Abstract
Extensive dysregulation of chromatin-modifying genes in dear cell renal cell carcinoma (ccRCC) has been uncovered through next-generation sequencing. However, a scientific understanding of the cross-talk between epigenetic and genomic aberrations remains limited. Here we identify three ccRCC epigenetic clusters, including a clear cell CpG island methylatorphenotype (C-CIMP) subgroup associated with promoter methylation of VEGF genes (FLT4, FLT1, and KDR). C-CIMP was furthermore characterized by silencing of genes related to vasculature development. Through an integrative analysis, we discovered frequent silencing of the his-tone H3 K36 methyltransferase NSD1 as the sole chromatin-modifying gene silenced by DNA methylation in ccRCC. Notably, tumors harboring NSD1 methylation were of higher grade and stage in different ccRCC datasets. NSD1 promoter methylation correlated with SETD2 somatic mutations across and within spatially distinct regions of primary ccRCC tumors. ccRCC harboring epigenetic silencing of NSD1 displayed a specific genome-wide methylome signature consistent with the NSD1 mutation methylome signature observed in Sotos syndrome. Thus, we concluded that epigenetic silencing of genes involved in angio-genesis is a hallmark of the methylator phenotype in ccRCC, implying a convergence toward loss of function of epigenetic writers of the H3K36 histone mark as a root feature of aggressive ccRCC.
Introduction
Clear cell renal cell carcinoma (ccRCC) represents the most frequently occurring subtype of renal cell carcinomas (1). ccRCC is often characterized by 3p loss and frequent mutation or methylation of the tumor suppressor gene VHL (2). The key roles of epigenetic inactivation of chromatin-remodeling genes have been uncovered through exome sequencing, revealing frequent mutations of PBRM1 (33%), BAP1 (15%), SETD2 (16%), and KDM5C (8%) genes (3–5). This is consistent with the notion that cancer is not only a genetic disease but also an epigenetic disease (6). The two main mechanisms defining epigenetic alterations in cancer are related to DNA methylation and histone modification (6). However, an understanding of the prognostic impact of DNA methylation aberrations in ccRCC remains limited.
A few studies have assessed the global scale of DNA methylation aberrations in ccRCC as well as the role of the polycomb repressive complex (PRC) in this setting (2, 7, 8). The CpG island methylator phenotype (CIMP) was initially defined by Arai and colleagues in a Japanese ccRCC cohort with 13.4% incidence (7). Another Japanese study used integrative analysis to investigate DNA methylation in almost 100 ccRCC cases and identified three rather than two ccRCC subgroups, which included a small subgroup that displayed high methylation levels (12.3%) (8). However, neither study used integrative analysis to identify sets of genes repressed by DNA methylation. In addition, neither study investigated whether there were coordinated changes in the methylome associated with somatic mutations. In light of reports revealing extensive genetic heterogeneity in ccRCC, it is important to determine the level of epigenetic heterogeneity in DNA methylation in ccRCC (9).
In the landmark paper of The Cancer Genome Atlas (TCGA), investigation of DNA methylation data found that promoter DNA methylation increases with cancer stage and grade (2). However, the relevance of DNA methylation for classifying ccRCC subtypes and especially the role of coordinated cancer-specific DNA methylation have not been determined. In an effort to make these determinations, we investigated ccRCC methylome datasets from TCGA and two independent cohorts.
Materials and Methods
Characteristics and statistical analysis of the discovery set of DNA methylation arrays
We analyzed DNA methylation of 271 primary ccRCC samples assessed by TCGA using the Infinium 450 K arrays (Supplementary Table S1). We performed hierarchical unsupervised clustering of probes located in promoter CGIs and described global correlations of DNA methylation with mRNA expression. We used the most variable probes for clustering analysis after excluding methylation probes found in normal kidney tissue with β value >0.2.
Definition of promoters
We defined promoters as regions located between—1,000 and +1,000 base pairs from the transcription start site. After excluding all probes with β values >0.2 in any normal kidney tissue sample (n = 161), we used 67,994 probes located in promoter CGIs for analysis on the Illumina HumanMeth450 K platform. We used the χ2 test and log-rank test to perform correlations between the identified DNA methylation clusters and clinicogenomic tumor features as well as overall survival. CGI was defined using Illumina Infinium HumanMethylation450 K annotation file.
Integrative analysis of DNA methylation and expression and histone
Chromatin immunoprecipitation-sequencing data.
We obtained chromatin immunoprecipitation-sequencing (ChlP-Seq) peak data for histone marks H3K4me3, H3K36me3, and H3K27me3 in the normal kidney cell line from UCSC ENCODE Histone Modification Tracks (https://genome.ucsc.edu/ENCODE/dataMatrix/encodeDataMatrixHuman.html). We extracted the histone data o f fetal kidney samples for H3K4me3, H3K36me3, and H3K27me3 from the Roadmap Epigenomics Project (http://www.roadmapepigenomics.org/).
Validation datasets
We validated our C-CIMP subgroup using the Infinium 27 K arrays in an independent dataset of 160 ccRCCs from TCGA (2). We applied supervised clustering with probes that were differentially methylated between the three groups obtained from Infinium 450 K arrays. In promoter CGI probes, 8,334 probes of 18,037 Promoter CGI probes in HM27 were present in the HM450 Promoter CGI probes. In addition, to explore the association between our 3 epi-clusters and response to sunitinib, which is standard first-line therapy for patients with metastatic ccRCC, we applied supervised clustering to the dataset from Beuselinck and colleagues (10). We assessed progression-free survival according to the subgroup classifications we established, as well as for selected genes.
To explore whether ccRCC cases with NSD1 methylation harbored a methylome alteration similar to that of Sotos syndrome, we performed supervised clustering of DNA methylation on ccRCC from TCGA training dataset (n = 271 cases) using the methylation signature associated with NSD1 mutations reported in Sotos syndrome by Berdasco and colleagues (11). We used Fisher exact test to evaluate the association between the epi-clusters and frequent somatic mutations in kidney tumors.
