Abstract
Ellis-van Creveld (EvC) syndrome is an autosomal recessive chondrodysplastic disorder. Affected patients present a wide spectrum of symptoms including short stature, postaxial polydactyly, and dental abnormalities. We previously disrupted Evc2, one of the causative genes for EvC syndrome, in mice using a neural crest-specific, Cre-mediated approach (i.e., P0-Cre, referred to as Evc2 P0 mutants). Despite the fact that P0-Cre predominantly targets the mid-facial region, we reported that many mid-facial defects identified in Evc2 global mutants are not present in Evc2 P0 mutants at postnatal day 8 (P8). In the current study, we used multiple Cre lines (P0-Cre and Wnt1-Cre, respectively), to specifically delete Evc2 in neural crest-derived tissues and compared the resulting mid-facial defects at multiple time points (P8 and P28, respectively). While both Cre lines indistinguishably targeted the mid-facial region, they differentially targeted the anterior portion of the skull base. By comprehensively analyzing the shapes of conditional mutant skulls, we detected differentially affected mid-facial defects in Evc2 P0 mutants and Evc2 Wnt1 mutants. Micro-CT analysis of the skull base further revealed that the Evc2 mutation leads to a differentially affected skull base, caused by premature closure of the intersphenoid synchondrosis (presphenoidal synchondrosis), which limited the elongation of the anterior skull base during the postnatal development of the skull. Given the importance of the skull base in mid-facial bone development, our results suggest that loss of function of Evc2 within the skull base secondarily leads to many aspects of the mid-facial defects developed by the EvC syndrome.
Keywords: EvC syndrome, Evc2, Limbin, ciliopathy, mid-facial defects, skull base, neural crest
Introduction
Ellis-van Creveld (EvC) syndrome is an autosomal recessive chondroectodermal dysplasia (McKusick et al., 1964). Affected individuals display a wide spectrum of symptoms including dwarfism, postaxial polydactyly, nail dysplasia, atrial septal or atrioventricular septal cardiovascular defects, and dental anomalies (McKusick et al., 1964; Baujat and Le Merrer, 2007). Genetic studies have allowed identification of two causative genes for EvC syndrome, EVC and EVC2, and homozygous mutations in either have been linked to two-thirds of EvC patients (Ruiz-Perez et al., 2003). Additionally, EvC syndrome has been characterized as a ciliopathy because the EVC and EVC2 proteins are intracellularly localized at the bottom of the primary cilium, where they form a heterotrimeric protein complex with SMO and transduce Hedgehog signaling (Dorn et al., 2012; Caparros-Martin et al., 2013). Our previous studies interestingly identified the bovine ortholog EVC2/LIMBIN as a causative gene for chondrodysplasia in Japanese brown cattle (Takeda et al., 2002). Recent studies also identify EVC2/LIMBIN mutations in dwarf Tyrolean Gray cattle (Murgiano et al., 2014). These two studies suggest an evolutionarily conserved function of EVC2 among different species.
Multiple studies have linked EvC syndrome with a wide range of craniofacial abnormalities including an enlarged skull, depressed nasal bridge, class II skeletal pattern with or without mandibular prognathism, and a skeletal open bite. Patients less frequently exhibit normocephaly with minimal facial defects and instances of class I skeletal patterns (Ellis and van Creveld, 1940; Eidelman et al., 1965; Prabhu et al., 1978; da Silva et al., 1980; Varela and Ramos, 1996). The variation of mid-facial defects reported in EvC patients prompted the generation of mouse models for EvC syndrome that facilitate characterization of affected mid-facial development and associated pathological mechanisms.
We and others have used both Evc and Evc2 mutant mice to characterize different aspects of abnormal development (Ruiz-Perez et al., 2007; Caparros-Martin et al., 2013; Zhang et al., 2015). Generating our own Evc2 mutant mice enabled us to characterize the pathological mechanisms leading to abnormal appendicular bone and hypomorphic enamel development (Zhang et al., 2015, 2016a,b). Our subsequent studies further demonstrated the expression of Evc2 in the mid-facial regions (Badri et al., 2016a) and mid-facial defects in Evc2 global mutant mice (Badri et al., 2016b). We most recently employed a conditional, Cre-mediated approach toward deleting Evc2 in a neural crest-specific manner. In a comparison between global and P0-Cre-mediated Evc2 mutants, we reported that at postnatal day 8 (P8): (1) Evc2 P0 mutants somewhat recapitulate craniofacial and tooth phenotypes exhibited by Evc2 global mutant mice and that (2) many mid-facial defects identified in the Evc2 global mutants are not present in the Evc2 P0 mutants (Kwon et al., 2018). This latter finding was surprising and warranted further study since P0-Cre predominantly targets the mid-facial region where Evc2 is, similarly, expressed.
To identify a pathological mechanism leading to the mid-facial defects in Evc2 mutant mice and EvC patients, we compared outcomes of Evc2 deletion within the neural crest-derived tissues of two similar, but not identical, neural crest-specific Cre lines: P0-Cre and Wnt1-Cre. Recombination efficiency and craniofacial deficiencies varied between mouse lines and facial region. Specifically, although both Cre lines showed indistinguishable recombination efficiency within the mid-facial bones, Wnt1-Cre-mediated excision of Evc2 resulted in a more robust mid-facial hypoplasia. Conversely, those two Cre lines showed differential recombination efficiencies within the anterior portion of the skull base. Analyses of these two mutant mice lines highlight the critical function of the skull base during postnatal mid-facial development and uncovers the pathological mechanisms whereby the shortened skull base, due to Evc2 loss of function within those bones, leads to characteristic EvC mid-facial defects.
Materials and Methods
Animal Model
Animals were maintained and used in compliance with the Institutional Animal Care and Use Committee (IACUC) of the University of Michigan in accordance with the National Institutes of Health Guidelines for Care and Use of Animals in research, and all experimental procedures were approved by the IACUC of the University of Michigan. Evc2 floxed mice used in this study were generated by our group and reported previously (Zhang et al., 2015). Neural crest-specific Evc2 mutant mice were generated by crossing Evc2 floxed mice either with Wnt1-Cre mice (Danielian et al., 1998) or with P0-Cre mice, C57BL/6J-Tg(P0-Cre)94Imeg (ID 148), which was provided by Dr. Kenichi Yamamura (Yamauchi et al., 1999). All mice were maintained in a mixed background of C57BL6/J and 129S6 and were crossed and maintained in our semi-closed mouse colony for at least 5 years.
Micro-CT (μCT)
Micro-CT scanning of fixed heads was performed at the University of Michigan using a Micro-CT Core (μCT100 Scanco Medical, Bassersdorf, Switzerland). Scan settings were as following: voxel size 12 μm, 55 kVp, 109 μA, 0.5 mm AL filter, and integration time 500 ms.
Cephalometric Analysis
Lateral radiographic films were taken according to previously published methods. The linear and angular cephalometric analyses followed the parameters outlined in a previous report (Engstrom et al., 1982) with certain modifications (Table 1). An additional landmark (Br) was placed on the bregma, the point of intersection between the sagittal and coronal sutures (see Supplementary Table S1). The distance from the bregma (Br) to the most posterior point of the skull (Po) was measured, denoted by the abbreviation Po-Br (see Supplementary Table S1). The angle ABrL/PoBrL was also measured between the Po-Br segment described above and the A-Br segment, which stretches the combined length of the nasal and frontal bones. This angle was used to emulate the concavity of the skull superficial to the cranial vault. Overall, 15 landmarks (Table 1) were placed in each sample and a total of 20 linear measurements and 15 angular measurements were taken (Table 1).
Table 1.
