Abstract
Across 8101 individuals in 46 villages, the proportion of Plasmodium spp. multiple clone infections (0%–53.8%) did not reflect prevalence by quantitative polymerase chain reaction (qPCR; 1.9%–38.4%), except for P. vivax in Solomon Islands (P < .001). Thus this parameter was not informative to identify transmission foci. In contrast, prevalence by microscopy and qPCR correlated well.
Keywords: malaria transmission, genotyping, multiple clone infection, prevalence, foci of transmission
Malaria transmission is often focal, with high levels of transmission being restricted to a few villages or even households [1, 2]. Although this offers the possibility for targeted control, in particular when transmission levels decline, the identification of such foci of transmission is challenging. Clinical cases identified through passive case detection might give indications of local outbreaks, but large population-wide surveys using sensitive diagnostics are needed to obtain a more complete picture by also identifying asymptomatic infections, which are likely to contribute to transmission.
However, population-wide surveys are time- and labor-intensive. Molecular data, such as the multiplicity of infection (MOI) assessed through genotyping of a representative smaller number of samples, might yield surrogate data on transmission intensity and asymptomatic prevalence. Multiple clone infections are expected to occur more frequently when transmission levels are higher, and it has been suggested to include MOI data to monitor changes in transmission intensity [3]. Indeed, lower proportions of P. falciparum multiple clone infections when transmission declined were found at large geographical scales, for example, in West Africa [4, 5] and over a 10-year period in Southeast Asia [6]. It is not known, however, how far these measures reflect prevalence of infection at small scales, for example, among villages, and how far they can help in identifying foci of transmission.
In the Southwest Pacific, transmission levels of P. falciparum and P. vivax have been high and decreased in recent years, with increased coverage of effective interventions [7, 8]. A recent country-wide study in Papua New Guinea (PNG) found P. falciparum prevalence at the provincial level to correlate moderately with mean MOI, whereas this was not the case for P. vivax [9]. To assess whether such relationships are stronger at smaller geographical scales, that is, at the village level, P. falciparum and P. vivax infections from 3 cross-sectional surveys covering 46 villages in PNG and Solomon Islands were genotyped, and the proportion of multiple clone infections was compared with prevalence. In addition, the correlation between prevalence by microscopy and quantitative polymerase chain reaction (qPCR) was assessed.
METHODS
Ethical approval for this study was obtained from the PNG IMR Institutional Review Board (approvals 1116/1204, 11.21/1206), the PNG Medical Research Advisory Committee (approvals 05.20, 12.06), the Solomon Islands National Health Research Ethics Committee (approval 12/022), and the WEHI Human Research Ethics Committee (approvals 12/01, 12/09). Informed written consent was obtained from all individuals or their legal guardians before sample collection.
Plasmodium vivax– and P. falciparum–positive samples from 3 previously described cross-sectional surveys in PNG and Solomon Islands were genotyped [7, 8]. Samples were collected from individuals aged 6 months to 80 years in 3 catchments (Malala, Mugil, Utu) in Madang 2010 (n = 2087) and Madang 2014 (n = 2531), containing 10 and 17 villages, respectively, and from 5 catchments (North Coast, South Coast, Bay, Anchor, Channel) including 19 villages in Solomon Islands in 2012 (n = 3501). Blood was collected into EDTA tubes using a 200-μL finger prick, and a Giemsa-stained slide was prepared for light microscopy. Plasmodium spp. were identified and quantified using qPCR assays with a sensitivity of 1 copy/μL [7, 8]. Plasmodium vivax– and P. falciparum–positive samples were genotyped by capillary electrophoresis of size-polymorphic markers: msp2 for P. falciparum, and msp1F3 and MS2 or MS16 for P. vivax [8].
Plasmodium vivax multiplicity of infection was defined as the maximum number of alleles detected by either marker, msp1F3 and MS2/MS16. The proportion of multiple clone infections and prevalence was compared using linear regression models. For the analysis at the village level, all villages with at least 10 infections genotyped were included. As frequency distributions of MOI were skewed, geometric mean MOI was calculated.
