Abstract
Asian rice (Oryza sativa) and African rice (Oryza glaberrima) are separately domesticated from their wild ancestors Oryza rufipogon and Oryza barthii, which are very sensitive to daylength. In the process of domestication, some traits that are favorable for the natural survival of wild rice such as seed dormancy and shattering have become favorable ones for human consumption due to the loss-of-function mutations in the genes that are underlying these traits. As a consequence, many genes that are related to these kinds of traits have been fixed with favorable alleles in modern cultivars by artificial selection. After domestication, Oryza sativa cultivars gradually spread to temperate and cool regions from the tropics and subtropics due to the loss of their photoperiod sensitivity. In this paper, we review the characteristics of domestication-related seed traits and heading dates in rice, including the key genes controlling these traits, the differences in allelic diversity between wild rice and cultivars, the geographic distribution of alleles, and the regulatory pathways of these traits. A comprehensive comparison shows that these genes contributed to rice domestication and its global expansion. In addition, these traits have also experienced parallel evolution by artificial selection on the homologues of key genes in other cereals.
Keywords: domestication, seed shattering, seed dormancy, flowering time, global expansion, parallel evolution
1. Introduction
A suite of common traits is selected during the domestication of crops, which are collectively known as a “domestication syndrome”. These traits make domesticated species different from their wild ancestors. In cereals, these traits include morphological traits such as larger grains, a loss of seed dispersal, increased apical dominance, more determinate growth, and physiological traits including seed dormancy, seed shattering, bitter substances in edible structures, photoperiod sensitivity, and synchronized flowering [1]. The seed-related traits of dormancy, seed shattering, and grain size determine which kinds of grain humans can choose for domestication, and the photoperiod-sensitive heading date limits the optimal growing region of rice. Rice is a model crop plant due to its small genome size, high-quality genome sequence, and efficient transformation [2]. In the last two decades, many genes that are related to domestication and global expansion have been isolated in rice. In this review, we summarize the genes controlling seed dormancy, seed shattering, grain size, and flowering time, which contribute to rice domestication and its global expansion. In addition, the homologues in other crops are also discussed to verify parallel evolution during domestication.
2. Seed Shattering in Rice
Wild rice seeds fall off after maturity to ensure their natural propagation in the natural environment. However, seed shattering causes yield loss for domesticated crop plants during harvest. The non-shattering trait is likely to undergo strong selection early in domestication and further facilitate the fixation of other domestication characters. Hence, the loss of seed shattering is considered to be direct ecological evidence for wild rice domestication.
2.1. Seed Shattering Genes Identified from Natural Variation
In rice, seed shattering is regulated by the formation of an abscission zone (AZ), which is composed of several layers of small and dense cytoplasmic cells in the joint between the lemma and pedicel [3]. Several shattering quantitative trait loci (QTLs) have been identified including SH4, encoding an Myb3 transcription factor, and qSH1, encoding a BELL1-like homeodomain protein (Table 1) [4,5,6]. The mutated alleles qsh1 and sh4 cause seed non-shattering owing to the absence of abscission layer formation (Table 1). The SHAT1 gene, which encodes an APETALA2 transcription factor, is identified from induced mutant through a 60Co γ-ray radiation and is required for seed shattering through specifying AZ development in rice. The expression of SHAT1 in the abscission layer is positively regulated by SH4, whose mutation results in incomplete development and the partial functioning of the AZ. qSH1 functions downstream of SHAT1 and SH4, and is involved in a positive feedback loop with SHAT1 and SH4 through the maintenance of SHAT1 and SH4 expression in the AZ, thereby promoting AZ differentiation [7].
Table 1.
Reported genes of domestication traits including seed shattering, seed dormancy, and grain size.
| Gene | MSU_LOC | RAP_LOC | Protein Category | Causative Mutation | References |
|---|---|---|---|---|---|
| SH4 | LOC_Os04g57530 | Os04g0670900 | Myb-like transcription factor | G/T substitution in exon | [4,5] |
| qSH1 | LOC_Os01g62920 | Os01g0848400 | BEL1-type transcription factor | G/T substitution in the 5′ UTR | [6] |
| Sdr4 | LOC_Os07g39700 | Os07g0585700 | Novel protein | 18-bp direct repeat | [8] |
| qSD7-1/qPC7 | LOC_Os07g11020 | Os07g0211500 | bHLH transcription factor | 14-bp deletion in exon | [9] |
| qSD1-2 | LOC_Os01g66100 | Os01g0883800 | Gibberellin-20 oxidase | 382-bp deletion | [10] |
| GS3 | Os03g0407400 | G protein γ subunit | C/A substitution in exon | [11,12] | |
| qLGY3/OsLG3b | LOC_Os03g11614 | Os03g0215400 | MADS transcription factor | Six SNPs in exon | [13,14] |
| GW5/qSW5 | LOC_Os05g09520 | Os05g0187500 | Plasma membrane | 1212-bp or 950-bp deletion in the promoter | [15,16,17,18] |
Bp: Base pair, SNP: Single nucleotide polymorphism; G/T: G substitutes for T; C/A: C substitutes for A; Myb-like: Myeloblastosis like; bHLH: Basic helix-loop-helix; MADS: MCM1, AGAMOUS, DEFICIENS, SRF.
2.2. Mutated Alleles Led to Seed Non-Shattering Domestication
Sh4 was detected using an F2 population derived from an indica-type cultivar and the wild rice progenitor Oryza nivara (an annual form of Oryza rufipogon). The nucleotide substitution guanine/ thymine (G/T) in the first exon of SH4 results in a mutated allele carried by all of the cultivars compared with the alleles in wild rice. The SH4 phylogeny, together with neutrality tests and coalescent simulations, suggested that sh4 had a single origin and was fixed to have a mutated allele by artificial selection during domestication [4,5,19], indicating its vital role in rice domestication. However, a near isogenic line carrying sh4 alleles showed strong seed-shattering behavior in the wild rice genetic background and weedy rice, indicating that other genes redundantly regulate abscission layer formation [20,21,22,23]. The interaction between the sh4 and qSH3 loci inhibits the formation of the abscission layer in rice [20,21,22,24,25]. The effect of qSH3 on seed shattering was weaker than that of qSH1 or sh4 when evaluated in the genetic background of cultivated rice [24,25]. qSH1 was detected using a mapping population derived from the japonica cultivar Nipponbare and indica cultivar Kasalath. The single nucleotide polymorphism (SNP) (G/T) in the 5’ regulatory region of qSH1 was conserved in japonica and highly associated with shattering in the temperate japonica subspecies, and has not been introgressed into indica subspecies [6,19].
2.3. The Parallel Evolution of the Non-Shattering Trait in Cereal Crops
Some seed shattering genes are identified from Asian rice including SH4 and qSH1, and some other genes are identified in African rice, including GL4 and ObSH3 [4,5,6,26,27]. GL4, encoding an Myb3 transcription factor, was identified in Oryza barthii, which is the ancestor of African cultivars (Oryza. glaberrima). A cytosine/thymine (C/T) substitution in the GL4 gene resulted in a premature stop codon and led to small seeds and the loss of seed shattering [26]. ObSH3, encoding a YABBY transcription factor, was identified from an F2 population derived from a cross between the African wild rice accession W1411 and the non-shattering African cultivar IRGC104165. Most accessions with both mutations gl4 and obsh3 occur in arid regions north of 11° N in African [26,27]. Their orthologues in either African rice or Asian rice have the conserved functions and have been subjected to selection. For example, OgSH1 and OgSH4, the orthologs of the Asian rice-shattering genes OsSh1 and SH4, also play important roles in controlling seed shattering in African rice. Ossh4 and Ogsh4 were selected in parallel during the domestication of African and Asian rice, respectively [4,5,27,28]. Therefore, artificial selection on common shattering genes resulted in non-shattering in Asian and African cultivars. Meanwhile, seed non-shattering due to the loss of the abscission layer also exists in other crops. Sh1, a homologue of OsSH1 in sorghum, also regulates seed shattering [29]; ZmSh1-1, ZmSh1-5.1, and ZmSh1-5.2, the homologues of ObSH3 in maize, have been verified to control seed shattering by QTL mapping with a large mapping population [27,29]. The Q gene, the homologue of SHAT1 in wheat, encodes a member of the APETALA2 family of transcription factors and confers the free-threshing character [30,31]. The same regulatory point mutation in rice Sh1 was also identified in an Sh1 orthologue gene in Brassica, and is responsible for the seed dispersal structures produced by natural selection [32]. Taken together, these findings indicate that parallel selection on non-shattering exists during sorghum, rice, and maize domestications.
