Abstract
The lipid composition of human meibomian gland secretions (meibum) has been analyzed using both targeted and untargeted mass spectrometric approaches, each of which has its advantages and disadvantages. Herein we report the results of shotgun lipidomic profiling of human meibum using a new approach that combines the advantages of targeted and untargeted analyses to yield highly sensitive and comprehensive profiles. Samples containing an estimated 7–13 µg (8–16 nL) of human meibum lipids were analyzed using MS/MSall, an untargeted approach for MS/MS. Using MS/MSall with ESI and successive polarity switching, we obtained tandem mass spectra in both modes at every 1 Da step for all ions in the m/z 200–1,200 range. In approximately 12 min, a total of 2 MS spectra and 2,000 MS/MS spectra were acquired for each sample, from which targeted analysis information was extracted. This approach allowed for the comprehensive and highly sensitive detection of meibum lipids, including species low in abundance. Altogether, more than 600 unique lipid molecular species were identified in meibum, 3 times more than previously reported in untargeted analyses of meibum samples. This untargeted MS and MS/MSall approach may be extended to other biological systems for the detection of lipids with sensitivity comparable to targeted analysis.
Keywords: eye, fatty acid, mass spectrometry, phospholipids, triglycerides, cholesteryl ester, diester
The tear film protects the surface of the eye and creates the smooth refractive surface required for optimal vision (1). Tear film is composed of three layers: mucin, aqueous, and lipid (1–3). The combined mucin and aqueous layers are sometimes referred to as the mucoaqueous gel layer (1, 4). The lipids in the outermost lipid layer originate from meibum, secretions from the meibomian glands situated posterior to the eyelashes on the eyelid margin (1). Changes in the composition and/or quantity of the lipid secretions from the meibomian glands lead to evaporative dry eye disease (DED) (5, 6), a disorder characterized by symptoms of dryness and visual disturbance that alter quality of life (7). DED affects nearly 5 million people aged 50 years and older in the United States and likely tens of millions more with less severe symptoms. The prevalence ranges from 5% to 50% worldwide depending on age and definition (7).
A clear understanding of the lipid composition of meibum would help to elucidate the mechanisms underlying DED, thereby facilitating the development of effective treatments. Meibum secretions have been shown to be almost exclusively composed of lipids (8). These lipids are predominantly neutral lipids and include the wax esters (WEs), cholesteryl esters (CEs), diesters, and triacylglycerols (TGs), as well as smaller amounts of polar lipids, such as FFAs and (O-acyl)-ω-hydroxy fatty acids (OAHFAs) (9–14). However, our knowledge of the exact lipid composition of meibum, including the specific isomeric species present and the identities of additional lipids present in low abundance, remains lacking (15).
Various mass spectrometric methods (9–14) have been applied to characterize the lipid composition of meibum. In 1981, Nicolaides et al. (10) were the first to report a comprehensive study of meibum. They reported the different classes of lipids present, along with the percentages of FA and fatty alcohol (FAl) moieties in each class. They followed up that study with a detailed analysis of the unusual lipid class diesters (16) and double-bond and chain patterns of meibum lipid moieties, including ω-hydroxy FAs and α,ω diols (17–19). However, their analysis lacked information regarding intact lipid molecules due to limitations of the technology available at the time (10). With the development of soft ionization methods in MS, several groups have characterized intact meibum lipids using a variety of approaches (9, 11–13). These approaches differ in the combinations of ionization, sample introduction, and detection methods. Ionization methods employed include atmospheric pressure chemical ionization (12) and ESI (9, 11, 13), and sample introduction methods include direct infusion (9, 11) and HPLC (12, 13). The use of direct infusion-based MS approaches for lipidome analysis is frequently referred to as shotgun lipidomics (20, 21). Common detection methods include multiple reaction monitoring (MRM) (11, 13), precursor ion scanning (PIS) (22), neutral loss scanning (NLS) (11), and full scanning (9, 23). The MRM, PIS, and NLS detection methods are considered targeted analyses, while full scanning is untargeted. MRM only detects lipids of interest characterized by specific pairs of precursor ions and product ions, and PIS and NLS only detect groups of lipids that share common product ions or neutral losses (11–13). In contrast, full scanning detects all lipids (9, 23). Each of these approaches has advantages and disadvantages, and the approach selected is often related to the particular mass spectrometer available. In addition to these mass spectrometric methods, spectroscopic methods for meibum analysis have also been reported. These methods focus more on the functional groups present in the lipids rather than the individual species present (24–26); therefore, a detailed discussion of these methods is beyond the scope of this study.
For the past several years, our group has been performing untargeted analyses of meibum lipids using direct-infusion, ESI, full-scan MS paired with prior MS/MS of lipids of interest in selected samples (6, 9, 15, 23). This approach has several advantages. First, direct infusion has the flexibility of using strong solvents, such as chloroform, to extract very hydrophobic lipids, including diesters of up to 79 carbon atoms (6, 9, 23). Chloroform can readily extract additives and polymers from plastics, which frequently impair the detection of neutral lipids, the major components of meibum. With the simple and plastic-free setup of the direct-infusion system, interference from these additives and polymers is minimized. In addition, in contrast to the continuous change in solution composition associated with HPLC separations, the solution composition remains the same across the acquisition period. Thus, it is feasible to correct for the ionization efficiency of different species with a limited number of internal standards (23) and, therefore, find subtle differences between sample groups (6). This method also allows sufficient time to perform MS/MS analysis of all ions of interest, in contrast to the time constraints due to the limited acquisition time encountered in HPLC-based methods (27). In addition to the advantages of direct infusion compared with atmospheric pressure chemical ionization, ESI is a gentle ionization method for neutral lipids, such as CEs, and in-source dissociation is typically minimal with appropriate instrument parameters (28). Finally, the full-scan detection provides a global view of all lipid components present in a spectrum (9, 23). By combining this direct-infusion, ESI, full-scan MS approach and MS/MS of lipids of interest, we have been able to identify most of the lipids present in meibum, including isomeric species of WEs (15). However, this type of data acquisition requires a stringent cleaning process; otherwise, ions from impurities will interfere with and sometimes could completely suppress the detection of lipids of interest.
The MS/MSall approach combines the comprehensiveness of full-scan MS and MS/MS analyses with the high-detection sensitivity for specific classes of lipids of interest associated with various pseudo-PIS (pPIS) and pseudo-NLS (pNLS) detection methods (29–33). In addition to the product ions analyzed in pPIS and pNLS, other product ion information is also available to confirm assignments. The MS/MSall method is performed on a high-resolution quadrupole TOF mass spectrometer, which minimizes the potential for false discovery. In contrast, a typical PIS instrument only detects precursor ions that generate a specific common product ion, and a typical NLS only detects pairs of precursor and product ions with a common difference that corresponds to the molecular weight of a neutral molecule (34). No information is obtained from other product ions when using PIS and NLS. Both of these methods are typically performed on low-resolution triple quadrupole mass spectrometers, and false discovery is possible, especially in the absence of front-end separation.