RESULTS
Across the 3 surveys, a total of 8101 individuals in 46 villages were sampled. By qPCR and microscopy, respectively, P. vivax prevalence was 12.7% and 7.0% in Madang in 2010, 19.7% and 2.7% in Madang in 2014, and 13.4% and 3.6% in Solomon Islands. P. falciparum prevalence was 18.7% and 7.4% in Madang in 2010, 9.0% and 2.7% in Madang in 2014, and 0.15% and 0.11% in the Solomon Islands [7, 8]. At the village level across all surveys, P. vivax prevalence by qPCR ranged from 3.0% to 38.5%, and P. falciparum prevalence ranged from 0% to 36.4%.
Eighty-three point six percent (515/616) of P. falciparum and 78.4% (968/1234) of P. vivax infections were successfully genotyped. Diversity of all genotyping markers was high (expected heterozygosity HE > 0.9), except for P. falciparum in the Solomon Islands, where all 5 isolates carried the same allele.
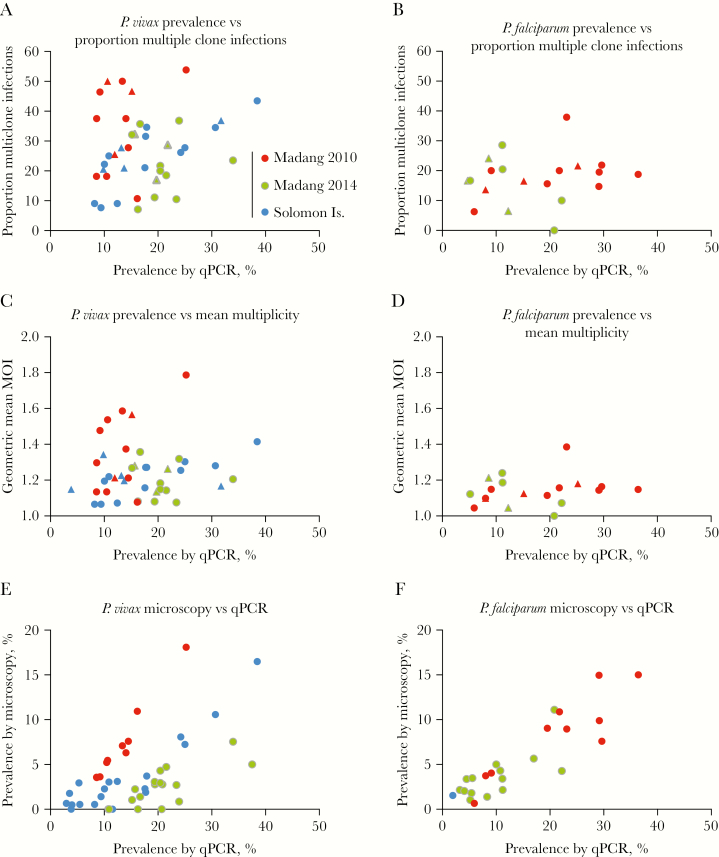
The proportion P. vivax multiple clone infections across villages (n = 32, prevalence: 8.2%–38.5%, 11–81 [median = 19] samples genotyped) ranged from 7.1% to 53.8% and was not correlated with prevalence by qPCR (P = .342) (Figure 1A). Likewise, no correlation was observed when data were analyzed at the catchment level (n = 11, P = .578). When surveys were assessed individually, no correlation was observed for Madang 2010 (n = 10, P = .476) or Madang 2014 (n = 10, P = .980). However, very high correlation was observed in Solomon Islands (n = 12, R2 = 0.876, P < .001). In line with results for the proportion of multiple clone infections, mean MOI (Figure 1C) was not correlated with prevalence in the entire data set (P = .354), at the catchment level (P = .766), in Madang 2010 (P = .147), or in Madang 2014 (P = .920), but it was strongly associated in Solomon Islands (P = .001).
Figure 1.