Bisulfite pyrosequendng
To estimate the frequency of NSD1 methylation, we applied bisulfite pyrosequendng to DNA extracted from 222 primary ccRCC spedmens and 10 adjacent normal kidney samples from Pitié-Salpêtrière hospital. All patients had previously provided informed consent for tumor collection and analysis. The study was approved by the ethical committee of the Pitié-Salpêtrière Hospital (IDF-6, lie de France). The collection and use of tissues followed procedures in accordance with the ethical standards formulated in the Dedaration of Helsinki. All cases were deidentified prior to analysis. We defined samples with NSD1 methylation as those with average methylation levels that were greater than those of normal kidney samples plus three SDs. We used Fisher exart test to determine assodations between methylation of NSD1 and dinicopathologic features; we considered a t test less than 0.05 as statistically significant. Analysis of NSD1 methylation consisted of performing two-step nested PCR. The primers of first Step F1 and R1 are as follows: primer F1—»GAGGGTAGGTGl I’lAGTGGA and primer R l—»CATΓCCCATCCCCΓCACCΓACCΓ. The primers of second step are as follows: primer F2—»TGGGGAGl’l iGGGTΓGTAAl’l’lAAGAT and primer R l—»CATΓCCCATCCCCΓCACCΓACCΓ. A calibration curve using in vitro methylated DNA (SssI enzyme), which is diluted with normal DNA then bisulfite treated and pyrosequenced, was performed. Data points were correrted accordingly.
NSD1 immunostaining
Whole slides from ccRCC with NSD1 promoter methylation (n = 10) and without (n = 10) were selerted. Ten normal kidney samples were also used as control. The slides were incubated with rabbit polydonal anti-NSDl antibody (ABE1009, Merck). The primary antibody was detected by using commerdally available detection kit (EnVisionTMFLEX+, Dako) following the manufacturer’s protocol. Slides were washed with Tris-buffered saline (TBS, 0.1 mol/L, pH = 7.4), 3–5 times after each step. Finally, the sections were counterstained with Mayer hematoxylin and mounted with Biomount (BIO-OPTICA). In the negative control tissue sections, the primary antibody was replaced by isotype spedfic nonimmune rabbit IgG. Tissue sections from normal kidney, were used as a positive control for NSD1 expression. The sections were evaluated by light microscopic examination on Olympus BX51 microscope. Each slide was evaluated for NSD1 immunostaining by using a semiquantitative scoring system of the percentage of positive neoplastic cells. NSD1 protein expression was evaluated by an expert pathologist (E. Compérat) who scored as positive if nudear reactivity was observed in tumor cells. The semiquantitative scale was based on the percent of immunoreactive neoplastic cells
Spatial heterogeneity of DNA methylation.
From an independent cohort of 20 primary ccRCC samples, we randomly selerted multiple cores within the primary tumor sample and performed pyrosequendng for NSD1 methylation. Overall, a median of 3 sections per primary kidney tumor were available.
Pathway analysis
We performed pathway analysis using the default settings for DAVID (https://david.ndfcrf.gov/; refs. 12, 13). Genes downregulated among the subgroups were defined as those with a fold-change ≤ 2, P < 0.05, and FDR < 0.05. Genes upregulated among the subgroups were defined as those with a fold-change ≥2, P < 0.05, and FDR < 0.05.
Analysis of histone status of H3K27me3 and H3K4me3 in fetal kidney and normal kidney tissue samples
To examine the histone modification profiles of mRNA genes for H3K4me3 and H3K27me3 in fetal kidney and normal kidney tissues, we analyzed the promoter regions of mRNA genes for overlap with histone mark enrichment peaks. Specifically, mRNA was defined as marked (associated with a specific histone mark) if the peak from ChlP-Seq data for a specific histone mark was located within ± 5 kb from the transcription start site (around promoter regions) for the mRNA.
Integrative analysis to identify epi-drivers
We used integrative analysis to investigate genes that are repressed through DNA methylation. For this purpose, we defined an arbitrary cutoff of gene expression with the value of fragments perkilobase of exon per million fragments mapped (FPKM) > 3 in unmethylated ccRCC; below this threshold, genes were considered to have low expression. We detected the genes and methylated probes that were significantly differentially expressed between the respective C-CIMP and no-CIMP epi-clusters, and then identified the significantly downregulated genes that were also differentially methylated. We used a starburst plot to visualize the results of the integrative analysis.
Analysis of SETD2 mutations
We used targeted hybrid capture-based next-generation sequencing in collaboration with Cancer Genetics, Inc., to detect VHL and SETD2 somatic mutations in cancer and adjacent normal tissue samples. Alignment, variant calling, and filtering, and annotation were performed essentially as described previously (14). All samples achieved >95% targets (2,400 total) with >95% of each target at >100× with a 5% variant allele frequency limit of detection or 2% for variants detected in ≥ 2 samples from the same patient.
Results
Identification of a CpG island methylator phenotype (CIMP) associated with patient outcome
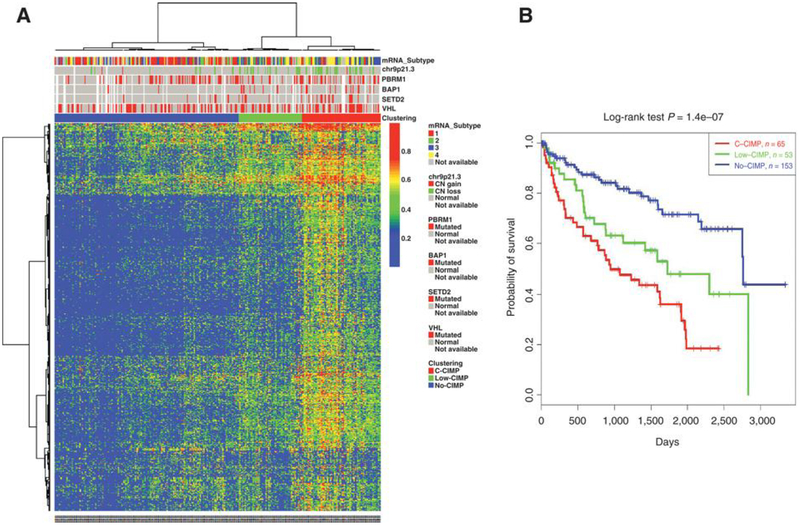
We analyzed DNA methylation patterns in a discovery set of 271 primary ccRCCs from TCGA that were assessed using the Illumina HumanMeth450 K platform. After excluding all probes with β values ≥0.2 in any of the normal kidney samples (π = 161), 67,994 probes remained; they were located in promoter CpG islands (CGI) with available DNA methylation data across all samples. Using the most variable probes, we performed unsupervised hierarchical clustering to identify ccRCC subgroups. We identified three robust DNA methylation epi-clusters (Fig. 1A). Epi-cluster Cl (n = 65; 21.8%) displayed markedly high DNA methylation levels reminiscent of the coordinated cancer-spedfic methylation seen in the CIMP of colorectal cancer; thus, we labeled this subgroup as having a clear cell CIMP (C-CIMP). The two other epi-clusters, C2 (n = 53; 17.8%) and C3 (n = 153; 51.4%), harbored low or no methylation levels, respectively. We thus respectively labeled the C2 and C3 epi-clusters as low-CIMP and no-CIMP subgroups.
