Abstract
Background
Lysine-specific demethylase 5B (KDM5B) is overexpressed in several types of cancer. However, the clinical significance of KDM5B expression in hepatocellular carcinoma (HCC) remains unclear. The aims of the present study were to examine the functional effects of KDM5B in the Hep3B cell line, the expression levels of KDM5B in human HCC tissues, and the association between KDM5B expression and clinical outcome in patients with HCC.
Material/Methods
Immunohistochemistry (IHC) and quantitative real-time polymerase chain reaction (qRT-PCR) were used to examine the expression levels of KDM5B in HCC tissues and adjacent normal liver tissues. In the HCC cell line, Hep3B, the effects of KDM5B on cell proliferation and migration, and KDM5B small interfering RNA (siRNA) were used to study KDM5B knockdown. Univariate and multivariate analysis assessed the prognostic role of KDM5B in HCC patients. Kaplan-Meier analysis and the log-rank test evaluated clinical outcomes.
Results
In the HCC cell line, Hep3B, KDM5B expression promoted promote tumor cell proliferation and colony formation. Increased expression of KDM5B in HCC tissues, compared with adjacent normal liver tissues, and was associated with larger tumor size, advanced TNM stage, and reduced overall survival in patients with HCC. Multivariate analysis identified KDM5B expression as an independent prognostic factor.
Conclusions
Increased expression of KDM5B was significantly correlated with poorer prognosis in patients with patients with HCC, indicating the possible potential of KDM5B as a novel clinical biomarker and therapeutic target.
MeSH Keywords: Carcinoma, Hepatocellular; Cell Proliferation; Prognosis
Background
Worldwide, hepatocellular carcinoma (HCC) is the main type of primary liver cancer and is the third most common cause of cancer-related death [1]. Current treatment options for patients with HCC include surgical resection, chemotherapy, radiotherapy and immunotherapy [2]. Despite the recent progress in early diagnosis and treatment, patients with HCC patients still have an aggressive clinical course and poor prognosis, especially from advanced-stage disease, with less than 6% of patients with HCC surviving more than 5 years [3]. Studies on the molecular signaling pathways involved in the pathogenesis of HCC have been undertaken, but the mechanisms underlying the development of HCC remain poorly understood [4]. Therefore, there is still a need to investigate the mechanisms involved in the pathogenesis of HCC and to identify potential prognostic biomarkers, more effective treatment strategies, as well as developing new therapeutic targets.
Epigenetic regulation plays multiple roles in a wide variety of physiological processes [5,6]. The homeostasis of epigenetic states is essential for the maintenance of chromatin structure and cell-specific functions [7]. Recent studies have shown that epigenetic alterations, including modifications in histones, are significantly associated with the development of several human malignancies, including osteosarcoma, glioma, and gastric adenocarcinoma [8,9]. As one of the most important types of histone modification, histone methylation contributes significantly to the regulation of chromatin structure and gene expression [10]. For example, lysine methylation on H3K4 is associated with transcriptional activation, whereas lysine methylation on H3K9 is associated with transcriptional repression [11]. It has now become acknowledged that alterations in histone methylation play important roles in the initiation and progression of cancer [12].
Lysine-specific demethylase 5 (KDM5), also known as JARID1, consists of four family members, KDM5A, KDM5B, KDM5C, and KDM5D [13]. The KDM5 family contains five conserved domains: the catalytic JmjC domain, N-terminal JmjN domain, ARID domain, PHD finger domain, and the C5CH2 domain [14]. It has been reported that tumor cells can utilize KDM5A to acquire chemotolerance to cytotoxic agents, such as cisplatin, which suggests a potential role of KDM5A in cancer therapeutics [15]. Although KDM5B is found to be primarily expressed in the normal testis, expression levels of KDM5B have been shown to be significantly upregulated in several human malignancies, including gastric carcinoma, glioma, and malignant osteosarcoma [16]. Also, KDM5B has been shown to have a role in tumor initiation and progression [17].
Therefore the aims of the present study were to examine the functional effects of KDM5B in the Hep3B human HCC cell line, the expression levels of KDM5B in human HCC tissues compared with adjacent normal liver tissues. The prognostic role of KDM5B expression was evaluated with univariate and multivariate analysis. Kaplan-Meier analysis and the log-rank test evaluated clinical outcomes associated with expression of KDM5B on tumor tissues from patients with HCC.
