Abstract
Despite decades of extensive drug discovery efforts, there are currently no targeted therapies approved to treat KRAS mutant cancers. In this review, we highlight the challenges and opportunities in targeting KRAS mutant tumors through inhibition of mitogen-activated protein kinase (MAPK) signaling with conformation-specific kinase inhibitors. Through structural analysis and mechanistic studies with BRAF and mitogen-activated protein kinase (MEK) inhibitors, we describe how kinase-dependent and -independent functions of MAPK signaling components regulate KRAS-driven tumorigenesis and how these insights can be used to treat RAS mutant cancers with small molecule kinase inhibitors.
MECHANISM OF NORMAL RAS/MAPK SIGNALING
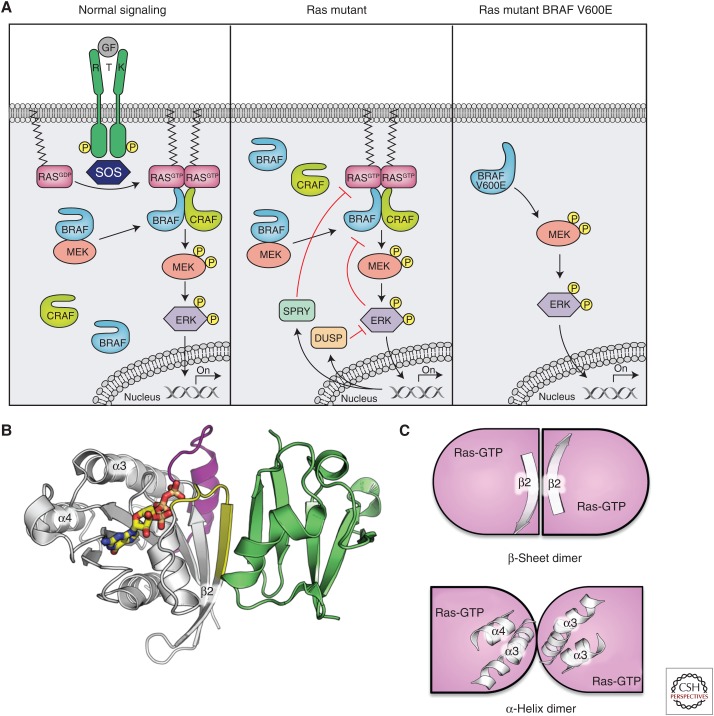
HRAS, KRAS, and NRAS comprise a highly conserved family of small GTPases that relay signals from upstream growth factor receptors to downstream effector pathways, including mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), and RAL guanine nucleotide dissociation stimulator (RAL-GDS). Ligand stimulation results in activation of the guanine nucleotide exchange factor son of sevenless (SOS) and facilitates exchange of the inactive guanosine diphosphate (GDP)-bound state of RAS to an active guanosine triphosphate (GTP)-bound state (Fig. 1A). This switch between inactive and active states involves structural rearrangement of the Switch I (SW1) and Switch II (SW2) regions in RAS, enabling RAS to adopt a conformation that interacts with the RAS-binding domain (RBD) of its downstream effectors (Fig. 1B) (Erijman and Shifman 2016). In the context of MAPK signaling, GTP-bound RAS recruits rapidly accelerated fibrosarcoma (RAF) family members (ARAF, BRAF, CRAF) from the cytosol to the plasma membrane through a direct and high affinity interaction with the RAF RBD (Fig. 1B). Structurally, the RBD has a ubiquitin-like fold and interacts with RAS by forming an extended β sheet in the vicinity of the switch regions (Nassar et al. 1995). This binding mode is common to most RAS–effector interactions.
Figure 1.