Figure 1.
Clustering of TCGA samples in ccRCC reveals a CpG Island methylator phenotype with poor outcome. A, Unsupervised hierarchical clustering for the most variable methylated probes in promoter CpG islands among 271 ccRCCs. The (β value) level of DNA methylation is represented by the color scale. Each column represents a sample; each row a probe set. The transcriptomic subtype in TCGA, the copy number variation at 9p23.1 locus (CDKN2A), somatic mutation status of four genes (PBRM1, BAP1, SETD2, and VHL) are indicated by red, green, and gray squares, with annotations in the legend. B, Kaplan-Meier curves showing distinct outcomes of patients according to the three subgroups of DNA methylation classification, with patients belonging to C-CIMP subgroup having the worst outcome.
We assessed whether tumors belonging to the C-CIMP subgroup were associated with distinct clinicopathologic tumor features, and found that C-CIMP tumors harbored higher pathologic Fuhrman grades (P < 10–5) and higher TNM stages (P < 10–5; Table 1). We then analyzed the association between our three subgroups of DNA methylation classification and overall survival (OS). We found that patients with C-CIMP had the worst overall survival when compared with the two other subgroups (P = 1.4 × 10–7; Fig. 1B). The median OS for patients with CCIMP tumors was 2.6 years [95% confidence interval (Cl), 1.94.4], which was significantly lower than the 4.7 years [95% Cl, 2.4-not reached (NR)] and 7.6 years [95% Cl, 7.5-NR] experienced by patients with low-CIMP and no-CIMP tumors, respectively (P = 1.4 × 10–7). Multivariate analysis revealed that CCIMP was not independently associated with poor overall survival when using other clinicopathologic features such as Fuhrman grade and TNM stage. This suggests an interplay between DNA methylation and known prognostic features in ccRCC.
Table 1.
Association between clinicopatholog¡c tumor features of ccRCCs in training and validation sets of TCGA and DNA methylation subgroups
| Training dataset (450 K) | Validation dataset (27 K) | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | No-CIMP (n = 53) |
Low-CIMP (n = 153) |
C-CIMP (n = 65) |
P | No-CIMP (n = 46) |
Low-CIMP (n = 75) |
C-CIMP (n = 39) |
P |
| Age (years) | 63 | 59 | 65 | 0.003 | 52 | 64 | 60.5 (37–85) | 0.0002 |
| Median (range) | (32–86) | (26–90) | (41–90) | (34–80) | (42–86) | |||
| Gender | 0.001 | 0.24 | ||||||
| Male | 42 | 66 | 50 | 25 | 41 | 32 | ||
| Female | 11 | 87 | 15 | 14 | 34 | 14 | ||
| Laterality | 0.28 | 0.31 | ||||||
| Right | 27 | 88 | 30 | 19 | 43 | 20 | ||
| Left | 26 | 65 | 35 | 20 | 32 | 26 | ||
| Histologic grade | <10e−5 | <10e−5 | ||||||
| G1 | 0 | 4 | 0 | 2 | 2 | 0 | ||
| G2 | 10 | 89 | 15 | 26 | 38 | 9 | ||
| G3 | 27 | 50 | 28 | 7 | 29 | 21 | ||
| G4 | 16 | 9 | 21 | 0 | 6 | 16 | ||
| Gx | 0 | 1 | 1 | 3 | 0 | 0 | ||
| Pathologic TNM stage | <10e−5 | <1.2e−5 | ||||||
| I | 12 | 103 | 11 | 30 | 37 | 12 | ||
| II | 6 | 13 | 5 | 4 | 11 | 7 | ||
| III | 21 | 25 | 21 | 5 | 20 | 12 | ||
| IV | 14 | 12 | 28 | 0 | 7 | 15 | ||
| Pathologic T | <0.0001 | 0.0002 | ||||||
| T1 | 14 | 103 | 12 | 30 | 37 | 13 | ||
| T2 | 8 | 16 | 9 | 4 | 11 | 9 | ||
| T3 | 29 | 33 | 39 | 5 | 27 | 22 | ||
| T4 | 2 | 1 | 5 | 0 | 0 | 2 | ||
| Pathologic N | 0.06 | 0.005 | ||||||
| N0 | 26 | 72 | 23 | 16 | 47 | 24 | ||
| N1 | 4 | 2 | 3 | 0 | 0 | 5 | ||
| Pathologic M | <0.0001 | <0.0001 | ||||||
| M0 | 40 | 141 | 38 | 39 | 67 | 31 | ||
| M1 | 13 | 12 | 27 | 0 | 8 | 15 | ||
Independent validation of C-CIMP subgroup
We then considered whether our classification of three ccRCC subtypes using 450 K arrays was also valid in an independent dataset of ccRCC that was assessed for DNA methylation using Infinium 27 K arrays. Supervised clustering for DNA methylation revealed three DNA methylation epi-clusters that were consistent with C-CIMP, low-CIMP, and no-CIMP subgroups (Supplementary Fig. S1A). Similar to the training set, the C-CIMP subgroup was enriched for tumors with higher pathologic Fuhrman grade (P < 10−5) and TNM stage (P < 1.2 × 10−5; Table 1). In addition, patients in the C-CIMP subgroup had the worst median OS when compared with that of the other subgroups (P = 0.0067); the median OS was 4.5 years (95% Cl, 3.4–6.2) for patients in the C-CIMP subgroup, 7.1 years (95% Cl, 6.4-NR) for those in the low-CIMP subgroup, and was not reached (95% Cl, 5.7-NR) forpatients in the no-CIMP subgroup (Supplementary Fig. S1B).
Methylation of VEGF receptor genes in C-CIMP subgroup
Overall, 13,439 of the 67,994 (19.8%) probes were differentially methylated between the C-CIMP and no-CIMP epi-clusters [false discovery rate (FDR) <0.05; Supplementary Fig. S2A], Using the most stringent criteria (average β-value in no-CIMP epi-cluster <0.2 and average β-value in C-CIMP epi-cluster >0.4), we identified 369 probe sets related to 242 genes that define C-CIMP (Supplementary Table S2). We then ranked the probes by decreasing adjusted P values and increasing β-value difference for probes with fold-change > 4 to identify the top hypermethylated probes within the C-CIMP subgroup. The highest ranked probes were related to the following genes: GALR1, FLT4, VWC2, SOX8, ASCL2, ASCL4, and CLEC2L.