In this study, we report the development of a method for analyzing meibum lipids that uses MS/MSall with successive switching between positive and negative ion modes. After a series of optimization steps, we increased the confidence for identifying individual lipid species, as well as the feasibility of analyzing nanoliter-volume lipid samples of human meibum. MS/MS spectra of all precursor ions in the m/z 200–1,200 range were acquired at every 1 Da step. Altogether, 2,000 high-quality MS/MS spectra were obtained, and extensive lipid composition information was extracted from these spectra. Acquisition of the data in successive positive and negative ion acquisition modes for the same solution minimized both the amount of sample required and the time spent on cleaning procedures (23).
MATERIALS AND METHODS
Chemicals
HPLC-grade chloroform (>99.9%, with amylene as the stabilizer), LC/MS-grade methanol (>99.9%), and ammonium hydroxide solution (25%, eluent additive for LC/MS, Fluka) were purchased from Sigma-Aldrich (St. Louis, MO). HPLC-grade ammonium acetate was purchased from Thermo Fisher Scientific (Norcross, GA).
Meibum samples
The study protocol was approved by the University of Alabama at Birmingham Institutional Review Board in accordance with the Declaration of Helsinki. Meibum samples were collected from each eye of healthy subjects as described previously (9, 23). Briefly, meibum samples were collected directly from meibomian gland orifices on the eyelid margin in a 32 mm 0.5 µl glass microcapillary tube (Drummond; Broomall, PA). The typical fill length of meibum samples collected in the tube was 0.5–1 mm, corresponding to approximately 8–16 nL or 7–13 μg (23).
MS analysis
The procedure was similar to that used in previous reports (9, 23). Briefly, each human meibum sample was directly dissolved in a chloroform-methanol solvent mixture (2:1; v/v) in a glass sample vial. No multiphase separation was performed because human meibum is almost exclusively composed of neutral lipids, with negligible amounts of other species (8). This approach has been reported by several groups (9, 12, 13) to minimize sample loss and contamination, particularly for analyzing such small amounts of samples (23). Typically, a volume of approximately 16 nL (13 µg) of meibum lipid sample was dissolved directly in 100 µL of the chloroform-methanol mixture. The resulting solution was then diluted 5-fold with methanol, along with ammonium acetate or ammonium hydroxide as the additive. No plastics were used in this work, with the exception of Teflon in the syringes.
The diluted working solution was directly infused into an ESI quadrupole TOF mass spectrometer (TripleTOF 5600; SCIEX, Concord, Canada) with a built-in syringe pump. MS and MS/MSall data were acquired. The same solution was successively analyzed in both modes. The MS signal was typically acquired for 3 min prior to MS/MSall acquisition. The MS/MSall acquisition time was 6 min in each ion mode. With MS/MSall, a total of 1,000 MS/MS spectra covering all precursor ions in the m/z 200–1200 range at every 1 Da step were acquired in each mode, yielding a total of 2,000 MS/MS spectra for both ion modes. A TripleTOF 5600 mass spectrometer has a resolving power of 30,000 (35) that can readily distinguish lipid peaks with subtle differences in m/z. A list of manufacturer’s predefined m/z values, including an appropriate mass defect (36), was used for both positive and negative ion modes to define the precursor ions for MS/MSall acquisition for general application (supplemental Table S1). Although the default m/z values may not exactly match the actual values of the precursor ions, they are sufficient for obtaining high mass accuracy product ions with unit resolution isolation. See supplemental data for other experimental details.
The spectra were processed using PeakView Software (SCIEX). Lipid species detected by MS analysis were identified manually by matching the m/z values (typically within 5 ppm) to a reference list of meibum lipid species or to related species (with different saturation levels and chain lengths) not on the list (9, 15, 22, 23, 37–39). To identify common lipid species such as CEs, SMs, and phosphatidylcholines (PCs), we used the peak list to query Lipidmaps (http://lipidmaps.org/tools/ms/lm_mass_form.php), typically using a limit of 0.005 Da. Assignments were verified with information from the corresponding tandem mass spectra when possible. The reference lipid list was created in-house from our previous MS and MS/MS spectra, combined with previously reported data (9, 15, 22, 23, 37–39). For MS/MSall spectra, we assigned peaks from pPIS spectra with the aid of the reference list mentioned above. When in doubt, we analyzed the corresponding full spectra to check the characteristic product ions or fragmentation patterns.
RESULTS
Use of ammonium hydroxide as an additive for high-sensitivity ESI-MS detection of both positive and negative ions
Most of our previous studies on MS analysis of meibum lipids used sodium iodide as the additive because of its ability to generate high-sensitivity, high-resolution, and high-stability lipid peaks (9, 23). However, sodiated ions are often difficult to dissociate into product ions and may, therefore, reduce the amount of information that can be collected in MS/MSall acquisition (29). Therefore, in the initial optimization steps, ammonium acetate, a common additive employed in ESI-MS analysis of neutral lipids (23), was used in positive ion mode. MS spectra acquired at different flow rates and temperatures were compared, and a flow rate of 7 µL/min and 250°C was found to be optimal for detecting meibum lipids.
MS/MSall acquisition requires a considerable amount of sample, and sample loss often occurs during each run due to the need to fill the tubing connections, wait for the flow to stabilize, and remove air bubbles in the syringes. It is also time-consuming to clean the experimental system setup after each run, which is required to minimize interference and suppression during full-spectrum analysis of neutral lipids (23). In the typical experimental setup for analysis in positive and negative ion modes, samples are prepared separately, with different additives, and run in different batches. Therefore, the sample preparation time, sample loss, and clean-up time are doubled. To overcome these limitations, sequential analysis of the same solution in both positive and negative ion modes would be ideal. Unfortunately, the additive ammonium acetate works poorly for detecting meibum lipids in negative-ion-mode MS analysis (9). In contrast, ammonium hydroxide is much more sensitive for detecting acidic lipids in the negative ion mode (9). Ideally, if ammonium hydroxide could also work for the positive ion mode, then the same sample solution containing the ammonium hydroxide additive could be used for both negative and positive ion modes. In this study, using optimized concentrations of the two additives, meibum lipids in ammonium hydroxide were detected in the positive ion mode with a level of sensitivity similar to that of lipids in ammonium acetate (Fig. 1). With ammonium hydroxide as the additive, we identified 360 lipid species collectively for both modes (Table 1, supplemental Table S2). As a result, we concluded that ammonium hydroxide could be used as the additive for both modes, saving not only samples but also time.
Fig. 1.
Comparison of two additives for ESI-MS analysis of meibum lipids in the positive ion mode. A: 0.025% ammonium hydroxide (NH4OH). B: 1 mM ammonium acetate (NH4Ac). The 0.025% ammonium hydroxide additive, ideal for negative ion mode detection, also works well in the positive ion mode and can therefore be used as the additive for both negative and positive ion detection.
TABLE 1.