RAS/mitogen-activated protein kinase (MAPK) pathway signaling and protein–protein interactions. (A) Pathway signaling showing normal, RAS mutant, and BRAF V600E mutant signaling. (B) Structure of the Ras-like small GTPase Rap1A (white) with Switch I (yellow) and Switch II (magenta) bound to the RAS-binding domain (RBD) domain (green) of CRAF (Protein Data Bank [PDB] code 1C1Y). (C) Cartoon of two proposed RAS dimers proposed by Muratcioglu et al. (2015) formed either by the β2 strand from Switch I, which forms a dimer analogous to the Ras–RBD interactions, or by mediated α3/α4.
More recent studies have suggested that RAS dimerizes via a region distal to the SW1 and SW2 regions (the α4–α5 region) (Fig. 1C). These dimers form at the membrane and may also be required for proper signaling activity, including RAF dimerization and activation (Lin et al. 2014; Spencer-Smith et al. 2017). Indeed, it has been proposed that upon GTP binding, the α4–α5 regions of RAS are positioned in close proximity and may promote RAF dimerization and activation. Lending support to this model is the fact that NS1, a monobody that selectively recognizes the α4–α5 region of RAS, prevents RAS dimerization and RAF activation (Spencer-Smith et al. 2017). Dimerization of RAF family members has been shown to induce kinase activity likely through a conformational change releasing the negative regulatory region (NRR) within the amino-terminal regions of RAF (Weber et al. 2001; Udell et al. 2011) and exposing the carboxy-terminal kinase domains. This results in phosphorylation and activation of RAF through an asymmetric allosteric mechanism (Garnett et al. 2005; Rajakulendran et al. 2009; Hu et al. 2013) that triggers mitogen-activated protein kinase (MEK) and extracellular signal-related kinase (ERK) phosphorylation.
Under physiological conditions, the duration and amplitude of MAPK signaling is regulated through a network of negative feedback loops mediated by ERK. ERK suppresses pathway signaling either directly through phosphorylation of upstream components such as SOS or CRAF or transcriptionally through the regulation of expression of MAPK phosphatases (dual-specificity phosphatase [DUSP] family members) and Sprouty family of proteins (Fig. 1A) (Dougherty et al. 2005; Owens and Keyse 2007; Pratilas et al. 2009). These negative feedback loops enable the pathway output to be tightly controlled under basal and stressed conditions. Pathway signaling is also tightly regulated by a variety of additional mechanisms, including phosphorylation and interaction with 14-3-3 proteins (both of which have been described extensively in previous reviews) as well as interaction with the scaffolding protein kinase suppressor of (activated) RAS ([KSR], see more below; Freeman et al. 2013; Lavoie and Therrien 2015).
RAF FAMILY DOMAIN ARCHITECTURE AND MECHANISM OF ASYMMETRIC ALLOSTERIC ACTIVATION
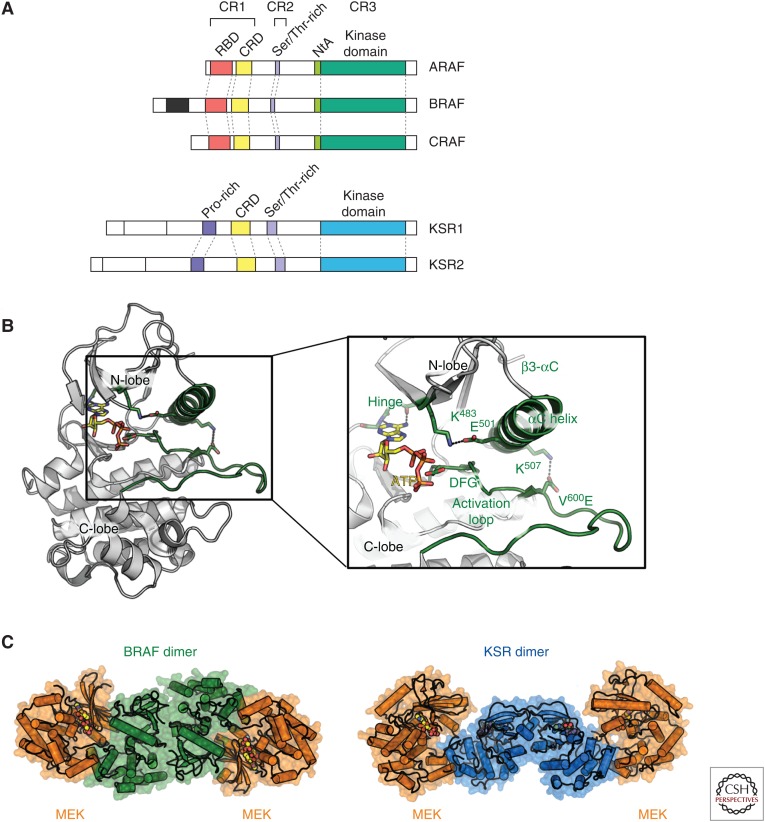
The protein domains of ARAF, BRAF, and CRAF are highly conserved, with each containing three conserved regions: CR1, CR2, and CR3 (Fig. 2A). CR1 contains the RBD and cysteine-rich domains (CRDs). CR2 contains a stretch of serine/threonine residues implicated in activation of RAF. Hence, both CR1 and CR2 together are commonly referred to as the NRR. CR3 is composed of the kinase domain and is held inactive via its interaction with the amino-terminal regulatory region (Lavoie and Therrien 2015). Interaction with RAS-GTP relieves the self-regulatory interaction between the NRR and kinase domains, such that RAF can dimerize with a second protomer and potentiate downstream signaling through the MEK-ERK pathway.