Material and Methods
Reagents
The anti-KDM5B (ab181089) and anti-beta actin (ab8227) antibodies were purchased from Abcam. The KDM5B small interfering RNA (siRNA) was purchased from ABM (Richmond, BC, Canada). The Lipofectamine 3000 Reagent was purchased from Invitrogen (Carlsbad, CA, USA). Other reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA), unless otherwise specified.
Patients and samples
This study was approved by the Ethics Committee of the Third Xiangya Hospital of Central South University. Written informed consent was obtained from all study participants. There were 152 pairs of formalin-fixed paraffin-embedded hepatocellular carcinoma (HCC) tissues and adjacent normal tissues selected from patients who underwent surgical resection in The Third Xiangya Hospital. Also, another 21 pairs of fresh HCC tissues together with adjacent normal liver tissues were obtained from the Department of Surgery, which were fresh-frozen with liquid nitrogen for mRNA analysis. All enrolled patients had no history of previous radiotherapy or chemotherapy and underwent followed-up until death or the end of the study, which ranged from between 3–59 months. All the specimens used in the present study underwent histopathology examination.
Immunohistochemistry (IHC) staining
Immunohistochemistry (IHC) staining for KDM5B was performed using a standard protocol, as previously described [18]. Briefly, 8 μm serial sections were cut onto glass slides, they were dried and then deparaffinized with xylene and rehydrated in alcohols. Microwave antigen retrieval was performed in citrate buffer at pH 6.0. Then, the sections were incubated overnight with the primary rabbit anti-human KDM5B antibody (1: 200 dilution) (Cat. No. ab181089) (Abcam). On the next day, the sections were washed and the localization of the primary antibody was visualized by using horseradish peroxidase (HRP)-conjugated IgG and 3,3′-diaminobenzidine (DAB) (brown) substrate. The negative control replaced the primary antibody with phosphate-buffered saline (PBS).
Evaluation of IHC staining
The stained slides were examined by light microscopy and scored by two independent investigators at a magnification of ×400. Six microscopic fields of each section were randomly selected. Staining intensity was divided into four grades, as follows: 1 (weak); 2 (moderate); 3 (strong). The percentage of positively-stained cells was scored as follows: 1 (<25%); 2 (25–50%); 3 (51–75%); 4 (>75%). The sum of intensity score and staining percentage score was defined as the final IHC score. To better evaluate the clinical significance of KDM5B in HCC, patients were divided into a high KDM5B expression group (IHC score ≥5) (N=80) and a low KDM5B expression group (IHC score <5) (N=70).
Western blot
The transfected cells were washed with PBS and lysed in cold lysis buffer. The cell lysates were centrifuged at 14,000×g at 4°C for 30 min. The supernatant was collected and denatured in the loading buffer. After protein quantification, equal amounts of total protein were separated using a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were immunoblotted with specific primary antibodies to KDM5B and β-actin at 4°C overnight and subsequently incubated with the secondary antibodies for a further 2 hours at room temperature. The protein bands were visualized with the enhanced chemiluminescence (ECL) substrate, using β-actin as the loading control.
RNA preparation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total mRNA was isolated from fresh-frozen specimens using Trizol reagent (Invitrogen, Carlsbad, USA), following a standard protocol [19]. RNA was reversely transcribed into cDNA by using the Primer-Script RT Enzyme Mix (Invitrogen, Carlsbad, CA, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the SYBR Premix Ex Tag (Takara, Japan) according to the manufacturer’s instructions.
GAPDH was used as the normal control, and the following primers were used:
KDM5B-Forward: 5′-CAGCCCGACGAGCAAAA-3′
KDM5B-Reverse: 5′-CGTTGTCTCCTCGGGTTCTATT-3′
GAPDH-Forward: 5′-TGCACCACCAACTGCTTAGC-3′
GAPDH-Reverse: 5′-GGCATGGACTGTGGTCATGAG-3′
Cell culture and transfection
The hepatocellular carcinoma (HCC) cell line, Hep3B was purchased from the American Type Culture Collection (ATCC) (Rockville, USA). The overexpression plasmid of KDM5B was synthesized by Genewiz (Suzhou, China) and verified by DNA sequencing. Hep3B cells were transfected with KDM5B plasmid or siRNA by using Lipofectamine 3000 Reagent following the manufacturer’s instructions [20].