The number of molecular species of each lipid class detected in human meibomian gland secretions using different analyses
| Number of Species Determined | ||||||
| MS/MSall | ||||||
| Lipid Class | MS | pPIS | Full MS/MS | |||
| Positive ion | CE | 28 | 369.352 ± 0.025 | 58 | — | |
| DE-I | 41 | 40 | — | |||
| WE | FA 16:0 | — | 257.248 ± 0.025 | 20 | — | |
| FA 17:0 | — | 271.263 ± 0.025 | 18 | — | ||
| FA 18:0 | — | 285.279 ± 0.025 | 22 | — | ||
| FA 16:1 | — | 255.232 ± 0.025 | 16 | — | ||
| FA 17:1 | — | 269.248 ± 0.025 | 14 | — | ||
| FA 18:1 | — | 283.263 ± 0.025 | 17 | — | ||
| FA 18:2 | — | 263.237 ± 0.025 | 23 | — | ||
| All | 71 | — | — | 163 | ||
| TG | FA 18:1 | — | 265.253 ± 0.025 | 14 | — | |
| All | 24 | — | — | — | ||
| DE-II | FA 18:1 | — | 265.253 ± 0.025 | 14 | — | |
| All | 33 | — | — | — | ||
| SM | 12 | 184.074 ± 0.025 | 17 | — | ||
| Glycerophosphocholine | 9 | 18 | — | |||
| LPC | 15 | 5 | — | |||
| Negative ion | OAHFA | FA 18:1 | — | 281.249 ± 0.025 | 23 | — |
| All | 67 | — | — | 196 | ||
| FFA | 59 | — | — | — | ||
| Cholesteryl sulfate | 1 | — | — | 1 | ||
| Sum | 360 | 319 | 360 | |||
| Total | 614 | |||||
The total number of species was calculated by adding the numbers of species highlighted in bold.
Unit isolation of precursor ions to obtain natural isotopic distributions in MS/MSall acquisition
In MS spectra of meibum lipids acquired with high-resolution MS, each lipid peak is composed of a set of isotopic peaks that includes the monoisotopic peak (M), the second (M+1) isotopic peak, the third (M+2) isotopic peak, and so on. The intensity of the M, M+1, and M+2 peaks for lipids usually decreases due to the natural abundance of C13 atoms and the total numbers of carbon atoms in the lipids (23, 40). Figure 2A shows a typical MS spectrum of meibum lipids detected in the negative ion mode, in which the isotopic distribution of a series of three OAHFA molecular species with different saturation levels can be seen. Because the intensity of the M+1 isotopic peak of each meibum lipid species is typically much lower than that of the corresponding monoisotopic peak, and the M+2 isotopic peak of each meibum lipid species is typically much lower than the corresponding M+1 peak (40), the three OAHFA species can be readily differentiated (9, 23). However, the low default setting for the isolation of precursor ions in MS/MSall acquisition skewed this natural isotopic distribution (Fig. 2B), making it difficult to assign the peaks and interpret the spectrum. To obtain the natural isotopic distribution (15, 41), we changed the isolation setting from low to unit; as a result, a typical isotopic distribution matching that observed in MS analysis was produced (Fig. 2C). The correspondence to the natural isotopic distribution not only makes spectrum interpretation less difficult but also minimizes false discovery by removing peaks from other species.
Fig. 2.
Effect of isolation efficiency of precursor ions for MS/MSall analysis. A: Full-scan MS spectrum of meibum lipids detected in the negative ion mode. B: TIC of the MS/MSall analysis with the low default isolation. C: TIC of the MS/MSall analysis with the unit isolation. The insets show that unit isolation is ideal because it yields isotopic distributions consistent with the full-scan MS spectrum, allowing for increased confidence in the assignment and minimization of false discovery. The m/z values of the precursor ions for the MS/MSall acquisition are based on a list of preset values considering the approximate mass defect. The PeakView program lists precursor ions with one-decimal place precision, although the spectra were acquired at high resolution.
MS/MSall and successive polarity switching for comprehensive and high-sensitivity shotgun lipidomics of small-volume samples
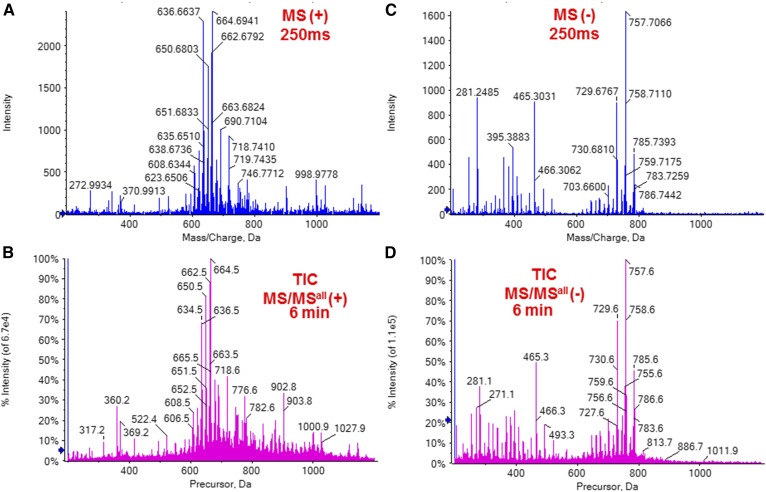
As discussed above, ammonium hydroxide was found to work efficiently as the additive in both positive and negative ion modes. The experimental conditions in the two modes, including the source temperature and flow rate, also needed to be as similar as possible. Constant temperature, as well as the stability of the spray, is essential for mass accuracy of TOF and detection sensitivity. After a series of tests, a flow rate of 7 µL/min and a temperature of 250°C were found to be optimal for both modes. Using these conditions, successive polarity switching was completed, and four acquisitions, including MS (+), MS/MSall (+), MS (−), and MS/MSall (−), were performed for the same sample solution. Comprehensive lipid profiles were obtained in approximately 12 min (Fig. 3). The quality of the data obtained after switching was assessed by running the same sample in subsequent replicates; the resulting spectra were identical.
Fig. 3.
Successive MS and MS/MSall acquisition of meibum with unit isolation for precursors. A: MS spectrum in the positive ion mode (250 ms acquisition). B: TIC of MS/MSall in the positive ion mode (6 min acquisition). C: MS spectrum in the negative ion mode (250 ms acquisition). D: TIC of MS/MSall in the positive ion mode (6 min acquisition).
A collision energy of 40 eV appeared to be optimal and showed the highest signal-to-noise ratio (S/N) for product ion peaks of almost all meibum lipids, except for PC, lysophosphatidylcholine (LPC), and SM, whose detection was more sensitive with a collision energy of 60 eV. Based on the representative MS/MS spectra extracted from the 12 min requisition (Fig. 4, Fig. 5), the characteristic product ions were applied to extract pPIS information from MS/MSall (Figs. 6–11). The pPIS identified molecular species of lipid classes previously reported to be present in meibum, including ω type I-St diesters (DE-Is) (9, 10, 14, 16, 23, 42) and CEs (9, 23, 43) (Fig. 6), WEs (9, 11, 13, 15, 23, 44) (Fig. 7), TGs (9, 11–13, 23) (Fig. 8), α,ω type II diesters (DE-IIs) (9, 10, 14, 16, 23, 45) (Fig. 9), and OAHFAs (9, 11, 13, 37, 44) (Fig. 10), as well as phospholipids, including lysophosphatidylcholine (LPC), PC, and SM (11, 13, 22) (Fig. 11). These phospholipids, which were negligible in previously reported untargeted MS analyses (9, 23), were detected with a high S/N (Fig. 11).
Fig. 4.
Representative MS/MS spectra extracted from MS/MSall acquisition. A: WE 44:1. B: WE 43:0. C: DE-II 68:3. D: SM d34:1. E: CE 24:0. F: PC 34:1. G: DE-I 50:2. H: OAHFA 50:2. The spectra in A–G were acquired in the positive ion mode; the spectrum in H was acquired in the negative ion mode. The characteristic product ions are labeled with asterisks. The combinations of the moieties for the major molecular species, including isomers, are shown. Ions corresponding to ammoniated, protonated, and deprotonated lipids are labeled as M+NH4+, M+H+, and M-H+, respectively. HO-FA, hydroxyl fatty acid; Chol, cholesteryl.