Description of landmarks and landmark-associated linear and angular measurements.
| Landmark | Description | ||
|---|---|---|---|
| A | The most anterior point on the nasal bone. | ||
| Bl | The intersection between the lingual surface of the lower incisors and the most anterior part of the lingual alveolar bone. | ||
| Br | The bregma, the point of intersection between the sagittal and coronal sutures. | ||
| Bu | A point on the premaxilla between jaw bone and the lingual surface of the upper lingual incisors. | ||
| E | The intersection between the frontal bone and the most superior-anterior point of the posterior limit of the ethmoid bone. | ||
| Id | The most inferior and anterior point on the alveolar process of the mandible. | ||
| Ii | The most prominent point between the incisal edges of the lower incisors. | ||
| Iu | The most prominent point between the incisal edges of the upper incisors. | ||
| Ml | The intersection between the mandibular alveolar bone and the mesial surface on the first molar. | ||
| Mn | A point in the deepest part of the antegonial notch curvature. | ||
| Mu | The intersection between the maxillary bone and the mesial surface of the upper first molar. | ||
| N | A point on the nasofrontal suture. | ||
| Po | The most posterior point on the cranial vault. | ||
| Pr | The most inferior and anterior point on the alveolar process of the premaxilla. | ||
| So | The intersection between the posterior border of the basisphenoid and the tympanic bulla. | ||
| Landmarks | Segment | Landmarks | Angle |
| A-N | Nasal bone length | PoEL/SoEL | Cranial vault to cranial base |
| A-Pr | Nasal bone height | ANL/SoEL | Nasal bone to cranial base |
| E-Mu | Viscerocranial height | ANL/PoEL | Nasal bone to cranial vault |
| E-Iu | Growth axis of upper face | ANL/PrNL | Nasal bone to premaxilla |
| E-Bu | Viscerocranial length (posterior to anterior incisors) | MuBuL/SoEL | Maxilla-premaxilla to cranial base |
| E-Pr | Viscerocranial length (anterior to anterior incisors) | MuBuL/PoEL | Maxilla-premaxilla to cranial vault |
| N-Pr | Relative position of pre-maxilla to cranium | PrEL/SoEL | Premaxilla to cranial base |
| Mu-Pr | Distance between molar and incisor in maxilla | BuEL/PoEL | Premaxilla to cranial vault |
| Mu-Bu | Palatal length | BuEL/SoEL | Upper incisors to cranial base |
| Mn-Id | Mandibular corpus length | IuEL/PoEL | Upper incisors to cranial vault |
| Ml-Bl | Mandibular lingual alveolar bone length | MuBuL/PrluL | Upper incisor inclination |
| Pr-Iu | Erupted upper incisor length | MlliL/ldLiL | Lower incisor inclination |
| Id-Ii | Erupted lower incisor length | ABrL/PoBrL | Angle of cranial vault |
| Ml-Ii | Distance between molar and incisor in mandible | PrEL/PoEL | Premaxilla to cranial vault |
| So-E | Length of anterior cranial base | ||
| Po-E | Neurocranial length | ||
| Po-Br | Length of posterior cranium | ||
| Po-Mu | The distance between the first molar to the most posterior point | ||
| Po-A | Total skull length | ||
Image Acquisition, Segmentation, and Surface Model Analysis
We performed shape comparisons of the nasal, frontal and parietal bones between controls and mutants (Evc2fl/fl; Wnt1-Cre). The surface model for each bone was generated based on the micro-CT data, using ITK-SNAP (open-source software developed by grants and contracts from the United States National Institutes of Health1). For shape comparisons, landmarks were placed on the individual bone surfaces by using “modules” developed in 3D Slicer (open-source software 2). 3D Slicer modules were then used to both superimpose the two bones based on the landmarks (CMF Registration) and to visualize shape differences (Shape Population Viewer). Model superimpositions for the mid-line region of the skulls were carried out using 3D Slicer with the following landmarks on the interparietal bones and basioccipital bones: right and left anterolateral tips of the interparietal bone, cross point between the median line and the line which connects left and right anterolateral tip of the interparietal bone, and posterior tip of the interparietal bone. Model superimpositions for nasal, frontal and parietal bones were carried out using 3D Slicer with landmarks on the right and left of anterolateral and posterior tips of each bone. Superimpositions were achieved by posterior registration of the two models. The length of the skull base and each indicated segment of the skull base were determined by assessing the linear distance between the most anterior midline point of and the most posterior point on the midline region of the skull base or the indicated segment of the skull base using a module program, Q3DC, in 3D Slicer.
Statistical Analysis
The Mann–Whitney U test was done by using SPSS21.0 to evaluate the linear and angular measurements between controls and mutants.
Results
Evc2 Mutation Within Neural Crest Cells Leads to Craniofacial Abnormalities
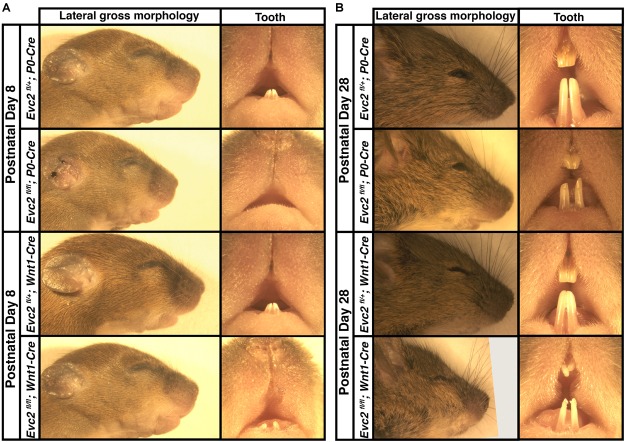
Our previous studies indicated that global Evc2 mutation led to retarded growth and significantly affected both body and head size (Zhang et al., 2015; Badri et al., 2016b). To better characterize the function of Evc2 in craniofacial development, we generated mutant mice with Evc2 deleted in the neural crest-derived cells. Both P0-Cre and Wnt1-Cre mediate nearly identical recombination efficiencies within the mid-facial region (Danielian et al., 1998; Yamauchi et al., 1999) (our own comparisons, data not shown). However, Evc2 deletion mediated by the P0-Cre (Evc2fl/fl; P0-Cre, abbreviated as Evc2 P0 mutants) leads to an overall shortened head and delayed mandibular incisor eruption at postnatal day 8 (P8) (Figure 1A). In Evc2 P0 mutants at P28, we observed less pronounced reductions in head and incisor lengths (Figure 1B). Evc2 deletions mediated by Wnt1-Cre (Evc2fl/fl; Wnt1-Cre, abbreviated as Evc2 Wnt1 mutants) did not affect head length at P8 but did lead to shortened head and hypomorphic incisor dimensions at P28 (Figures 1A,B). Unlike Evc2 global mutant mice, there was no apparent neonatal or postnatal death observed in either Evc2 P0 or Evc2 Wnt1 mutant groups at the above time points. Furthermore, in both types of Evc2 cKO, at P8 and P28, we observed insignificant gender-specific differences (Supplementary Table S1), particularly in the parameters characterizing mid-facial defects. Lack of gender-specific observations is consistent with previous studies and we, therefore, did not match genders in our comparative analyses (Vora et al., 2015; Wei et al., 2017). Therefore, we combined both genders for the rest of our comparisons.
FIGURE 1.
Gross morphologies of heads from Evc2 P0 mutants and Evc2 Wnt1 mutants at P8 (A) and P28 (B).
