Abstract
Long noncoding RNA (lncRNA) H19 is abundantly expressed in fetal liver. Its expression is significantly diminished in adult healthy liver but is re‐induced in chronic liver diseases, including cholestasis. In this study, we developed a new method with combined in situ hybridization (ISH) and immunofluorescence (IF) colabeling to establish an H19 expression profile with both parenchymal and nonparenchymal cell‐specific markers in the livers of cholestatic mouse models and patients with cholestasis. H19RNA+ cells showed no colocalization with biliary epithelial cell marker cytokeratin 19 (CK19)+ cholangiocytes but were immediately adjacent to biliary structures in bile duct ligation (BDL), 3,5‐diethoxycarbony1‐1,4‐dihydrocollidine (DDC), and multidrug‐resistant gene 2 knockout (Mdr2–/–) mouse models and in human primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) liver specimens. In contrast, double‐positive H19RNA+/sex‐determining region Y (SRY)‐box 9 (SOX9)+ ductal progenitor cells, H19RNA+/hepatocyte nuclear factor 4α (HNF4α)+ hepatocytes, H19RNA+/F4/80+ Kupffer cells, HNF4α+/SOX9+ hybrid hepatocytes, as well as triple‐positive H19RNA+/HNF4α+/SOX9+ periportal hepatocytes were identified. In addition, H19RNA could not be detected in mesenchymal cell marker desmin+ cells. Furthermore, H19RNA was predominately detected in cytoplasm with a small amount at the interspace with neighboring cells. Conclusion: H19RNA is localized in HNF4α+ periportal hepatocytes, SOX9+ ductal progenitor cells, and F4/80+ Kupffer cells but not in CK19+ cholangiocytes and desmin+ stellate cells in cholestatic livers.
Abbreviations
- BDL
bile duct ligation
- CK19
cytokeratin 19
- CoH
canals of Hering
- DAPI
4´,6‐diamidino‐2‐phenylindole
- DDC
3,5‐diethoxycarbony1‐1,4‐dihydrocollidine
- DR
ductular reaction
- EpCAM
epithelial cell adhesion molecule
- FFPF
formalin‐fixed, paraffin‐embedded
- HNF4α
hepatocyte nuclear factor 4α
- IF
Immunofluorescence
- ISH
in situ hybridization
- lncRNA
long noncoding RNA
- LSC
liver stem cell
- Mat H19–/–
maternal H19‐deleted mice
- Mdr2–/–
multidrug‐resistant gene 2 knockout mice
- Pat H19–/–
paternal H19‐deleted mice
- PBC
primary biliary cholangitis
- PBS
phosphate‐buffered saline
- PH
partial hepatectomy
- PSC
primary sclerosing cholangitis
- SOX9
sex‐determining region Y (SRY)‐box 9
Long noncoding RNAs (lncRNAs), defined as a nonprotein‐coding transcript of more than 200 nucleotides in length, are highly abundant in mammalian species.1 It has been estimated there are 14,880 lncRNA transcripts in the human genome,2 but the number could be much higher.3 Even with lower expression levels compared with messenger RNA,2, 4 lncRNAs play a vital role in various cellular processes in both physiologic and pathologic conditions.5 They hold important roles in regulating the expression and function of global‐ or local‐coding genes, and their aberrant expression could lead to diseases in diverse organs.6
H19 is a paternal‐imprinted lncRNA that is 2.3 kb in length; it is highly expressed in embryonic liver but markedly reduced after birth and in adult liver.7 Due to its high expression level in fetal liver, it is postulated that H19 is an important gene in regulating liver development, but the mechanism remains elusive. Liver is the only organ with a strong ability to regenerate.8 After partial hepatectomy (PH), H19 was increased in hepatocytes isolated from experimental mouse liver,9 indicating that the re‐activated H19 was associated with promoting liver regeneration. Recent studies have also observed increased H19 expression in patients with chronic liver diseases, including liver cancer,10 cholestatic liver injury,7 liver fibrosis and cirrhosis,11 PBC, and PSC.12 Overexpression of H19 in mouse hepatocytes augmented liver injury induced by BDL and DDC. These observations indicate an important role of H19 in liver disease.
Despite recent progress on the hepatic function of H19, its cell‐type‐specific expression profile in liver remains controversial. Current knowledge about H19 expression is based on quantitative reverse‐transcription polymerase chain reaction (PCR) analysis in isolated primary cells from animal models. A major limitation of this method is that the isolated primary cells are likely contaminated with neighboring cells, thereby producing false positives. Indeed, discrepant results have been reported. H19RNA was highly expressed in isolated hepatocytes at day 3 after PH in one study,9 whereas another study reported that H19RNA was mainly expressed in primary cholangiocytes and was not expressed in hepatocytes or Kupffer cells.13 There is an urgent need to develop a new method to precisely localize the subcellular distribution of H19, especially in human liver specimens.
In this study, we established a novel in situmethod to directly detect H19RNA and colocalize it with cell‐type‐specific markers in mouse models of cholestatic liver fibrosis and in human PBC and PSC specimens. We found that H19RNA was not expressed in CK19+ cholangiocytes but was exclusively expressed in cytoplasm that partially overlapped with HNF4α+ hepatocytes, SOX9+ progenitor cells, and F4/80+ Kupffer cells in periportal areas; it was also found at the interspace between neighboring cells or translocated into the nucleus under severe cholestatic conditions. Our findings are of critical importance to guide future research to further understand the role of H19 in liver metabolic diseases.
Materials and Methods
ANIMALS AND HUMAN LIVER SPECIMENS
Mouse models included maternal H19‐deleted mice (Mat–/–) with paternal H19‐deleted mice (Pat–/–) as control, as described.12, 14 Mice were provided with free access to a standard rodent chow diet (Harlan No. 2018) and water and housed in a temperature‐controlled (23°C) and pathogen‐free facility with a 12‐hour light/dark cycle. BDL surgeries were performed on 6‐week‐old male mice for 1 week as described.15 Cholestatic liver injury was also induced with a DDC‐containing diet as described.16 Mdr2–/– mice have been described.17 Animal studies complied with the guidelines of the Institutional Animal Care and Use Committee of the University of Connecticut.
The coded liver specimens PBC and PSC were obtained through the Liver Tissue Cell Distribution System (National Institutes of Health contract no. HSN276201200017C) as described.12 Because we did not ascertain individual identities associated with the samples, the Institutional Review Board for Human Research at the University of Connecticut determined that this project was not research involving human subjects.