Fig. 5.
Representative structures of the lipids detected in meibum. In A, B, C, E, G, and H, the chain structures (iso-methyl, anteiso-methyl, or straight) and double-bond positions were based on the most abundant structures of these moieties reported in references 10 and 16–19; the combinations of these moieties were based on references 9, 11, 13, 15, 23, 37, 38, 42, 43, and 44.
Fig. 6.
pPIS spectrum of m/z 369.352 ± 0.025 extracted from MS/MSall analysis. Two classes of Chol moiety-containing lipids, i.e., CEs and DE-Is, were detected. The two numbers labeling each peak, separated by a colon, represent the total number of carbon atoms in the lipid ion, excluding the Chol moiety, and the number of double bonds. HO-FA, hydroxyl fatty acid; Chol, cholesteryl.
Fig. 11.
pPIS of m/z 184.074 ± 0.025 extracted from MS/MSall analysis. PC and SM peaks are shown along with LPC peaks in the inset. The two numbers labeling each peak, separated by a colon, represent the total number of carbon atoms and the number of double bonds in the acyl chains of PCs, SMs, or LPCs. The letter “d” or “t” in front of the numbers for SMs indicate di- or trihydroxy bases, respectively.
Fig. 7.
pPIS spectra of m/z 283.263 ± 0.025 (A) and m/z 271.263 ± 0.025 (B) extracted from MS/MSall analysis. The peaks correspond to FA 18:1-based WEs and FA 17:0-based WEs, respectively. The spectra were acquired in the positive ion mode, and only the part of the spectra that correspond to these WEs is shown. The two numbers labeling each peak, separated by a colon, represent the total number of carbon atoms and the number of double bonds in the FAl moiety of the WE ion.
Fig. 8.
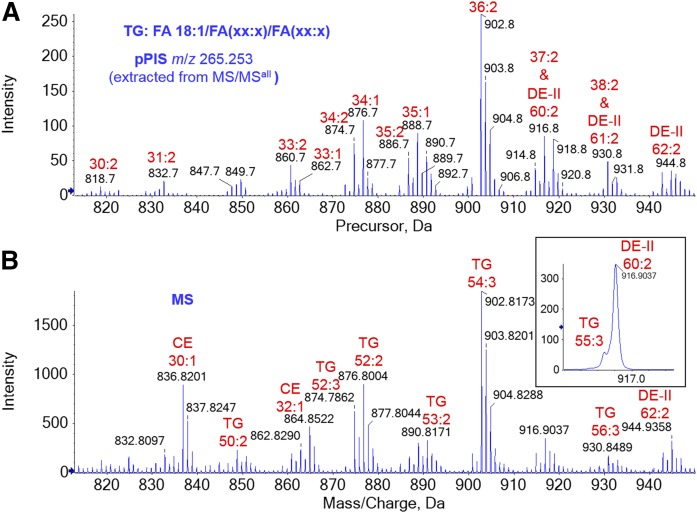
Detection of FA 18:1-based TGs in human meibum. A: pPIS spectrum of m/z 265.253 in the m/z 815–950 range extracted from MS/MSall analysis. B: The corresponding MS spectrum. Almost all of the peaks (A) correspond to FA 18:1-based TGs. The inset in B shows that a small peak of TG 55:3 overlaps with the peak of DE-II 60:2. The two numbers labeling each TG peak, separated by a colon, represent the total number of carbon atoms and the number of double bonds in the other two fatty acyl chains of the TG ion (A) and in all three fatty acyl chains of the TG ion (B). Peaks associated with CEs and DE-IIs are labeled in B.
Fig. 9.
Detection of FA 18:1-based DE-IIs in human meibum. A: pPIS spectrum of m/z 265.253 ± 0.025 in the m/z 910–1,042 range extracted from the same MS/MSall analysis described in Fig. 7. B: The corresponding MS spectrum. Almost all of the peaks (A) correspond to FA 18:1-based DE-IIs. The inset in B shows that the intensities of TG 56:3 and DE-II 61:3 are similar. The corresponding mass errors are 0.5 and −1.3 ppm. The two numbers labeling each DE-II peak, separated by a colon, represent the total number of carbon atoms and the number of double bonds in the DE-II ion, excluding the FA18:1 moiety (A), and in the whole DE-II ion (B). TG peaks are labeled in B.
Fig. 10.
pPIS spectrum of m/z 281.249 ± 0.025 extracted from MS/MSall analysis. The peaks correspond to FA 18:1-based OAHFAs. The spectrum was acquired in the negative ion mode. The two numbers labeling each OAHFA peak, separated by a colon, represent the total number of carbon atoms and the number of double bonds in OAHFA ions, excluding the FA18:1 moiety.
DISCUSSION
Use of ammonium hydroxide as an additive for high-sensitivity ESI-MS detection of both positive and negative ions
The incorporation of MS/MSall into lipidomic analysis requires a more extended infusion time and, therefore, more sample volume. To minimize the required amount of sample and the extensive system cleaning time required for successfully detecting these meibum lipids, we acquired the data in both positive and negative ion modes under similar experimental conditions for the same solution. The use of ammonium hydroxide as the additive was found to work efficiently for both modes. The detection of lipids in the negative ion mode with ammonium hydroxide as the additive is not surprising, but the ability to detect positive ions with the ammonium hydroxide additive is somewhat unexpected. Ammonium hydroxide is basic, and compounds tend to lose protons under basic conditions. Consequently, acids (such as acetic acid and formic acid) or acidic salts (such as ammonium acetate) are typically used to promote the formation of protonated adducts for ESI-MS detection in the positive ion mode (46). It is true that under these acidic solutions compounds can form protonated/ammoniated ions more readily than in basic solutions, such as ammonium hydroxide. However, because electrochemical reactions and gas phase formation occur during ESI, the behavior of these compounds likely differs from that expected in the solution phase under typical conditions. The high-sensitivity detection of meibum lipids in the positive ion mode in the presence of ammonium hydroxide may result from excess protons formed in the electrochemical reaction of water during ESI. In this electrochemical reaction, water molecules are reduced by electrons into oxygen molecules and protons (46). The excess protons are then attached to ammonia to form ammonium ions, which in turn form cation adducts with meibum lipids. Similarly, the sensitivity for detecting negative ions by ESI-MS has also been reported to be increased in the presence of weak acids, a condition under which compounds would not be expected to be in the deprotonated form in the solution phase, which should make these compounds difficult to detect (47). These reports suggest factors other than pH play key roles in the ionization process.
Unit isolation of precursor ions for obtaining natural isotopic distributions in MS/MSall acquisition
Analyzing isotopic peak distributions is essential for identifying unknown species, as well as differentiating between peaks of distinct species (48). This aspect is particularly true for lipids, as there are often a series of peaks corresponding to lipids of varying saturation levels with differences of two, four, or more hydrogen atoms between the corresponding molecules. The peaks associated with increasingly unsaturated lipid species often overlap with the M+2, M+4, and other isotopic peaks of less unsaturated lipids (40). Interestingly, for meibum samples, the intensity of the monoisotopic peaks for most of the more unsaturated lipids (lower m/z) is much lower than that of the corresponding less unsaturated lipids (higher m/z). Even if not, the intensities of their corresponding M+2 isotopic peaks, which are often much lower than their corresponding monoisotopic peaks, are usually still of much lower intensity compared with that of lipids with less unsaturation (9, 23). As a result, it is easy to differentiate between peaks of different lipid species.