Colony formation assay
Transfected cells were seeded into 60 mm wells at a density of 500 cells per well and cultured for 10 days. The colonies were then fixed with methanol for 15 min and stained with crystal violet for 20 min, and colonies containing more than 60 cells were counted. All the experiments were performed in triplicate [21].
Cell proliferation assay
A cell proliferation assay was performed using the Cell Counting Kit-8 (CCK-8) assay (Dojindo; Japan). Briefly, 100 μl of cell suspension (5×103 cells per well) were incubated in 96-well plates and cultured for designated time points, with 10 μl CCK-8 solution added to each well of the plate and incubated at 37°C for 1 hour. The absorbance was measured at a wavelength of 450 nm using a microplate reader. All the experiments were performed in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS version 19.0 (IBM, New York, NY, USA). The relationship between KDM5B expression and clinical characteristics were assessed using a chi-squared (χ2) test. The overall survival (OS) curves of patients with HCC were plotted using Kaplan-Meier analysis. Statistical validation of independent prognostic factors was performed with multivariate analysis. Student’s t-test was used to analyze the results of cell experiments. P<0.05 was considered to be statistically significant.
Results
Expression of KDM5B was increased in human hepatocellular carcinoma (HCC) tissues
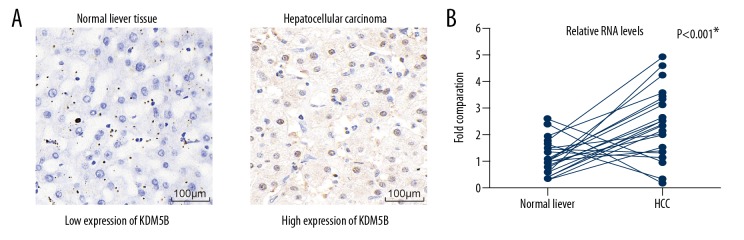
To investigate the role of KDM5B in HCC, we first examined its protein level in HCC tissues together with adjacent normal liver tissues by IHC staining. KDM5B was highly expressed in HCC tissues compared with adjacent normal tissues (Figure 1A). The mRNA levels of KDM5B in a further 21 pairs of fresh frozen HCC tissues compared with adjacent normal liver tissues were compared using quantitative real-time polymerase chain reaction (qRT-PCR) assay (Figure 1B). The mRNA level of KDM5B was significantly increased in HCC tissues when compared with adjacent normal liver tissues (P<0.001). These results indicated that the KDM5B was highly expressed in HCC tissues and might be involved in the progression of HCC.
Figure 1.
Analysis of KDM5B expression in tissue from patients with hepatocellular carcinoma (HCC). (A) Photomicrographs of the immunohistochemical (IHC) staining for KDM5B in liver tissues. The left panel shows a representative image of low expression of KDM5B in normal adjacent liver tissues; the right panel shows a representative image of high expression of KDM5B in HCC tumor tissues. Magnification ×400. (B) mRNA levels of KDM5B were examined in HCC tissues together with adjacent normal liver tissues by quantitative real-time polymerase chain reaction (qRT-PCR). * P<0.001 by Student’s t-test.
Increased expression levels of KDM5B indicated poorer clinical outcome in patients with HCC
To investigate the potential role of KDM5B in HCC, using the results from immunohistochemistry (IHC) and tissue staining for KDM5B expression, 72 patients were in the low KDM5B expression group (IHC score <5), and the other 80 HCC patients were in the high KDM5B expression group (IHC score ≥5). Correlations between KDM5B expression and patient clinical outcome were made (Table 1). Increased expression levels of KDM5B expression were significantly associated with increased tumor size (P<0.001) and advanced TNM stage (P=0.012). However, no significant correlations were identified between KDM5B expression and patient age, gender, serum alpha-fetoprotein (AFP), tumor number, histopathological grade, and portal vein invasion (all P>0.05).
Table 1.