Using DAVID to perform gene ontology analysis, we found that the C-CIMP subgroup was enriched for genes related to Homeobox (P = 2 × 10−10), developmental proteins (P = 1.3 × 10−5), chordate embryonic development (P = 6.4 × 10−5), and neuroactive ligand-receptor interaction (P = 2.4 × 10–13). Of the 16 markers of CIMP previously identified in ccRCC (7), 8 belong to the set of 242 genes that define C-CIMP: FAM150A, ZFP42, ASCL2, RIMS4, TRH, ZNF154, GRM6, and KHDRBS2.
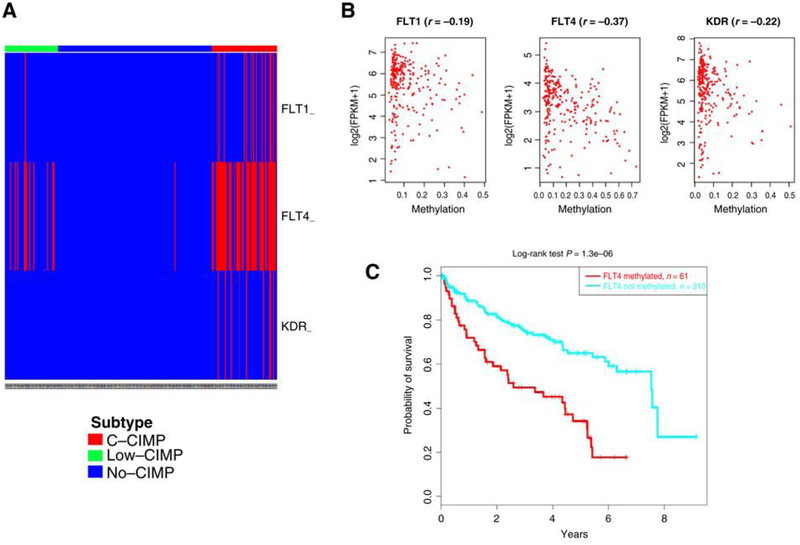
To determine the set of genes regulated epigenetically in CCIMP, we explored genes that show gains in DNA methylation in the C-CIMP subgroup (average β-value ≥ 0.25) compared with the no-CIMP subgroup (average β-value <0.2; FDR< 0.05), as well as downregulation of their expression (FDR<0.05). Amongthese, we identified 75 probes related to 34 gene promoters (Supplementary Fig. S2B). DAVID analysis revealed that those genes were enriched for VEGF receptors (P = 2.9 × 10−5) and vasculature development (P = 6.7 × 10−5). VEGF receptor genes FLT4, FLT1, and KDR were methylated in 22.5%, 6.2%, and 4.4% of ccRCC samples, respectively (Fig. 2A). Importantly, the highest inverse correlation between DNA methylation and expression was related to the FLT4 gene (Fig. 2B). Among genes associated with angio-genesis, FLT4 was also associated with poor patient outcome (Fig. 2C); this was not the case for FLT1 and KDR genes (not shown).
Figure 2.
Charting methylation of VEGF receptors in ccRCC. A, Heatmap for methylation of FLT4, KDR, and FLTÌ in TCGA ccRCC data. B, Correlation of methylation in VEGF receptor genes and expression assessed by RNA-seq. C, Kaplan-Meier curves for overall survival of patients according to FLT4 methylation status.
We then considered whether the overall gene expression patterns of the C-CIMP subgroup differed from those of the no-CIMP subgroup. We found that 377 genes were significantly down-regulated and 477 genes were significantly upregulated in the CCIMP subgroup as compared with the no-CIMP subgroup. The differentially downregulated genes were related to vasculature development (P = 6.2 × 10−10), cell adhesion (P = 7.8 × 10−9) and cell migration (P = 9.1 × 10−8). Most of the significantly upregulated genes were related to mitosis (P = 2 × 10−19), cell division (P = 1.8 × 10−11), and regulation of cell proliferation (P = 3.3 × 10−6). Gene set enrichment analysis confirmed the activation of the mitosis and hypoxia pathways in this setting, which is consistent with VEGF receptor inactivation.
Association between copy number alterations and somatic mutations in C-CIMP subgroup
To determine recurrent alterations in the C-CIMP subgroup, we analyzed statistically significant copy number variations in this subgroup as compared with the other subgroups. We identified frequent deletions in chromosomes 9p21.3 and 9p23, which are known to be associated with aggressive ccRCC (Fig. 1A). We also identified a frequent gain in chr8q24.22, which we previously identified as being associated with aggressive ccRCC using long noncoding RNA (IncRNA) subtype classification (15). Of note, chr8q24.22 contains the following overexpressed IncRNAs: PVΓ1, RP11–47304.5, and RP11–62901.2. We then looked at the IncRNAs differentially expressed between the three clusters and identified PVT1 as overexpressed in C-CIMP, which is consistent with 8q gain. These data suggest that MYC might be activated in the C-CIMP subgroup through PVT 1 expression.
Finally, we investigated the association between somatic mutations and subgroups of DNA methylation and found that the CCIMP and low-CIMP subgroups harbored increased mutational load as compared with the no-CIMP subgroup (P = 0.0006; Supplementary Fig. S3A). In addition, C-CIMP was associated with increased mutational rates of BAP1 (P = 8.6 × 10−6) and SEΓD2 (P = 0.002) genes (Supplementary Fig. S3B).