The same isotopic distribution would be expected for MS/MSall acquisition. However, the use of the low default setting for the isolation of precursor ions, intended to increase detection sensitivity, skewed the isotopic distribution of the product ion peaks in the total ion chromatogram (TIC) from MS/MSall acquisition. This skewed isotopic distribution is due to the isolation of multiple ions of different m/z values, making it difficult to locate the monoisotopic peak and to differentiate between peaks of lipids with different saturation levels (Fig. 2B). As a result, the product ions attributed to the set precursor ion could originate from multiple precursor ions of different m/z values, which could readily lead to false discovery.
For this study, we obtained the isotopic patterns for product ion peaks in the TIC chromatograms of the MS/MSall acquisition as expected for the natural isotopic distribution, suggesting that the contribution from neighboring peaks was negligible and that reliable identification of lipids could be achieved. However, setting the isolation to the unit resolution decreased the detection sensitivity, making the detection of neutral lipids in meibum more difficult. To increase the detection sensitivity and S/N, careful handling of samples and the system becomes even more critical (23). An alternative approach is to increase the detection time. Current work is ongoing to address this issue.
Meibum lipid profiles
By optimizing the isolation window and experimental conditions for both modes of acquisition, more information was obtained regarding the composition of meibum lipids. The overall profiles of the classes of lipids present in meibum are consistent with previous reports (9, 11–13).
Based on the MS/MSall data, it is straightforward to identify lipid classes that contain only one characteristic product ion, such as CEs (9, 43) and DE-Is (9, 42, 49). From a pPIS spectrum of m/z 369.3516, at least 58 CEs of different molecular weights were detected (Fig. 6, supplemental Table S2, supplemental Figs. S1–S6), which corresponds to more species than that reported in previous studies (10, 13, 23, 43), including the 56 CEs identified by targeted analysis (11). In the high m/z region of the same spectrum, at least 40 cholesteryl-containing diesters, or DE-Is (9, 16, 45), were detected (Fig. 6, supplemental Table S2, supplemental Figs. S7–S11), which doubles the previously reported number (23). The assignments of these two lipid classes were further supported by data from the full MS/MS spectra (supplemental Figs. S1–S11) from the MS/MSall acquisition.
WEs differ from CEs and cholesteryl-containing diesters because they do not generate a common product ion for all species within the same class (38). The characteristic product ions from the dissociation of WE ions differ depending on the FA moiety present. A protonated FA corresponding to the FA moiety in a WE is usually detected, and this peak is often accompanied by associated ions corresponding to one or two water molecule losses for unsaturated WEs (38). For example, FA 18:1-based WEs generate product ions of m/z 283.2632, 265.2526, and 247.2420, while FA 17:0-based WEs generate a product ion of m/z 271.2632. Two representative series of WEs, FA 18:1- and 17:0-based WEs, are shown in Fig. 7. Because the most abundant FA moiety and the most abundant odd-chain FA moiety present in WEs in meibum are FA 18:1 and FA 17:0, respectively (10, 15), the corresponding FA 18:1- and 17:0-based WE profiles are of the most interest. The four most abundant WE species contain the FAl moieties 24:0, 25:0, 26:0, and 26:1 (Fig. 7A). The same top four species were observed for FA 18:2-, 16:1-, 17:1-, 16:0-, and 18:0-based WE profiles (supplemental Figs. S12–S16), consistent with previous reports using different approaches (11, 15). However, the approach presented in this report provides information that is more comprehensive in scope. Unlike the MRM-targeted analysis, which only monitors a limited number of predefined transits, our MS/MSall analysis detected all potential precursor ions containing all corresponding product ions. With pPIS data extracted from the MS/MSall acquisition, at least 17 WEs that contain an FA 18:1 moiety were detected (Fig. 7A, supplemental Table S3), compared with only 8 FA 18:1-containing WEs that were reported (11). Similarly, at least 18 WEs that contain the FA 17:0 moiety were detected (Fig. 7B), compared with a previous report of 8 (11). Many more WEs containing other FA moieties were also detected in our study compared with literature reports (supplemental Figs. S12–S16). Importantly, unlike MRM analyses in which only several pairs of the precursor ion/product ions were monitored, full-scan MS/MS spectra were obtained using MS/MSall acquisition in our study, which could verify assignments, thereby minimizing false discovery due to random matches. The use of MS/MSall provides a more robust method for characterizing WEs, including isomeric species (15, 23). By examining the 1,000 MS/MS spectra and considering the characteristic fragmentation pattern of WEs (38), 163 WE molecular species were detected altogether (supplemental Table S4, supplemental Fig. S17, supplemental Fig. S18), doubling the number reported in our recent study (15).
Similar to WEs, TG (9, 50, 51) and DE-II (9, 45) molecular species generated different characteristic product ions, i.e., protonated FAs and associated water-loss ions that depended on the identity of the FA moiety (9, 16) (Fig. 4G, supplemental Fig. S19, supplemental Fig. S20). The presence of these different product ions complicated the analysis of these isomers relative to lipid classes that contain a unique product ion, such as CEs. Compared with WEs, obtaining detailed isomer information for TGs and DE-IIs based on MS/MS spectra is even more complicated because TGs and DE-IIs contain more moieties than WEs. WEs contain one fatty acyl moiety and one FAl moiety, while TGs contain three fatty acyl moieties and DE-IIs contain two fatty acyl moieties with a diol moiety between them. These fatty acyl moieties belong to the same family and are thus theoretically interchangeable. Therefore, there are a greater number of potential combinations within the intact lipid molecules based on the detected ions. A detailed analysis of the isomer combinations is beyond the scope of this study. In this report, we only show peaks corresponding to FA 18:1-based TGs and DE-IIs, which are close in the regions of the pPIS spectra and partially overlap (Figs. 8A, 9A), making it difficult to directly determine the identities. However, by comparing these spectra with the corresponding MS spectra (Figs. 8B, 9B), we can assign the peaks with high confidence. We determined that the numbers of molecular species of FA 18:1-based TGs (Fig. 8A) and FA 18:1-based DE-IIs (Fig. 8B) were 14 and 20, respectively (supplemental Table S3). In contrast to other lipids that contain ultra-long carbon (≥24) chain FAs or FAls, the composition of TGs is quite typical in terms of carbon chain length (<24) (40, 50). Note that the number of carbon atoms and double bonds in Fig. 8A is the sum of the two FA moieties. The detection of TG (9, 11, 13, 23) and DE-II (6, 9, 23, 42, 45) molecular species in meibum has previously been reported; however, our understanding of the fatty acyl composition of these lipids is still limited. The combination of MS/MSall and MS increases the confidence of the peak assignment and provides a systematic means for obtaining more information about the structures of these molecules.