Effects of KDM5B in HCC patients.
| Variables | Cases | KDM5B expression | P value | |
|---|---|---|---|---|
| (n=152) | Low (n=72) | High (n=80) | ||
| Age (years) | 0.570 | |||
| ≤55 | 85 | 42 | 43 | |
| >55 | 67 | 30 | 37 | |
| Sex | 0.520 | |||
| Female | 33 | 14 | 19 | |
| Male | 119 | 58 | 61 | |
| Serum AFP | 0.813 | |||
| ≤400 U/mL | 50 | 23 | 27 | |
| >400 U/mL | 102 | 49 | 53 | |
| Tumor number | 0.058 | |||
| Single | 98 | 52 | 46 | |
| Multiple | 54 | 20 | 34 | |
| Tumor size | <0.001* | |||
| ≤5.0 cm | 61 | 40 | 21 | |
| >5.0 cm | 91 | 32 | 59 | |
| Pathological grade | 0.730 | |||
| Grade 1 | 12 | 7 | 5 | |
| Grade 2 | 101 | 47 | 54 | |
| Grade 3 | 39 | 18 | 21 | |
| Portal vein invasion | 0.264 | |||
| Negative | 103 | 52 | 51 | |
| Positive | 49 | 20 | 29 | |
| TNM stage | 0.012* | |||
| TNM I–II | 60 | 26 | 24 | |
| TNM III–IV | 92 | 36 | 56 | |
Statistically significant by chi-square test.
Increased expression levels of KDM5B were correlated with poor prognosis in patients with HCC
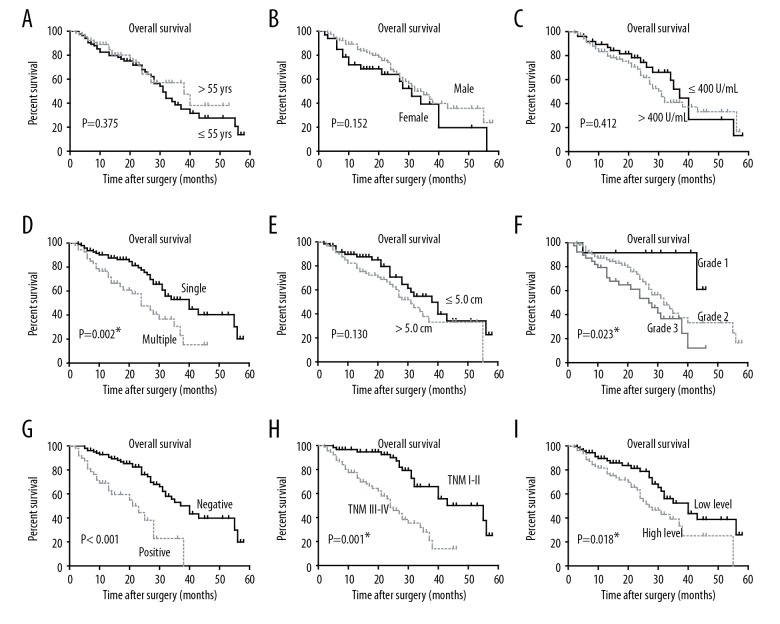
Kaplan-Meier analysis assessed the role of KDM5B expression on overall survival (OS) in patients with HCC and evaluated by the log-rank test (Figure 2). Patients with HCC who expressed higher protein levels of KDM5B had a reduced mean overall survival time (30.1±2.7 months) compared with patients with HCC who expressed lower protein levels of KDM5B (38.5±2.7 months) (P=0.018) (Figure 2I; Table 2). Some conventional prognostic factors were correlated with the overall survival time in patients with HCC, such as the number of tumor nodules, histopathological grade, portal vein invasion, and TNM stage (all P<0.005) (Figure 2; Table 2).
Figure 2.
Analysis of the overall survival of patients with patients with hepatocellular carcinoma (HCC). The overall survival (OS) curves plotted by Kaplan–Meier analysis and assessed by log-rank test, based on patient age (A), gender (B), serum alpha-fetoprotein (AFP) (C), tumor number (D), tumor size (E), pathological grade (F), portal vein invasion (G), TNM stage (H), and KDM5B protein level (I), respectively. * P<0.05 by log-rank test.
Table 2.
Overall survival of HCC patients.