H19 ISH COMBINED WITH IF STAINING
Liver samples were processed in formalin for 48 hours followed by embedding in paraffin. We cut 4‐µm sections for staining and performed H19 ISH following instructions in the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics). For combined staining, pretreatment durations were optimized based on different cell markers. IF staining was performed before incubating liver sections with 4',6‐diamidino‐2‐phenylindole (DAPI). Briefly, liver sections were washed with distilled water and phosphate‐buffered saline (PBS) twice each, followed by blocking in 10% goat serum in PBS with 0.1% Triton X‐100, then incubated with primary antibodies against different cell types (Table 1) at 4°C overnight. On the following day, liver sections were incubated with fluorescent‐conjugated goat anti‐mouse/rabbit secondary antibody (Table 1) at room temperature for 1 hour after three washes in PBS with 0.1% Triton X‐100. After another three washes, liver sections were stained with DAPI and mounted. Images were taken under a confocal microscope using fluorescence microscopy (Leica SP8).
Table 1.
ANTIBODIES USED FOR IMMUNOHISTOCHEMISTRY STAINING
| Antibody | Source/Catalog#/dilution |
|---|---|
| CK19 | Abcam/ab52625/1:50 |
| EpCAM | Abcam/ab71916/1:50 |
| HNF4α (for mouse) | Abcam/ab41898/1:40 |
| HNF4α (for human) | Abcam/ab92378/1:50 |
| SOX9 | Abcam/ab185966/1:50 |
| F4/80 | Abcam/ab15200/1:50 |
| Desmin | Abcam/ab15200/1:40 |
| Alexa647 conjugated goat anti‐rabbit | Thermo Fisher/A‐21245/1:500 |
| Alexa594 conjugated goat anti‐mouse | Thermo Fisher/A‐11005/1:500 |
| Alexa594 conjugated goat anti‐rabbit | Thermo Fisher/A‐11037/1:500 |
| TSA plus fluorescein (for H19) | PerkinElmer/NEL741E001KT/1:750 |
Abbreviation: TSA, tyramide signal amplification.
RNA ISOLATION AND REAL‐TIME PCR
Total RNAs from human specimens were isolated by TRIzol (Ambion), and the complementary DNA (cDNA) library was synthesized using the High‐Capacity cDNA Reverse Transcription Kit (Thermo Fisher) according to the manufacturer's protocol.18 Universal SYBR Green (Bio‐Rad Laboratories) was used for quantitative PCR.16 Primers used for human H19 were TGGTGCACTTTACAACCACTG (forward) and ATGGTGTCTTTGATGTTGGGGC (reverse); human glyceraldehyde 3‐phosphate dehydrogenase was used for normalization.
STATISTICAL ANALYSIS
Data were expressed as mean ± SEM and represented by triplicate assays. Statistical analysis was performed using one‐way analysis of variance among multiple groups and the Student t test between two groups.19 P < 0.05 was considered statistically significant.
Results
H19RNA IS PREDOMINATELY EXPRESSED IN CYTOPLASM AND CAN BE SECRETED INTO NEIGHBORING CELLS
H19 is a paternal‐imprinted gene; therefore, Pat–/– were used as wild‐type mice and Mat–/– were used as H19 knockout mice (negative controls). Because H19RNA is barely detectable in adult normal liver, we could perform H19 staining in sham mice. H19 was induced in cholestasis7; thus, BDL liver sections were used for the initial analysis. We first tested RNA quality in formalin‐fixed paraffin‐embedded (FFPF) mouse liver sections by using both negative‐mouse bacterial gene dapB and positive‐mouse peptidylprolyl isomerase B probes supplied from the RNAScope Assay kit following the manufacturer's instructions. No green fluorescence signals were detected using the negative‐control probe, whereas strong punctate green dots were detected by the positive‐control probe (Supporting Fig. S1). In addition, no H19RNA was detected in Mat–/–‐BDL and Mat–/–‐sham liver (Supporting Fig. S2). In contrast, H19RNA‐positive cells were detected in Pat–/–‐BDL liver in fibrosis areas (Supporting Fig. S3A, yellow arrow) and were also found close to the portal vein (Supporting Fig. S3B, yellow arrow) adjacent to the bile duct (Supporting Fig. S3B, red arrow). We note that H19RNA was exclusively expressed in cytoplasm with a punctate staining pattern (Supporting Fig. S4A, yellow arrow). It was also found between cells (Supporting Fig. S4B, orange arrow), indicating a likelihood of secretion into neighboring cells. Future studies will determine how the H19 secretion occurs.
H19RNA IS NOT EXPRESSED IN CK19+OR EPITHELIAL CELL ADHESION MOLECULE+ CHOLANGIOCYTES BUT MAY RESIDE IN CANALS OF HERING LINED BY HEPATOCYTES IN THE BDL MOUSE MODEL
Based on prior findings that H19 promoted BDL‐induced bile duct epithelial cell (i.e., cholantiocyte) proliferation,20 we first costained H19RNA with the biliary epithelial cell marker CK19. Immunostaining for CK19 labeled a single cell, cell cluster, or string surrounding the bile ducts in response to bile duct damage (ductular reaction [DR]) (Fig. 1A). However, none of the H19RNA+(green)‐positive cells overlapped with CK19+ (red)‐positive cholangiocytes. Interestingly, H19RNA+ cells were found at the interphase of bile duct and hepatocyte plates in small numbers near the bile ducts and were clearly distinguishable from neighboring hepatocytes. These cells did not reside in but were immediately adjacent to biliary structures (Fig. 1, yellow arrow). The characteristic oval cell response was identified by the expansion of CK19+ cells in periportal regions. Some of these cells were next to H19RNA+ cells (Fig. 1B, pink circle; Supporting Fig. S5). In addition, a cluster of H19RNA+ cells was observed in other areas distant from bile ducts (Fig. 1C, white circle).
Figure 1.