Figure 2.
Domain organization and structure of RAF family members. (A) Domain organization of ARAF, BRAF, CRAF, and kinase suppressor of (activated) RAS (KSR) with features of interest labeled. (B) Model of BRAF kinase domain (based on chain B form PDB code 4MNE) with adenosine triphosphate (ATP) docked in and V600E mutation modeled. Regions of interest are labeled and colored green. (C) Comparison of BRAF-mitogen-activated protein kinase (MEK) heterotetramer (left, in green and orange, respectively; PDB code 1MNE) and the KSR-MEK heterotetramer (right, in blue and orange, respectively; PDB code 2Y4I).
Although the molecular details of the interaction between the NRR and the RAF kinase domain are yet to be determined, the structure of the RAF kinase domain has been studied in detail both to better understand the mechanism of normal and oncogenic kinase activation as well as to support structure-based drug design of pathway-specific inhibitors. Structures of BRAF and CRAF kinase domains show a typical bi-lobed architecture of kinases with an adenosine triphosphate (ATP) (or inhibitor)-binding site at the interface between the amino (N-lobes) and carboxy-terminal lobes (C-lobes) (Wan et al. 2004; Hatzivassiliou et al. 2010). Key features include the activation loop (A-loop), the αC-helix, the catalytic salt bridge between K483 and E501 (in BRAF), the DFG motif, and the hinge region between the N- and C-lobes. All of these features contribute to the normal catalytic role of kinases and most are affected by the transition between the active and inactive conformation of RAF. On activation of RAF, the A-loop forms an extended platform for substrate binding while the αC-helix moves “in” toward the active site leading the formation of the catalytic K483–E501 salt bridge while the DFG motif is positioned such that the hydrophobic phenylalanine contributes to the hydrophobic core (Fig. 2B).
Immediately before the kinase domain is a short stretch of amino acids termed the amino-terminal acidic region (NtA; residues 446SSDD449 in BRAF). This region is constitutively phosphorylated in BRAF in addition to possessing two negatively charged residues (Lavoie and Therrien 2015). In CRAF, the sequence (338SSYY341) lacks the negatively charged residues and is only phosphorylated on recruitment to the plasma membrane. Phosphorylation of these residues in CRAF has been shown to enhance dimerization and RAF kinase activity (Hu et al. 2013). This amino acid sequence difference may account for the increased intrinsic activity of BRAF relative to CRAF (Emuss et al. 2005). The phosphorylated residues in the NtA are typically disordered in structures of BRAF, indicating that the mechanism of activation may rely on destabilizing the repressive effects of the NRR. When they are resolved, they reside close to R509 and have been suggested to help form the proper dimerization interface. On release of the NRR region by interaction with RAS, the kinase domains dimerize stabilizing the active conformation of BRAF.