As reported previously, OAHFAs also generate different characteristic deprotonated FA product ions (9, 37) (Fig. 4H). The pPIS spectrum of m/z 281.25 clearly shows a series of FA 18:1 moiety-containing OAHFAs (Fig. 10). Approximately 23 FA 18:1-containing OAHFAs were detected in this analysis (supplemental Table S3), which exceeds the 16 and 17 FA 18:1-containing OAHFAs previously reported in targeted MRM analyses (11, 13) and approaches the 42 FA 18:1-based OAHFAs reported in a study dedicated to the analysis of OAHFAs and FAs (37). Importantly, full-scan MS/MS spectra were obtained using MS/MSall acquisition in our study, allowing for the verification of assignments and thereby minimizing the rate of false discovery due to random matches. By examining the potential MS/MS spectra and considering the characteristic fragmentation pattern for OAHFAs (9, 37, 39), 196 OAHFA molecular species were detected altogether (supplemental Table S4, supplemental Fig. S24, supplemental Fig. S25).
With the increased sensitivity of pPIS using this untargeted MS/MSall method, species low in abundance, including LPC, PC, and SM, could be detected with high sensitivity. These species generate a common characteristic product ion with the theoretical m/z 184.0733 (52, 53). SM and PC were estimated to account for only 18 ± 5 ppm (22), 0.006 ± 0.003% (11), ∼0.6% (54), or below the detection limit in meibum in previous studies (9, 12). We recently detected peaks corresponding to phospholipids in MS spectra based on accurate mass (55); however, the S/N of those peaks was low. In this study, the pPIS spectrum of m/z 184.07 detected these LPCs, PCs, and SMs with a high S/N (Fig. 11, supplemental Table S3, supplemental Figs. S21, S22), and this lipid profile matched many phospholipids identified in single-stage MS (supplemental Table S2). Cholesteryl sulfate, previously reported to be present in tears (54), was also detected in both MS and MS/MS (supplemental Table S2, supplemental Fig. S23, supplemental Fig. S26).
The peak pattern in the low m/z region (m/z 670–820), including the assignment of some dihydro-SMs, is consistent with the reported peak pattern of PCs and SMs in meibum lipids (22) and is significantly different from tears (22, 54), suggesting different sources for these classes of lipids in meibum and tears. Interestingly, a series of new peaks that also yielded the characteristic product ion of m/z 184.07, including m/z 831, 845, 859, 873, and 901, were observed in this study. These peaks appear to match trihydroxy long-chain base SMs (56–59) and are expected to contain acyl chains of 24 to >30 carbon atoms, considering that the typical base of SMs contains 18 carbon atoms. The acyl chains of these putative trihydroxy SMs appear to match the chain lengths of fatty acyl moieties of CEs and FAl moieties of WEs. There might be some connection in the biosynthesis of lipids of these different classes in meibum. Studies in our lab are ongoing to confirm the identity of these peaks.
In addition to the characterization of individual classes of lipids, the lipid profiles can also be compared between classes. When comparing the lipid profiles of the major components of each class (Figs. 6, 7, 10; supplemental Figs. S12–S16), there appear to be two subgroups of carbon chain moieties. One group is composed of ultra-long carbon chains, mainly saturated or monounsaturated, including 24:0, 25:0, 26:0, and 27:0 (15, 23), and the other group is composed of even longer carbon chains, mainly monounsaturated, including 30:1, 32:1, and 34:1 (9). The presence of these two subgroups of carbon chains is consistent with a previous report (43). These two subgroups of lipids likely play different roles in the function of tear film and warrant further study.
All of the lipid classes described above were detected from pPIS extracted from the MS/MSall acquisition. Theoretically, pNLS could also be applied for the detection of some lipids, e.g., neutral loss of m/z 299.2824 for FA 18:1-based TGs. However, we found that the sensitivity and S/N of pNLS are usually not as good as those of pPIS, likely due to the fact that the neutral loss is calculated from the default m/z value of the precursor ion (supplemental Table S1). The default m/z of the precursor ions is intended for application to different samples. However, these values usually differ from the actual values. For example, the default m/z for ammoniated TG 54:3 is m/z 902.752, which differs from the theoretical value m/z 902.8172. As a result, to extract pNLS, the tolerance for the neutral loss has to be set much wider than necessary, thereby decreasing the S/N.
One limitation of the data presented in this study is the lack of internal standards. In the absence of internal standards, it is unclear whether and how much the ionization efficiency differs for either different classes of lipids or lipids with different saturation levels within the same class. As a result, it is difficult to compare the amounts of the lipids based solely on the peak intensities. However, under our experimental conditions, the ionization efficiency was found to be independent of chain length for WE standards of the same saturation level, and the ionization efficiency of monounsaturated WE standards was approximately twice that of their saturated counterparts (data not shown). These observations are consistent with our previous data obtained using a different mass spectrometer (Q-TOF II; Waters, Milford, MA), although the influence of saturation was slight in that report (23). Based on these results, we would predict that for WEs of the same saturation level, their relative amounts would be proportional to their peak intensities; for WEs of different saturation levels, a good estimate of their relative amounts would require a comparison of the peak intensities after a correction. To perform the correction, the peak intensity should be divided by a coefficient depending on the level of saturation: 1 for saturated, 2 for monounsaturated, and 3 for diunsaturated. A similar trend would be expected for CEs and other neutral lipids in this study (23). In terms of polar lipids, because ionization efficiency is predominantly determined by their head groups (60), the relative amounts of the lipids within the same class would be proportional to the corresponding peak intensities as long as the concentrations are low enough to minimize lipid-lipid interaction (21). Accurate determination of the relative amounts of these beyond these estimates would require spiking internal standards into the samples (23), which is beyond the scope of this study.
CONCLUSIONS
Shotgun lipidomics with successive switching between acquisition polarity modes has been reported in a previous study using an LTQ Orbitrap mass spectrometer (61). High mass accuracy was achieved in that study, and 331 and 222 lipid species were detected from total extracts of bovine heart and human blood plasma, respectively. Those lipid species were detected in 7 min based on MS spectra collected in both positive and negative ion modes, and the identified species were independently validated by MS/MS. In contrast, in this study, 2,000 MS/MS spectra were acquired in both modes in less than 12 min. Including some additional lipids identified in the MS analysis, such as FAs (Table 1, supplemental Table S2, supplemental Fig. S23), more than 600 unique lipid molecules were identified in meibum (Table 1), representing three times more lipids than were previously reported using an untargeted approach to analyze lipids in meibum (23). These spectra were of a high resolution that was enabled by a TOF mass analyzer, which makes it possible to identify the lipid species with high confidence. Several optimization steps not only increased the confidence in lipid assignment and relative quantification but also increased the sample throughput. Only a fraction of the information has been extracted from these 2,000 MS/MS spectra, and we expect that the further development of bioinformatic approaches will aid in the extraction of more information. Untargeted MS and MS/MSall is a promising comprehensive method for detecting lipids that could also be extended to other biological systems for comprehensively detecting lipids with sensitivity comparable to targeted analyses.