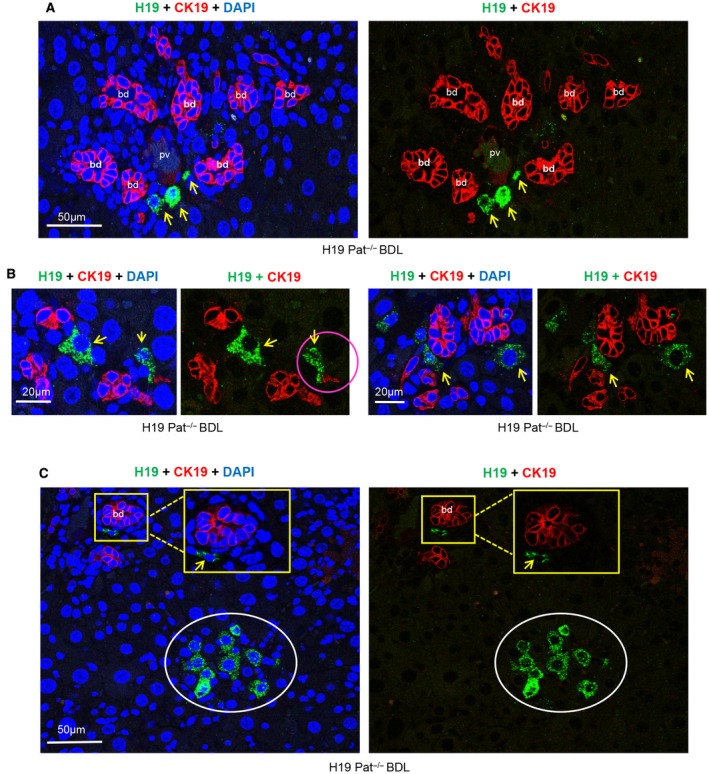
H19RNA is not expressed in CK19+ cholangiocytes but in periportal cells of close proximity in BDL‐induced cholestasis liver fibrosis. Pat H19–/– mice livers were harvested 1 week after BDL, and FFPF sections were used for combined ISH+IF costaining. (A,B) Representative images of H19RNA and CK19 colabeling are shown; (A) lower magnification (scale bar, 50 µm); (B) higher magnification (scale bar, 20 μm). H19RNA was expressed in cells close to the biliary structure and DR. Red, CK19; green, H19RNA; blue, DAPI; yellow arrow, H19RNA+ cells; pink circle, H19RNA+ cells in close vicinity to CK19+ cells. (C) A cluster of H19RNA+ cells was observed in hepatocyte plates distinct from CK19‐labeled bile ducts (scale bar, 50 µm). White circle, cluster of H19RNA+ cells; yellow square, enlarged images. Abbreviations: bd, bile duct; pv, portal vein.
Epithelial cell adhesion molecule (EpCAM) is another cholangiocyte marker that is expressed around the portal vein in wild‐type mice.21 EpCAM stain of human hepatitis B cirrhosis showed cytoplasmic staining of biliary structures.22 DRs, inclusive of intermediate cells, showed some cytoplasmic staining but also membranous staining. The hepatocytes surrounding the DRs showed extensive membranous staining. Interestingly, we observed membranous staining of EpCAM in the DRs (Supporting Fig. S6). Consistent with results from CK19 colabeling, H19RNA+ cells did not overlap but rather were in close proximity to EpCAM+ cells.
Canals of Hering (CoH) are considered a niche of hepatic progenitor cells. CoH begin in the lobules, are lined partially by cholangiocytes and partly by hepatocytes, and conduct bile from bile canaliculi to terminal bile ducts in portal tracts.23 The cholangiocytes of the CoH can be highlighted by CK19 and EpCAM. Because H19RNA+ cells either contacted duct cells (Fig. 1B, pink arrow) or were located proximately 1 cell diameter away but did not overlap with CK19+ and EpCAM+cells, we postulate that H19 may reside in CoH lined by hepatocytes or in progenitor cells expressing a different marker.
H19RNA IS IDENTIFIED IN SOX9+ PROGENITOR CELLS IN THE BDL MOUSE MODEL
Based on the above observation, we next determined whether H19 is expressed in progenitor cells in mouse liver. SOX9 is a relatively broad ductal progenitor marker. It is considered primarily active in bile duct cells and bipotential hepatobiliary progenitors. Our results showed that SOX9 was highly expressed in adjacent biliary ductules (Fig. 2A, left and middle; Supporting Fig. S7, white arrow). Interestingly, dual immunostaining with SOX9 and H19RNA identified a small population (~6%) of double H19RNA+/SOX9+ cells located in the distal end of DRs (Fig. 2A, right; Supporting Fig. S8, red circle), despite the majority of SOX9+ cells showing H19RNA–. We also observed single H19RNA+ (yellow circle) and single SOX9+ (blue circle) cells. All H19RNA+ cells were spatially restricted to areas directly adjacent to the portal veins and ductal cells.
Figure 2.
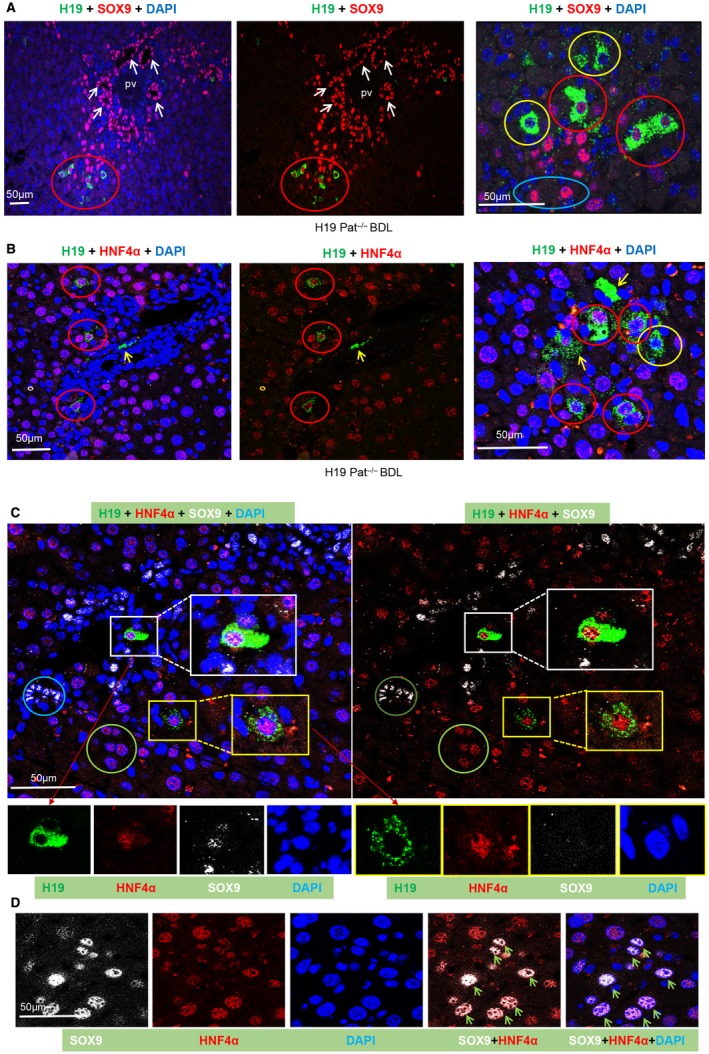
H19RNA is expressed in SOX9+ progenitor cells, or HNF4α+ hepatocytes, or SOX9+/HNF4α+ cells in BDL mice. (A‐C) Representative images of H19RNA and (A) SOX9, (B) HNF4α, or (C) SOX9+HNF4α colabeling are shown at lower magnification (scale bar, 50 µm) (left and middle panels) and higher magnification (scale bar, 20 µm) (right panel). (D) Colocalization of HNF4α and SOX9 are shown in different single and merged channels (scale bar, 50 µm). Red, SOX9 (A) and HNF4α (B‐D); green, H19RNA; blue, DAPI; yellow arrows, H19RNA+ cells; green arrows, SOX9 and HNF4α double‐positive cells; red circle, H19 and HNF4α double‐positive cells; yellow circle, H19 single‐labeling cells; blue circle, SOX9 single‐labeling cells; green circle, HNF4α single‐labeling cell; white square, H19RNA, SOX9, and HNF4α triple‐labeling cells shown in different single enlarged channels; yellow square, H19RNA and HNF4α double‐labeling cells shown in different single enlarged channels.