BRAF and CRAF dimers can exist as homodimers (Farrar et al. 1996; Luo et al. 1996) and heterodimers (Mizutani et al. 2001), with heterodimers contributing to the majority of kinase activity within the cell (Rushworth et al. 2006). Recent work has shown that RAF activity is allosterically regulated via the side-to-side dimer configuration that positions the αC-helix in a catalytically active orientation (Rajakulendran et al. 2009). Removal of the regulatory amino terminus results in a constitutively active kinase, as does the induction of RAF dimerization through mutation of an interface residue (Rajakulendran et al. 2009). Furthermore, kinase-dead BRAF can activate CRAF in trans (Wan et al. 2004; Rushworth et al. 2006; Heidorn et al. 2010). A similar transactivation mechanism of CRAF by BRAF has been reported with certain ATP-competitive BRAF inhibitors (Hatzivassiliou et al. 2010; Heidorn et al. 2010; Poulikakos et al. 2010).
Further arguing for an asymmetric dimer state, the distant RAF family member KSR can also activate RAF activity. KSR was originally identified through RAS synthetic lethal genetic screens in Drosophila melanogaster and Caenorhabditis elegans (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995). In these studies, KSR was shown to function downstream from RAS regulating RAS/MAPK signaling through direct interactions with RAF, MEK, and ERK family members. Interestingly, the KSR proteins are most closely related to the RAF kinase family, with the carboxy-terminal region of KSR containing a pseudokinase domain, a region similar to canonical kinase domains yet lacking a critical catalytic lysine residue (Fig. 2A). Hence, KSR lacks kinase activity but has been shown to regulate RAF activity through a dimerization-dependent mechanism involving its pseudokinase domain (Roy et al. 2002; Rajakulendran et al. 2009; Hu et al. 2011). Given its role as a scaffolding protein, KSR expression levels have been shown to modulate the duration and amplitude of MAPK signaling (Therrien et al. 1996). Moderate levels of KSR expression enhance MAPK signaling, whereas high levels of KSR expression appear to function in a dominant negative manner by dampening pathway signaling likely through a sequestering mechanism (Kortum and Lewis 2004). Biochemical and structural studies have shown that KSR can dimerize with RAF and MEK proteins through its pseudokinase domain (Rajakulendran et al. 2009; Brennan et al. 2011). Dimerization of KSR with BRAF through a side-to-side dimer interface has been shown to enhance RAF kinase activity (Brennan et al. 2011).
MECHANISM OF ONCOGENIC RAS/MAPK SIGNALING
Oncogenic mutations in BRAF and KRAS circumvent the highly regulated activation process through a variety of mechanisms. The precise mechanism of MAPK pathway activation is dependent on the specific underlying genetic mutation. For example, the activating BRAF-V600E mutation or BRAF-β3-αC deletion, which is found in nearly 70% of melanomas and a small subset of pancreatic adenocarcinomas, respectively, predisposes BRAF to an active αC-helix-“in” conformation that is independent of RAS activation and BRAF dimerization for activity (Wan et al. 2004; Poulikakos et al. 2011; Foster et al. 2016b). In contrast, noncanonical BRAF mutations are not necessarily highly activating and, in some cases, abrogate kinase activity completely (i.e., BRAF-D594V). In this context, the mutant BRAF requires dimerization with CRAF for MAPK activity (Wan et al. 2004; Yao et al. 2015). Interestingly, noncanonical BRAF mutations are enriched in lung cancers (∼5%–7% of lung adenocarcinomas) and are mutually exclusive with KRAS mutations, suggesting that there may be a tissue-specific preference for this alteration (Foster et al. 2016a).
