Google Scholar]
- 49. Mabashi-Asazuma H., Sohn B. H., Kim Y. S., Kuo C. W., Khoo K. H., Kucharski C. A., Fraser M. J. Jr, and Jarvis D. L. (2015) Targeted glycoengineering extends the protein N-glycosylation pathway in the silkworm silk gland. Insect Biochem. Mol. Biol. 65, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walski T., Van Damme E. J., Smargiasso N., Christiaens O., De Pauw E., and Smagghe G. (2016) Protein N-glycosylation and N-glycan trimming are required for postembryonic development of the pest beetle Tribolium castaneum. Sci. Rep. 6, 35151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hård K., Van Doorn J. M., Thomas-Oates J. E., Kamerling J. P., and Van der Horst D. J. (1993) Structure of the Asn-linked oliogsaccharides of apolipophorin III from the insect Locusta migratoria. Carbohydrate-linked 2-aminoethylphosphonate as a constituent of a glycoprotein. Biochemistry 32, 766–775 [DOI] [PubMed] [Google Scholar]
- 52. Aoki K., Perlman M., Lim J. M., Cantu R., Wells L., and Tiemeyer M. (2007) Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J. Biol. Chem. 282, 9127–9142 [DOI] [PubMed] [Google Scholar]
- 53. Yan S., Jin C., Wilson I. B. H., and Paschinger K. (2015) Comparisons of Caenorhabditis fucosyltransferase mutants reveal a multiplicity of isomeric N-glycan structures. J. Proteome Res. 14, 5291–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hykollari A., Balog C. I., Rendić D., Braulke T., Wilson I. B. H., and Paschinger K. (2013) Mass spectrometric analysis of neutral and anionic N-glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. J. Proteome Res. 12, 1173–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kimura M., Kimura Y., Tsumura K., Okihara K., Sugimoto H., Yamada H., and Yonekura M. (2003) 350-kDa royal jelly glycoprotein (apisin), which stimulates proliferation of human monocytes, bears the β1-3galactosylated N-glycan: analysis of the N-glycosylation site. Biosci. Biotechnol. Biochem. 67, 2055–2058 [DOI] [PubMed] [Google Scholar]
- 56. Kimura M., Hama Y., Tsumura K., Okihara K., Sugimoto H., Yamada H., and Kimurai Y. (2002) Occurrence of GalNAcβ1–4GlcNAc unit in N-glycan of royal jelly glycoprotein. Biosci. Biotechnol. Biochem. 66, 1985–1989 [DOI] [PubMed] [Google Scholar]
- 57. Maes E., Garenaux E., Strecker G., Leroy Y., Wieruszeski J. M., Brassart C., and Guerardel Y. (2005) Major O-glycans from the nest of Vespula germanica contain phospho-ethanolamine. Carbohydr. Res. 340, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 58. Scott N. E., Nothaft H., Edwards A. V., Labbate M., Djordjevic S. P., Larsen M. R., Szymanski C. M., and Cordwell S. J. (2012) Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287, 29384–29396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McConville M. J., Collidge T. A., Ferguson M. A., and Schneider P. (1993) The glycoinositol phospholipids of Leishmania mexicana promastigotes. Evidence for the presence of three distinct pathways of glycolipid biosynthesis. J. Biol. Chem. 268, 15595–15604 [PubMed] [Google Scholar]
- 60. Mackinnon F. G., Cox A. D., Plested J. S., Tang C. M., Makepeace K., Coull P. A., Wright J. C., Chalmers R., Hood D. W., Richards J. C., and Moxon E. R. (2002) Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol. Microbiol. 43, 931–943 [DOI] [PubMed] [Google Scholar]
- 61. Roberts W. L., Santikarn S., Reinhold V. N., and Rosenberry T. L. (1988) Structural characterization of the glycoinositol phospholipid membrane anchor of human erythrocyte acetylcholinesterase by fast atom bombardment mass spectrometry. J. Biol. Chem. 263, 18776–18784 [PubMed] [Google Scholar]
- 62. Cabrera G., Salazar V., Montesino R., Tambara Y., Struwe W. B., Lugo E. L., Harvey D. J., Antoine L., Rincon M., Domon B., Mendez Martinez M. D., Portela M., Gonzalez-Hernandez A., Triguero A., Duran R., Lundberg U., Vonasek E., and Gonzalez L. J. (2015) Structural characterization and biological implications of sulfated N-glycans in a serine protease from the neotropical moth Hylesia metabus (Cramer [1775]) (Lepidoptera: Saturniidae). Glycobiology 26, 230–250 [DOI] [PubMed] [Google Scholar]
- 63. De Waard P., Koorevaar A., Kamerling J. P., and Vliegenthart J. F. G. (1991) Structure determination by 1H NMR spectroscopy of (sulfated) sialylated N-linked carbohydrate chains released from porcine thyroglobulin by peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase-F. J. Biol. Chem. 266, 4237–4243 [PubMed] [Google Scholar]
- 64. Green E. D., and Baenziger J. U. (1988) Asparagine-linked oligosaccharides on lutropin, follitropin, and thyrotropin. I. Structural elucidation of the sulfated and sialylated oligosaccharides on bovine, ovine, and human pituitary glycoprotein hormones. J. Biol. Chem. 263, 25–35 [PubMed] [Google Scholar]
- 65. Kawashima H., and Fukuda M. (2012) Sulfated glycans control lymphocyte homing. Ann. N.Y. Acad. Sci. 1253, 112–121 [DOI] [PubMed] [Google Scholar]