H19RNA IS IDENTIFIED IN HNF4α+ HEPATOCYTES IN THE BDL MOUSE MODEL
Hepatocytes are the chief functional cells of the liver, and roughly 80% of the mass of the liver is contributed by hepatocytes. A follow‐up question is whether H19 is expressed in hepatocytes. We therefore tested HNF4α, a well‐established hepatocyte marker. On double staining for H19 and HNF4α, we found ~77.5% of H19RNA+ (excluding secreted H19RNA) expressed in HNF4α+ cells that were adjacent to the DRs (Fig. 2B; Supporting Fig. S9, red circle). Most mature HNF4α+ hepatocytes were not colabeled with H19RNA; thus, H19 was only expressed in a specific population of hepatocytes that were also positive for HNF4α. In addition, a small number of cells showed H19RNA+ but HNF4α– (Fig. 2B, right panel, yellow circle). H19RNA+ signals were also observed between two neighboring cells (Fig. 2B, right panel, yellow arrow), indicating its secretion.
IDENTIFYING TRIPLE‐POSITIVE H19RNA+, HNF4α+, andSOX9+ PERIPORTAL HEPATOCYTES IN THE BDL MOUSE MODEL
Because we observed double‐labeled H19RNA+ cells with either SOX9 or HNF4α, we colabeled those three genes using three colors (green [H19], white [SOX9], and red [HNF4α]) (Fig. 2C). In addition to single SOX9+ (brown circle) and single HNF4α+ (green circle) cells and double H19+/HNF4α+ (yellow square) cells, triple H19+/HNF4α+/SOX9+ hepatocytes (Fig. 2C, white square) were identified. Moreover, many double SOX9+/HNF4α+ hepatocytes were found located at the interphase of bile duct and hepatocyte plates (Fig. 2D, green arrow; Supporting Fig. S10, blue square). The majority of positive cells were only single labeled with either HNF4α or SOX9. On the other hand, we did not observe single H19RNA+or H19RNA+/HNF4α+ hybrid hepatocytes surrounding the central veins.
H19RNA IS IDENTIFIED IN F4/80+ KUPFFER CELLS IN THE BDL MOUSE MODEL
Kupffer cells are the resident macrophages in the liver. Because BDL altered functional activities of Kupffer cells, we determined H19 co‐expression with the macrophage marker F4/80. By colabeling with H19, we found that H19RNA was colocalized with F4/80 in cytoplasm and stained as punctate dots in a number of cells (Fig. 3A; Supporting Fig. S11A). Interestingly, in some cells, both H19RNA and F4/80 were completely overlapped in a vesicle structure that was attached to or immediately adjacent to the nucleus (Fig. 3B; Supporting Fig. S11B, white arrowhead). We also observed single H19RNA+ (yellow arrow) and single F4/80+ (red arrow) cells (Supporting Fig. S12). Because F4/80 can mark both resident Kupffer cells and infiltrating macrophages, H19 could be expressed in both cell populations. The results are in agreement with a recent report showing that hepatic overexpression of H19 enriched immune cell populations in liver and spleen.12
Figure 3.
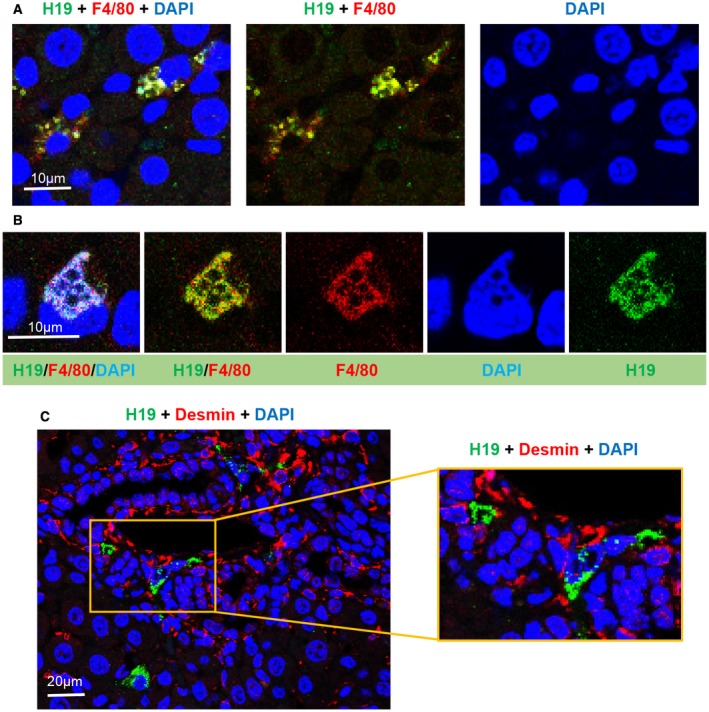
H19RNA is colocalized with F4/80 in Kupffer cells but not with desmin in stellate cells in BDL mice. (A) Representative images of H19RNA and F4/80 costaining are shown at a 10‐μm scale. (B) Enlarged double‐labeling images are shown in different single and merged channels (scale bar, 10 μm). (C) H19RNA and desmin costaining showed no overlapping localization (scale bar, left 20 µm and right 10 µm); square in right panel shows enlarged image with no colocalization. Red, F4/80 (A,B) and desmin (C); green, H19RNA; blue, DAPI.