3. Seed Dormancy in Rice
Seed dormancy is a special period in the plant life cycle when seed germination is unable to proceed under a combination of environmental factors that are normally suitable for the germination of non-dormant seeds. Dormancy is a mechanism in wild species to prevent germination during unsuitable external conditions. Excessive seed dormancy is not a desired trait in crops. Some degree of dormancy is desired to prevent pre-harvest sprouting. Hence, domesticated crops have undergone selection against dormancy. Abscisic acid (ABA) and gibberellin (GA) are two major regulators of seed dormancy and germination. Abscisic acid positively regulates dormancy induction and maintenance, while GA promotes seed germination [33,34].
3.1. Seed Dormancy Genes Identified from Natural Variation
Many QTLs affecting seed dormancy or germination-related traits have been mapped in rice [35,36,37,38,39,40,41], but only a few QTLs have been cloned in rice (Table 1), such as Sdr4, encoding a novel protein; qSD7-1/Rc, encoding a bHLH transcription factor; and qSD1-2, encoding OsGA20ox2 [8,9,10,42]. Sdr4 acts as a seed dormancy-specific regulator that is under the control of OsVP1, which is a global positive regulator of seed maturation through its effect on ABA signaling and the regulation of the expression of OsDOG1L-1, which is a positive regulator of dormancy in rice [8]. qSD7-1/Rc controls dormancy and pigment traits by regulating ABA and the flavonoid biosynthetic pathways, respectively [9]. However, qSD1-2 regulates seed dormancy by controlling the seed GA level [10]. A recent study demonstrated that the antagonistic relationship between the ABA and GA metabolic pathways regulates the switch of cereal seeds between dormancy and germination [43].
3.2. Mutated Alleles Led to Seed Dormancy Domestication
The 18-bp (base pair) direct repeat sequence variation resulting from a double-strand cleavage and repair event in Sdr4 substantially contributes to the differences in seed dormancy between japonica (Nipponbare) and indica (Kasalath) cultivars. A sequence analysis of 59 cultivars and 46 accessions of O. rufipogon revealed that the Sdr4-n sequence that causes reduced dormancy was not found in any wild rice accessions, and that the Sdr4-n in indica cultivars is introgressed from japonica rice. Sdr4-n appears to have been produced through at least two mutation events from the closest O. rufipogon allele among the examined accessions. Sdr4-k and Sdr4-k′ in indica were inherited from these subgroups of the wild ancestor [8]. qSD7-1/qPC7 controls the seed dormancy/pericarp color in weedy red rice. The dormancy-enhancing alleles qSD7-1/qPC7 was differentiated into two groups that are generally associated with the tropical and temperate ecotypes of weedy rice. qSD7-1/qPC7 may contribute the most to weed adaptation [9]. A loss-of-function mutation in qSD1-2 enhances seed dormancy and results in semi-dwarfism, which has been used to develop high-yield, semi-dwarf varieties worldwide. The sd1 mutant originally occurred in an O. rufipogon population and in weedy rice. The allelic distribution of qSD1-2/OsGA20ox2 was found to be associated with the subspeciation of indica and japonica rice [44]. However, there is no evidence that the primitive indica-specific and japonica-specific alleles are functionally differentiated at the presumably domestication-related locus of qSD1-2/OsGA20ox2 [35].
3.3. The Parallel Evolution of Seed Dormancy in Cereals
Recently, many QTLs affecting seed dormancy or germination-related traits have been identified in plant species such as barley [45,46,47] and wheat [40,48,49,50]. However, most seed dormancy QTLs have not been finely mapped. Currently, it is difficult to infer the identities of these QTLs. Based on synteny analysis, HvGA20ox has been suggested as the candidate of a seed dormancy QTL in barley [47]. The orthologues of OsSdr4 in wheat, namely TaSdr, in which a single-nucleotide mutation causes a distinct phenotype in seed germination, have been demonstrated to be key regulators of pre-harvest sprouting in wheat [51]. Seed dormancy genes are probably under parallel selection in cereals, which needs to be further demonstrated by testing the function of more homologues of seed dormancy genes in other cereal crops in the future.
4. Grain Size in Rice
Plant seeds are major sources of human nutrition and are the major means of crop propagation. The grain size of wild rice is significantly smaller than that of cultivated rice [1]. Early in domestication, farmers preferred to select larger seeds to increase grain yield and obtain more food during domestication [52]. Therefore, seed size has always been subjected to selection.
4.1. Grain Size Genes Associated with Domestication in Rice
Dozens of major QTLs for grain size have been molecularly characterized, and their regulatory roles in determining grain size or weight have been explored. The beneficial alleles gs3 [11,12], gw5 [15,16,17,18], GW8 [53,54], gif1 [55], GLW7 [56], qLGY3/OsLG3b [13,14,57], GL7/GW7/SLG7 [54,58,59], and gl4 [26] were artificially selected in modern rice varieties during domestication. However, the beneficial alleles qgl3.1 [60,61,62], tgw6 [63], GS2/GL2/OsGRF4/PT2 [64,65,66,67,68], GS5 [69], GW6a [70], tgw3 [71,72,73], and GS9 [74] are rare alleles, which would have mutated recently, and thus were not widely selected by breeders due to limited time.
4.2. Mutated Alleles Contributing to Domestication
GS3 contains four putative domains, and the organ size regulation (OSR) domain is necessary and sufficient for its function as a negative regulator [11,12]. GS3 has undergone positive selection in cultivars [75,76]. A cytosine/adenine (C/A) mutation in the second exon is associated with large grain size and classifies global rice collections into long-grain and short-grain groups. C/A variation exists in the wild rice gene bank [75,77,78], and the C/A variation in wild rice may have resulted from the gene flow from cultivated rice. However, this mutation does not play a similar role in wild rice [77]. The short-grain alleles of GS3 have multiple independent origins, because farmers and early breeders imposed artificial selection, favoring short seeds [79]. The long-grain allele gs3 likely originated from a japonica-like ancestor, and was subsequently introduced into indica by introgression [77]. However, a haplotype network and phylogenetic analyses showed that the japonica and indica haplotypes evolved independently [80].
qLGY3/OsLG3b encodes the MADS-domain transcription factor OsMADS1, and it regulates grain size by interacting with the Gγ subunits GS3 and DEP1. Six SNPs in the OsLG3b region led to alternative splicing, which was associated with grain length, resulting in increases in both the grain quality and yield potential of rice [13,14]. Haplotype analysis revealed that the long-grain allele of OsLG3b might have arisen after the domestication of tropical japonica, and then spread to the subspecies indica or temperate japonica by natural crossing and artificial selection, which is similar to the events related to the GS3 gene that lead to the improvement of tropical japonica [14].
GW5/qSW5 encodes a plasma membrane-associated protein with IQ calmodulin-binding motifs, and is a novel positive regulator of brassinosteroid (BR) signaling that controls grain width and weight through the proteasomal degradation pathway [15,16,17,18]. A 1212-bp deletion (DEL2) in japonica varieties and a 950-bp deletion (DEL1) in indica varieties in the promoter region of qSW5 are confirmed to be the causal mutations that led to increases in both grain width and grain weight, and have strong correlations with grain width in rice. Both the DEL1 and DEL2 deletions likely originated in different wild rice accessions during rice domestication, and were enriched by artificial selection, as well as the propagation of cultivation and natural crosses, and eventually became widely utilized by rice breeders [15,16,17,18]. A nucleotide diversity analysis showed that qSW5 has high nucleic acid polymorphism in both cultivated and wild rice populations, and it has been subjected to positive selection for genetic improvement [81].
4.3. The Parallel Evolution of Grain Size in Cereals
The GS3-homologous gene ZmGS3 in maize contains the same conserved domain as GS3. Correlation analysis shows that the SNP located in the fifth exon is significantly correlated with the kennel length of maize [82]. The TaGW2-6A of a GW2 homologue in wheat and the ZmGW2-CHR4 and ZmGW2-CHR5 of GW2 homologues in maize are highly associated with grain width and grain weight in a large germplasm collection [83,84]. The orthologue of rice GS5 in wheat, TaGS5-3A-T, is significantly associated with larger grain size and a higher thousand-kernel weight [85]. In contrast to the evolutionary model of GS5 in rice, TaGS5-3A-T is positively selected in Chinese wheat breeding, and undergoes selection during each polyploidization event [85,86]. Among the 15 seed size genes previously identified to be under selection in rice or maize, 12 orthologues in sorghum have been under selection during domestication [87]. The major grain size genes in rice and their homologues in other cereal crops have large effects on grain weight; therefore, they have been easily selected based on trait performance. Therefore, grain size has undergone parallel evolution in cereals.