| Variables | Patients | Overall survival months | P value | |
|---|---|---|---|---|
| (n=152) | 3-year OS | Mean ±S.D. | ||
| Age (years) | 0.375 | |||
| ≤55 | 85 | 38.6% | 32.9±2.4 | |
| >55 | 67 | 57.2% | 35.0±2.9 | |
| Sex | 0.152 | |||
| Female | 33 | 39.3% | 29.6±3.8 | |
| Male | 119 | 47.0% | 35.8±2.3 | |
| Serum AFP | 0.412 | |||
| ≤400 U/mL | 50 | 52.1% | 35.5±3.2 | |
| >400 U/mL | 102 | 41.0% | 33.4±2.3 | |
| Tumor number | 0.002* | |||
| Single | 98 | 53.0% | 38.4±2.4 | |
| Multiple | 54 | 30.6% | 24.9±2.3 | |
| Tumor size | 0.130 | |||
| ≤5.0 cm | 61 | 54.7% | 37.2±2.9 | |
| >5.0 cm | 91 | 37.9% | 32.3±2.5 | |
| Pathological grade | 0.023* | |||
| Grade 1 | 12 | 91.7% | 41.7±3.3 | |
| Grade 2 | 101 | 41.2% | 34.9±2.5 | |
| Grade 3 | 39 | 36.7% | 25.9±2.5 | |
| Portal vein invasion | <0.001* | |||
| Negative | 103 | 53.0% | 38.7±2.2 | |
| Positive | 49 | 22.8% | 20.9±2.2 | |
| TNM stage | <0.001* | |||
| TNM I–II | 60 | 65.8% | 43.5±2.6 | |
| TNM III–IV | 92 | 27.8% | 24.8±1.8 | |
| KDM5B level | 0.018* | |||
| Low | 72 | 52.4% | 38.5±2.7 | |
| High | 80 | 39.1% | 30.1±2.7 | |
Statistically significant by log-rank test.
Multivariate analysis using a Cox hazard regression model was used to test the independent effects of KDM5B on the overall survival of patients with HCC (Table 3). All the factors that showed statistical significance in univariate analysis were enrolled in the Cox regression model, including tumor number, histopathological grade, portal vein invasion, TNM stage, and KDM5B levels. Accordingly, KDM5B expression was identified as an independent prognostic factor (HR=1.686; 95% CI, 1.076–2.642) (P=0.023). Also, the presence of portal vein invasion (P=0.015), and advanced TNM stage (P=0.044) were also found to be independent factors of reduced overall survival in patients with HCC.
Table 3.
Multivariate analysis of HCC patients.
| Variables | HR | 95% CI | P value |
|---|---|---|---|
| Tumor number (multiple vs. single) | 1.167 | 0.573–2.376 | 0.670 |
| Pathological grade (grade 3 vs. 1/2) | 1.330 | 0.770–2.300 | 0.307 |
| Portal vein (positive vs. negative) | 2.311 | 1.177–4.536 | 0.015* |
| TNM stage (III/IV vs. I–II) | 1.369 | 1.015–3.753 | 0.044* |
| KDM5B level (high vs. low) | 1.686 | 1.076–2.642 | 0.023* |
Statistically significant by Cox regression model.
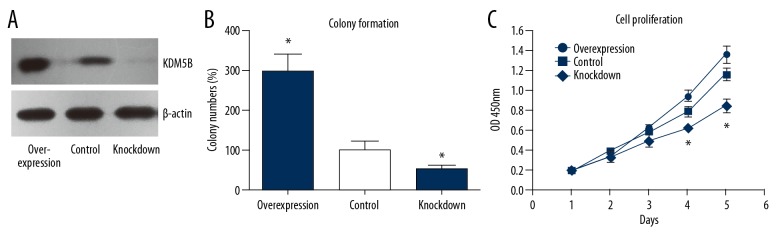
Increased expression of KDM5B promoted colony formation and cell proliferation of the Hep3B human HCC cell line
Overexpression or knockdown of KDM5B expression in the human HCC cell line, Hep3B, and the transfection efficiency was confirmed by Western blotting (Figure 3A). The characteristics of different transfected cells were investigated. KDM5B overexpression enhanced colony formation and cell proliferation when compared with KDM5B knockdown Hep3B cells (Figure 3B, 3C). These data suggested that KDM5B might contribute to the progression of human HCC by enhancing tumor cell growth.
Figure 3.
KDM5B promoted colony formation and proliferation of the hepatocellular carcinoma (HCC) cell line Hep3B. (A) Western blot results showed transfection efficiency of KDM5B plasmids and KDM5B-small interfering RNA (siRNA) in the hepatocellular carcinoma (HCC) cell line, Hep3B. (B) The results of colony formation assays performed in KDM5B overexpressing or siRNA knocked down cells. (C) Cell proliferation ability was tested in KDM5B overexpressing or in the siRNA knocked down cells.