H19RNA IS NOT EXPRESSED IN DESMIN+ STELLATE CELLS IN THE BDL MOUSE MODEL
Hepatic stellate cells are collagen‐producing cells in the liver that are responsible for fibrogenesis in chronic liver diseases. Encouraged by the above results that H19RNA was detected in F4/80 Kupffer cells, we next assessed its expression in hepatic stellate cells. Direct fluorescence of H19RNA (green) with immunostaining for the mesenchymal cell marker desmin (red) showed no colocalization of either gene (Fig. 3C). Prior studies revealed co‐IF of CK19 and desmin of ductular structures within the portal field.24 We found high labeling efficiency of desmin+ periportal mesenchymal cells in DRs. Colabeling with α‐smooth muscle action (α‐SMA) also failed to detect the presence of H19RNA in α‐SMA+ cells (not shown). Together these results argue against a contribution from resident mesenchymal cells to H19RNA+ progenitor cells.
H19RNA IS NOT EXPRESSED IN CK19+ CHOLANGIOCYTES IN DDC OR Mdr2 –/– MOUSE MODELS OF BILIARY LIVER FIBROSIS
The liver toxin DDC‐enriched diet is commonly used as a chemically induced chronic cholestatic liver injury model with a strong expansion of atypical ductal proliferation near the periportal areas.24 To expand our findings from the above BDL model, we examined H19RNA in DDC‐fed mouse liver. Similarly, H19RNA did not overlap but was in close proximity to CK19+ cells (Fig. 4A; Supporting Fig. S13).
Figure 4.
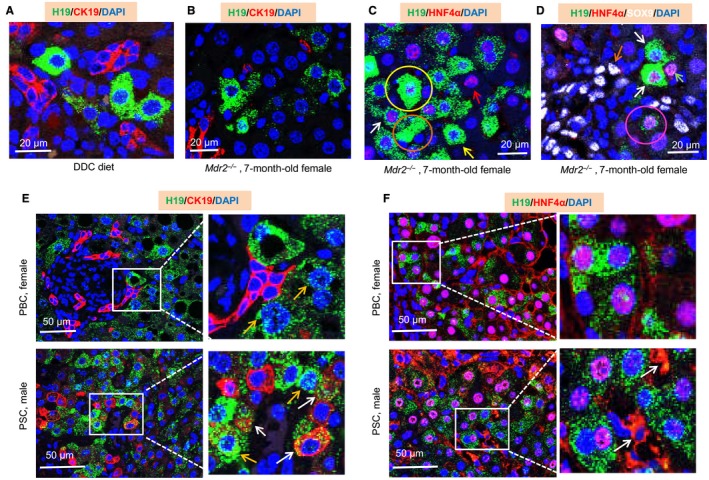
H19RNA is not expressed in CK19+ cholangiocytes but is expressed in HNF4α+ hepatocytes in mouse DDC and Mdr2 (Abcb4)–/– models and in human PBC and PSC liver specimens. (A) Representative image of H19RNA with CK19 in DDC mouse liver. (B) Representative image of H19RNA with CK19 in 7‐month‐old female Mdr2 –/– mouse liver. (C) Representative image of H19RNA with HNF4α in 7‐month‐old female Mdr2 –/– mouse liver. Yellow arrow, H19RNA single‐labeling cell; red arrow, HNF4α single‐labeling cell; white arrow, H19RNA and HNF4α double‐labeling cell; yellow circle, remarkable induction of H19RNA that overlaid with nucleus; brown circle, secreted H19RNA. (D) Representative image of H19RNA with HNF4α and SOX9 in 7‐month‐old female Mdr2 –/– mouse liver. White arrow, H19RNA and HNF4α double‐labeling cell; brown arrow, SOX9 single‐labeling cell; green arrow, HNF4α single‐labeling cell; pink circle, H19RNA, HNF4α, and SOX9 triple‐labeling cell. (E,F) Representative images of H19RNA with (E) CK19 and (F) HNF4α in female PBC and male PSC liver specimens. Orange arrow, H19RNA detected in nucleus; white arrow, H19RNA dispersing into neighboring (E) CK19+ cells and (F) cytoplasmic HNF4α cells. Scale bar, 20 µm (A‐D), 50 µm (E,F).
Mdr2 (Abcb4)–/– mice spontaneously develop hepatic lesions resembling PSC, representing one of the best characterized biliary fibrosis models of cholangiopathy.25 We examined H19RNA in 3‐month‐old male Mdr2–/– and 7‐month‐old female Mdr2–/–livers. H19RNA was barely detectable in 3‐month‐old male Mdr2–/–liver (Supporting Fig. S14). In contrast, H19RNA was highly induced in cells adjacent to the DRs (Supporting Fig. S15A) and in periportal regions (Supporting Fig. S15B) in 7‐month‐old female Mdr2–/– liver. Consistent with the results in BDL and DDC livers, H19RNA was not expressed in CK19+cholangiocytes in 7 month‐old female Mdr2–/–liver (Fig. 4B; Supporting Fig. S16).
H19RNA IS EXPRESSED IN HNF4α+HEPATOCYTES IN THE Mdr2 –/– MOUSE MODEL
In addition, single H19RNA+ (yellow arrow), single HNF4α+(red arrow), and double H19+/HNF4α+ (white arrow; 70.8% of H19RNA in HNF4α‐positive hepatocytes) periportal hepatocytes were identified in 7‐month‐old female Mdr2–/– liver (Fig. 4C; Supporting Fig. S17). We found that the H19RNA signal overlaid with the nucleus in some cells, likely due to its remarkable induction (Supporting Fig. S17, yellow circle). An extracellular H19RNA signal was also observed (orange circle), indicative of its secretion.
Because triple H19RNA+/HNF4α+/SOX9+ periportal hepatocytes were observed in the BDL mouse model as presented above, we costained these three genes in 7‐month‐old female Mdr2–/– liver. SOX9+ (white) cells were abundantly observed around the DRs (Fig. 4D; Supporting Fig. S18). However, we barely detected triple‐positive H19RNA+/HNF4α+/SOX9+ (pink circle indicates the cell with weak staining) or double‐positive H19RNA+/SOX9+ and HNF4α+/SOX9+ periportal hepatocytes in Mdr2–/– liver. The results suggest a differential expression regulation of the bipotential hybrid HNF4α+/SOX9+ cells in BDL and Mdr2–/– models.