5. Flowering and Adaptation in Rice Expansion
Rice flowering time is determined by internal genetic factors and external environmental factors such as daylength, temperature, drought, nutrients, and biotic stresses. The molecular mechanism of rice flowering has been well characterized and has been recently reviewed [88]. Many genes are included in the review, but here we mainly focused on the genes that have been isolated from natural variations and are related to rice adaptation. O. rufipogon is mainly in a limited tropical region nearby and has been domesticated to the cultivar O. sativa in the Yangtze River valley region in China [78,89]. After a long process of domestication and improvement, rice has been successfully grown worldwide as a global crop, indicating that the rich variation in heading date genes has allowed cultivated rice to adapt to different environments, especially the changing daylength at different latitudes [90]. Rice is a short-day plant. Rice flowering is repressed when the daylength is longer than 13.5 h [91]. In tropical regions, the daylength is less than 13.5 h, and the daily temperature is high, which ensures that rice can grow all year. However, in other regions, such as temperate regions, rice only grows in the short and warm summer, when the daylength is long. Therefore, during rice expansion, cultivars have gradually adapted to the long daylength at high latitudes. Here, we will review the genes that are responsible for rice adaptation to various daylengths, trace the nucleotide changes, and try to reveal how cultivars expanded into diverse regions.
5.1. Heading Date Genes Identified from Natural Variation
Cultivars have abundant variation in flowering time [92], indicating that many heading date genes are responsible for rice flowering. Dozens of genes have been mapped for heading date in rice [93]. Eighteen QTLs (Hd1-Hd18) were detected with the different populations that have been derived from crosses between Nipponbare and Kasalath and between Koshihikari and Hayamasari, and most of the QTLs have been cloned [94,95,96,97]. Hd1, the first cloned flowering gene from natural variation in rice, is the orthologue of CONSTANT, which encodes a transcription factor with a zinc-finger domain and a CONSTANS, CO-like, and TOC1 (CCT) domain. This gene is a bifunctional regulator that promotes flowering in short days and delays flowering in long days [98]. Hd2, which is also named Ghd7.1/OsPRR37/DTH7, encodes a transcription factor with a pseudoreceiver domain and a CCT domain. This gene delays flowering under long days [99,100,101]. qHd3 includes two heading date genes, namely Hd3a and Hd3b (Hd17/OsELF3/EF7) [102,103,104]. Hd3a encodes one of the mobile flowering signal florigens in rice, and RFT1 encodes another florigen. Both genes belong to the phosphatidylethanolamine-binding protein (PEBP) gene family [105,106,107,108]. Hd3b, which is also named OsELF3, Hd17, and EF7, is the homologue of ELF3, which is a component of the Arabidopsis circadian clock [104]. Hd4/Ghd7, encoding a transcription factor with a CCT domain, delays flowering on long days [109]. Hd5/Ghd8/DTH8/LHD1 is a member of the heme-associated proteins 3 (HAP3) family [110,111,112]. Hd6 and Hd16/EL1/EF7 encode the subunits of casein kinase II and casein kinase I separately [113,114,115]. Hd9/DTH3/OsMADS50, the homologue of SOC1 in rice, is a member of the MADS-box gene family [116]. Hd18 encodes an amine oxidase domain-containing protein and is the homologue of Arabidopsis flowering locus D [97]. With other bi-parentally derived populations, additional heading date QTLs have been identified and cloned such as Ehd1, DTH2, and Ehd4 [117,118,119]. Ehd1 encodes a B-type response regulator and promotes flowering independently of Hd1. DTH2 also encodes a transcription factor with a zinc-finger domain and CCT domain, and delays flowering on long days. Ehd4 encodes a CCCH-type zinc finger transcription factor and promotes flowering. Interestingly, when QTL analyses were performed using F2 populations from crosses between Koshihikari and 12 cultivars originating from various regions in Asia, most of the major QTLs were identified, including Hd1, Hd6, Hd16, Hd17, Ghd7, Ghd7.1, Ghd8, Hd3a, and RFT1 [120,121,122]. In addition, many of the minor QTLs were identified with advanced populations [122]. Genome-wide association analysis studies on the basis of diverse germplasm collections identified some of the flowering genes that were detected in bi-parental mapping populations [123,124,125]. These results strongly suggest that these genes largely determine the variations in the heading date of rice.
5.2. Regulatory Networks of Flowering
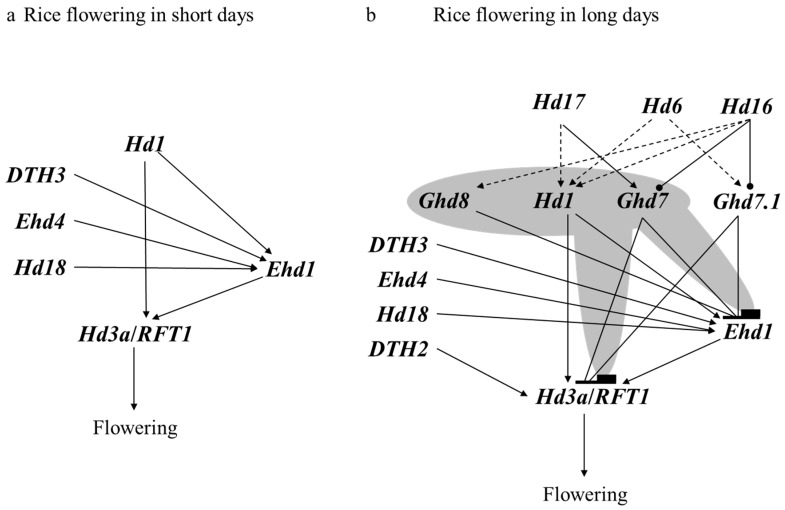
Combined with the latest research findings, a draft regulatory network of how these QTLs/genes coordinate to determine rice flowering time is suggested (Figure 1). The key regulators of flowering are the florigens Hd3a and RFT1, which are expressed in leaves, and then move to the shoot to accelerate flowering [105,106,107,108]. Hd1 only triggers the expression of florigen genes directly in leaves, but it can switch to repress the expression of florigen genes when Ghd7 and Ghd8 are present in long days [126,127,128,129,130]. Ehd1 also triggers the expression of florigen genes independently of Hd1; moreover, it is regulated by Ghd7, Ghd8, Ghd7.1, Ehd4, DTH2, DTH3, and other flowering genes [101,109,111,112,116,118]. In addition, other regulators that are dependent on these major genes have been identified as contributing to flowering. Hd17 is a component of the circadian clock that delays flowering, depending on Hd1 [104]. Hd6 and Hd16 encode kinase and directly phosphorylate GHD7 and GHD7.1, and interact genetically with Ghd7, Ghd7.1, Ghd8, and Hd1 to further delay flowering [113,114,131]. Even more extreme, there are two combinations that lead to non-flowering in natural long days. One is the combination of functional Hd1, Ghd7, and Ghd8 in Zhenshan97, while another includes non-functional ehd1 and rft1 in both the Nona Bokra background and a recombinant inbred line derived from Guangluai 4 and Taichung 65 [126,132,133]. Taken together, these studies show that cultivars with different combinations of known flowering genes can exhibit extensive variation in the heading date, from earlier flowering to extremely later flowering or even non-flowering on long days. On short days, only a few genes such as Hd1, Ehd4, DTH3, and Hd18 still promote flowering as they do on long days, while most others have weak effects or even no effect such as Ghd7, Ghd7.1, Ghd8, Hd17, and DTH2 [100,104,109,111,116,117,118,127,128,129].
Figure 1.
The regulatory network of rice flowering. Lines with a triangle indicate active transcriptional regulation; lines with a cap indicate a repression of transcriptional regulation; lines with a dot indicate phosphorylation; dotted lines indicate genetic interaction; and the dark background indicates that Hd1 interacts with Ghd8 and Ghd7 to largely repress Ehd1 and Hd3a/RFT1 expression.