- 66. Liedtke S., Geyer H., Wuhrer M., Geyer R., Frank G., Gerardy-Schahn R., Zahringer U., and Schachner M. (2001) Characterization of N-glycans from mouse brain neural cell adhesion molecule. Glycobiology 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 67. Kjellén L., and Lindahl U. (1991) Proteoglycans: structures and interactions. Annu. Rev. Biochem. 60, 443–475 [DOI] [PubMed] [Google Scholar]
- 68. Praissman J. L., Willer T., Sheikh M. O., Toi A., Chitayat D., Lin Y. Y., Lee H., Stalnaker S. H., Wang S., Prabhakar P. K., Nelson S. F., Stemple D. L., Moore S. A., Moremen K. W., Campbell K. P., and Wells L. (2016) The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife 5, e14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dennis R. D., Antonicek H., Wiegandt H., and Schachner M. (1988) Detection of the L2/HNK-1 carbohydrate epitope on glycoproteins and acidic glycolipids of the insect Calliphora vicina. J. Neurochem. 51, 1490–1496 [DOI] [PubMed] [Google Scholar]
- 70. Blank S., Bantleon F. I., McIntyre M., Ollert M., and Spillner E. (2012) The major royal jelly proteins 8 and 9 (Api m 11) are glycosylated components of Apis mellifera venom with allergenic potential beyond carbohydrate-based reactivity. Clin. Exp. Allergy. 42, 976–985 [DOI] [PubMed] [Google Scholar]
- 71. Hayashi T., Takamatsu N., Nakashima T., and Arita T. (2011) Immunological characterization of honey proteins and identification of MRJP 1 as an IgE-binding protein. Biosci. Biotechnol. Biochem. 75, 556–560 [DOI] [PubMed] [Google Scholar]
- 72. Rendić D., Klaudiny J., Stemmer U., Schmidt J., Paschinger K., and Wilson I. B. H. (2007) Towards abolition of immunogenic structures in insect cells: characterization of a honey-bee (Apis mellifera) multi-gene family reveals both an allergy-related core α1,3-fucosyltransferase and the first insect Lewis-histo-blood-group-related antigen-synthesizing enzyme. Biochem. J. 402, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Petitou M., Casu B., and Lindahl U. (2003) 1976–1983, a critical period in the history of heparin: the discovery of the antithrombin binding site. Biochimie 85, 83–89 [DOI] [PubMed] [Google Scholar]
- 74. Izvolsky K. I., Shoykhet D., Yang Y., Yu Q., Nugent M. A., and Cardoso W. V. (2003) Heparan sulfate-FGF10 interactions during lung morphogenesis. Dev. Biol. 258, 185–200 [DOI] [PubMed] [Google Scholar]
- 75. Yeh J. C., Hiraoka N., Petryniak B., Nakayama J., Ellies L. G., Rabuka D., Hindsgaul O., Marth J. D., Lowe J. B., and Fukuda M. (2001) Novel sulfated lymphocyte homing receptors and their control by a Core1 extension β1,3-N-acetylglucosaminyltransferase. Cell 105, 957–969 [DOI] [PubMed] [Google Scholar]
- 76. Roseman D. S., and Baenziger J. U. (2000) Molecular basis of lutropin recognition by the mannose/GalNAc-4-SO4 receptor. Proc. Natl. Acad. Sci. U.S.A. 97, 9949–9954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cebo C., Durier V., Lagant P., Maes E., Florea D., Lefebvre T., Strecker G., Vergoten G., and Zanetta J.-P. (2002) Function and molecular modeling of the interaction between human interleukin 6 and Its HNK-1 oligosaccharide ligands. J. Biol. Chem. 277, 12246–12252 [DOI] [PubMed] [Google Scholar]
- 78. Chou D. K., Evans J. E., and Jungalwala F. B. (2001) Identity of nuclear high-mobility-group protein, HMG-1, and sulfoglucuronyl carbohydrate-binding protein, SBP-1, in brain. J. Neurochem. 77, 120–131 [DOI] [PubMed] [Google Scholar]
- 79. Pilzweger C., and Holdenrieder S. (2015) Circulating HMGB1 and RAGE as clinical biomarkers in malignant and autoimmune diseases. Diagnostics 5, 219–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Garlanda C., Bottazzi B., Bastone A., and Mantovani A. (2005) Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Ann. Rev. Immunol. 23, 337–366 [DOI] [PubMed] [Google Scholar]
- 81. Martinez de la Torre Y., Fabbri M., Jaillon S., Bastone A., Nebuloni M., Vecchi A., Mantovani A., and Garlanda C. (2010) Evolution of the pentraxin family: the new entry PTX4. J. Immunol. 184, 5055–5064 [DOI] [PubMed] [Google Scholar]
- 82. Shrive A. K., Metcalfe A. M., Cartwright J. R., and Greenhough T. J. (1999) C-reactive protein and SAP-like pentraxin are both present in Limulus polyphemus haemolymph: crystal structure of Limulus SAP. J. Mol. Biol. 290, 997–1008 [DOI] [PubMed] [Google Scholar]
- 83. Gaunitz S., Jin C., Nilsson A., Liu J., Karlsson N. G., and Holgersson J. (2013) Mucin-type proteins produced in the Trichoplusia ni and Spodoptera frugiperda insect cell lines carry novel O-glycans with phosphocholine and sulfate substitutions. Glycobiology 23, 778–796 [DOI] [PubMed] [Google Scholar]
- 84. Harnett M. M., Kean D. E., Boitelle A., McGuiness S., Thalhamer T., Steiger C. N., Egan C., Al-Riyami L., Alcocer M. J., Houston K. M., Gracie J. A., McInnes I. B., and Harnett W. (2008) The phosphorycholine moiety of the filarial nematode immunomodulator ES-62 is responsible for its anti-inflammatory action in arthritis. Ann. Rheum. Dis. 67, 518–523 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometric data in mzXML format are appended in a supplementary zip file.