H19RNA IS NOT EXPRESSED IN MATURE CK19+ CHOLANGIOCYTES IN HUMAN PBC OR PSC LIVER SPECIMENS
PBC and PSC are two main human cholangiopathies.26 PBC is a chronic progressive cholestatic liver disease characterized by inflammatory obliteration of the intrahepatic bile ducts, leading to fibrosis and ultimately to cirrhosis complicated by liver failure or hypertension.27 PSC is another chronic liver disease characterized by intrahepatic and extrahepatic duct inflammation and fibrosis, is associated with inflammatory bowel diseases, and ultimately leads to biliary cirrhosis in advanced stages.28 To determine sex disparity, we determined H19RNA in both male and female PBC and PSC liver specimens (Supporting Table S1). Interestingly, the levels of H19RNA were significantly elevated in female but not male PBC and in male but not female PSC, as compared to normal controls (Supporting Fig. S19). Based on these results, we selected two female PBC and two male PSC samples with high H19RNA expression for further analysis. We first tested RNA quality in FFPF human liver sections by using both negative human bacterial gene dapB and positive human peptidylprolyl isomerase B probes. No green fluorescence signals were detected using the negative‐control probe, whereas strong punctate green dots were detected by the positive‐control probe (Supporting Fig. S20).
As expected, H19RNA was markedly induced in female PBC livers (Supporting Figs. S21 and S22). Despite being located in close proximity, H19RNA did not overlap with mature CK19+ cholangiocytes in bile ducts (Fig. 4E, top panel). H19RNA was diffuse in the nucleus (orange arrow), which could indicate either nuclear translocation from cytoplasm or newly synthesized RNAs that have not been completely translocated into cytoplasm due to the extremely high expression of H19RNA.
Similarly, H19RNA was markedly induced in male PSC livers and did not overlap with mature CK19+ cholangiocytes in biliary structures (Supporting Figs. S23 and S24). Some H19RNA signals, however, were dispersed into individual neighboring cells positive for CK19+ (Fig. 4E, bottom panel, white arrow). These cells may represent the newly regenerated immature cholangiocytes from DRs or may represent bipotential liver progenitor cells.
H19RNA IS EXPRESSED IN HNF4α+ HEPATOCYTES IN HUMAN PBC AND PSC LIVER SPECIMENS
H19RNA was found expressed in HNF4α+ hepatocytes in both female PBC and male PSC liver specimens (76.6% and 74.5%, respectively) (Fig. 4F; Supporting Fig. S25). In addition to its nuclear localization in hepatocytes, HNF4α was highly expressed in the cytoplasmic compartment in DR and fibrotic areas (white arrow). Prior studies showed cytoplasmic expression of HNF4α messenger RNA in ductular hepatocytes in massive necrosis areas.29 The subcellular localization of HNF4α could be associated with its phosphorylation status.30 Because HNF4α is a critical master transcriptional regulator of liver function, its cytoplasmic retention will affect the expression of its target genes and consequently hepatic homeostasis during cholestatic liver injury. Taken together, H19RNA was colocalized with HNF4α protein in mature hepatocytes in PBC and PSC specimens.
Discussion
The conventional RNA ISH technique is difficult to perform and lacks sensitivity and specificity, especially in measuring RNA that is in low abundance.31 Fluorescent labeling is a sensitive method broadly used in research and has the advantage of a multiple‐label possibility for detecting more than one target signal simultaneously. The recently developed RNAscope employs new strategies in designing probes and amplifying signals32; however, this technique can only detect RNA. Zimmerman et al.33 set up a combined IF and fluorescent ISH labeling approach in Drosophila ovaries by performing IF before fluorescent ISH. However, this approach was complicated and took 5 days to complete. Our new method used the advantage of RNAscope to obtain high‐quality H19RNA signals and followed this by high‐resolution IF staining of protein markers; this was easy to perform, had a shorter duration time, and was reproducible with very reliable outcomes.
Using the above new method, we directly visualized the localization of H19RNA in cholestatic livers. We found the following: 1) H19RNA was predominately detected in the cytoplasm of cells but also at the interspace with neighboring cells under severe cholestatic conditions; 2) in contrast, H19RNA was not detected in CK19+ cholangiocytes but likely resides in CoH lined by hepatocytes; 3) H19RNA was partially colocalized with HNF4α+ hepatocytes, SOX9+ progenitor cells, and F4/80+ Kupffer cells in periportal areas; and 4) H19RNA was not expressed in desmin+ stellate cells. Importantly, we identified significant enrichments of H19RNA in hepatocytes in a subset of female PBC and male PSC livers, signifying the importance of H19 in human biliary liver fibrosis.
Liver contains two major types of cells: parenchymal cells, which include hepatocytes and cholangiocytes, and nonparenchymal cells, which include sinusoidal endothelial, Kupffer, and stellate cells.34 Hepatic bile acid homeostasis is tightly controlled by transcriptional regulators.17 Dysregulation of bile synthesis or bile flow results in cholestasis and liver injury, which are often accompanied with striking morphologic changes.31 In particular, periportal bile ductules and CoH are constantly injured by inflammatory cells, resulting in CK19‐positive reactions. Cells in DRs are phenotypically heterogeneous, expressing transcription factors as well as surface and cytoplasmic markers, including CK19 and SOX9, for stem/progenitors. Recent studies indicate that EpCAM could serve as a hepatic stem/progenitor cell‐specific marker.35 In liver biopsy specimens from patients with chronic hepatitis B and C, EpCAM+hepatocytes were rare in early stages of disease, became increasingly prominent in later stages in parallel with the emergence of DRs, and were consistently arrayed around the periphery of cords of CK19+ hepatobiliary cells of the DRs with which they shared EpCAM expression.22 In human liver, hepatic stem/progenitor cells can be isolated by immune selection for EpCAM.22 In the present study, we did not identify H19RNA+ cells overlapping with either CK19+ or EpCAM+ ductal cells, suggesting that H19RNA is not expressed in this cell population that composes or generates cholangiocytes. Interestingly, H19RNA was not found in all segments of the biliary tree but in areas near small ducts, suggesting its unique expression activation by unknown signaling molecules.