Evc2 P0 Mutants Showed More Aspects of Mid-Facial Defects Than Evc2 Wnt1 Mutants at P8
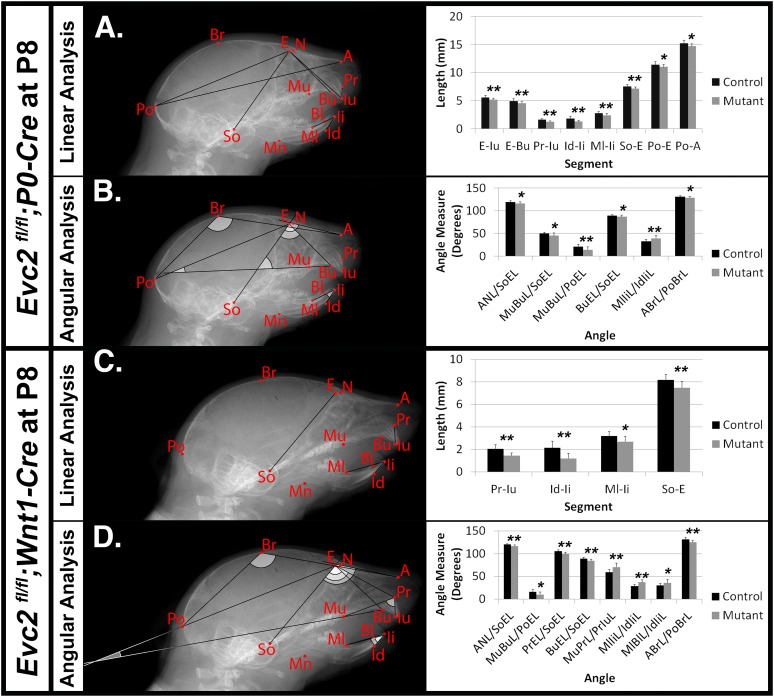
To characterize the abnormal craniofacial development observed in both Evc2 P0 mutants and Evc2 Wnt1 mutants, we examined the linear and angular dimensions from lateral cephalometric radiographs. In Evc2 P0 mutants at P8, the lateral cephalometric analysis indicated abnormalities in the incisors, skull base, and mid-facial regions. We specifically found decreases in growth axes of the upper face (E-Iu), viscerocranial length (E-Bu), erupted upper incisor length (Pr-Iu), erupted lower incisor length (Id-Ii), distance between the first molar and incisor in mandible (Ml-Ii), length of anterior cranial base (So-E), neurocranial length (Po-E), the distance between the first molar to the most posterior point (Po-Mu), and total skull length (Po-A), respectively (Figure 2A and Table 2). These phenotypic observations recapitulate many of the mid-facial defects of the Evc2 global mutants that we previously reported (Badri et al., 2016a). Additionally, we found a shallow mid-facial region in the Evc2 P0 mutants, as evidenced by decreased angle measures in nasal bone to cranial base (ANL/SoEL), maxilla-premaxilla to cranial base (MuBuL/SoEL), maxilla-premaxilla to cranial vault (MuBuL/PoEL), upper incisors to cranial base (BuEL/SoEL), and angle of cranial vault (ABrL/PoBrL), respectively (Figure 2B and Table 2).
FIGURE 2.
Differential mid-facial defects between Evc2 P0 mutants and Evc2 Wnt1 mutants at P8. Lateral X-ray cephalogram indicates the linear (A) and angular (B) measurements with significant differences between controls and Evc2 P0 mutants. Lateral X-ray cephalogram indicates the linear (C) and angular (D) measurements with significant differences between controls and Evc2 Wnt1 mutants. Graphs indicate the linear and angular values of the measurement with significant differences (N = 7; ∗p < 0.05; and ∗∗p < 0.01).
Table 2.
Summary of phenotypic spectrums of Evc2 global mutants, P0 mutants and Wnt1 mutants.
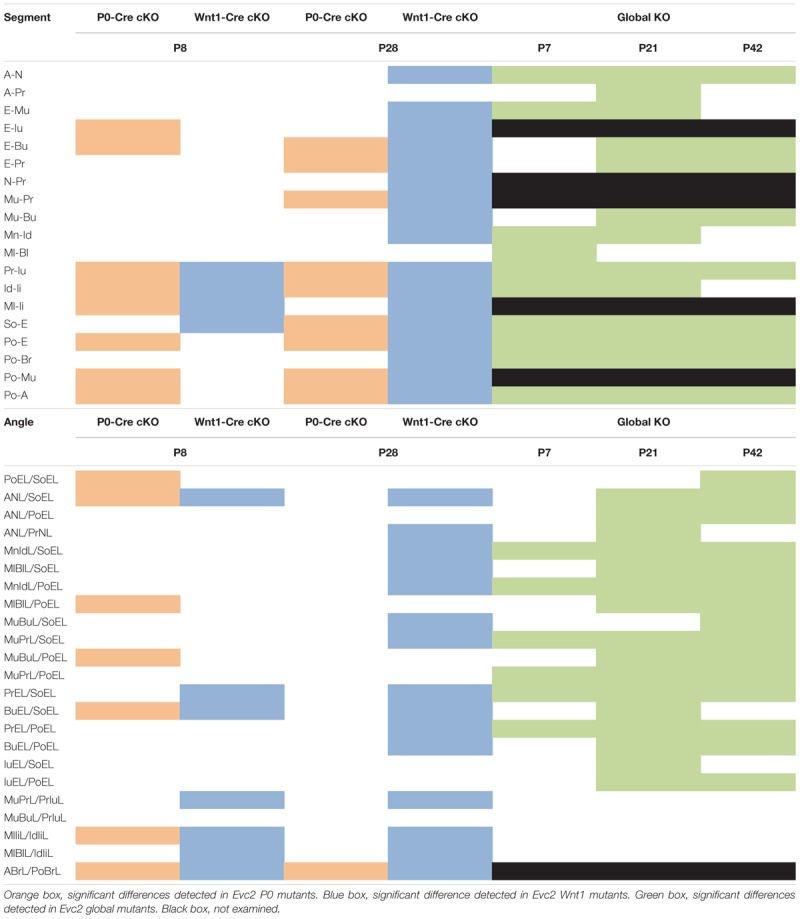 |
Similar analyses of the Evc2 Wnt1 mutants revealed abnormalities limited to the incisors and the skull base. We detected decreases in erupted upper incisor (Pr-Iu), erupted lower incisor (Id-Ii), and distance between the first molar and incisor in the mandible (Ml-Ii), respectively (Figure 2C and Table 2). Similar to Evc2 P0 mutants, these phenotypic observations recapitulate some characteristics of Evc2 global mutants (Badri et al., 2016a,b). Additionally, we also found a shortened skull base in the Evc2 Wnt1 mutants, as evidenced by decreased length in the anterior skull base (So-E) (Figure 2C). Although most of the linear measurements showed no significant differences between control and Evc2 Wnt1 mutants, we observed a slanted mid-facial region in the Evc2 Wnt1 mutants, supported by the following decreased angle measures: the nasal bone to the skull base (ANL/SoEL), the maxilla-premaxilla to the skull base (MuBuL/SoEL), the premaxilla to the cranial base (PrEL/SoEL), the premaxilla to the cranial vault (BuEL/PoEL), and the angle of the cranial vault (ABrL/PoBrL) (Figure 2D). In summary (Table 2), at P8 both Evc2 P0 mutants and Evc2 Wnt1 mutants showed fewer aspects of mid-facial defects compared to Evc2 global mutants. Of the two types of conditional mutants, Evc2 P0 mutants showed more severe and widespread abnormalities compared to Evc2 Wnt1 mutants at P8 (Table 2).