Histone marks of C-CIMP-assodated gene promoters are similar to those of no-CIMP and low-CIMP subgroups
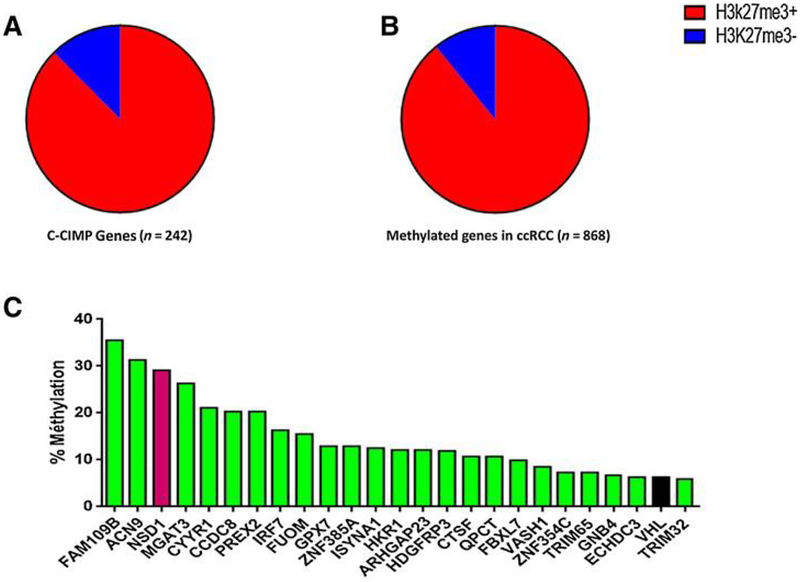
Of the 242 highly methylated genes in the C-CIMP subgroup, 212 (87.6%) were marked by H3K27me3 in fetal kidney samples (Fig. 3A), as compared with 5157 genes (51.1%) marked by the PRC in fetal kidney samples (P< 0.0001). Likewise, 204 of the 242 (84.3%) genes were marked by H3K27me3 in the normal renal samples as compared with2,363 genes (23.4%) marked by PRCin the normal kidney samples (P < 0.0001). These data highly indicate that genes that are silenced through DNA methylation in ccRCC are not random and are marked by the PRC in normal kidney tissue. Consistent with these data, 162 of 242 genes showed no expression level in ccRCC (Supplementary Table S2). Based upon cutoff (FDR = 0.25), 15 genes were suppressed, 6 were increased, and 221 of 242 genes did not see gene expression change (Supplementary Table S2).
Figure 3.
Correlation between DNA methylation and polycomb mark. A, Distribution of CCIMP genes marked by H3K27me3 in fetal kidney samples. B, Distribution of genes that gain DNA methylation in ccRCC according to H3K27me3 mark status in fetal kidney samples. C, Twenty-five genes identified as frequently methylated and repressed in ccRCC. Rate of Λ/SD7 methylation (red) is high compared with that of VHL (black).
We investigated genes that gained DNA methylation in more than 5% of ccRCC samples; we identified 868 genes, including 194 genes that are the hallmark of the C-CIMP subgroup. Of note, 774 (89.2%) and 684 (78.8%) of those genes were, respectively, marked by H3K27me3 in the fetal kidney (Fig. 3B) and the normal kidney tissue, suggesting that these features are not defining characteristics of the C-CIMP subgroup but are features of ccRCC.
Identification of NSD1 methylation as an epi-driver of ccRCC
We investigated the set of genes that gained DNA methylation in at least 5% of ccRCCs and are downregulated. Using integrative analysis, we identified 108 gene candidates that we believe represent the most important epi-drivers in ccRCC. VHL was among those genes and was methylated in 6.2% of our total samples, which is consistent with TCGA analysis and validates the accuracy of our approach.
To identify the genes that might act as tumor suppressor genes, we looked for genes harboring only the H3k4me3 histone mark in the fetal kidney tissue and that were without any H3K27me3 mark. This was based on a recent finding that tumor suppressor genes harbor broad H3K4me3 peaks in samples of normal tissue (16). Of 108 genes, 25 were identified, including VHL (Fig. 3C). We found two other members of the ubiquitin conjugation pathway that had not been demonstrated to be involved in renal carcinogenesis, namely FBXL7 (10%) and TRIM32 (5.9%). We also discovered NSD1, a SET domain histone methyltransferase that primarily dimethylates nucleosomal histone H3 lysine 36 (H3K36). NSD1 was methylated in 29.1% of ccRCCs. Correlation with clinicopathologic features identified a higher rate of NSD1 methylation in metastatic versus localized ccRCC cases (52% vs. 16%, P < 0.0001) as well as in tumors with Fuhrman grades III—ΓV (28% vs. 16%, P = 0.02).
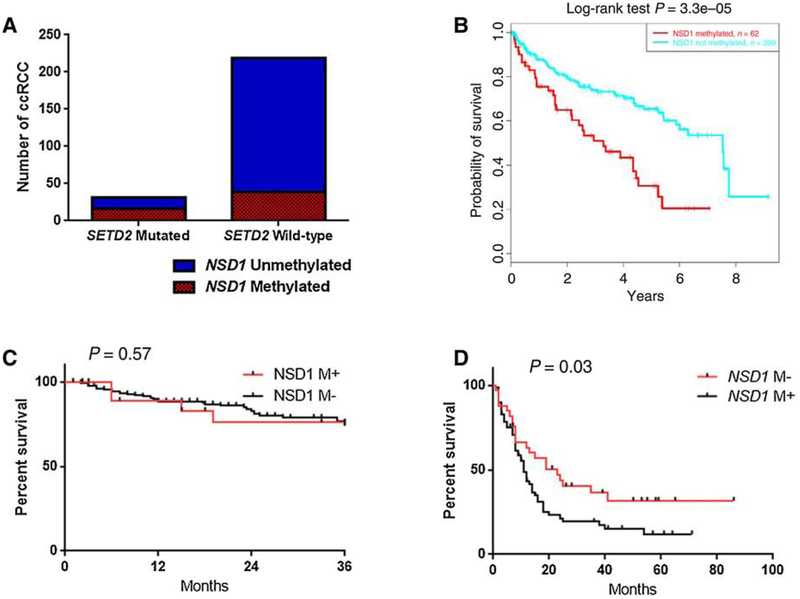
We then considered whether there was a correlation between NSD1 methylation and SEΓD2 mutations using the TCGA training set. Strikingly, 16 of31 (51.6%) ccRCC cases from TCGA with SETD2 mutations harbored concomitant NSD1 methylation as compared with 39 of 219 (17.8%) ccRCC cases with no SETD2 mutations (P < 0.0001; Fig. 4A). We thus conduded that ccRCC converges to alter the methylation status of H3K36 through multiple aberrations of the pathway by silendng several enzymes, induding SETD2 and NSD1. To assess the impact of alterations in NSD1 methylation in TCGA dataset (450 K), we analyzed the OS of 2 71 patients with ccRCC and found that NSD1 methylation was assodated with poor OS (P = 3.3 × 10−5; Fig. 4B).
Figure 4.
Prognostic impact of NSDΊ methylation and correlation with SETD2 mutation. A, Association between NSDΊ methylation and SETD2 somatic mutations in TCGA dataset shows high rate of NSDΊ methylation in tumors with SETD2 mutations. B, Kaplan-Meier curves for overall survival according to NSDΊ methylation in TCGA cohort (450 K). C, Kaplan-Meier curves for recurrence-free survival according to NSDΊ methylation in Pitié-Salpêtrière cohort. D, Progression-free survival according to NSDΊ methylation in patients with ccRCC treated with sunitinib (Beuselinck cohort).