Similar to the mechanism of noncanonical BRAF mutations, activating KRAS mutations drive MAPK signaling through CRAF and BRAF dimerization and CRAF activation (Garnett et al. 2005; Dumaz et al. 2006; Rushworth et al. 2006). Mutations within the KRAS family member represent the second most frequent alteration in cancer, found in 20% of all patients, and 90% of pancreatic, 43% of colorectal, and 32% of lung adenocarcinomas (Cerami et al. 2012; Gao et al. 2013) with mutations typically occurring at codons 12, 13, or 61. These mutations result in reduced intrinsic and GTPase-activating protein (GAP)-mediated GTPase activity, rendering KRAS in a constitutively active, GTP-bound form. Activated RAS can then signal to its downstream effector pathways driving cell growth, proliferation, and survival in the absence of external signaling cues.
More recently, it has been reported that in the context of KRAS mutant tumors, BRAF is in a preassembled, inactive complex with MEK (Fig. 1A) (Haling et al. 2014). MEK phosphorylation results in rapid dissociation of the complex and kinetic studies with epidermal growth factor (EGF) stimulation (in wild-type cells) show that the complex can be reformed on accumulation of inactive MEK. Intriguingly, a cocrystal structure of the active, heterotetrameric complex of BRAF with MEK has been determined in the presence of a MEK inhibitor G-573 and ATP (Haling et al. 2014). In this structure, BRAF side-to-side dimers reside at the heart of the tetrameric complex, with BRAF adopting an active conformation and mediating a face-to-face interaction with MEK (Fig. 2C). Small molecule inhibitors of RAF or MEK can perturb this complex depending on their binding mode. Interestingly, unlike the KSR-MEK complex previously reported, the BRAF-MEK complex is high in affinity and appears to favor BRAF in its active conformation. Furthermore, structural studies show that the BRAF-MEK and KSR-MEK interfaces are largely overlapping such that MEK and KSR cannot interact with the BRAF kinase domain simultaneously (Haling et al. 2014). This suggests that KSR is more likely to function as a scaffolding protein that may transactivate RAF family members through an allosteric, side-to-side dimerization mechanism rather than through recruitment of MEK as a substrate for RAF.
These mechanistic insights into BRAF and KRAS mutant tumors underscore the fact that BRAF has both catalytic and noncatalytic functions in regulating MAPK signaling. In the context of KRAS mutant tumors, BRAF resides in the cytosol in a dormant complex with MEK and additional signaling cues are required to form a transient, active heterotetrameric complex with CRAF that results in MEK phosphorylation. In contrast, BRAF-V600E and BRAF deletions activate BRAF in the absence of dimerization (Fig. 1). Finally, noncanonical BRAF mutations (including kinase-dead oncogenic mutations) constitutively dimerize with CRAF and allosterically activate CRAF through a side-to-side dimerization mechanism. Taken together, regulation of RAF dimerization is the rate-limiting step in pathway activity and oncogenic mutations in BRAF either abrogate the requirement for dimerization for kinase activity or result in constitutive dimers independent of KRAS as in the case of the noncanonical BRAF mutations. In KRAS mutant tumors, additional studies need to be conducted to understand how the preassembled MEK-BRAF complex is primed for activity.
TARGETING THE MAPK PATHWAY TO TREAT RAS MUTANT TUMORS
Several lines of evidence support the notion that targeting the MAPK effector pathway may be beneficial for KRAS mutant patients. First, CRAF but not BRAF has been shown to be required for KRAS driven signaling (Dumaz et al. 2006) as well as for the initiation of KRAS driven lung tumorigenesis in a genetically engineered KRAS driven lung cancer mouse model (Blasco et al. 2011; Karreth et al. 2011). In this KRAS genetically engineered mouse model, depletion of CRAF and not BRAF increased survival and decreased tumor incidence. Second, of the effector pathways downstream from RAS, KRAS and BRAF mutations appear to be mutually exclusive of one another, suggesting redundant functions. In contrast, mutations in other effector pathways such as PI3K can co-occur significantly with KRAS mutations (as exemplified in colorectal cancer [CRC]), suggesting nonoverlapping functions. Third, a recent genome-wide clustered regularly interspaced short palindromic repeat (CRISPR) dropout screen (Wang et al. 2017) of 14 acute myeloid leukemia (AML) cell lines with a subset of KRAS mutant and KRAS wild-type lines identified CRAF as a top KRAS synthetic lethal hit and not an essential gene, again suggesting that of the effector pathways downstream from KRAS, the MAPK pathway, namely, the RAF family members, remain critical for KRAS-driven tumorigenesis. Thus, of the downstream effector pathways, the MAPK pathway continues to be the most compelling therapeutic target in the context of RAS mutant cancers.