Evc2 Wnt1 Mutants Showed More Aspects of Mid-Facial Defects Than Evc2 P0 Mutants at P28
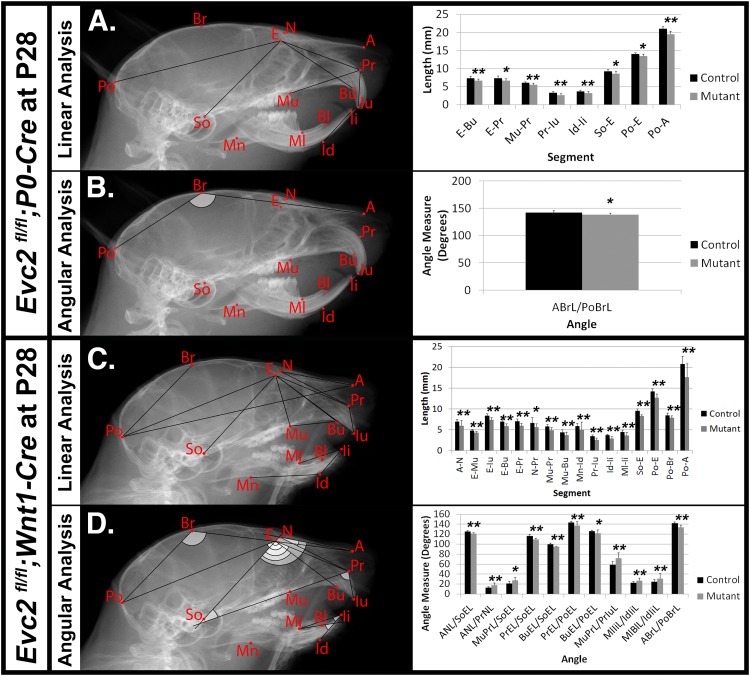
Mouse cranial growth rate of peaks and reaches 80% of its adult length within the first month (Vora et al., 2015; Wei et al., 2017). We therefore performed similar lateral cephalometric analyses at P28 but, unlike analyses at P8, observed more widespread craniofacial deficiencies in Evc2 Wnt1 mutants compared to Evc2 P0 mutants. In Evc2 P0 mutants we found significant decreases in: viscerocranial length (E-Bu and E-Pr), distance between molar and incisor in maxilla (Mu-Pr), erupted upper incisor length (Pr-Iu), erupted lower incisor length (Id-Ii), length of anterior skull base (So-E), neurocranial length (Po-E), distance between the first molar to the most posterior point (Po-Mu), and total skull length (Po-A) (Figure 3A). The only significant angle difference was a decreased angle of cranial vault (ABrL/PoBrL) in the Evc2 P0 mutants (Figure 3B).
FIGURE 3.
Differential mid-facial defects between Evc2 P0 mutants and Evc2 Wnt1 mutants at P28. Lateral X-ray cephalogram indicates the linear (A) and angular (B) measurements with significant differences between controls and Evc2 P0 mutants. Lateral X-ray cephalogram indicates the linear (C) and angular (D) measurements with significant differences between controls and Evc2 Wnt1 mutants. Graphs indicate the linear and angular values of the measurement with significant differences (N = 7; ∗p < 0.05; and ∗∗p < 0.01).
In contrast to the Evc2 P0 mutants, Evc2 Wnt1 mutants at P28 displayed a more severe and widespread decreases in their craniofacial linear measurements. We found significant decreases in nasal bone length (A-N), nasal bone height (E-Mu), viscerocranial length (E-Bu), and (E-Pr), relative position of pre-maxilla to cranium (N-Pr), distance between molar and incisor in maxilla (Mu-Pr), palatal length (Mu-Bu), length of anterior cranial base (So-E), neurocranial length (Po-E), length of posterior cranium (Po-Br), and total skull length (Po-A). Similar to Evc2 Wnt1 mutants at P8, we also observed shortened erupted upper (Pr-Iu) and lower (Id-Ii) incisors, and decreased distance between molar and incisor in mandible (Ml-Ii) (Figure 3C). Consistent with the slanted mid-facial region in the gross morphologic observations, we also detected decreased angle measures of nasal bone to cranial base (ANL/SoEL), premaxilla to cranial base (PrEL/SoEL), premaxilla to cranial vault (BuEL/PoEL), and the angle of the cranial vault (ABrL/PoBrL), respectively (Figure 3D). Overall, Evc2 Wnt1 mutants showed severely affected mid-facial bones and a more slanted mid-facial region at P28 when compared to both control and Evc2 P0 mutant counterparts.
In Table 2, a series of linear and angular defects detected in Evc2 P0 mutants and Evc2 Wnt1 mutants (present study) are compared with those found in Evc2 global mutants (Badri et al., 2016a,b). At P7/P8, comparisons in linear measurements revealed that all tooth-associated abnormalities detected in Evc2 P0 mutants and Evc2 Wnt1 mutants were also detected in the Evc2 global mutants. Additionally, nasal bone length (A-N), viscerocranial height (E-Mu), and mandibular lingual alveolar bone length (Ml-Bl) were significantly shorter in the Evc2 global mutants than in controls, whereas these characteristics were normal in both Evc2 Wnt1 mutants and Evc2 P0 mutants. In the angular measurements, we found that lower incisor inclination (MlIiL/IdIiL) and upper incisor inclination (MuPrL/PrIuL) were increased in the Evc2 P0 mutants and Evc2 Wnt1 mutants but not in the Evc2 global mutants. At P21/P28, all linear measurement differences detected in Evc2 Wnt1 mutants were detected in Evc2 global mutants. Of these, decreased palatal length (Mu-Bu) and length of posterior cranium (Po-Br) were not detected in the Evc2 P0 mutants. These results reflect subtle phenotypic dissimilarities between mutants at P28 that are not apparent at P8. In the P28 angular measurements, we found that defective nasal bone to cranial vault (ANL/PoEL), maxilla-premaxilla to cranial base (MuBuL/PoEL), and upper incisor to cranial vault in Evc2 P0 mutants, but not in the Evc2 Wnt1 mutants, whereas defective upper incisor inclination (MuPrL/PrIuL) and lower incisor inclination (MlIiL/IdIiL) was observed in the Evc2 Wnt1 mutants but not in the Evc2 global mutants. Except for the decreased angle of cranial vault (ABrL/PoBrL), there were no other angular differences detected in the Evc2 P0 mutants.