5.3. Diverse Alleles of Flowering Genes in Wild Rice and Cultivars
Oryza rufipogon has limited genetic and nucleotide diversity for a partial outcrossing species [89,134]. Here, we summarize the functional diversities of known flowering genes in wild rice and cultivars (Table 2). Some key flowering genes have already diversified into functional (strong) and non-functional alleles in wild rice such as Hd1, Hd6, Hd16, and RFT1, and some genes such as Ehd4, Hd17, and Hd3a have generated weak alleles. However, no non-functional alleles have been identified for the strong photoperiod sensitivity genes Ghd7, Ghd7.1, Ghd8, DTH2, and DTH3 in wild rice. These results indicate that the functional mutated alleles of some flowering genes exist in wild rice, but only in rare wild rice accessions. In cultivars, all of the flowering genes have several kinds of mutant alleles with a weak effect or no effect, in addition to the pre-existing alleles in wild rice. The newly generated alleles are enriched in high-latitude regions, because they are artificially selected to be grown under long-day conditions due to their weak or lack of photoperiod sensitivity [104,117]. Both Ghd7 and Ghd7.1 are sensitive to photoperiod. The alleles with strong effects are preserved from wild rice, but the indica and japonica alleles independently originated from different wild rice accessions [100,135], which indicates that indica–japonica differentiation has already occurred in wild rice. This notion is also supported by an evolutionary analysis of a major reproductive barrier regulator, S5 [136]. Weak alleles or non-functional alleles of Ghd7 and Ghd7.1 were then generated in japonica and indica in parallel. Thus, the retention of their pre-existing genetic variants in ancestral species and the acquisition of mutations after domestication have increased the natural variation in the heading date.
Table 2.
Functional and non-functional alleles of flowering genes in cultivars and wild rice.
| Gene Name | MSU_LOC | RAP_LOC | Cultivars | Wild Rice |
|---|---|---|---|---|
| DTH2 | LOC_Os02g49230 | Os02g0724000 | F/N | F [117] |
| Ehd4 | LOC_Os03g02160 | Os03g0112700 | F/W | F/W [118] |
| DTH3/OsMADS50 | LOC_Os03g03070 | Os03g0122600 | F/N | F [116] |
| Hd6 | LOC_Os03g55389 | Os03g0762000 | F/N | F/N [140] |
| Hd16/EL1 | LOC_Os03g57940 | Os03g0793500 | F/W | F/W [114] |
| ELF3/Hd17/EF7 | LOC_Os06g05060 | Os06g0142600 | F/W | F/W [104] |
| RFT1 | LOC_Os06g06300 | Os06g0157500 | F/N | F/N [133] |
| Hd3a | LOC_Os06g06320 | Os06g0157700 | F/N | F/W a |
| Hd1 | LOC_Os06g16370 | Os06g0275000 | F/N | F/N [126] |
| Ghd7/Hd4 | LOC_Os07g15770 | Os07g0261200 | F/N | F [126] |
| Ghd7.1/Hd2/OsPRR37/DTH7 | LOC_Os07g49460 | Os07g0695100 | F/N | F [100] |
| Ghd8/Hd5/DTH8 | LOC_Os08g07740 | Os08g0174500 | F/N | F [126] |
| Ehd1 | LOC_Os10g32600 | Os10g0463400 | F/N | Unknown |
| Hd18 | LOC_Os08g04780 | Os08g0143300 | F/W | Unknown |
a. the wild rice data query from http://ecogems.ncpgr.cn/ with 602942292 and 602942293 sites; MSU_LOC: LOC number from MSU; RAP_LOC: LOC number from RAP; F: Functional; N: Non-functional; W: Weak functional.
An analysis of the eco-geographical distribution patterns of flowering genes showed that flowering repressors are present in low-latitude regions at high frequencies, while the activators (except Hd1) are present in high-latitude regions at high frequencies. As an important flowering signal integrator, the exception—Hd1—has an eco-geographical distribution pattern that is well explained by its bifunctionality. When Hd1 is combined with the non-functional alleles ghd7 and ghd8, Hd1 promotes flowering under either long-day or short-day conditions [128]. When Hd1 is combined with Ghd7 and Ghd8, Hd1 interacts with the repressors of Ghd7 and Ghd8 under long-day conditions, and in turn greatly delays the flowering of this genotype, which is mainly grown in tropical regions. In addition, the interactions between Hd6, Hd16, and Ghd7, Ghd8, Ghd7.1, Hd1 significantly increase the effects of these genes on the heading date [114,137,138]. These genes likely work together and may be the components of a large complex. Therefore, these interactions should be taken into consideration when developing a cultivar for a local region. With the help of humans/breeders, cultivars with different gene combinations are grown in optimized ecological regions, including high-latitude and low-latitude regions, to generate the maximum rice product [126]. However, for the flowering activators such as RFT1, the defective allele would be limited to the tropical region, and functional alleles would move to northern regions or the regions with double-cropping seasons such as those where early rice is grown, where the daylength is long throughout the cropping season [132,133]. Similarly, the functional Hd18 with a minor effect is present in Northeastern China at a higher frequency [139].
5.4. The Parallel Evolution of Key Flowering Genes in Cereals
Plants are classified into three types according to their responses to photoperiods: short-day plants such as rice, maize, and sorghum; long-day plants such as wheat and barley; and day-neutral plants such as tomato [141]. When grown under the same daylength conditions, short-day and long-day plants show opposite responses. However, accessions with different photoperiod response-related genes exhibit various photoperiod responses. We have described how Asian rice expands into different latitudes from its limited region of origin. The other crops such as wheat, maize, and sorghum also adapt to local photoperiod conditions and consequently expand to different latitudes.
The major determinant of long-day response in barley has been cloned as photoperiod-H1 (Ppd-H1) in a colinear region of Ghd7.1. In spring barley, an amino acid change in the CCT domain caused a reduced photoperiod response, which extended the growth period of barley, allowing it to adapt to the long growing seasons and produce higher yields in Western Europe and North America [142]. Different mutations in SbPRR37, the Ghd7.1 homologue in sorghum, also reduce the photoperiod sensitivity and cause sorghum to flower earlier, which is critical for the cultivation of this tropical crop in temperate regions worldwide [143]. BvBTC1, the homologue of Ghd7.1 in sugar beet (Beta vulgaris), is a master switch distinguishing annual from biennial. The loss-of-function BvBTC1 allele confers a reduced photoperiod sensitivity, later flowering, and bienniality [144]. Taken together, these findings indicate that Ghd7.1 and its homologues are key factors in expanding the cultivation of cereals. However, no identical mutant alleles were found in different crops, indicating that mutations occurred after species differentiation, but not from the common ancestor. Ghd7 is another important major determinant of photoperiod response and adaptation in rice. ZmCCT9 and ZmCCT10 are homologues of Ghd7 in maize. Insertions of transposable elements in the promoters of both genes caused a change in mRNA expression, and thus contributed to maize adaptation to higher latitudes after maize was domesticated from southern Mexico [145]. Mutations of EAM8, the homologue of Hd17, facilitate adaptation to a short growing season in barley and expand the geographic range of this species [146]. In sorghum, besides Ma1/SbPRR37 and EAM8, Ma6, which is the homologue of Ghd7, and FT, which is the homologue of Hd3a and RFT1 in sorghum, also regulates photoperiod flowering, and collectively with other genes contribute to its adaptation to diverse environments [147]. In addition, the parallel domestication of Hd1 was elucidated in sorghum, foxtail millet, and rice [148]. Thus, the photoperiod response genes Ghd7.1, Ghd7, Hd1, ELF3, Hd3a, and their homologues underwent parallel selection, and the loss of function of these genes helped crops extend their ranges into higher latitudes from the original range in the tropics or subtropics.