Analysis of NSD1 methylation status in two independent cohorts
We then assessed NSD1 methylation using pyrosequendng in an independent cohort of 222 ccRCCs and 10 samples of normal kidney tissue adjacent to kidney tumors from the Pitié-Salpêtrière cohort. Median methylation in the normal kidney samples was 1.04% (SD = 1.39%). Using a stringent cutoff to define NSD1 methylation (above the methylation rate of normal tissue plus 3 SDs), 26 of 222 (11.7%) ccRCC samples harbored NSD1 methylation. Consistent with TCGA cohort, tumors with NSD1 methylation were more often metastatic (n = 6/22; 27.2%) as compared with those without NSD1 methylation (n = 16/200; 8%; P = 0.03); although there was no difference between NSD1 methylated cases in terms of tumor size (P = 0.57). In addition, NSD1 methylated tumors displayed higher Fuhrman grades of III—ΓV (n = 15/26; 57.7%) as compared with those without NSD1 methylation (n = 66/196; 33.6%; P = 0.007). With a median follow-up of 21 months, median recurrence-free survival was not different between the patients with and those without NSD1 methylation (P = 0.57; Fig. 4C).
To assess whether NSD1 was methylated in a third independent cohort, we used the Beuselinck study, which investigated patients with metastatic ccRCC who were treated with sunitinib as a first-line treatment (10). We found that NSD1 was methylated in 67.6% of metastatic ccRCCs (n = 69/102), which is consistent with the 52% rate that we discovered in metastatic ccRCCs from TCGA. Methylation of FLT4, KDR, and FLT1 (13/107) had also been found, respectively, in 40.8%, 11.8%, and 12.1% of patients with metastatic ccRCC who were treated with sunitinib (10). Importantly, among patients treated with sunitinib, VEGF receptor methylation of FLT4, KDR, and FLT1 was not assodated with progression-free survival (Supplementary Fig. S4A-S4C), but NSD1 methylation was assodated with the lowest progression-free survival (P = 0.03; Fig. 4D). The C-CIMP subgroup was not assodated with the response to sunitinib in the Beuselinck study (Supplementary Fig. S5A and S5B).
NSD1 promoter methylation and NSD1 immunostaining
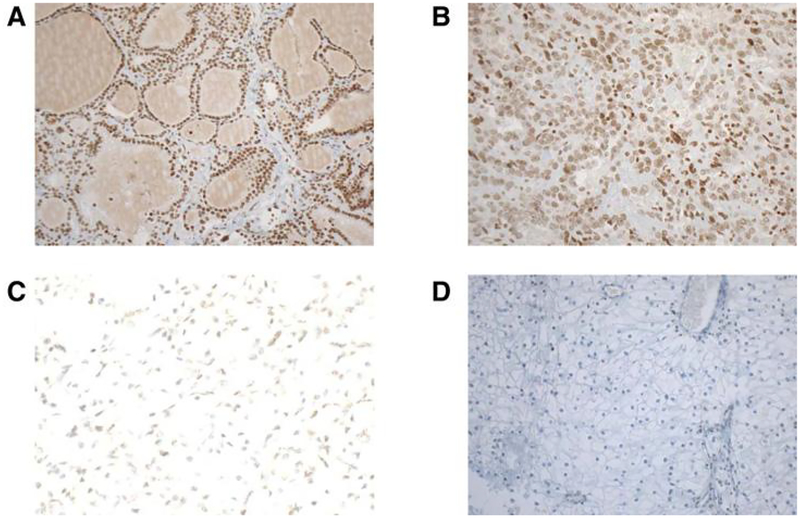
To assess the correlation between epigenetic silendng of NSD1 gene and the protein levels in ccRCCs assessed by IHC, 10 ccRCC tumor samples with NSD 1 promoter methylation were compared with 10 ccRCC tumors without NSD1 methylation; ten normal adjacent kidneys were also assessed. Of note, we observed 100% NSD1 expression both in normal kidneys (n = 10) and in clear cell RCC without NSD1 methylation (n = 10; Fig. 5A and B). Conversely, in NSD1-methylated ccRCC cases, NSD1 expression was completely lost in 5 cases, reduced (10%–50%) in 3 cases and normal in 2 cases (Fig. 5C and D).
Figure 5.
Representative images of IHC staining for NSDΊ protein on whole slides of ccRCCs. A and B, Microphotographs are from a ccRCC sample positive for NSDΊ expression. A, Original magnification, ×ΊO. B, Original magnification, ×20.C,Weakstaining in one ccRCC case with NSDΊ promoter methylation. Original magnification, ×20. D, Negative staining in one ccRCC case with NSDΊ promoter methylation. Original magnification, ×ΊO.
Association between NSD1 promoter methylation and the ccRCC methylome
We then considered whether NSD1 epigenetic silencing through DNA methylation was associated with a specific genome-wide methylome signature of ccRCC, as it is the case for NSD1 mutations in Sotos syndrome (17). We used the DNA methylation signature associated with NSD1 mutations as reported for the Sotos syndrome (17) and applied it to theTCGA cohort of 271 ccRCCs assessed by Infinium 450 K arrays. We obtained two epi-clusters, namely clusters 1 and 2 (Supplementary Fig. S6A). Strikingly, cluster 1 was highly enriched for tumors harboring NSD1 methylation (55.7%; n = 44/79) as compared with 9.4% (n = 18/192) in cluster 2 (P = 2.7 × 10−15; HR = 12; 95% Cl, 6.0–24.9). Cluster 1 was also enriched for tumors harboring SETD2 mutations (27.4%; n = 20/73) relative to 6.2% (n = 11/177) in cluster 2 (P = 2.3 × 10−5; HR = 5.6; 95% Cl, 2.4–14). We found that ccRCCs with the NSD1 mutation genome-wide methylome signature were associated with poor overall survival as compared with those without the signature (P = 0.0021) ; Supplementary Fig. S6B).