Loss of Evc2 Expression in Neural Crest-Derived Cells Severely Affects Head Shape and Mid-Facial Bone Shapes
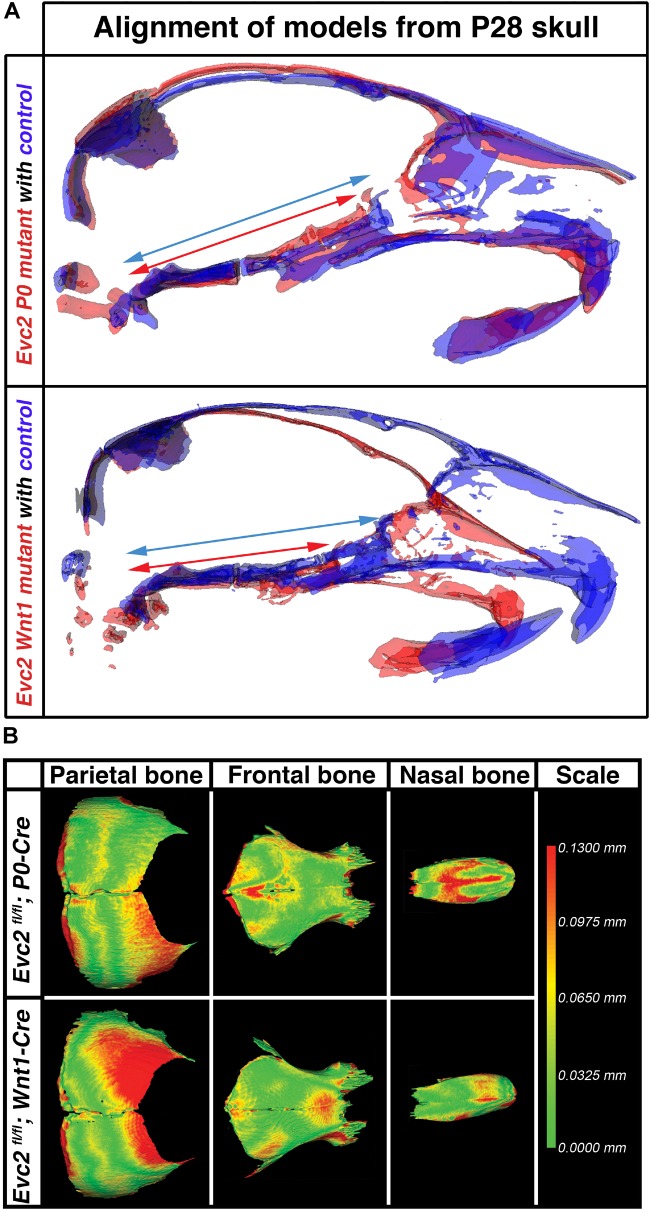
Particularly at P28, Evc2 P0 mutants showed very limited angular defects. To better appreciate the impacts of Evc2 mutation on the overall skull shape at P28, we: (1) generated 3D surface models from micro-CT data for the midline portion of the controls, Evc2 P0 mutants, and Evc2 Wnt1 mutants at P28 and (2) registered the skulls at the most posterior region (Figure 4A). Registration of each pair of skulls revealed nearly no shape differences in the interparietal and basioccipital bones (i.e., tissues negative for neither P0-Cre nor Wnt1-Cre expression). When we individually superimposed nasal, frontal, and parietal, respectively, bones, respectively, using Evc2 P0 mutants and Evc2 Wnt1 mutants with corresponding controls, we did not see noticeable size differences (Figure 4B). This suggests that there was no overt overall growth retardation since general growth retardation would affect all bones. Morphometric comparison, however, revealed mutant-specific structural differences. Evc2 Wnt1 mutants had a more shortened and downward-angled facial region than Evc2 P0 mutants had (Figure 4A). Since the skull base connects the facial region with the posterior part of the skull, the extent of skull base linear differences may directly result in angular measurement differences in Evc2 Wnt1 and Evc2 P0 mutants, respectively. To visualize the potential shape or scaling differences within each individual bone, we digitally extracted nasal, frontal, and parietal bones separately and superimposed control surface models with corresponding mutant samples (Figure 4B). Superimpositions of individual bones revealed shape differences in all nasal, frontal, and parietal bones (Figure 4B). Most strikingly, in the parietal bones (non-neural crest derived), we detected deviations in the anterior regions, in which the mutant surfaces “stuck out” of the control surfaces (red color in Figure 4B). Evc2 Wnt1 mutants have more severely affected parietal bones than Evc2 P0 mutants. Three other mutant skulls from each mutant group showed similar patterns when compared with littermate controls (Supplementary Figure S1). When we compared four littermate controls within the Evc2 P0 mutant group or four littermate controls within the Evc2 Wnt1 mutant group using the same standard, almost all areas showed green color indicating that surface structures of those bones are highly consistent among controls (Supplementary Figure S1, one pair of comparison for each group is shown). The defective skull bones in Evc2 mutants likely reflect the shortened skull base and resultant shortened and downward-angled mid-facial region. Therefore, the more severely affected parietal bones in the Evc2 Wnt1 mutants (compared to Evc2 P0 mutants) are likely due to the more severely affected skull base in these mutants, as previously discussed (Table 2).
FIGURE 4.
Evc2 cKO mutants demonstrate skull shape and mid-facial shape differences. (A) Mid-sagittal planes were generated based on the micro-CT scans of the controls (Blue), Evc2 P0 mutants (red) and Evc2 Wnt1 mutants (red). Models of skulls from mutants and corresponding controls were then superimposed at the occipital bones of the skull. Blue and red arrows are spanning the entire regions of the skull bases in controls and mutants, respectively. (B) Nasal, frontal, and parietal bones from P28 Evc2 P0 mutants and Evc2 Wnt1 mutants were superimposed with corresponding skull bones from P28 controls. Pictures shown represent comparisons within indicated mutants with corresponding controls. Color indicates the distances the mutants bone surface protruding out of the control surface at the indicated region.
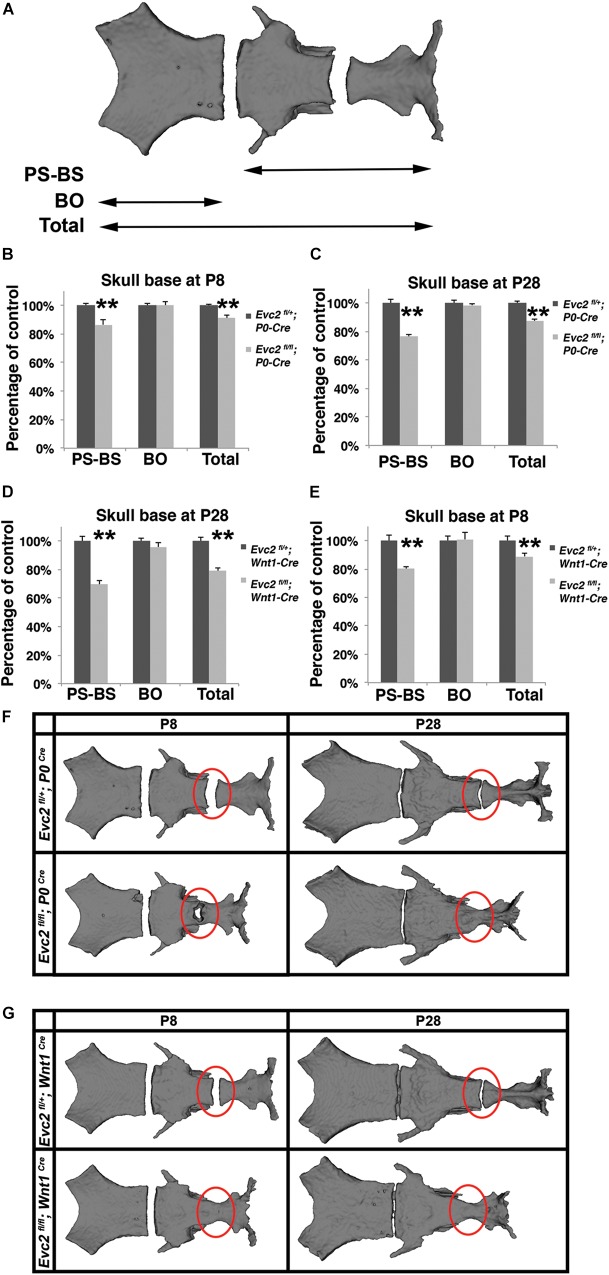
The Differential Early Fusion of the Presphenoidal Synchondrosis Leads to the Differentially Shortened Skull Base in the Evc2 P0 and Evc2 Wnt1 Mutants
The skull base is a unique structure that connects multiple bones in the skull and has a significant influence on facial growth, particularly in the mid-facial region (Nie, 2005). Our studies demonstrated that Evc2 is expressed in both intersphenoidal synchondrosis (ISS) and the spheno-occipital synchondrosis (SOS) (Supplementary Figure S2). Quantification of the skull base length (described in Figure 5A) revealed that, compared to controls, skull bases from Evc2 Wnt1 mutants are 20% shorter while skull bases from Evc2 P0 mutants are only 10% shorter at P28 (Figures 5B–E, N = 5, p < 0.01). Elongation of the skull base is supported by the endochondral ossification within the ISS and the SOS. To investigate whether the abnormal ISS or SOS is the cause of the affected skull base elongation in either Evc2 P0 or Evc2 Wnt1 mutants, we generated surface models from the skull bases of Evc2 P0 and Evc2 Wnt1 mutants, respectively. At P28, we observed premature fusion of ISS in both Evc2 P0 and Evc2 Wnt1 mutants (Figures 5B,C). However, at P8, we observed premature fusion of ISS in Evc2 Wnt1 mutants and partial fusion of ISS in the Evc2 P0 mutants (Figures 5B,C). These results suggest that the premature fusion of the ISS in Evc2 P0 mutants happens between P8 and P28, while the premature fusion of ISS in Evc2 Wnt1 happens before P8 (Figure 5C).