Our results argue against a recent study by Li et al.36 showing that H19RNA is mainly expressed in primary cholangiocytes and that cholangiocyte‐derived exosomal H19 promotes cholestatic liver injury by suppressing the small heterodimer partner in hepatocytes. A potential explanation for this discrepancy could be the contamination of CK19+/H19RNA–cholangiocytes from CK19–/H19RNA+neighboring cells during cholangiocyte isolation, as it would be technically challenging to separate both cell populations due to their close proximity. H19 was also reported to be induced by bile acids in the mouse small cholangiocyte cell line (MSC).12 MSC was derived from cholangiocytes isolated from small ducts that may have acquired certain characteristic changes with stem‐like features during immortalization that are not present in primary cholangiocytes. Removing cells from their in vivo environment may also confer properties of plasticity not representative of in vivo biology, which likely contributes to the difference in results. Cell signaling can take place either through direct cell–cell contacts or through the action of secreted signaling molecules. In paracrine signaling, a molecule released by one cell acts on neighboring target cells. Due to the close vicinity of H19RNA+ cells to CK19+ cholangiocytes, we postulate that H19 may act as a local signaling molecule to regulate the behavior of nearby cholangiocytes. It should also be noted that liver H19 is highly induced in female Mdr2–/– mice, an animal model of human PSC; however, it is highly induced in PBC female and PSC male livers. The results suggest that the sex dimorphic regulation of H19 may be different in mice and humans. The pathways that contribute to increased H19 expression in PBC and PSC remain to be determined in future studies.
Most hepatic injuries, such as surgical PH, result in a progenitor‐independent proliferative response of preexisting mature cells. However, other forms of liver damage can induce progenitor‐dependent liver regeneration. This process is considered to involve the emergence of a new epithelial cell type not present in normal liver, termed “hepatic oval cell.” The cellular origin of oval cells is unknown, but it is hypothesized that oval cells are the progeny of resident adult hepatic stem cells.35 Liver stem cells (LSCs) are viewed as bipotential nonhepatocyte precursors of highly proliferative progenitor cells that can differentiate into either hepatocytes or biliary epithelial cells. However, using genetic fate tracing in adult mice fed a choline‐deficient ethionine‐supplemented diet, Schaub et al.37 failed to provide evidence for hepatocytes derived from LSCs or other nonhepatocyte cell types, which argues against the notion that LSCs replenish lost hepatocytes during liver regeneration. Conflicting results have been reported about the identity of LSCs. Recent reports in adult mice showed two distinct subpopulations of hepatocytes, namely “Axin2+” cells located pericentrally24 and “SOX9+ hybrid periportal hepatocytes” located periportally within the liver lobule.25
Interestingly, we identified a very small population of triple‐positive H19RNA+/HNF4α+/SOX9+ hepatocytes immediately adjacent to biliary structures that were located within the periportal mesenchyme. Periportal mesenchyme may harbor cells capable of hepatocyte differentiation. The small number of H19RNA+ hepatocytes may represent a newly identified reservoir of stem cells within the SOX9+ stem/progenitor cell niches and derived from HNF4α+‐expressing hepatocytes. Indeed, double HNF4α+/SOX9+ hybrid periportal hepatocytes evidently exist and could be the source of triple H19RNA+/HNF4α+/SOX9+ hepatocytes. We also noted single H19RNA+ cells that did not express either HNF4α or SOX9. Our findings suggest that H19RNA+ cells may not represent a single cell type but a heterogeneous population of hepatic cells. Additional studies are required to determine whether these cells have intrinsic “stem cell‐like” potential and whether they represent liver's transit‐amplifying cells or intermediates between hepatic stem cells and hepatoblasts or between hepatoblasts and adult parenchyma. H19RNA also showed co‐expression with F4/80‐expressing cells, suggesting periportal nonparenchymal cell types of unknown potential and function present during cholestasis induced DRs.
During liver maturation, multiple stem cell niches persist in specific anatomical locations within adult liver, including biliary tree stem/progenitor cells in peribiliary glands along extrahepatic and large intrahepatic bile ducts and hepatic stem/progenitors in or near CoH.23 It would be interesting to determine the origin of H19RNA+ cells during liver development in future studies using lineage tracing. Identifying new factors that stimulate proliferation and differentiation of hepatic stem cells in response to liver injury are of interest for the development of cell‐based therapies in cholestatic liver fibrosis.
In conclusion, we successfully developed a new combined ISH+IF costaining method that is simple and reliable for determining cell‐type‐specific H19 localization. By costaining with hepatic cell‐specific markers, we uncovered that H19RNA is not expressed in CK19+ cholangiocytes but in HNF4α+ hepatocytes immediately adjacent to biliary structures in different cholestatic mouse models and human specimens. We also revealed that H19RNA can be secreted into neighboring cells, providing a solid foundation to investigate the mechanisms by which H19 regulates cholangiocyte proliferation and liver fibrosis, likely through cell–cell communication in a paracrine fashion. Future studies are warranted to identify endogenous signaling pathways that induce H19 expression during the progression of cholestasis.
Author names in bold designate shared co‐first authorship.
Supporting information
Acknowledgment
The Leica SP8 confocal microscope is supported by the National Institutes of Health grant S10OD016435 awarded to Akiko Nishiyama at the University of Connecticut.
Supported by the National Institutes of Health (grants DK104656, ES025909, AA024935, AA026322 to L.W.), a Veterans Affairs Merit Award (1I01BX002634 to L.W.), and the Yale Liver Center (DK34989 and DK25636 to J.L.B.).
Potential conflict of interest: Nothing to report.