Discussion
Post-translational demethylation of lysine residues on histone tails is an important chromatin modification that is mediated by specific subfamilies of lysine demethylases (KDMs) and has roles in many cellular processes [22]. Mutations in the KDM5A gene in humans are associated with chronic inflammatory diseases [23,24]. Reduction of the expression of KDM5C has been shown to be associated with some neurodegenerative diseases [25]. Recent studies have shown that overexpression or mutations of KDMs are associated with the initiation and progression of several human cancers [26]. For example, overexpression of KDM4C is expressed in renal cell carcinoma [27]. Inactivation of mutations of the KDM5D gene has been shown to promote the progression of prostate cancer [28]. Emerging evidence from published studies has shown that members of the KDM5 family are involved in tumor development and progression, and may serve as novel cancer therapeutic targets [14,29].
KDM5B, which belongs to the KDM5 family, can function as a transcriptional suppressor by specifically removing methyl residues from lysine 4 of histone 3 (H3K4), and consequently suppresses gene transcription [30]. A recently published study has shown that KDM5B exhibited tumorigenic activity in a variety of human cancer types [31]. Several studies have shown that KDM5B has a role in both tumor initiation and progression, and that overexpression of KDM5B has been reported in several human malignancies, suggesting that KDM5B may be required for cancer cell development [32,33]. However, the potential role of KDM5B in the pathogenesis of HCC remains poorly understood.
In the present study, we demonstrated that KDM5B expression was significantly increased in liver tissues containing HCC tissues when compared with adjacent normal liver tissues by immunohistochemistry (IHC) and quantitative real-time polymerase chain reaction (qRT-PCR). Also, the associations between KDM5B expression and the clinical characteristics of patients with HCC who were included in this study were evaluated. The increased expression of KDM5B was significantly associated with larger tumor size and advanced TNM stage in patients with HCC. Also, using multivariate analysis, increased expression of KDM5B was identified as an independent prognostic factor for patients with HCC. Finally, in vitro studies using the human HCC cell line, Hep3B, was used to investigate the potential effect of KDM5B on HCC cells, which showed that overexpression of KDM5B in Hep3B cells promoted colony formation and tumor cell proliferation, which were reversed by knockdown of KDM5B. These findings support that increased expression of KDM5B was associated with a more aggressive clinicopathological outcome for patients with HCC, possibly by promoting tumor cell growth.
Previously published studies support the findings of the present study. KDM5B expression has been shown to be significantly upregulated in glioma tissues and associated with poor clinical outcomes in glioma patients [34]. KDM5B has been shown to promote the progression of thyroid cancer through direct suppression of p21 [35]. In gastric cancer cells, KDM5B expression was shown to increase cell proliferation and invasion [36]. Also, KDM5B was previously identified as a potential oncoprotein, as the inhibition of KDM5B activity was shown to suppress tumorigenesis in vivo in tumor-bearing mouse models [37]. Therefore, the increased expression of the KDM5B protein is significantly associated with the pathogenesis of malignant tumors. Further studies are needed to support these findings and to investigate the underlying molecular mechanisms for the roles of KDM5B in human malignancy, including HCC, to determine potential clinical applications.
Conclusions
The findings of this study showed that KDM5B was highly expressed in tissues from patients with hepatocellular carcinoma (HCC), and expression levels were significantly correlated with the poor clinicopathological features of tumors in patients with HCC. Also, using multivariate analysis, upregulation of KDM5B was identified as an independent negative prognostic factor for patients with HCC. These findings indicated that KDM5B requires further investigation as a potential novel prognostic biomarker in HCC.
Footnotes
Conflict of interest
None.