FIGURE 5.
Evc2 mutants have defective skull bases. (A) Diagram of measurements in labels (B–E). Quantification of skull base length at P8 (B,D) and P28 (C,E). The length of the skull base was determined by assessing the linear distance shown in label (A) (N = 5; ∗∗p < 0.01 comparing to controls). Surface models of skull bases were generated based on micro-CT scans of P28 (F) and P8 samples (G). Red circles indicating the ISS or fused ISS.
The anterior regions of the skull base are derived from neural crest cells and are the active sites for postnatal skull base elongation (Nie, 2005). To examine if differential skull base lengths in Evc2 P0 mutants and Evc2 Wnt1 mutants result from differential Cre recombination efficiency in the anterior part of the skull base, we examined the Cre recombined cells in the anterior region of the skull base. At E18.5, while P0-Cre leads to sporadic recombination within the chondrocytes in the anterior part of the skull base, Wnt1-Cre leads to nearly 100% recombination in the chondrocytes in the same region (Figure 6). These results suggest that (1) Evc2 function within the ISS is critical for maintaining the ISS and supporting the elongation of skull base; and that (2) differential Evc2 deletion efficiency mediated by P0-Cre and Wnt1-Cre leads to differential impacts upon skull base elongation, which secondarily affects differential outcomes in mid-facial abnormalities between P0-Cre and Wnt1-Cre mutants.
FIGURE 6.
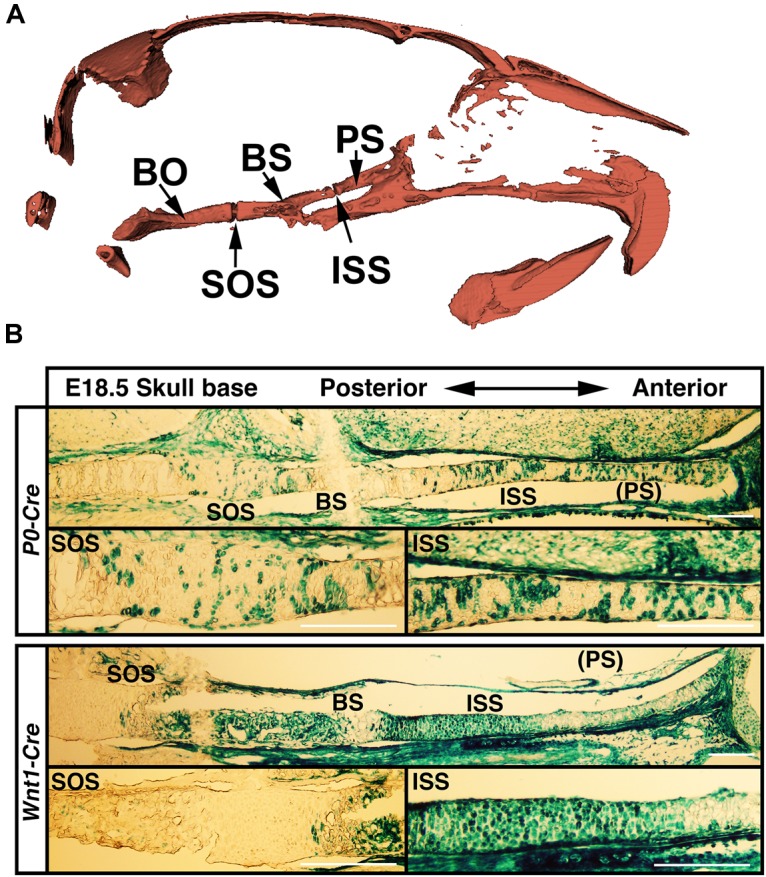
(A) Diagram to skull base structure. (B) Differential Cre-dependent recombination in the skull bases of P0 and Wnt1 Cre lines. Indicated Cre mice were crossed with Rosa26-LacZ Cre reporter mice, followed by beta-galactosidase activity staining to visualize the Cre recombinant cells in the skull base. PS, presphenoid bone; ISS, intersphenoidal synchondrosis; BS, basisphenoid bone; SOS, spheno-occipital synchondrosis. Scale bar = 100 μm.
Discussion
Craniofacial abnormalities have been reported in EvC patients (Ellis and van Creveld, 1940; McKusick et al., 1964; Eidelman et al., 1965; Prabhu et al., 1978). Despite our recent studies describing mouse models that recapitulate the craniofacial abnormalities of EvC patients (Badri et al., 2016b), the underlying pathophysiological mechanisms of these abnormalities remain largely unknown. In this study, we deleted Evc2 in the neural crest derived cells using P0-Cre and Wnt1-Cre. While both of these Cre lines equally target the mid-facial region, they differentially target the anterior region of the skull base. Comprehensive analysis of these two types of Evc2 mutant lines allows us to delineate the craniofacial abnormalities due to Evc2 loss of function in the neural crest derived tissues and the secondary abnormalities due to the affected skull base. The results here are potentially applicable to understand postnatal mid-facial development in other types of ciliopathies.
Compared to their global mutant counterparts, we believe that two types of Evc2 cKO mice, Evc2 P0 mutants and Evc2 Wnt1 mutants, are great tools in helping to elucidate the pathological mechanism leading to the mid-facial defects. Our phenotypic analysis at P8 and P28 indicated that mid-facial defects were recapitulated in Evc2 Wnt1 mutants, but to a less extent in Evc2 P0 mutants. The Evc2 P0 mutants show a smaller head (Figure 1A, 2nd row) and differences in some of the parameters (Figures 2A,B) at P8, which are consistent with our previous report (Kwon et al., 2018). However, many of the differences are compensated by P28 (Figure 1B, second row and Figures 3A,B). In contrast, the Evc2 Wnt1 mutants show comparable head size at P8 but become smaller at P28 (Figures 1A,B, fourth row), and more parameters show differences at P28 than P8 (Figures 2C,D, 3C,D). Those facts strongly suggest that tissues that are specifically positive for Wnt1-Cre mediated recombination play a critical role for post-natal mid-facial development. Considering the impacts of shortened skull base on the mid-facial region as shown in Figure 4A and that loss of Evc2 leads to early fusion of ISS in Evc2 Wnt1 mutants, we believe that the differential Evc2 deletion efficiency mediated by P0-Cre and Wnt1-Cre at the anterior region of the skull base is a major reason leading to the differential mid-facial defects in these two types of Evc2 cKO mice.
Differential phenotypic differences between Evc2 global mutants, Evc2 P0 mutants and Evc2 Wnt1 mutants suggest that different pathological mechanisms lead to various craniofacial defects due to Evc2 mutations. For example, viscerocranial length (E-Bu and E-Pr), erupted upper incisor length (Pr-Iu), erupted lower incisor length (Id-Ii), cranial base length (So-E) are due to the Evc2 loss of function within the neural crest cells. In contrast, the nasal bone to cranial base (ANL/SoEL), nasal bone to premaxilla (ANL/PrNL), premaxilla to cranial base (PrEL/SoEL), upper incisors to cranial base (BuEL/SoEL), premaxilla to cranial vault (PrEL/PoEL), premaxilla to cranial vault (BuEL/PoEL), upper incisor inclination (MuPrL/PrIuL), lower incisor inclination and (MlIiL/IdIiL), are due to the unique spatial activity of the Wnt1-Cre driver. Since the skull base connects the posterior and mid-facial regions, the shortened skull base is a well-known cause of mid-facial defects (Nie, 2005; Vora et al., 2015). Therefore, our data suggest that the differential Cre recombination efficiency mediated by the Wnt1-Cre and P0-Cre lineage cells within the anterior part of the skull base leads to the differential impacts on the pre-mature fusion of the ISS, which connects the presphenoid bone and the basisphenoid bone, and thus the length of the skull base. Those structural changes secondarily lead to the shortened and downward angled facial defects observed more prominently in the Evc2 Wnt1 mutants.