USING MAPK INHIBITORS IN BRAF AND RAS MUTANT CANCERS
Significant efforts have been undertaken to find small molecule inhibitors to selectively target the MAPK pathway in malignancies. BRAF and MEK inhibitors have been approved to treat BRAF mutant cancers with dozens of other RAF, MEK, and ERK inhibitors either still in preclinical research stage or early stage clinical trials (Table 1). These drugs bind to a diverse array of kinase conformational states and have been shown to exhibit efficacy in mutation-specific contexts. Despite extensive clinical efforts, the only approved MAPK therapies to-date are the BRAF inhibitors vemurafenib and dabrafenib and the MEK inhibitors trametinib and cobimetinib, all of which have been approved only for BRAF V600E/K metastatic melanomas (Kim et al. 2014).
Table 1.
BRAF, MEK, and ERK inhibitors with their binding modes defined with structures shown below
| Molecule | Target | Binding mode | αC-helix “in” or “out” |
|---|---|---|---|
| GDC-0879 | BRAF | Type I | “in” |
| Vemurafenib | BRAF | Type I.5 | “out” |
| Dabrafenib | BRAF | Type I.5 | “out” |
| PLX-8394 | BRAF | Type I.5 | “out” |
| Sorafenib | BRAF | Type II | “in” |
| AZ-628 | BRAF | Type II | “in” |
| Trametinib | MEK | Class I | “out” |
| Cobimetinib | MEK | Class I | “out” |
| GDC-0623 | MEK | Class II | “out” |
| G-573 | MEK | Class II | “out” |
| CH5126766 | MEK | Class II | “out” |
| GDC-0994 | ERK | Type I | “in” |
| SCH772984 | ERK | Type I | “in” |
MEK, Mitogen-activated protein kinase; ERK, extracellular signal-related kinase.
CLASSES OF RAF INHIBITORS
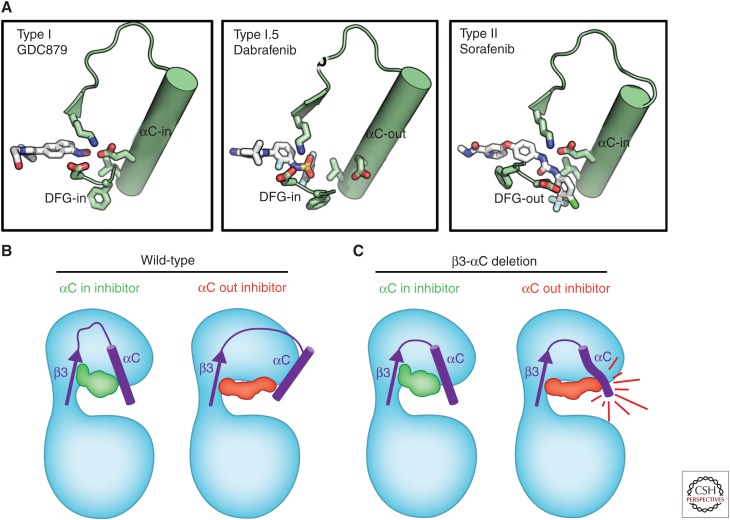
RAF inhibitors can be grouped into several classes based on their molecular mechanism of action, all of which are ATP-competitive (Fig. 3). Type I inhibitors such as GDC-0879, which bind to a similar site as ATP, make interactions with the hinge region and favor an active conformation of the A-loop, αC-helix, DFG motif and permit formation of the K483–E501 catalytic salt bridge (Hatzivassiliou et al. 2010). In contrast, the DFG-“out” (also known as type II) inhibitors such as sorafenib, AZ-628, and LY3009120 bind to RAF such that the F within the DFG motif is oriented toward solvent as opposed to being buried in the hydrophobic core of the protein (Wan et al. 2004; Peng et al. 2015; Foster et al. 2016b). However, the αC-helix remains in the “in” position with formation of the catalytic salt bridge. Type I.5 inhibitors have an intermediate effect. They typically occupy both the ATP-binding site as well as extend toward the αC-helix, perturbing its conformation and forcing it into varying degrees of an “out” conformation in which the catalytic salt bridge is disrupted.