Source of support: This study was supported by the grant from Science and Technology Agency of Hunan Province (No. 2013FJ6071)
References
- 1.Mortezaee K. Human hepatocellular carcinoma: Protection by melatonin. J Cell Physiol. 2018 doi: 10.1002/jcp.26586. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 2.Kudo M. Combination cancer immunotherapy in hepatocellular carcinoma. Liver Cancer. 2018;7(1):20–27. doi: 10.1159/000486487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kudo M. Lenvatinib may drastically change the treatment landscape of hepatocellular carcinoma. Liver Cancer. 2018;7(1):1–19. doi: 10.1159/000487148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Tian Y, Chen H, et al. Key signaling pathways, genes and transcription factors associated with hepatocellular carcinoma. Mol Med Rep. 2018;17(6):8153–60. doi: 10.3892/mmr.2018.8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grabiec AM, Potempa J. Epigenetic regulation in bacterial infections: Targeting histone deacetylases. Crit Rev Microbiol. 2018;44(3):336–50. doi: 10.1080/1040841X.2017.1373063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itabashi E, Osabe K, Fujimoto R, Kakizaki T. Epigenetic regulation of agronomical traits in Brassicaceae. Plant Cell Rep. 2018;37(1):87–101. doi: 10.1007/s00299-017-2223-z. [DOI] [PubMed] [Google Scholar]
- 7.Kameda T, Imamura T, Nakashima K. Epigenetic regulation of neural stem cell differentiation towards spinal cord regeneration. Cell Tissue Res. 2018;371(1):189–99. doi: 10.1007/s00441-017-2656-2. [DOI] [PubMed] [Google Scholar]
- 8.Kagohara LT, Stein-O’Brien GL, Kelley D, et al. Epigenetic regulation of gene expression in cancer: Techniques, resources and analysis. Brief Funct Genomics. 2018;17(1):49–63. doi: 10.1093/bfgp/elx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Leary K, Shia A, Schmid P. Epigenetic regulation of EMT in non-small cell lung cancer. Curr Cancer Drug Targets. 2018;18(1):89–96. doi: 10.2174/1568009617666170203162556. [DOI] [PubMed] [Google Scholar]
- 10.Daskalaki MG, Tsatsanis C, Kampranis SC. Histone methylation and acetylation in macrophages as a mechanism for regulation of inflammatory responses. J Cell Physiol. 2018;233(9):6495–507. doi: 10.1002/jcp.26497. [DOI] [PubMed] [Google Scholar]
- 11.Feng B, Zhu Y, Su Z, et al. Basil polysaccharide attenuates hepatocellular carcinoma metastasis in rat by suppressing H3K9me2 histone methylation under hepatic artery ligation-induced hypoxia. Int J Biol Macromol. 2018;107(Pt B):2171–79. doi: 10.1016/j.ijbiomac.2017.10.088. [DOI] [PubMed] [Google Scholar]
- 12.Fu LN, Tan J, Chen YX, Fang JY. Genetic variants in the histone methylation and acetylation pathway and their risks in eight types of cancers. J Dig Dis. 2018;19(2):102–11. doi: 10.1111/1751-2980.12574. [DOI] [PubMed] [Google Scholar]
- 13.Nie Z, Shi L, Lai C, et al. Structure-based design and discovery of potent and selective KDM5 inhibitors. Bioorg Med Chem Lett. 2018;28(9):1490–94. doi: 10.1016/j.bmcl.2018.03.083. [DOI] [PubMed] [Google Scholar]
- 14.Taylor-Papadimitriou J, Burchell J. JARID1/KDM5 demethylases as cancer targets? Expert Opin Ther Targets. 2017;21(1):5–7. doi: 10.1080/14728222.2017.1263616. [DOI] [PubMed] [Google Scholar]
- 15.Leadem BR, Kagiampakis I, Wilson C, et al. A KDM5 inhibitor increases global H3K4 trimethylation occupancy and enhances the biological efficacy of 5-Aza-2′-Deoxycytidine. Cancer Res. 2018;78(5):1127–39. doi: 10.1158/0008-5472.CAN-17-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He R, Kidder BL. H3K4 demethylase KDM5B regulates global dynamics of transcription elongation and alternative splicing in embryonic stem cells. Nucleic Acids Res. 2017;45(11):6427–41. doi: 10.1093/nar/gkx251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumber A, Nuzzi A, Hookway ES, et al. Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem Biol. 2017;24(3):371–80. doi: 10.1016/j.chembiol.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Xu Y, Zhang Q, et al. Prognostic significance of TBL1XR1 in predicting liver metastasis for early stage colorectal cancer. Surg Oncol. 2017;26(1):13–20. doi: 10.1016/j.suronc.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Tan W, Pan M, Liu H, et al. Ergosterol peroxide inhibits ovarian cancer cell growth through multiple pathways. Onco Targets Ther. 2017;10:3467–74. doi: 10.2147/OTT.S139009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou S, Du P, Wang P, et al. Significance of MNK1 in prognostic prediction and chemotherapy development of epithelial ovarian cancer. Clin Transl Oncol. 