References
- 1. Zhang L, Yang Z, Trottier J, Barbier O, Wang L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 2017;65:604‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775‐1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol 2011;3:a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Z, Cappello T, Wang L. Emerging role of microRNAs in lipid metabolism. Acta Pharm Sin B 2015;5:145‐150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Z, Ross RA, Zhao S, Tu W, Liangpunsakul S, Wang L. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol Commun 2017;1:513‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non‐coding RNAs. Nucleic Acids Res 2012;40:6391‐6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep 2016;6:20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol 2013;3:485‐513. [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto Y, Nishikawa Y, Tokairin T, Omori Y, Enomoto K. Increased expression of H19 non‐coding mRNA follows hepatocyte proliferation in the rat and mouse. J Hepatol 2004;40:808‐814. [DOI] [PubMed] [Google Scholar]
- 10. Chen X, Yamamoto M, Fujii K, Nagahama Y, Ooshio T, Xin B, et al. Differential reactivation of fetal/neonatal genes in mouse liver tumors induced in cirrhotic and non‐cirrhotic conditions. Cancer Sci 2015;106:972‐981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esposti DD, Hernandez‐Vargas H, Voegele C, Fernandez‐Jimenez N, Forey N, Bancel B, et al. Identification of novel long non‐coding RNAs deregulated in hepatocellular carcinoma using RNA‐sequencing. Oncotarget 2016;7:31862‐31877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song Y, Liu C, Liu X, Trottier J, Beaudoin M, Zhang L, et al. H19 promotes cholestatic liver fibrosis by preventing ZEB1‐mediated inhibition of epithelial cell adhesion molecule. Hepatology 2017;66:1183‐1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li X, Liu R, Yang J, Sun L, Zhang L, Jiang Z, et al. The role of long noncoding RNA H19 in gender disparity of cholestatic liver injury in multidrug resistance 2 gene knockout mice. Hepatology 2017;66:869‐884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu C, Yang Z, Wu J, Zhang L, Lee S, Shin DJ, et al. Long noncoding RNA H19 interacts with polypyrimidine tract‐binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology 2018;67:1768‐1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y, Xu N, Xu J, Kong B, Copple B, Guo GL, et al. E2F1 is a novel fibrogenic gene that regulates cholestatic liver fibrosis through the Egr‐1/SHP/EID1 network. Hepatology 2014;60:919‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smalling RL, Delker DA, Zhang Y, Nieto N, McGuiness MS, Liu S, et al. Genome‐wide transcriptome analysis identifies novel gene signatures implicated in human chronic liver disease. Am J Physiol Gastrointest Liver Physiol 2013;305:G364‐G374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cai SY, Mennone A, Soroka CJ, Boyer JL. Altered expression and function of canalicular transporters during early development of cholestatic liver injury in Abcb4‐deficient mice. Am J Physiol Gastrointest Liver Physiol 2014;306:G670‐G676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou T, Zhang Y, Macchiarulo A, Yang Z, Cellanetti M, Coto E, et al. Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J Biol Chem 2010;285:24871‐24881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tran M, Lee SM, Shin DJ, Wang L. Loss of miR‐141/200c ameliorates hepatic steatosis and inflammation by reprogramming multiple signaling pathways in NASH. JCI. Insight 2017;2;pii:96094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. LeSage G, Glaser S, Alpini G. Regulation of cholangiocyte proliferation. Liver 2001;21:73‐80. [DOI] [PubMed] [Google Scholar]
- 21. Okabe M, Tsukahara Y, Tanaka M, Suzuki K, Saito S, Kamiya Y, et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development 2009;136:1951‐1960. [DOI] [PubMed] [Google Scholar]
- 22. Yoon SM, Gerasimidou D, Kuwahara R, Hytiroglou P, Yoo JE, Park YN, et al. Epithelial cell adhesion molecule (EpCAM) marks hepatocytes newly derived from stem/progenitor cells in humans. Hepatology 2011;53:964‐973. [DOI] [PubMed] [Google Scholar]
- 23. Lanzoni G, Cardinale V, Carpino G. The hepatic, biliary, and pancreatic network of stem/progenitor cell niches in humans: a new reference frame for disease and regeneration. Hepatology 2016;64:277‐286. [DOI] [PubMed] [Google Scholar]
- 24. Mariotti V, Strazzabosco M, Fabris L, Calvisi DF. Animal models of biliary injury and altered bile acid metabolism. Biochim Biophys Acta 2018;1864:1254‐1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kulkarni SR, Soroka CJ, Hagey LR, Boyer JL. Sirtuin 1 activation alleviates cholestatic liver injury in a cholic acid‐fed mouse model of cholestasis. Hepatology 2016;64:2151‐2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo Z, Jegga AG, Bezerra JA. Gene‐disease associations identify a connectome with shared molecular pathways in human cholangiopathies. Hepatology 2017; doi: 10.1002/hep.29504. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beuers U, Gershwin ME, Gish RG, Invernizzi P, Jones DE, Lindor K, et al. Changing nomenclature for PBC: From 'cirrhosis' to 'cholangitis'. Clin Res Hepatol Gastroenterol 2015;39:e57‐e59. [DOI] [PubMed] [Google Scholar]
- 28. Rossi RE, Conte D, Massironi S. Primary sclerosing cholangitis associated with inflammatory bowel disease: an update. Eur J Gastroenterol Hepatol 2016;28:123‐131. [DOI] [PubMed] [Google Scholar]
- 29. Hakoda T, Yamamoto K, Terada R, Okano N, Shimada N, Suzuki T, et al. A crucial role of hepatocyte nuclear factor‐4 expression in the differentiation of human ductular hepatocytes. Lab Invest 2003;83:1395‐1402. [DOI] [PubMed] [Google Scholar]
- 30. Song Y, Zheng D, Zhao M, Qin Y, Wang T, Xing W, et al. Thyroid‐stimulating hormone increases HNF‐4alpha phosphorylation via cAMP/PKA pathway in the liver. Sci Rep 2015;5:13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mahmood R, Mason I. In‐situ hybridization of radioactive riboprobes to RNA in tissue sections. Methods Mol Biol 2008;461:675‐686. [DOI] [PubMed] [Google Scholar]
- 32. Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin‐fixed, paraffin‐embedded tissues. J Mol Diagn 2012;14:22‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zimmerman SG, Peters NC, Altaras AE, Berg CA. Optimized RNA ISH, RNA FISH and protein‐RNA double labeling (IF/FISH) in Drosophila ovaries. Nat Protoc 2013;8:2158‐2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kmiec Z. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol 2001;161:III‐XIII, 1‐151. [DOI] [PubMed] [Google Scholar]
- 35. Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, et al. Identification of microRNA‐181 by genome‐wide screening as a critical player in EpCAM‐positive hepatic cancer stem cells. Hepatology 2009;50:472‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li X, Liu R, Huang Z, Gurley EC, Wang X, Wang J, et al. Cholangiocyte‐derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology 2018; doi: 10.1002/hep.29838. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khan FM, Komarla AR, Mendoza PG, Bodenheimer HC Jr, Theise ND. Keratin 19 demonstration of canal of Hering loss in primary biliary cirrhosis: "minimal change PBC"? Hepatology 2013;57:700‐707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