Figure 3.
BRAF inhibitor-binding modes. (A) Cartoon images of BRAF wild-type and β3-αC deletion mutants bound to type I, type I.5, and type II inhibitors, including the effect on αC-helix and the catalytic salt bridge. (B) Cartoon representations of BRAF bound to type I (left), type I.5 (center), and type II inhibitors (right), including the effect on the αC-helix and the catalytic salt bridge.
All of these inhibitors prevent the ability of RAF to phosphorylate MEK. However, the differences in RAF conformation, affinity for ATP, and interactions with partners results in different cellular consequences. For instance, all classes of RAF inhibitors preferentially inhibit BRAF-V600E in cells. This is attributed to the fact that the V600E mutations result in a ∼10-fold decrease in ATP KM (reduced affinity for ATP), similar to what has been reported for epidermal growth factor receptor (EGFR) mutations, thereby sensitizing this specific mutant oncoprotein to ATP-competitive inhibitors (Hatzivassiliou et al. 2010). In contrast, type II (but not type I.5) inhibitors are able to inhibit tumors harboring noncanonical BRAF mutations that are constitutively dimerized with CRAF (Yao et al. 2015). In this context, BRAF-CRAF dimerization conformationally restricts the αC-helix from shifting to accommodate the type I.5 inhibitor-binding mode, and hence RAF dimer inhibitors (which are typically type II inhibitors) have been shown to be effective toward RAF dimers (Foster et al. 2016b; Karoulia et al. 2016). In yet another context, BRAF-β3-αC deletions are resistant to type I.5 inhibitors because the αC-helix is conformationally restricted from moving as a result of the deletion in the β3-αC loop (Foster et al. 2016b). However, because the conformation caused by these deletions is similar to conformation induced by dimerization (shift of the αC-helix “in”), this class of mutation is sensitive to type II inhibitors. Interestingly, whereas both the BRAF-V600E and BRAF-β3-αC deletions do not require dimerization for activity, they have distinct requirements for inhibition. In the case of BRAF-V600E, a type I.5 inhibitor can inhibit its activity effectively because the C-helix remains conformationally flexible and able to “shift” on inhibitor binding; however, deletion of the β3-αC loop restricts the C-helix from this same movement with type I.5 inhibitors (Fig. 3).
The approved BRAF inhibitors vemurafenib and dabrafenib are both type I.5 inhibitors, although the extent of the interaction with the αC-helix results in differences in their activity (Joseph et al. 2010; Zhang et al. 2015; Foster et al. 2016b). Vemurafenib was derived from a fragment-based drug discovery approach and, along with the B-cell lymphoma 2 (Bcl-2) inhibitor venetoclax, are examples of this approach leading to approved therapeutics (Erlanson et al. 2016). In particular, the fragment lead for vemurafinib was found in a high-concentration biochemical assay-based screen of a 20,000 library of small (150–300 Da) scaffold-like compounds. Extensive structure-based drug design was used in the process of discovering vemurafenib. A highly engineered version of BRAF with improved expression, purification, and crystallization properties was developed to support this effort (Bollag et al. 2012).