2017;19(9):1107–16. doi: 10.1007/s12094-017-1646-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Fan H, Zou Q, et al. TEAD 4 exerts pro-metastatic effects and is negatively regulated by miR6839-3p in lung adenocarcinoma progression. J Cell Mol Med. 2018;22(7):3560–71. doi: 10.1111/jcmm.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng Y, Li H, Liu C, et al. Jumonji domain-containing protein family: The functions beyond lysine demethylation. J Mol Cell Biol. 2018 doi: 10.1093/jmcb/mjy010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong SY, Kim W, Lee HR, Kim HJ. The histone demethylase KDM5A is required for the repression of astrocytogenesis and regulated by the translational machinery in neural progenitor cells. FASEB J. 2018;32(2):1108–19. doi: 10.1096/fj.201700780R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shokri G, Doudi S, Fathi-Roudsari M, et al. Targeting histone demethylases KDM5A and KDM5B in AML cancer cells: A comparative view. Leuk Res. 2018;68:105–11. doi: 10.1016/j.leukres.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Vallianatos CN, Farrehi C, Friez MJ, et al. Altered gene-regulatory function of KDM5C by a novel mutation associated with autism and intellectual disability. Front Mol Neurosci. 2018;11:104. doi: 10.3389/fnmol.2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamurrad S, Hatch HAM, Drelon C, et al. A drosophila model of intellectual disability caused by mutations in the histone demethylase KDM5. Cell Rep. 2018;22(9):2359–69. doi: 10.1016/j.celrep.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Fernandez L, Carmona FD, Lopez-Mejias R, et al. Cross-phenotype analysis of Immunochip data identifies KDM4C as a relevant locus for the development of systemic vasculitis. Ann Rheum Dis. 2018;77(4):589–95. doi: 10.1136/annrheumdis-2017-212372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arseneault M, Monlong J, Vasudev NS, et al. Loss of chromosome Y leads to down regulation of KDM5D and KDM6C epigenetic modifiers in clear cell renal cell carcinoma. Sci Rep. 2017;7:44876. doi: 10.1038/srep44876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brier AB, Loft A, Madsen JGS, et al. The KDM5 family is required for activation of pro-proliferative cell cycle genes during adipocyte differentiation. Nucleic Acids Res. 2017;45(4):1743–59. doi: 10.1093/nar/gkw1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui G, Liu D, Li W, et al. Original research: miR-194 inhibits proliferation and invasion and promotes apoptosis by targeting KDM5B in esophageal squamous cell carcinoma cells. Exp Biol Med (Maywood) 2017;242(1):45–52. doi: 10.1177/1535370216662712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson C, Velupillai S, Tumber A, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol. 2016;12(7):539–45. doi: 10.1038/nchembio.2087. [DOI] [PubMed] [Google Scholar]
- 32.Tarhonskaya H, Nowak RP, Johansson C, et al. Studies on the interaction of the histone demethylase KDM5B with tricarboxylic acid cycle intermediates. J Mol Biol. 2017;429(19):2895–906. doi: 10.1016/j.jmb.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Song C, Ding Y, et al. Transcriptional regulation of JARID1B/KDM5B histone demethylase by ikaros, histone deacetylase 1 (HDAC1), and casein kinase 2 (CK2) in B-cell acute lymphoblastic leukemia. J Biol Chem. 2016;291(8):4004–18. doi: 10.1074/jbc.M115.679332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, An Q, Guo RX, et al. miR424-5p functions as an anti-oncogene in cervical cancer cell growth by targeting KDM5B via the Notch signaling pathway. Life Sci. 2017;171:9–15. doi: 10.1016/j.lfs.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Han S, Peng R, et al. Depletion of histone demethylase KDM5B inhibits cell proliferation of hepatocellular carcinoma by regulation of cell cycle checkpoint proteins p15 and p27. J Exp Clin Cancer Res. 2016;35:37. doi: 10.1186/s13046-016-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao J, Zou JH, Li CY, Zheng GQ. miR-194 inhibits gastric cancer cell proliferation and tumorigenesis by targeting KDM5B. Eur Rev Med Pharmacol Sci. 2016;20(21):4487–93. [PubMed] [Google Scholar]
- 37.Zhou Q, Obana EA, Radomski KL, et al. Inhibition of the histone demethylase Kdm5b promotes neurogenesis and derepresses Reln (reelin) in neural stem cells from the adult subventricular zone of mice. Mol Biol Cell. 2016;27(4):627–39. doi: 10.1091/mbc.E15-07-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]