6. Conclusions
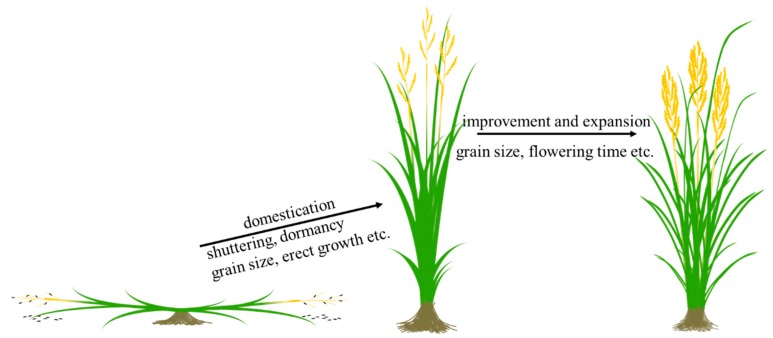
As described above, many important genes contribute to domestication in rice by loss of function. Moreover, many homologues in other cereals have conserved functions, which indicates that parallel evolution plays an important role in the “domestication syndrome”. Parallel evolution provides us with the chance to examine the function of homologues in other cereals once a major gene for such traits has been identified. In general, domestication syndrome genes are divided into two types according to their distribution in cultivars and wild rice. The genes of type one control prostrate growth (not included in this review), seed dormancy, and seed shattering. Loss-of-function mutations in these genes are required for rice domestication, because these mutations allow cultivars to grow more easily and produce more grains initially. In contrast, the genes of type two have diverse functional alleles in cultivars, but some strong functional alleles in cultivars come directly from wild rice, and weak or non-functional alleles are generated in cultivars such as genes for grain size and flowering time. These genes contribute to rice domestication but play more important roles in rice global expansion and genetic improvement to develop modern rice. In brief, the domestication of wild traits such as prostrate growth, seed dormancy, and seed shattering is the sudden result of loss-of-function mutations, and adaptive traits determining rice expansion such as photoperiod sensitivity are the continuing results of the mutations from strong alleles to weak alleles, and then to non-functional alleles (Figure 2).
Figure 2.
Two-step selection from wild rice to modern cultivars.
Funding
This work was supported by grants from the National Special Program for Research of Transgenic Plant of China (2011ZX08009-001-002), National Key Research and Development Program of China (2016YFD0100301), and National Natural Science Foundation of China (31601283) and the Natural Science Foundation of Hubei province China (2015CFA006).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Doebley J.F., Gaut B.S., Smith B.D. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Izawa T., Shimamoto K. Becoming a model plant: The importance of rice to plant science. Trends Plant Sci. 1996;1:95–99. doi: 10.1016/S1360-1385(96)80041-0. [DOI] [Google Scholar]
- 3.Roberts J.A., Elliott K.A., Gonzalez-Carranza Z.H. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 2002;53:131–158. doi: 10.1146/annurev.arplant.53.092701.180236. [DOI] [PubMed] [Google Scholar]
- 4.Li C., Zhou A., Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 5.Lin Z.W., GriYth M.E., Li X., Zhu Z., Tan L., Fu Y., Zhang W., Wang X., Xie D., Sun C. Origin of seed shattering in rice (Oryza sativa L.) Planta. 2007;226:11–20. doi: 10.1007/s00425-006-0460-4. [DOI] [PubMed] [Google Scholar]
- 6.Konishi S., Izawa T., Lin S.Y., Ebana K., Fukuta Y., Sasaki T., Yano M. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y., Lu D., Li C., Luo J., Zhu B.F., Zhu J., Shangguan Y., Wang Z., Sang T., Zhou B., et al. Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell. 2012;24:1034–1048. doi: 10.1105/tpc.111.094383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto K., Takeuchi Y., Ebana K., Miyao A., Hirochika H., Hara N., Ishiyama K., Kobayashi M., Ban Y., Hattori T., et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA. 2010;107:5792–5797. doi: 10.1073/pnas.0911965107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu X.Y., Foley M.E., Horvath D.P., Anderson J.V., Feng J., Zhang L., Mowry C.R., Ye H., Suttle J.C., Kadowaki K., et al. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics. 2011;189:1515–1524. doi: 10.1534/genetics.111.131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye H., Feng J., Zhang L., Zhang J., Mispan M.S., Cao Z., Beighley D.H., Yang J., Gu X.Y. Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 2015;169:2152–2165. doi: 10.1104/pp.15.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., Li X., Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 12.Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., Li X., Zhang Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA. 2010;107:19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q., Han R., Wu K., Zhang J., Ye Y., Wang S., Chen J., Pan Y., Li Q., Xu X., et al. G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018;9:852. doi: 10.1038/s41467-018-03047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J., Miao J., Zhang Z., Xiong H., Zhu X., Sun X., Pan Y., Liang Y., Zhang Q., Abdul Rehman R.M., et al. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 2018 doi: 10.1111/pbi.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 16.Weng J., Gu S., Wan X., Gao H., Guo T., Su N., Lei C., Zhang X., Cheng Z., Guo X., et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 17.Duan P., Xu J., Zeng D., Zhang B., Geng M., Zhang G., Huang K., Huang L., Xu R., Ge S., et al. Natural variation in the promoter of GS5 contributes to grain size diversity in rice. Mol. Plant. 2017;10:685–694. doi: 10.1016/j.molp.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Liu J., Chen J., Zheng X., Wu F., Lin Q., Heng Y., Tian P., Cheng Z., Yu X., Zhou K., et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants. 2017;3:17043. doi: 10.1038/nplants.2017.43. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L.B., Zhu Q., Wu Z.Q., Ross-Ibarra J., Gaut B.S., Ge S., Sang T. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009;184:708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 20.Htun T.M., Inoue C., Chhourn O., Ishii T., Ishikawa R. Effect of quantitative trait loci for seed shattering on abscission layer formation in Asian wild rice Oryza rufipogon. Breed. Sci. 2014;64:199–205. doi: 10.1270/jsbbs.64.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue C., Htun T.M., Inoue K., Ikeda K., Ishii T., Ishikawa R. Inhibition of abscission layer formation by an interaction of two seed-shattering loci, sh4 and qSH3, in rice. Genes Genet. Syst. 2015;90:1–9. doi: 10.1266/ggs.90.1. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa R., Nishimura A., Htun T.M., Nishioka R., Oka Y., Tsujimura Y., Inoue C., Ishii T. Estimation of loci involved in non-shattering of seeds in early rice domestication. Genetica. 2017;145:201–207. doi: 10.1007/s10709-017-9958-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y., Ellstrand N.C., Lu B.R. Sequence polymorphisms in wild, weedy, and cultivated rice suggest seed-shattering locus sh4 played a minor role in Asian rice domestication. Ecol. Evol. 2012;2:2106–2113. doi: 10.1002/ece3.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onishi K., Horiuchi Y., Ishigoh-Oka N., Takagi K., Ichikawa N., Maruoka M., Sano Y. A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed. Sci. 2007;57:7–16. doi: 10.1270/jsbbs.57.7. [DOI] [Google Scholar]
- 25.Onishi K., Takagi K., Kontani M., Tanaka T., Sano Y. Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome. 2007;50:757–766. doi: 10.1139/G07-051. [DOI] [PubMed] [Google Scholar]
- 26.Wu W., Liu X., Wang M., Meyer R.S., Luo X., Ndjiondjop M.N., Tan L., Zhang J., Wu J., Cai H., et al. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants. 2017;3:17064. doi: 10.1038/nplants.2017.64. [DOI] [PubMed] [Google Scholar]
- 27.Lv S., Wu W., Wang M., Meyer R.S., Ndjiondjop M.N., Tan L., Zhou H., Zhang J., Fu Y., Cai H., et al. Genetic control of seed shattering during African rice domestication. Nat. Plants. 2018;4:331–337. doi: 10.1038/s41477-018-0164-3. [DOI] [PubMed] [Google Scholar]
- 28.Wang M., Yu Y., Haberer G., Marri P.R., Fan C., Goicoechea J.L., Zuccolo A., Song X., Kudrna D., Ammiraju J.S., et al. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 2014;46:982–988. doi: 10.1038/ng.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z., Li X., Shannon L.M., Yeh C.T., Wang M.L., Bai G., Peng Z., Li J., Trick H.N., Clemente T.E., et al. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 2012;44:720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons K.J., Fellers J.P., Trick H.N., Zhang Z., Tai Y.S., Gill B.S., Faris J.D. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172:547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Belcram H., Gornicki P., Charles M., Just J., Huneau C., Magdelenat G., Couloux A., Samain S., Gill B.S., et al. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl. Acad. Sci. USA. 2011;108:18737–18742. doi: 10.1073/pnas.1110552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnaud N., Lawrenson T., Ostergaard L., Sablowski R. The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. Curr. Biol. 2011;21:1215–1219. doi: 10.1016/j.cub.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Kucera B., Cohn M.A., Leubner-Metzger G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005;15:281–307. doi: 10.1079/SSR2005218. [DOI] [Google Scholar]