Spatial heterogeneity o f NSD1 in primary ccRCC tumors
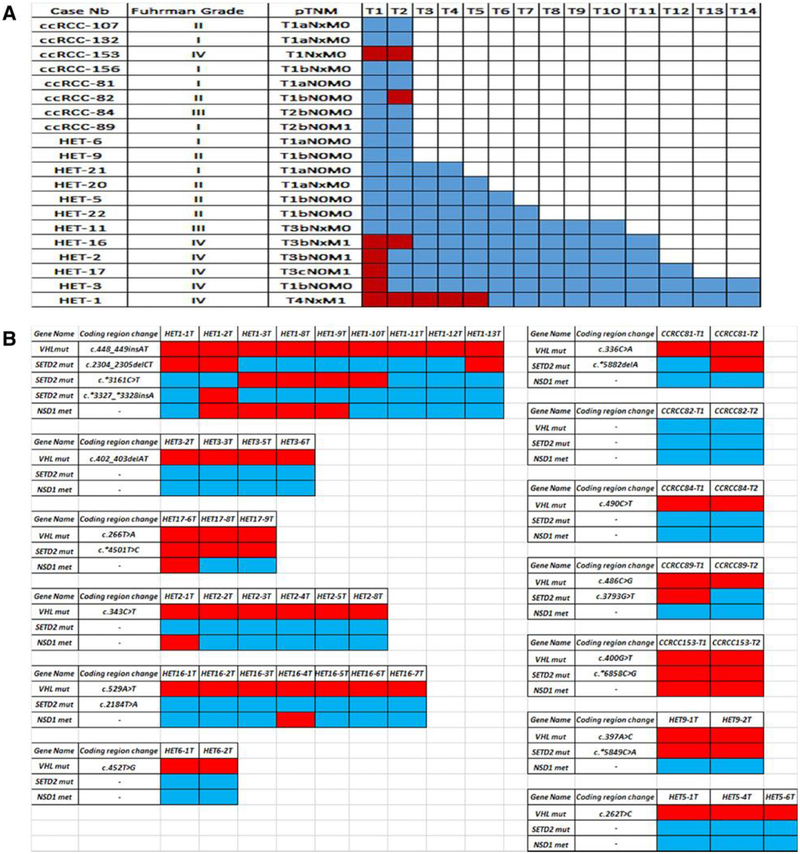
As ccRCC is a genetically heterogeneous disease, we analyzed NSD1 in a fourth independent set of 20 primary ccRCCs from the Pitié-Salpêtrière cohort. The median number of core sections analyzed for NSD1 methylation was 3 per primary tumor (range: 2–13), with a total of 114 successfully analyzed. We found heterogeneity of NSD1 methylation in at least one section of the primary tumor in 7 of 20 samples (35%; Fig. 6A). That was far higher than the approximately 11.7% rate observed in the analysis of one tumor core. Strikingly, 100% (n = 6/6) of the ccRCC cases that harbored NSD1 methylation were grade IV tumors as compared with none of the cases that were grade I—III tumors (P <0.0001). In addition, 80% (n = 4/5) of metastatic ccRCCs harbored NSD1 methylation as compared with 13.3% (n = 3/15) of the cases with no metastasis at diagnosis (P = 0.03), which is consistent with the data on 222 primary ccRCC cases.
Figure 6.
Heterogeneity of NSD1 methylation and SETD2 somatic mutation. A, Heterogeneity of NSD1 methylation in 20 primary ccRCCs. Each column represents a section; each row a primary ccRCC sample. Red, NSD1 methylation; blue, no DNA methylation. B, Association between NSD1 methylation and SETD2 mutations in a cohort of 13 primary ccRCCs. Red, methylation of NSD1 and mutations of SETD2\ blue, unmethylated NSD1 and wild-type SETD2. Annotation according to NM_000551 for VHL and NM_014159 for SETD2.
We then decided to explore the association between SETD2 mutations and NSD1 methylation in 13 ccRCC cases for which material was available. Of 114 samples assessed for DNA methylation, 47 sections related to 13 cases were also assessed for SETD2 mutations (Fig. 6B). Strikingly, we observed a high rate of mutation of SETD2 (46.1%, n = 6/13), with mutational convergence of SETD2 in different loci observed in one case (HET-1). Consistent with our analytic results on the data obtained from TCGA, we found an association between SETD2 mutations and NSD1 methylation (P = 0.0049).
Discussion
To our knowledge, this work represents the first integrative analysis showing frequent epigenetic silencing of NSD1 as the sole methylated and repressed H3K36 methyltransferase in ccRCC. Strikingly, compared with localized ccRCC, metastatic ccRCC harbored high rates of NSD1 methylation that ranged from 27.2% in Pitié-Salpêtrière cohort to 67.6% in Beuselinck cohort; differences regarding NSD1 methylation rates might be related to distinct distribution of clinicopathologic tumor features. Consistent with data showing heterogeneity of SETD2 mutations in ccRCC with convergence toward mutations in the SWI-SNF complex (18), our analysis uncovered an association between SETD2 somatic mutations and NSD1 methylation in two different cohorts, suggesting that loss of H3K36me3 in ccRCC might occur through crosstalk between the inactivation of NSD1 and SETD2. Although descriptive, our study suggests an epigenetic drift toward inactivating the H3K36 pathway in metastatic ccRCC. Further explorations using animal models should be undertaken to examine whether the initiating event of metastasis in the context of ccRCC is SETD2 mutation or NSD1 methylation. Future studies are also needed to clarify genetic hierarchy and temporal epigenetic changes in the clonal history of renal carcinomas.
Consistent with the requirement of a minimum of three distinct ∞res for accurate tumor genotyping (19), we herein demonstrated heterogeneity of NSD1 methylation with the rate of heterogeneity increasing with the number of cores analyzed, reaching a rate of 100% in metastatic cases. Although not causative, ccRCCs with NSD1 methylation harbor the genome-wide methylome signature of Sotos syndrome (17), suggesting that the silencing of this histone methyltransferase affects genes involved in cellular morphogenesis.
Several studies have reported a possible oncogenic role for NSD1 in acute myeloid leukemia through the cryptic NLΓP98-NSD1 fusion t(5;ll)(q 35;p l5.5) (20). Other reports have implied that NSD1 functions as a tumor suppressor gene, as we observed for ccRCC (20). Indeed, NSD1 silencing through CpG island-promoter hypermethylation has been frequently observed in neuroblastomas and gliomas and has predicted poor patient outcomes (11). Restoration of NSD1 expression in neuroblastoma and glioma cell lines led to decreased cell proliferation, which is consistent with the tumor-suppressive effect of NSD1 (11). This is also consistent with the genomic analysis of squamous cell carcinomas of the head and neck, and endometrial and gastric adenocarcinomas that revealed recurrent loss-of-function mutations in NSD1 in approximately 10% of cases (21, 22). Most importantly, NSD1 hypermethylation was a predictor of poor outcome in ccRCC. These findings highlight the importance of NSD1 epigenetic inactivation in ccRCCs, which, concomitantly with SETD2 mutations, leads to a disrupted histone methylation landscape.