Supplementary Material
Acknowledgments
The authors thank Landon Wilson for technical assistance.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- DED
- dry eye disease
- DE-I
- ω type I-St diester
- DE-II
- α,ω type II diester
- FAl
- fatty alcohol
- LPC
- lysophosphatidylcholine
- MRM
- multiple reaction monitoring
- NLS
- neutral loss scanning
- OAHFA
- (O-acyl)-ω-hydroxy fatty acid
- PC
- phosphatidylcholine
- PIS
- precursor ion scanning
- pNLS
- pseudo neutral loss scanning
- pPIS
- pseudo precursor ion scanning
- S/N
- signal-to-noise ratio
- TG
- triacylglycerol
- TIC
- total ion chromatogram
- WE
- wax ester
This work was supported by National Institutes of Health Grants S10 RR027822 and P30 EY003039. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Willcox M. D. P., Argüeso P., Georgiev G. A., Holopainen J. M., Laurie G. W., Millar T. J., Papas E. B., Rolland J. P., Schmidt T. A., Stahl U., et al. 2017. TFOS DEWS II Tear Film Report. Ocul. Surf. 15: 366–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holly F. J., and Lemp M. A.. 1977. Tear physiology and dry eyes. Surv. Ophthalmol. 22: 69–87. [DOI] [PubMed] [Google Scholar]
- 3.Wolff E. 1946. The muco-cutaneous junction of the lid margin and the distribution of the tear fluid. Trans. Ophthalmol. Soc. U. K. 66: 291–308. [Google Scholar]
- 4.Cher I. 2008. A new look at lubrication of the ocular surface: fluid mechanics behind the blinking eyelids. Ocul. Surf. 6: 79–86. [DOI] [PubMed] [Google Scholar]
- 5.Foulks G. N. 2007. The correlation between the tear film lipid layer and dry eye disease. Surv. Ophthalmol. 52: 369–374. [DOI] [PubMed] [Google Scholar]
- 6.Chen J., Keirsey J. K., Green K. B., and Nichols K. K.. 2017. Expression profiling of nonpolar lipids in meibum from patients with dry eye: a pilot study. Invest. Ophthalmol. Vis. Sci. 58: 2266–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stapleton F., Alves M., Bunya V. Y., Jalbert I., Lekhanont K., Malet F., Na K. S., Schaumberg D., Uchino M., Vehof J., et al. 2017. TFOS DEWS II Epidemiology Report. Ocul. Surf. 15: 334–365. [DOI] [PubMed] [Google Scholar]
- 8.Linton R. G., Curnow D. H., and Riley W. J.. 1961. The meibomian glands: an investigation into the secretion and some aspects of the physiology. Br. J. Ophthalmol. 45: 718–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Green-Church K. B., and Nichols K. K.. 2010. Shotgun lipidomic analysis of human meibomian gland secretions with electrospray ionization tandem mass spectrometry. Invest. Ophthalmol. Vis. Sci. 51: 6220–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolaides N., Kaitaranta J. K., Rawdah T. N., Macy J. I., Boswell F. M. 3rd, and Smith R. E.. 1981. Meibomian gland studies: comparison of steer and human lipids. Invest. Ophthalmol. Vis. Sci. 20: 522–536. [PubMed] [Google Scholar]
- 11.Brown S. H., Kunnen C. M., Duchoslav E., Dolla N. K., Kelso M. J., Papas E. B., Lazon de la Jara P., Willcox M. D., Blanksby S. J., and Mitchell T. W.. 2013. A comparison of patient matched meibum and tear lipidomes. Invest. Ophthalmol. Vis. Sci. 54: 7417–7424. [DOI] [PubMed] [Google Scholar]
- 12.Butovich I. A., Uchiyama E., and McCulley J. P.. 2007. Lipids of human meibum: mass-spectrometric analysis and structural elucidation. J. Lipid Res. 48: 2220–2235. [DOI] [PubMed] [Google Scholar]
- 13.Lam S. M., Tong L., Yong S. S., Li B., Chaurasia S. S., Shui G., and Wenk M. R.. 2011. Meibum lipid composition in Asians with dry eye disease. PLoS One. 6: e24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathers W. D., and Lane J. A.. 1998. Meibomian gland lipids, evaporation, and tear film stability. Adv. Exp. Med. Biol. 438: 349–360. [DOI] [PubMed] [Google Scholar]
- 15.Chen J., Green K. B., and Nichols K. K.. 2016. Compositional analysis of wax esters in human meibomian gland secretions by direct infusion electrospray ionization mass spectrometry. Lipids. 51: 1269–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolaides N., and Santos E. C.. 1985. The di- and triesters of the lipids of steer and human meibomian glands. Lipids. 20: 454–467. [DOI] [PubMed] [Google Scholar]
- 17.Nicolaides N., and Ruth E. C.. 1982. Unusual fatty acids in the lipids of steer and human meibomian Gland excreta. Curr. Eye Res. 2: 93–98. [DOI] [PubMed] [Google Scholar]
- 18.Nicolaides N., Santos E. C., and Papadakis K.. 1984. Double-bond patterns of fatty acids and alcohols in steer and human meibomian gland lipids. Lipids. 19: 264–277. [DOI] [PubMed] [Google Scholar]
- 19.Nicolaides N., Santos E. C., Papadakis K., Ruth E. C., and Muller L.. 1984. The occurrence of long chain alpha, omega-diols in the lipids of steer and human meibomian glands. Lipids. 19: 990–993. [DOI] [PubMed] [Google Scholar]
- 20.Han X., and Gross R. W.. 2003. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44: 1071–1079. [DOI] [PubMed] [Google Scholar]
- 21.Han X., and Gross R. W.. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 22.Saville J. T., Zhao Z., Willcox M. D., Ariyavidana M. A., Blanksby S. J., and Mitchell T. W.. 2011. Identification of phospholipids in human meibum by nano-electrospray ionisation tandem mass spectrometry. Exp. Eye Res. 92: 238–240. [DOI] [PubMed] [Google Scholar]
- 23.Chen J., Green K. B., and Nichols K. K.. 2013. Quantitative profiling of major neutral lipid classes in human meibum by direct infusion electrospray ionization mass spectrometry. Invest. Ophthalmol. Vis. Sci. 54: 5730–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borchman D., Foulks G. N., Yappert M. C., Kakar S., Podoll N., Rychwalski P., and Schwietz E.. 2010. Physical changes in human meibum with age as measured by infrared spectroscopy. Ophthalmic Res. 44: 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borchman D., Sledge S., Michiel H., Dennis E., Gerlach D., and Bhola R.. 2015. Lipid hydrocarbon chain conformation of surface lipid films by Raman spectroscopy and the rate of evaporation. Invest. Ophthalmol. Vis. Sci. 56: 1643. [Google Scholar]
- 26.Shrestha R. K., Borchman D., Foulks G. N., Yappert M. C., and Milliner S. E.. 2011. Analysis of the composition of lipid in human meibum from normal infants, children, adolescents, adults, and adults with meibomian gland dysfunction using (1)H-NMR spectroscopy. Invest. Ophthalmol. Vis. Sci. 52: 7350–7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang K., and Han X.. 2011. Accurate quantification of lipid species by electrospray ionization mass spectrometry—meet a key challenge in lipidomics. Metabolites. 1: 21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J., Green-Church K. B., and Nichols K. K.. 2011. Author response: on the presence of (O-acyl)-omega-hydroxy fatty acids and their esters in human meibomian gland secretions. Invest. Ophthalmol. Vis. Sci. 52: 1894–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simons B., Kauhanen D., Sylvanne T., Tarasov K., Duchoslav E., and Ekroos K.. 2012. Shotgun lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites. 2: 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao F., McDaniel J., Chen E. Y., Rockwell H., Lynes M. D., Tseng Y. H., Sarangarajan R., Narain N. R., and Kiebish M. A.. 2016. Monoacylglycerol analysis using MS/MSALL quadruple time of flight mass spectrometry. Metabolites. 