- 34.Finkelstein R., Reeves W., Ariizumi T., Steber C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 35.Gu X.Y., Kianian S.F., Foley M.E. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166:1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marzougui S., Sugimoto K., Yamanouchi U., Shimono M., Hoshino T., Hori K., Kobayashi M., Ishiyama K., Yano M. Mapping and characterization of seed dormancy QTLs using chromosome segment substitution lines in rice. Theor. Appl. Genet. 2012;124:893–902. doi: 10.1007/s00122-011-1753-y. [DOI] [PubMed] [Google Scholar]
- 37.Magwa R.A., Zhao H., Xing Y. Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.) BMC Genet. 2016;17:28. doi: 10.1186/s12863-016-0340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee G.A., Jeon Y.A., Lee H.S., Hyun D.Y., Lee J.R., Lee M.C., Lee S.Y., Ma K.H., Koh H.J. New genetic loci associated with preharvest sprouting and its evaluation based on the model equation in rice. Front. Plant Sci. 2017;8:1393. doi: 10.3389/fpls.2017.01393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y., Xie Y., Cai J., Liu C., Zhu H., Jiang R., Zhong Y., Zhang G., Tan B., Liu G., et al. Substitution mapping of QTLs controlling seed dormancy using single segment substitution lines derived from multiple cultivated rice donors in seven cropping seasons. Theor. Appl. Genet. 2017;130:1191–1205. doi: 10.1007/s00122-017-2881-9. [DOI] [PubMed] [Google Scholar]
- 40.Liu S., Sehgal S.K., Li J., Lin M., Trick H.N., Yu J., Gill B.S., Bai G. Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics. 2013;195:263–273. doi: 10.1534/genetics.113.152330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W., Xu L., Bai X.F., Xing Y.Z. Quantitative trait loci for seed dormancy in rice. Euphytica. 2011;178:427–435. doi: 10.1007/s10681-010-0327-4. [DOI] [Google Scholar]
- 42.Gu X.Y., Liu T., Feng J., Suttle J.C., Gibbons J. The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice. Plant Mol. Biol. 2010;73:97–104. doi: 10.1007/s11103-009-9555-1. [DOI] [PubMed] [Google Scholar]
- 43.Tuan P.A., Kumar R., Rehal P.K., Toora P.K., Ayele B.T. Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 2018;9:668. doi: 10.3389/fpls.2018.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asano K., Yamasaki M., Takuno S., Miura K., Katagiri S., Ito T., Doi K., Wu J., Ebana K., Matsumoto T., et al. Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl. Acad. Sci. USA. 2011;108:11034–11039. doi: 10.1073/pnas.1019490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao W., Clancy J.A., Han F., Prada D., Kleinhofs A., Ullrich S.E. Molecular dissection of a dormancy QTL region near the chromosome 7 (5H) L telomere in barley. Theor. Appl. Genet. 2003;107:552–559. doi: 10.1007/s00122-003-1281-5. [DOI] [PubMed] [Google Scholar]
- 46.Hori K., Sato K., Takeda K. Detection of seed dormancy QTL in multiple mapping populations derived from crosses involving novel barley germplasm. Theor. Appl. Genet. 2007;115:869–876. doi: 10.1007/s00122-007-0620-3. [DOI] [PubMed] [Google Scholar]
- 47.Li C., Ni P., Francki M., Hunter A., Zhang Y., Schibeci D., Li H., Tarr A., Wang J., Cakir M., et al. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genomics. 2004;4:84–93. doi: 10.1007/s10142-004-0104-3. [DOI] [PubMed] [Google Scholar]
- 48.Flintham J., Adlam R., Bassoi M., Holdsworth M., Gale M. Mapping genes for resistance to sprouting damage in wheat. Euphytica. 2002;126:39–45. doi: 10.1023/A:1019632008244. [DOI] [Google Scholar]
- 49.Kulwal P.L., Singh R., Balyan H.S., Gupta P.K. Genetic basis of pre-harvest sprouting tolerance using single-locus and two-locus QTL analyses in bread wheat. Funct. Integr. Genomics. 2004;4:94–101. doi: 10.1007/s10142-004-0105-2. [DOI] [PubMed] [Google Scholar]
- 50.Cabral A.L., Jordan M.C., McCartney C.A., You F.M., Humphreys D.G., MacLachlan R., Pozniak C.J. Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat (Triticum aestivum L.) BMC Plant Biol. 2014;14:340. doi: 10.1186/s12870-014-0340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Miao X., Xia X., He Z. Cloning of seed dormancy genes (TaSdr) associated with tolerance to pre-harvest sprouting in common wheat and development of a functional marker. Theor. Appl. Genet. 2014;127:855–866. doi: 10.1007/s00122-014-2262-6. [DOI] [PubMed] [Google Scholar]
- 52.Sundaresan V. Control of seed size in plants. Proc. Natl. Acad. Sci. USA. 2005;102:17887–17888. doi: 10.1073/pnas.0509021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., Zeng R., Zhu H., Dong G., Qian Q., et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 54.Wang S., Li S., Liu Q., Wu K., Zhang J., Wang S., Wang Y., Chen X., Zhang Y., Gao C., et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015;47:949–954. doi: 10.1038/ng.3352. [DOI] [PubMed] [Google Scholar]
- 55.Wang E., Wang J., Zhu X., Hao W., Wang L., Li Q., Zhang L., He W., Lu B., Lin H., et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- 56.Si L., Chen J., Huang X., Gong H., Luo J., Hou Q., Zhou T., Lu T., Zhu J., Shangguan Y., et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 57.Yu J., Xiong H., Zhu X., Zhang H., Li H., Miao J., Wang W., Tang Z., Zhang Z., Yao G., et al. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 2017;15:28. doi: 10.1186/s12915-017-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., Fang Y., Zeng L., Xu E., Xu J., et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015;47:944–948. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y., Miao J., Gu H., Peng X., Leburu M., Yuan F., Gu H., Gao Y., Tao Y., Zhu J., et al. Natural variations in SLG7 regulate grain shape in rice. Genetics. 2015;201:1591–1599. doi: 10.1534/genetics.115.181115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X.J., Wang J.F., Huang J., Lan H.X., Wang C.L., Yin C.F., Wu Y.Y., Tang H.J., Qian Q., Li J.Y., et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA. 2012;109:21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Z., He H., Zhang S., Sun F., Xin X., Wang W., Qian X., Yang J., Luo X. A Kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice. J. Integr. Plant Biol. 2012;54:979–990. doi: 10.1111/jipb.12008. [DOI] [PubMed] [Google Scholar]
- 62.Qi P., Lin Y.S., Song X.J., Shen J.B., Huang W., Shan J.X., Zhu M.Z., Jiang L., Gao J.P., Lin H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishimaru K., Hirotsu N., Madoka Y., Murakami N., Hara N., Onodera H., Kashiwagi T., Ujiie K., Shimizu B., Onishi A., et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 64.Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., Xiao Y., Hu B., Liu L., Wang H., et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants. 2015;2:15195. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- 65.Duan P., Ni S., Wang J., Zhang B., Xu R., Wang Y., Chen H., Zhu X., Li Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants. 2015;2:15203. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- 66.Hu J., Wang Y., Fang Y., Zeng L., Xu J., Yu H., Shi Z., Pan J., Zhang D., Kang S., et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 67.Li S., Gao F., Xie K., Zeng X., Cao Y., Zeng J., He Z., Ren Y., Li W., Deng Q., et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016;14:2134–2146. doi: 10.1111/pbi.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun P., Zhang W., Wang Y., He Q., Shu F., Liu H., Wang J., Wang J., Yuan L., Deng H. OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 2016;58:836–847. doi: 10.1111/jipb.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., Shao D., Xu C., Li X., Xiao J., et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 70.Song X.J., Kuroha T., Ayano M., Furuta T., Nagai K., Komeda N., Segami S., Miura K., Ogawa D., Kamura T., et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. USA. 2015;112:76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu Z., Lu S.J., Wang M.J., He H., Sun L., Wang H., Liu X.H., Jiang L., Sun J.L., Xin X., et al. A Novel QTL qTGW3 encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant. 2018;11:736–749. doi: 10.1016/j.molp.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 72.Xia D., Zhou H., Liu R., Dan W., Li P., Wu B., Chen J., Wang L., Gao G., Zhang Q., et al. GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol. Plant. 2018;11:754–756. doi: 10.1016/j.molp.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 73.Ying J.Z., Ma M., Bai C., Huang X.H., Liu J.L., Fan Y.Y., Song X.J. TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant. 2018;11:750–753. doi: 10.1016/j.molp.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 74.Zhao D.S., Li Q.F., Zhang C.Q., Zhang C., Yang Q.Q., Pan L.X., Ren X.Y., Lu J., Gu M.H., Liu Q.Q. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. commun. 2018;9:1240. doi: 10.1038/s41467-018-03616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang C., Chen S., Yu S. Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice. Theor. Appl. Genet. 2011;122:905–913. doi: 10.1007/s00122-010-1497-0. [DOI] [PubMed] [Google Scholar]