Correlation between the proportion of multiple clone infections and prevalence by quantitative polymerase chain reaction (qPCR) among 46 villages in the South Pacific for Plasmodium vivax (A) and Plasmodium falciparum (B), between geometric mean multiplicity of infection and prevalence by qPCR (C, D) and between prevalence by light microscopy and qPCR (E, F). Each dot represents 1 village. In (A, B, C, and D), values for each catchment are given as triangles.
For P. falciparum, ≥10 genotyped infections were available from 15 villages in the PNG surveys (prevalence: 5.2%–36.4%, 12–55 [median = 32] samples genotyped). No correlation between prevalence and the proportion of multiple clone infection was observed for the entire data set (P = .640) (Figure 1B), the Madang 2010 survey only (n = 10, P = .286), the Madang 2014 survey only (n = 5, P = .205), or at the catchment level (n = 6, P = .678). Mean MOI (Figure 1D) was not correlated with prevalence in the entire data set (P = .603), at the catchment level (P = .694), in Madang 2010 (P = .341), or in Madang 2014 (P = .233).
Across villages, prevalence by qPCR and microscopy was moderately correlated for P. vivax (R = 0.575, P < .001) (Figure 1E) and highly correlated for P. falciparum (R = 0.941, P < .001) (Figure 1F). The relatively lower correlation for P. vivax was mainly caused by the Madang 2014 survey, where only 13.7% (68/496) of infections were microscopy-positive. Excluding this survey, correlation was high (R = 0.8075, P < .001). There were no villages with P. falciparum detected by qPCR but not by microscopy, whereas this was the case in 6/46 villages for P. vivax.
DISCUSSION
Genotyping using length-polymorphic markers is frequently applied in epidemiological studies to determine MOI as a surrogate marker for transmission intensity. Typing infections diagnosed by qPCR in >8000 individuals in 46 villages in the South Pacific found no correlation between the proportion of multiple clone infections or mean MOI and prevalence by qPCR at the village level in 2 out of 3 surveys for P. vivax, and in both surveys for P. falciparum. Genotyping was thus not informative to identify villages with a large number of individuals infected, where control activities should be scaled up. In contrast, prevalence by light microscopy correlated well with prevalence by qPCR.
The P. vivax results from Solomon Islands were in stark contrast to the results of both species in PNG, with a high correlation between prevalence and the proportion of multiple clone infections and mean MOI. The difference to PNG is particularly intriguing as P. vivax prevalence was similar in all surveys. Genotyping a panel of 9 microsatellite markers has shown that in Solomon Islands, infections within households and villages are closely related [10]. This suggests that transmission occurring at small scales and higher transmission might result directly in more people carrying multiple clone infections. Comparable studies from PNG, that is, assessing relatedness at the household or village level, are not available. Country-wide studies that included samples from the Madang area found no structure of P. vivax populations [11], suggesting transmission occurring across wider scales.
A number of factors affect the lack of correlation between MOI and prevalence. Even at low transmission levels, a few households might be at high risk of infection, and several clones might be transmitted among them [12]. Thus, despite very low overall population prevalence, the proportion of multiple clone infections might be high. In the case of P. vivax, 80% of blood-stage infections in PNG are caused by relapses [13], and thus are not directly related to recent mosquito-to-human transmission levels. Even at low transmission levels, genetically diverse inocula might be frequent, which can result in heterologous relapse during ongoing blood-stage infection, and thus in multiple clone infection. Importantly, population prevalence might not directly reflect mosquito-to-human transmission levels. Previous work in PNG has shown that a 10-fold increase in the entomological inoculation rate (EIR) would only result in approximately a 10%–30% increase in prevalence [14]. Studies comparing EIR with multiplicity and prevalence will be required to assess whether spatial differences in EIR could explain differences in MOI among villages. Finally, the genotyping methods used, though widely applied in epidemiological studies, likely miss low-density clones and thus underestimate multiplicity [15].
It appears that a detailed understanding of EIR, transmission levels at even smaller scales, and possibly additional factors are needed to interpret patterns of multiplicity of infection at the population level. Given these complexities, assessing multiplicity is of limited benefit to inform malaria control programs on transmission foci and asymptomatic prevalence at the village level.