6: E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao F., McDaniel J., Chen E. Y., Rockwell H. E., Drolet J., Vishnudas V. K., Tolstikov V., Sarangarajan R., Narain N. R., and Kiebish M. A.. 2017. Dynamic and temporal assessment of human dried blood spot MS/MSALL shotgun lipidomics analysis. Nutr. Metab. (Lond.). 14: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasain J. K., Wilson L., Hoang H. D., Moore R., and Miller M. A.. 2015. Comparative lipidomics of caenorhabditis elegans metabolic disease models by SWATH non-targeted tandem mass spectrometry. Metabolites. 5: 677–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokol E., Ulven T., Færgeman N. J., and Ejsing C. S.. 2015. Comprehensive and quantitative profiling of lipid species in human milk, cow milk and a phospholipid-enriched milk formula by GC and MS/MSALL. Eur. J. Lipid Sci. Technol. 117: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Hoffmann E. 1996. Tandem mass spectrometry: a primer. J. Mass Spectrom. 31: 129–137. [Google Scholar]
- 35.Andrews G. L., Simons B. L., Young J. B., Hawkridge A. M., and Muddiman D. C.. 2011. Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600). Anal. Chem. 83: 5442–5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sleno L. 2012. The use of mass defect in modern mass spectrometry. J. Mass Spectrom. 47: 226–236. [DOI] [PubMed] [Google Scholar]
- 37.Mori N., Fukano Y., Arita R., Shirakawa R., Kawazu K., Nakamura M., and Amano S.. 2014. Rapid identification of fatty acids and (O-acyl)-omega-hydroxy fatty acids in human meibum by liquid chromatography/high-resolution mass spectrometry. J. Chromatogr. A. 1347: 129–136. [DOI] [PubMed] [Google Scholar]
- 38.Chen J., Green K. B., and Nichols K. K.. 2015. Characterization of wax esters by electrospray ionization tandem mass spectrometry: double bond effect and unusual product ions. Lipids. 50: 821–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hancock S. E., Ailuri R., Marshall D. L., Brown S. H. J., Saville J. T., Narreddula V. R., Boase N. R., Poad B. L. J., Trevitt A. J., Willcox M. D. P., et al. 2018. Mass spectrometry-directed structure elucidation and total synthesis of ultra-long chain (O-acyl)-omega-hydroxy fatty acids. J. Lipid Res. 59: 1510–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X., and Gross R. W.. 2001. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100. [DOI] [PubMed] [Google Scholar]
- 41.Rockwood A. L., Kushnir M. M., and Nelson G. J.. 2003. Dissociation of individual isotopic peaks: predicting isotopic distributions of product ions in MSn. J. Am. Soc. Mass Spectrom. 14: 311–322. [DOI] [PubMed] [Google Scholar]
- 42.Butovich I. A., Borowiak A. M., and Eule J. C.. 2011. Comparative HPLC-MS analysis of canine and human meibomian lipidomes: many similarities, a few differences. Sci. Rep. 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butovich I. A. 2009. Cholesteryl esters as a depot for very long chain fatty acids in human meibum. J. Lipid Res. 50: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butovich I. A., Wojtowicz J. C., and Molai M.. 2009. Human tear film and meibum. Very long chain wax esters and (O-acyl)-omega-hydroxy fatty acids of meibum. J. Lipid Res. 50: 2471–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butovich I. A., Lu H., McMahon A., and Eule J. C.. 2012. Toward an animal model of the human tear film: biochemical comparison of the mouse, canine, rabbit, and human meibomian lipidomes. Invest. Ophthalmol. Vis. Sci. 53: 6881–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cech N. B., and Enke C. G.. 2001. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 20: 362–387. [DOI] [PubMed] [Google Scholar]
- 47.Wu Z., Gao W., Phelps M. A., Wu D., Miller D. D., and Dalton J. T.. 2004. Favorable effects of weak acids on negative-ion electrospray ionization mass spectrometry. Anal. Chem. 76: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kind T., and Fiehn O.. 2010. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2: 23–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalužíková A., Vrkoslav V., Harazim E., Hoskovec M., Plavka R., Buděšínský M., Bosáková Z., and Cvačka J.. 2017. Cholesteryl esters of ω-(O-acyl)-hydroxy fatty acids in vernix caseosa. J. Lipid Res. 58: 1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAnoy A. M., Wu C. C., and Murphy R. C.. 2005. Direct qualitative analysis of triacylglycerols by electrospray mass spectrometry using a linear ion trap. J. Am. Soc. Mass Spectrom. 16: 1498–1509. [DOI] [PubMed] [Google Scholar]
- 51.Murphy R. C., James P. F., McAnoy A. M., Krank J., Duchoslav E., and Barkley R. M.. 2007. Detection of the abundance of diacylglycerol and triacylglycerol molecular species in cells using neutral loss mass spectrometry. Anal. Biochem. 366: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godzien J., Ciborowski M., Martínez-Alcázar M. P., Samczuk P., Kretowski A., and Barbas C.. 2015. Rapid and reliable identification of phospholipids for untargeted metabolomics with LC-ESI-QTOF-MS/MS. J. Proteome Res. 14: 3204–3216. [DOI] [PubMed] [Google Scholar]
- 53.Liebisch G., Drobnik W., Lieser B., and Schmitz G.. 2002. High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin. Chem. 48: 2217–2224. [PubMed] [Google Scholar]
- 54.Lam S. M., Tong L., Duan X., Petznick A., Wenk M. R., and Shui G.. 2014. Extensive characterization of human tear fluid collected using different techniques unravels the presence of novel lipid amphiphiles. J. Lipid Res. 55: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J., Keirsey J., Basso K., and Nichols K. K.. 2015. Differentially expressed non-polar lipids in human meibum of dry eye disease. Invest. Ophthalmol. Vis. Sci. 56: 342. [Google Scholar]
- 56.Morrison W. R. 1971. Trihydroxy long-chain bases in bovine milk sphingomyelin. FEBS Lett. 19: 63–64. [DOI] [PubMed] [Google Scholar]
- 57.Karlander S. G., Karlsson K. A., Leffler H., Lilja A., Samuelsson B. E., and Steen G. O.. 1972. The structure of sphingomyelin of the honey bee (Apis mellifera). Biochim. Biophys. Acta. 270: 117–131. [DOI] [PubMed] [Google Scholar]
- 58.Karlsson K. A., Samuelsson B. E., and Steen G. O.. 1973. Detailed structure of sphingomyelins and ceramides from different regions of bovine kidney with special reference to long-chain bases. Biochim. Biophys. Acta. 316: 336–362. [DOI] [PubMed] [Google Scholar]
- 59.Breimer M. E. 1975. Distribution of molecular species of sphingomyelins in different parts of bovine digestive tract. J. Lipid Res. 16: 189–194. [PubMed] [Google Scholar]
- 60.Han X., and Gross R. W.. 1994. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. USA. 91: 10635–10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuhmann K., Almeida R., Baumert M., Herzog R., Bornstein S. R., and Shevchenko A.. 2012. Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J. Mass Spectrom. 47: 96–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.