- 76.Fan C., Yu S., Wang C., Xing Y. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 2009;118:465–472. doi: 10.1007/s00122-008-0913-1. [DOI] [PubMed] [Google Scholar]
- 77.Takano-Kai N., Jiang H., Kubo T., Sweeney M., Matsumoto T., Kanamori H., Padhukasahasram B., Bustamante C., Yoshimura A., Doi K., et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics. 2009;182:1323–1334. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang X., Kurata N., Wei X., Wang Z.X., Wang A., Zhao Q., Zhao Y., Liu K., Lu H., Li W., et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takano-Kai N., Jiang H., Powell A., McCouch S., Takamure I., Furuya N., Doi K., Yoshimura A. Multiple and independent origins of short seeded alleles of GS3 in rice. Breed. Sci. 2013;63:77–85. doi: 10.1270/jsbbs.63.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh N., Singh B., Rai V., Sidhu S., Singh A.K., Singh N.K. Evolutionary insights based on SNP haplotypes of red pericarp, grain size and starch synthase genes in wild and cultivated rice. Front. Plant Sci. 2017;8:972. doi: 10.3389/fpls.2017.00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., Li C., Zhu C., Lu T., Zhang Z., et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 82.Li Q., Yang X., Bai G., Warburton M.L., Mahuku G., Gore M., Dai J., Li J., Yan J. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 2010;120:753–763. doi: 10.1007/s00122-009-1196-x. [DOI] [PubMed] [Google Scholar]
- 83.Su Z., Hao C., Wang L., Dong Y., Zhang X. Identification and development of a functional marker of TaGW2 associated with grain weight in bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 2011;122:211–223. doi: 10.1007/s00122-010-1437-z. [DOI] [PubMed] [Google Scholar]
- 84.Li Q., Li L., Yang X., Warburton M.L., Bai G., Dai J., Li J., Yan J. Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 2010;10:143. doi: 10.1186/1471-2229-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma L., Li T., Hao C., Wang Y., Chen X., Zhang X. TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 2016;14:1269–1280. doi: 10.1111/pbi.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang S., Yan X., Wang Y., Liu H., Cui D., Chen F. Haplotypes of the TaGS5-A1 gene are associated with thousand-kernel weight in chinese bread wheat. Front. Plant Sci. 2016;7:783. doi: 10.3389/fpls.2016.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tao Y., Mace E.S., Tai S., Cruickshank A., Campbell B.C., Zhao X., Van Oosterom E.J., Godwin I.D., Botella J.R., Jordan D.R. Whole-genome analysis of candidate genes associated with seed size and weight in sorghum bicolor reveals signatures of artificial selection and insights into parallel domestication in cereal crops. Front. Plant Sci. 2017;8:1237. doi: 10.3389/fpls.2017.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cho L.H., Yoon J., An G. The control of flowering time by environmental factors. Plant J. 2017;90:708–719. doi: 10.1111/tpj.13461. [DOI] [PubMed] [Google Scholar]
- 89.Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 2007;58:3091–3097. doi: 10.1093/jxb/erm159. [DOI] [PubMed] [Google Scholar]
- 90.Xiong Z.Y., Zhang S.J., Ford-Lloyd B.V., Jin X., Wu Y., Yan H.X., Liu P., Yang X., Lu B.R. Latitudinal distribution and differentiation of rice germplasm: Its implications in breeding. Crop Sci. 2011;51:1050–1058. doi: 10.2135/cropsci2010.07.0431. [DOI] [Google Scholar]
- 91.Itoh H., Nonoue Y., Yano M., Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- 92.Vergara B.S., Chang T.-T. The Flowering Response of the Rice Plant to Photoperiod. A Review of the Literature. International Rice Research Institute; Los Baños, Laguna, Philippines: 1985. [Google Scholar]
- 93.Hori K., Matsubara K., Yano M. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor. Appl. Genet. 2016;129:2241–2252. doi: 10.1007/s00122-016-2773-4. [DOI] [PubMed] [Google Scholar]
- 94.Matsubara K., Kono I., Hori K., Nonoue Y., Ono N., Shomura A., Mizubayashi T., Yamamoto S., Yamanouchi U., Shirasawa K., et al. Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theor. Appl. Genet. 2008;117:935–945. doi: 10.1007/s00122-008-0833-0. [DOI] [PubMed] [Google Scholar]
- 95.Lin H., Yamamoto T., Sasaki T., Yano M. Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor. Appl. Genet. 2000;101:1021–1028. doi: 10.1007/s001220051576. [DOI] [Google Scholar]
- 96.Yano M., Kojima S., Takahashi Y., Lin H., Sasaki T. Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 2001;127:1425–1429. doi: 10.1104/pp.010710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shibaya T., Hori K., Ogiso-Tanaka E., Yamanouchi U., Shu K., Kitazawa N., Shomura A., Ando T., Ebana K., Wu J. Hd18, encoding histone acetylase related to Arabidopsis FLOWERING LOCUS D, is involved in the control of flowering time in rice. Plant Cell Physiol. 2016;57:1828–1838. doi: 10.1093/pcp/pcw105. [DOI] [PubMed] [Google Scholar]
- 98.Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koo B.H., Yoo S.C., Park J.W., Kwon C.T., Lee B.D., An G., Zhang Z., Li J., Li Z., Paek N.C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant. 2013;6:1877–1888. doi: 10.1093/mp/sst088. [DOI] [PubMed] [Google Scholar]
- 100.Yan W., Liu H., Zhou X., Li Q., Zhang J., Lu L., Liu T., Liu H., Zhang C., Zhang Z., et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013;23:969–971. doi: 10.1038/cr.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao H., Jin M., Zheng X.M., Chen J., Yuan D., Xin Y., Wang M., Huang D., Zhang Z., Zhou K., et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA. 2014;111:16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Monna L., Lin X., Kojima S., Sasaki T., Yano M. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor. Appl. Genet. 2002;104:772–778. doi: 10.1007/s00122-001-0813-0. [DOI] [PubMed] [Google Scholar]
- 103.Kojima S., Takahashi Y., Kobayashi Y., Monna L., Sasaki T., Araki T., Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 104.Matsubara K., Ogiso-Tanaka E., Hori K., Ebana K., Ando T., Yano M. Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 2012;53:709–716. doi: 10.1093/pcp/pcs028. [DOI] [PubMed] [Google Scholar]
- 105.Taoka K., Ohki I., Tsuji H., Furuita K., Hayashi K., Yanase T., Yamaguchi M., Nakashima C., Purwestri Y.A., Tamaki S., et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- 106.Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- 107.Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 108.Komiya R., Yokoi S., Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- 109.Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 110.Dai X., Ding Y., Tan L., Fu Y., Liu F., Zhu Z., Sun X., Sun X., Gu P., Cai H., et al. LHD1, an allele of DTH8/Ghd8, controls late heading date in common wild rice (Oryza rufipogon) J. Integr. Plant Biol. 2012;54:790–799. doi: 10.1111/j.1744-7909.2012.01166.x. [DOI] [PubMed] [Google Scholar]
- 111.Yan W.H., Wang P., Chen H.X., Zhou H.J., Li Q.P., Wang C.R., Ding Z.H., Zhang Y.S., Yu S.B., Xing Y.Z., et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant. 2011;4:319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- 112.Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H., Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takahashi Y., Shomura A., Sasaki T., Yano M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA. 2001;98:7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hori K., Ogiso-Tanaka E., Matsubara K., Yamanouchi U., Ebana K., Yano M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013;76:36–46. doi: 10.1111/tpj.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kwon C.T., Yoo S.C., Koo B.H., Cho S.H., Park J.W., Zhang Z., Li J., Li Z., Paek N.C. Natural variation in Early flowering1 contributes to early flowering in japonica rice under long days. Plant Cell Environ. 2014;37:101–112. doi: 10.1111/pce.12134. [DOI] [PubMed] [Google Scholar]