Similar to the appendicular bones, the skull base develops through endochondral ossification, wherein the cartilage primordium forms first, followed by mineral deposition. The proliferation and maturation of chondrocytes at the synchondrosis in the skull base is quite important for skull base elongation (Nie, 2005). In this study, we found loss of function of Evc2 results in premature closure of the ISS before P8 in Evc2 global mutants and in Evc2 Wnt1 mutants. We previously reported that loss of function of Evc2 results in severe chondrodysplastic dwarfism; however, we never observed premature closure of appendicular growth plates through adult stages (Zhang et al., 2016a). Those data suggest that a function of Evc2 to maintain cartilage is different between appendicular growth plates and synchondroses. A potential mechanism leading to the premature fusion of the ISS is a subject under active investigation.
In this study, we used both P0-Cre and Wnt1-Cre to specifically delete Evc2 in the neural crest derived cells. Despite largely overlapping targeted regions within the facial regions of P0-Cre and Wnt1-Cre (Danielian et al., 1998; Yamauchi et al., 1999) (our own comparisons, data not shown), small differences have been noticed between those Cre lines by multiple studies (Wang et al., 2011; Liu et al., 2012; Suzuki et al., 2013; Chen et al., 2017). Particularly, our recent studies demonstrated that both P0-Cre and Wnt1-Cre differentially target neural crest cells emerging at the anterior, middle and posterior portions of the head during embryogenesis (Chen et al., 2017). Previously, we reported a neural crest-specific disruption of TGF-beta activating kinase 1 (Tak1) using Wnt1-Cre and P0-Cre, respectively (Yumoto et al., 2013; Liu et al., 2018). While Tak1 P0 mutants do not show overt craniofacial phenotypes at birth (Liu et al., 2018), Tak1 Wnt1 mutants develop cleft palate secondarily to the micrognathia (Yumoto et al., 2013). This is another example that Wnt1-Cre and P0-Cre result in distinctive phenotypes. In the current study, we identified that P0-Cre and Wnt1-Cre differentially target the anterior regions of the skull base, and we took advantage of those differences to identify the pathophysiological mechanism leading to the mid-facial defects in Evc2 mutant mice. However, there is a formal possibility that the differential Cre activity within the unknown types of cells or tissues contributing to the differential mid-facial defects in Evc2 P0 mutants and Wnt1 mutants. We will further investigate an involvement of the premature fusion of the ISS to the mid-facial defects in our mutant mouse models.
Wnt1-Cre is known to target the brain (Danielian et al., 1998), while P0-Cre is not expressed in the central nervous system. However, it is unlikely that the differential mid-facial defects observed in the Evc2 Wnt1 mutants and Evc2 P0 mutants are secondary effects of the differential Cre targeting within the brain. Firstly, the mid-facial region is not in direct contact with the brain and thus brain development should not have an impact on the development of the mid-facial region. Secondly, despite the fact that Evc2 is a positive regulator of Hedgehog signaling, there are no brain developmental defects observed in patients or mice. Recent studies also suggest that the transgenic construct in Wnt1-Cre mouse line transiently elevates WNT signaling during craniofacial development (Lewis et al., 2013). It is unlikely that this is the reason leading to differential mid-facial defects observed in the Evc2 Wnt1 mutants and Evc2 P0 mutants. Our analysis demonstrated that no overt head shape differences between Evc2fl/+ and Evc2fl/+; Wnt1-Cre, suggesting the transiently elevated WNT signaling due to Wnt1-Cre did not lead to craniofacial abnormalities.
By specifically deleting Evc2 in neural crest-derived tissues, we were able to generate a mouse model of human EvC that recapitulates the mid-facial defects associated with EvC syndrome. Multiple analyses at different developmental stages confirm the critical role of the skull base during face elongation and development within the first month and demonstrate that the mid-facial defects in EvC syndrome are largely due to the premature fusion of the ISS in the skull base. Examination and targeting the shortened skull is therefore a suggested therapeutic option for the mid-facial defect in EvC patients.
Author Contributions
AK, KL, YMi, and HZ conceived this study. AK, KL, and HZ performed the analysis. AK, KL, MY, ACOR, YMo, LC, YMi, and HZ contributed to data interpretation and critical revision of the manuscript. AK, KL, YMi, and HZ drafted the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Dr. Taylor N. Snider and Mr. Nikko Simmons for their initial contribution to the 3D-image analyses and Drs. Hera Kim-Berman and Xiaoxi Wei for constructive comments of this manuscript.
Funding. This study was supported by the National Institutes of Health (R01DE019527 to YMo, R01DE020843 to YMi, and R03 DE027456 to HZ). Research reported in this publication was supported in part by a core grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (P30 AR069620 to Karl Jepsen, PI; David H. Kohn, Core Director). The micro-CT core at the School of Dentistry, University of Michigan was funded in part by NIH/NCRR S10RR026475-0.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01484/full#supplementary-material
Superimposition of each skull bones within the Evc2 P0 group. Comparisons of two controls (Evc2 P0 C vs. C) and three additional pairs of control and mutant (Evc2 P0 C vs. m) are shown. (B) Superimposition of each skull bones within the Evc2 Wnt1 group. Comparisons of two controls (Evc2 Wnt1 C vs. C) and three additional pairs of control and mutant (Evc2 Wnt1 C vs. m) are shown.
Evc2 expression in synchondrosis in the skull base. Beta-galactosidase activity from the lacZ knock-in allele of Evc2 locus was used as a surrogate of the expression of Evc2. The skull bases from Evc2lacZ/+ at P10 were stained with X-gal followed by eosin counter staining. Robust staining (blue) was detected in the ISS and the SOS. Evc2+/+ littermates were used as negative controls.
Lateral X-ray cephalogram indicates the linear and angular measurements within P0-Cre group at P8 and P28 and within Wnt1-Cre group at P8 and P28. Within each group, the potential gender specific differences were evaluated by comparing control male with control females; and mutant males and mutant females. Occasional significant differences observed were marked in yellow color.