Patients with localized ccRCC of the C-CIMP subtype displayed poor outcomes, but this association was not independent from other clinicopathologic parameters, suggesting that aggressive tumors might acquire epigenetic aberrations during their evolution. Of note, genes that gain DNA methylation in CIMP were in majority repressed and marked by H3K27me3 in normal kidneys consistent with previous findings (23). This is also consistent with the observation that Polycomb Group (PcG) targets in embryonic stem cells (ESC) are 12-fold more likely to become methylated in cancer (24). Strikingly, the key feature of C-CIMP was methylation of VEGF receptor genes, in particular FLT4, as a key feature of C-CIMP. Inverse correlation between FLT4 methylation and expression was observed to be consistent with repression of these receptors. Tumors with C-CIMP seem to rely on a high mitotic rate, which may be associated with aggressiveness. Despite the methylation of those receptors, response to sunitinib treatment was similar in ccRCC cases without methylation of FLT4, KDR, or FLT1 genes, which may result from heterogeneous methylation of these receptors. This provides a rationale for targeting ccRCC using a hypomethylating inhibitor in association with a VEGF inhibitor, as recently shown in vitro using several cell lines (25).
Predicting the response to VEGF inhibitors has been an important goal in cancer research (14). Whether these agents work via an effect on tumor cells or the environment remains unclear. Resistance to sunitinib treatment could not be predicted through our classification of ccRCC into three subgroups, nor was it associated with the methylation of a VEGF receptor. The heterogeneous nature of the methylation of VEGF receptors, as we showed for FLT4, might limit the capacity of such measurement categories to predict treatment sensitivity. Epigenetic therapy might work in a large population of patients with ccRCCs that harbor these aberrations. Finally, tumors with C-CIMP showed enrichment with BAP1 and SETD2 mutations consistent with Sato dataset (8). Consistent with these data, Tiedemann and colleagues showed that SETD2-depleted cell lines exhibit a DNA hypermethylation phenotype coinciding with ectopic gains in H3K36me3 (26). Likewise, SETD2-mutant primary ccRCC, papillary RCCs, and lung adenocarcinomes all demonstrated a DNA hypermethylation phenotype that segregated tumors by SETD2 genotype (26). Mechanistic data are further needed to clarify the link between H3K36 methylation and CpG island methylation.
In summary, our study provides evidence about the involvement of alterations of NSD1 concomitantly with SETD2 in metastatic ccRCC. Epigenetic heterogeneity of NSD1 methylation seems to mirror SETD2 mutational heterogeneity, leading to a convergence toward alterations in the kidney epigenetic machinery. Thus, targeting the H3K3 6 pathway may represent a potential avenue in the management of patients with an aggressive ccRCC phenotype.
Supplementary Material
Acknowledgments
We would like to thank FONDAΉON AVEC and Magali Kemaleguen for her technical help in performing NSD1 methylation calibration curve.
Grant Support
This work was supported in part by grants from Fondation AVEC and the Genitourinary Cancers Program and BISR of the CCSG shared resources at MD Anderson Cancer Center.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
J. Houldsworth is a vice-president of Research and Development at Cancer Genetics Inc. and has ownership interest (including patents) in Cancer Genetics, Inc. J.-P. Spano is a consultant/advisory board member for Roche, MSD, Pfizer, Gilead, and Novartis. G.G. Malouf reports receiving a commercial research grant from Pfizer and Novartis and is a consultant/advisory board member for BMS, Novartis, and Pfizer. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Shuch B, Amin A, Armstrong AJ, Eble JN, Ficarra V, Lopez-Beltran A, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol 2015;67:85–97. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 2012;44:751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011;469:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brugarolas J PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer J 2013;19:324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baylin SB, Jones PA. A decade of exploring the cancer epigenome-biological and translational implications. Nat Rev Cancer 2011;11: 726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arai E, Chiku S, Mori T, Gotoh M, Nakagawa T, Fujimoto H, et al. Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis 2012;33:1487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013;45:860–7. [DOI] [PubMed] [Google Scholar]
- 9.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beuselinck B, Job S, Becht E, Karadimou A, Verkarre V, Couchy G, et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin Cancer Res 2015;21: 1329–39. [DOI] [PubMed] [Google Scholar]
- 11.Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sd U S A 2009;106:21830–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang da W, Sherman BT, Lempidd RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempidd RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nudeic Adds Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fay AP, de Velasco G, Ho TH, Van Allen EM, Murray B, Albiges L, et al. Whole-exome sequendng in two extreme phenotypes of response to VEGF-targeted therapies in patients with metastatic dear cell renal cell cardnoma. J Natl Compr Cane Netw 2016;14:820–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malouf GG, Zhang J, Yuan Y, Comperat E, Roupret M, Cussenot O, et al. Characterization of long non-coding RNA transcriptome in dear-cell renal cell cardnoma by next-generation deep sequencing. Mol Oncol 2014;9: 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, et al. Broad H3K4me3 is assodated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet 2015;47:1149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choufani S, Cytrynbaum C, Chung BH, Turinsky AL, Grafodatskaya D, Chen YA, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun 2015;6:10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of dear cell renal cell cardnomas defined by multiregion sequendng. Nat Genet 2014;46:225–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sankin A, Hakimi AA, Mikkilineni N, Ostrovnaya I, Silk MT, Liang Y, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med 2014;3:1485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vougiouklakis T, Hamamoto R, Nakamura Y, Saloura V. The NSD family of protein methyltransferases in human cancer. Epigenomics 2015;7: 863–74. [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007;39:232–6. [DOI] [PubMed] [Google Scholar]
- 24.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet 2007;39:157–8. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Hwang J, Jeong H, Song HJ, Shin J, Hur G, et al. Promoter methylation status of VEGF receptor genes: a possible epigenetic biomarker to antidpate the efficacy of intracellular-acting VEGF-targeted drugs in cancer cells. Epigenetics 2012;7:191–200. [DOI] [PubMed] [Google Scholar]
- 26.Tiedemann RL, Hlady RA, Hanavan PD, Lake DF, Tibes R, Lee JH, et al. Dynamic reprogramming of DNA methylation in SEΓD2-deregulated renal cell carcinoma. Oncotarget 2016;7:1927–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.