Acknowledgments
Disclaimer. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by the NIH International Centers of Excellence in Malaria Research (19 AI089686), the TransEpi consortium funded by the Bill & Melinda Gates Foundation, and National Health and Medical Research Council (NHMRC) Project Grant funding (1021544). This work was made possible through Victorian State Government Operational Infrastructure Support and the Australian Government NHMRC Independent Research Institute Infrastructure Support Scheme (IRIISS). C.K. was supported by a fellowship from the Swiss National Science Foundation (P2BSP3_151880). L.J.R. was supported by an National Health and Medical Research Council Early Career Fellowship (1016443). I.M. is supported by an NHMRC Senior Research Fellowship.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Bousema T, Stresman G, Baidjoe AY, et al. The impact of hotspot-targeted interventions on malaria transmission in rachuonyo South district in the Western Kenyan highlands: a cluster-randomized controlled trial. PLoS Med 2016; 13:e1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mogeni P, Williams TN, Omedo I, et al. Detecting malaria hotspots: a comparison of rapid diagnostic test, microscopy, and polymerase chain reaction. J Infect Dis 2017; 216:1091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tusting LS, Bousema T, Smith DL, Drakeley C. Measuring changes in Plasmodium falciparum transmission: precision, accuracy and costs of metrics. Adv Parasitol 2014; 84:151–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bei AK, Niang M, Deme AB, et al. Dramatic changes in malaria population genetic complexity in Dielmo and Ndiop, Senegal, revealed using genomic surveillance. J Infect Dis 2018; 217:622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mobegi VA, Loua KM, Ahouidi AD, et al. Population genetic structure of Plasmodium falciparum across a region of diverse endemicity in West Africa. Malar J 2012; 11:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nkhoma SC, Nair S, Al-Saai S, et al. Population genetic correlates of declining transmission in a human pathogen. Mol Ecol 2013; 22:273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waltmann A, Darcy AW, Harris I, et al. High rates of asymptomatic, sub-microscopic Plasmodium vivax infection and disappearing plasmodium falciparum malaria in an area of low transmission in solomon Islands. PLoS Negl Trop Dis 2015; 9:e0003758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koepfli C, Ome-Kaius M, Jally S, et al. Sustained malaria control over an 8-year period in Papua New Guinea: the challenge of low-density asymptomatic plasmodium infections. J Infect Dis 2017; 216:1434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fola AA, Harrison GLA, Hazairin MH, et al. Higher complexity of infection and genetic diversity of Plasmodium vivax than Plasmodium falciparum across all malaria transmission zones of Papua New Guinea. Am J Trop Med Hyg 2017; 96:630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waltmann A, Koepfli C, Natacha T, et al. Long-term sustained malaria control leads to inbreeding and fragmentation of Plasmodium vivax populations. bioRxiv. [Google Scholar]
- 11. Jennison C, Arnott A, Tessier N, et al. Plasmodium vivax populations are more genetically diverse and less structured than sympatric Plasmodium falciparum populations. PLoS Negl Trop Dis 2015; 9:e0003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karl S, White MT, Milne GJ, et al. Spatial effects on the multiplicity of Plasmodium falciparum infections. PLoS One 2016; 11:e0164054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robinson LJ, Wampfler R, Betuela I, et al. Strategies for understanding and reducing the Plasmodium vivax and Plasmodium ovale hypnozoite reservoir in Papua New Guinean children: a randomised placebo-controlled trial and mathematical model. PLoS Med 2015; 12:e1001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith T, Hii JL, Genton B, et al. Associations of peak shifts in age–prevalence for human malarias with bednet coverage. Trans R Soc Trop Med Hyg 2001; 95:1–6. [DOI] [PubMed] [Google Scholar]
- 15. Lerch A, Koepfli C, Hofmann NE, et al. Development of amplicon deep sequencing markers and data analysis pipeline for genotyping multi-clonal malaria infections. BMC Genomics 2017; 18:864. [DOI] [PMC free article] [PubMed] [Google Scholar]