References
- Badri M. K., Zhang H., Ohyama Y., Venkitapathi S., Alamoudi A., Kamiya N., et al. (2016a). Expression of Evc2 in craniofacial tissues and craniofacial bone defects in Evc2 knockout mouse. Arch. Oral Biol. 68 142–152. 10.1016/j.archoralbio.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badri M. K., Zhang H. H., Ohyama Y., Venkitapathi S., Kamiya N., Takeda H., et al. (2016b). Ellis van creveld2 is required for postnatal craniofacial bone development. Anat. Rec. 299 1110–1120. 10.1002/ar.23353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baujat G., Le Merrer M. (2007). Ellis-van creveld syndrome. Orphanet J. Rare Dis. 2:27. 10.1186/1750-1172-2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparros-Martin J. A., Valencia M., Reytor E., Pacheco M., Fernandez M., Perez-Aytes A., et al. (2013). The ciliary Evc/Evc2 complex interacts with Smo and controls Hedgehog pathway activity in chondrocytes by regulating Sufu/Gli3 dissociation and Gli3 trafficking in primary cilia. Hum. Mol. Genet. 22 124–139. 10.1093/hmg/dds409 [DOI] [PubMed] [Google Scholar]
- Chen G. Q., Ishan M., Yang J. W., Kishigami S., Fukuda T., Scott G., et al. (2017). Specific and spatial labeling of P0-Cre versus Wnt1-Cre in cranial neural crest in early mouse embryos. Genesis 55:e23034. 10.1002/dvg.23034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva E. O., Janovitz D., de Albuquerque S. C. (1980). Ellis-van Creveld syndrome: report of 15 cases in an inbred kindred. J. Med. Genet. 17 349–356. 10.1136/jmg.17.5.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8 1323–1326. 10.1016/S0960-9822(07)00562-3 [DOI] [PubMed] [Google Scholar]
- Dorn K. V., Hughes C. E., Rohatgi R. (2012). A Smoothened-Evc2 complex transduces the hedgehog signal at primary cilia. Dev. Cell 23 823–835. 10.1016/j.devcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidelman E., Odont D., Rosenzweig K. A. (1965). Ellis-van creveld syndrome; report of a case. Oral Surg. Oral Med. Oral Pathol. 20 174–179. 10.1016/0030-4220(65)90185-4 [DOI] [PubMed] [Google Scholar]
- Ellis R. W., van Creveld S. (1940). A syndrome characterized by ectodermal dysplasia, polydactyly, chondro-dysplasia and congenital morbus cordis: report of three cases. Arch. Dis. Child. 15 65–84. 10.1136/adc.15.82.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom C., Linde A., Thilander B. (1982). Craniofacial morphology and growth in the rat. Cephalometric analysis of the effects of a low calcium and vitamin D-deficient diet. J. Anat. 134(Pt 2), 299–314. [PMC free article] [PubMed] [Google Scholar]
- Kwon E. K., Louie K., Kulkarni A., Yatabe M., Ruellas A. C. O., Snider T. N., et al. (2018). The role of Ellis-Van creveld 2(EVC2) in mice during cranial bone development. Anat. Rec. 301 46–55. 10.1002/ar.23692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A. E., Vasudevan H. N., O’Neill A. K., Soriano P., Bush J. O. (2013). The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev. Biol. 379 229–234. 10.1016/j.ydbio.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. X., Komatsu Y., Mishina Y., Mistretta C. M. (2012). Neural crest contribution to lingual mesenchyme, epithelium and developing taste papillae and taste buds. Dev. Biol. 368 294–303. 10.1016/j.ydbio.2012.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Hayano S., Pan H., Inagaki M., Ninomiya-Tsuji J., Sun H., et al. (2018). Compound mutations in Bmpr1a and Tak1 synergize facial deformities via increased cell death. Genesis 56:e23093. 10.1002/dvg.23093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick V. A., Egeland J. A., Eldridge R., Krusen D. E. (1964). Dwarfism in the Amish I. The Ellis-Van creveld syndrome. Bull. Johns Hopkins Hosp. 115 306–336. [PubMed] [Google Scholar]
- Murgiano L., Jagannathan V., Benazzi C., Bolcato M., Brunetti B., Muscatello L. V., et al. (2014). Deletion in the EVC2 gene causes chondrodysplastic dwarfism in Tyrolean Grey cattle. PLoS One 9:e94861. 10.1371/journal.pone.0094861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X. (2005). Cranial base in craniofacial development: developmental features, influence on facial growth, anomaly, and molecular basis. Acta Odontol. Scand. 63 127–135. 10.1080/00016350510019847 [DOI] [PubMed] [Google Scholar]
- Prabhu S. R., Daftary D. K., Dholakia H. M. (1978). Chondroectodermal dysplasia (Ellis-van Creveld syndrome): report of two cases. J. Oral Surg. 36 631–637. [PubMed] [Google Scholar]
- Ruiz-Perez V. L., Blair H. J., Rodriguez-Andres M. E., Blanco M. J., Wilson A., Liu Y. N., et al. (2007). Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development 134 2903–2912. 10.1242/dev.007542 [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez V. L., Tompson S. W., Blair H. J., Espinoza-Valdez C., Lapunzina P., Silva E. O., et al. (2003). Mutations in two nonhomologous genes in a head-to-head configuration cause Ellis-van Creveld syndrome. Am. J. Hum. Genet. 72 728–732. 10.1086/368063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Yoshizaki K., Kobayashi T., Osumi N. (2013). Neural crest-derived horizontal basal cells as tissue stem cells in the adult olfactory epithelium. Neurosci. Res. 75 112–120. 10.1016/j.neures.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Takeda H., Takami M., Oguni T., Tsuji T., Yoneda K., Sato H., et al. (2002). Positional cloning of the gene LIMBIN responsible for bovine chondrodysplastic dwarfism. Proc. Natl. Acad. Sci. U.S.A. 99 10549–10554. 10.1073/pnas.152337899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela M., Ramos C. (1996). Chondroectodermal dysplasia (Ellis-van Creveld syndrome): a case report. Eur. J. Orthod. 18 313–318. 10.1093/ejo/18.1.313 [DOI] [PubMed] [Google Scholar]
- Vora S. R., Camci E. D., Cox T. C. (2015). Postnatal ontogeny of the cranial base and craniofacial skeleton in male C57BL/6J mice: a reference standard for quantitative analysis. Front. Physiol. 6:417. 10.3389/fphys.2015.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. K., Komatsu Y., Mishina Y. (2011). Potential contribution of neural crest cells to dental enamel formation. Biochem. Biophys. Res. Commun. 415 114–119. 10.1016/j.bbrc.2011.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Thomas N., Hatch N. E., Hu M., Liu F. (2017). Postnatal craniofacial skeletal development of female C57BL/6NCrl mice. Front. Physiol. 8:697. 10.3389/fphys.2017.00697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Abe K., Mantani A., Hitoshi Y., Suzuki M., Osuzu F., et al. (1999). A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev. Biol. 212 191–203. 10.1006/dbio.1999.9323 [DOI] [PubMed] [Google Scholar]
- Yumoto K., Thomas P. S., Lane J., Matsuzaki K., Inagaki M., Ninomiya-Tsuji J., et al. (2013). TGF-beta-activated kinase 1 (Tak1) mediates agonist-induced Smad activation and linker region phosphorylation in embryonic craniofacial neural crest-derived cells. J. Biol. Chem. 288 13467–13480. 10.1074/jbc.M112.431775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kamiya N., Tsuji T., Takeda H., Scott G., Rajderkar S., et al. (2016a). Elevated fibroblast growth factor signaling is critical for the pathogenesis of the dwarfism in Evc2/Limbin mutant mice. PLoS Genet. 12:e1006510. 10.1371/journal.pgen.1006510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Takeda H., Tsuji T., Kamiya N., Kunieda T., Mochida Y., et al. (2016b). Loss of function of Evc2 in dental mesenchyme leads to hypomorphic enamel. J. Dent. Res. 96 421–429. 10.1177/0022034516683674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. H., Takeda H., Tsuji T., Kamiya N., Rajderkar S., Louie K., et al. (2015). Generation of Evc2/Limbin global and conditional KO mice and its roles during mineralized tissue formation. Genesis 53 612–626. 10.1002/dvg.22879 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Superimposition of each skull bones within the Evc2 P0 group. Comparisons of two controls (Evc2 P0 C vs. C) and three additional pairs of control and mutant (Evc2 P0 C vs. m) are shown. (B) Superimposition of each skull bones within the Evc2 Wnt1 group. Comparisons of two controls (Evc2 Wnt1 C vs. C) and three additional pairs of control and mutant (Evc2 Wnt1 C vs. m) are shown.
Evc2 expression in synchondrosis in the skull base. Beta-galactosidase activity from the lacZ knock-in allele of Evc2 locus was used as a surrogate of the expression of Evc2. The skull bases from Evc2lacZ/+ at P10 were stained with X-gal followed by eosin counter staining. Robust staining (blue) was detected in the ISS and the SOS. Evc2+/+ littermates were used as negative controls.
Lateral X-ray cephalogram indicates the linear and angular measurements within P0-Cre group at P8 and P28 and within Wnt1-Cre group at P8 and P28. Within each group, the potential gender specific differences were evaluated by comparing control male with control females; and mutant males and mutant females. Occasional significant differences observed were marked in yellow color.