- 116.Bian X.F., Liu X., Zhao Z.G., Jiang L., Gao H., Zhang Y.H., Zheng M., Chen L.M., Liu S.J., Zhai H.Q., et al. Heading date gene, dth3 controlled late flowering in O. Glaberrima Steud. by down-regulating Ehd1. Plant Cell Rep. 2011;30:2243–2254. doi: 10.1007/s00299-011-1129-4. [DOI] [PubMed] [Google Scholar]
- 117.Wu W., Zheng X.M., Lu G., Zhong Z., Gao H., Chen L., Wu C., Wang H.J., Wang Q., Zhou K., et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA. 2013;110:2775–2780. doi: 10.1073/pnas.1213962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qu L.-J., Gao H., Zheng X.-M., Fei G., Chen J., Jin M., Ren Y., Wu W., Zhou K., Sheng P., et al. Ehd4 encodes a novel and oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 2013;9:e1003281. doi: 10.1371/journal.pgen.1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ebana K., Shibaya T., Wu J., Matsubara K., Kanamori H., Yamane H., Yamanouchi U., Mizubayashi T., Kono I., Shomura A., et al. Uncovering of major genetic factors generating naturally occurring variation in heading date among Asian rice cultivars. Theor. Appl. Genet. 2011;122:1199–1210. doi: 10.1007/s00122-010-1524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Itoh H., Wada K.C., Sakai H., Shibasaki K., Fukuoka S., Wu J., Yonemaru J.I., Yano M., Izawa T. Genomic adaptation of flowering-time genes during the expansion of rice cultivation area. Plant J. 2018;94:895–909. doi: 10.1111/tpj.13906. [DOI] [PubMed] [Google Scholar]
- 122.Hori K., Nonoue Y., Ono N., Shibaya T., Ebana K., Matsubara K., Ogiso-Tanaka E., Tanabata T., Sugimoto K., Taguchi-Shiobara F., et al. Genetic architecture of variation in heading date among Asian rice accessions. BMC Plant Biol. 2015;15:115. doi: 10.1186/s12870-015-0501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Han Z.M., Zhang B., Zhao H., Ayaad M., Xing Y.Z. Genome-wide association studies reveal that diverse heading date genes respond to short and long day lengths between indica and japonica rice. Front. Plant Sci. 2016;7:1270. doi: 10.3389/fpls.2016.01270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ye J., Niu X., Yang Y., Wang S., Xu Q., Yuan X., Yu H., Wang Y., Wang S., Feng Y., et al. Divergent Hd1, Ghd7, and DTH7 alleles control heading date and yield potential of japonica rice in northeast China. Front. Plant Sci. 2018;9:35. doi: 10.3389/fpls.2018.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., Li W., Guo Y., Deng L., Zhu C. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 126.Zhang J., Zhou X., Yan W., Zhang Z., Lu L., Han Z., Zhao H., Liu H., Song P., Hu Y. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 2015;208:1056–1066. doi: 10.1111/nph.13538. [DOI] [PubMed] [Google Scholar]
- 127.Du A., Tian W., Wei M., Yan W., He H., Zhou D., Huang X., Li S., Ouyang X. The DTH8-Hd1 module mediates day-length-dependent regulation of rice flowering. Mol. Plant. 2017;10:948–961. doi: 10.1016/j.molp.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 128.Zhang Z.Y., Hu W., Shen G.J., Liu H.Y., Hu Y., Zhou X.C., Liu T.M., Xing Y.Z. Alternative functions of Hd1 in repressing or promoting heading are determined by Ghd7 status under long-day conditions. Sci. Rep. 2017;7:5388. doi: 10.1038/s41598-017-05873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nemoto Y., Nonoue Y., Yano M., Izawa T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016;86:221–233. doi: 10.1111/tpj.13168. [DOI] [PubMed] [Google Scholar]
- 130.Goretti D., Martignago D., Landini M., Brambilla V., Gomez-Ariza J., Gnesutta N., Galbiati F., Collani S., Takagi H., Terauchi R., et al. Transcriptional and post-transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 2017;13:e1006530. doi: 10.1371/journal.pgen.1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kwon C.T., Koo B.H., Kim D., Yoo S.C., Paek N.C. Casein kinases I and 2α phosphorylate Oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Mol. Cells. 2015;38:81–88. doi: 10.14348/molcells.2015.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhao J., Chen H., Ren D., Tang H., Qiu R., Feng J., Long Y., Niu B., Chen D., Zhong T. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa) New Phytol. 2015;208:936–948. doi: 10.1111/nph.13503. [DOI] [PubMed] [Google Scholar]
- 133.Ogiso-Tanaka E., Matsubara K., Yamamoto S.-i., Nonoue Y., Wu J., Fujisawa H., Ishikubo H., Tanaka T., Ando T., Matsumoto T. Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PLoS ONE. 2013;8:e75959. doi: 10.1371/journal.pone.0075959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu W., Shahid M.Q., Bai L., Lu Z., Chen Y., Jiang L., Diao M., Liu X., Lu Y. Evaluation of genetic diversity and development of a core collection of wild rice (Oryza rufipogon Griff.) populations in China. PLoS ONE. 2015;10:e0145990. doi: 10.1371/journal.pone.0145990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lu L., Shao D., Qiu X., Sun L., Yan W., Zhou X., Yang L., He Y., Yu S., Xing Y. Natural variation and artificial selection in four genes determine grain shape in rice. New Phytol. 2013;200:1269–1280. doi: 10.1111/nph.12430. [DOI] [PubMed] [Google Scholar]
- 136.Chen J., Ding J., Ouyang Y., Du H., Yang J., Cheng K., Zhao J., Qiu S., Zhang X., Yao J. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica–japonica hybrids in rice. Proc. Natl. Acad. Sci. USA. 2008;105:11436–11441. doi: 10.1073/pnas.0804761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yamamoto T., Lin H., Sasaki T., Yano M. Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics. 2000;154:885–891. doi: 10.1093/genetics/154.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ogiso E., Takahashi Y., Sasaki T., Yano M., Izawa T. The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 2010;152:808–820. doi: 10.1104/pp.109.148908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li X., Sun Y., Tian X., Ren Y., Tang J., Wang Z., Cheng Y., Bu Q. Comprehensive identification of major flowering time genes and their combinations, which determined rice distribution in Northeast China. Plant Growth Regul. 2018;84:593–602. doi: 10.1007/s10725-017-0364-2. [DOI] [Google Scholar]
- 140.Yamane H., Ito T., Ishikubo H., Fujisawa M., Yamagata H., Kamiya K., Ito Y., Hamada M., Kanamori H., Ikawa H., et al. Molecular and evolutionary analysis of the Hd6 photoperiod sensitivity gene within genus oryza. Rice. 2008;2:56–66. doi: 10.1007/s12284-008-9019-2. [DOI] [Google Scholar]
- 141.Nakamichi N. Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol. 2014;56:594–604. doi: 10.1093/pcp/pcu181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. The Pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- 143.Murphy R.L., Klein R.R., Morishige D.T., Brady J.A., Rooney W.L., Miller F.R., Dugas D.V., Klein P.E., Mullet J.E. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA. 2011;108:16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pin P.A., Zhang W., Vogt S.H., Dally N., Buttner B., Schulze-Buxloh G., Jelly N.S., Chia T.Y., Mutasa-Gottgens E.S., Dohm J.C., et al. The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 2012;22:1095–1101. doi: 10.1016/j.cub.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 145.Huang C., Sun H., Xu D., Chen Q., Liang Y., Wang X., Xu G., Tian J., Wang C., Li D., et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA. 2018;115:E334–E341. doi: 10.1073/pnas.1718058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Faure S., Turner A.S., Gruszka D., Christodoulou V., Davis S.J., von Korff M., Laurie D.A. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. USA. 2012;109:8328–8333. doi: 10.1073/pnas.1120496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li X., Guo T., Mu Q., Li X., Yu J. Genomic and environmental determinants and their interplay underlying phenotypic plasticity. Proc. Natl. Acad. Sci. USA. 2018;115:6679–6684. doi: 10.1073/pnas.1718326115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu H., Liu H., Zhou L., Zhang Z., Zhang X., Wang M., Li H., Lin Z. Parallel domestication of the heading date 1 gene in cereals. Mol. Biol. Evol. 2015;32:msv148. doi: 10.1093/molbev/msv148. [DOI] [PubMed] [Google Scholar]