Most type I and type I.5 inhibitors, including the clinically approved drugs, have been shown preclinically to induce paradoxical activation of MAPK signaling in RAS mutant cells or cells with elevated RTK signaling (Hatzivassiliou et al. 2010; Poulikakos et al. 2010). In some contexts, these inhibitors have been shown to increase cellular proliferation both in vitro and in vivo (Hatzivassiliou et al. 2010). Paradoxical activation is believed to be the consequence of drug-induced dimerization of the drug-bound BRAF protomer with a second RAF protomer (i.e., CRAF), which results in increased CRAF kinase activity (which is RAS-GTP dependent) and downstream signaling through MEK and ERK. At high concentrations of the drugs, both protomers of the dimer (i.e., BRAF and CRAF) are occupied and the paradoxical activation is inhibited. It has been hypothesized that the significant incidence of cutaneous squamous cell carcinoma and keratoacanthomas (∼20% of patients) observed clinically with BRAF-V600E melanoma patients treated with BRAF inhibitors is attributed to paradoxical activation (Anforth et al. 2012a,b; Su et al. 2012). In support of this, these paradox-induced skin tumors show a high incidence of HRAS mutations, consistent with the preclinical observations demonstrating enhanced growth of RAS mutant tumors on treatment with vemurafenib (Anforth et al. 2012a; Callahan et al. 2012; Oberholzer et al. 2012). These clinical findings have raised concerns that secondary RAS-driven tumors may be accelerated by this class of drugs.
Two main approaches have been explored to address the deleterious effects of paradoxical activation. Type I.5 binders have been discovered that induce even greater effects on the conformation of the αC-helix such that BRAF/CRAF dimerization can be more effectively inhibited (Zhang et al. 2015). This class of compounds, also called “paradox busters,” still effectively inhibits BRAF-V600E tumors but do not induce paradoxical activation. The best characterized paradox buster PLX-8394 has been shown to block BRAF-V600E activity without inducing paradoxical activation in RAS mutant tumors (Zhang et al. 2015). Although the structural basis of this compared with other type I.5 inhibitors is not clear, structural studies suggest that these sulfonamide-containing type I.5 inhibitors may interact with Leu505 within the C-helix, which may result in greater dynamics in this region, thus disfavoring the dimeric structure. Unfortunately, despite the lack of paradoxical activation, paradox busters still show no efficacy in KRAS mutant tumors as they are not able to bind to preexisting RAF dimers. Alternatively, type II pan-RAF inhibitors have been shown to bind to BRAF/CRAF heterodimers and thereby prevent induction of MAPK signaling while remaining effective at inhibiting BRAF-V600E-dependent signaling. In this context, the type II inhibitors will still induce BRAF and CRAF dimerization in RAS mutant tumors, but paradoxical activation is not observed owing to occupancy of both promoters simultaneously (Nakamura et al. 2013; Peng et al. 2015).
Despite the significant progress that has been made in developing inhibitors that selectively target BRAF, several of which have been clinically approved for treating BRAF-V600 mutant cancers, these approaches have resulted in little success with KRAS mutant cancers. Some hope may lie in recent reports of type II RAF dimer inhibitors demonstrating efficacy in KRAS mutant tumors; however, the dose required for efficacy in KRAS mutant tumors is significantly greater than that required for efficacy in BRAF-V600E models, raising concerns around the therapeutic utility of such an approach as a single agent (Peng et al. 2015). Several recent reports have also described the combination of RAF inhibitors, with MEK inhibitors showing efficacy in RAS mutant tumors (Lamba et al. 2014; Lito et al. 2014). In this context, it has been shown that MEK inhibitors disable the negative feedback loops upstream of RAF, resulting in RAF kinase activation and increased sensitivity to RAF kinase inhibition. It remains to be seen whether the combination of RAF and MEK inhibitors will be successful clinically in KRAS mutant tumors.
CLASSES OF MEK INHIBITORS
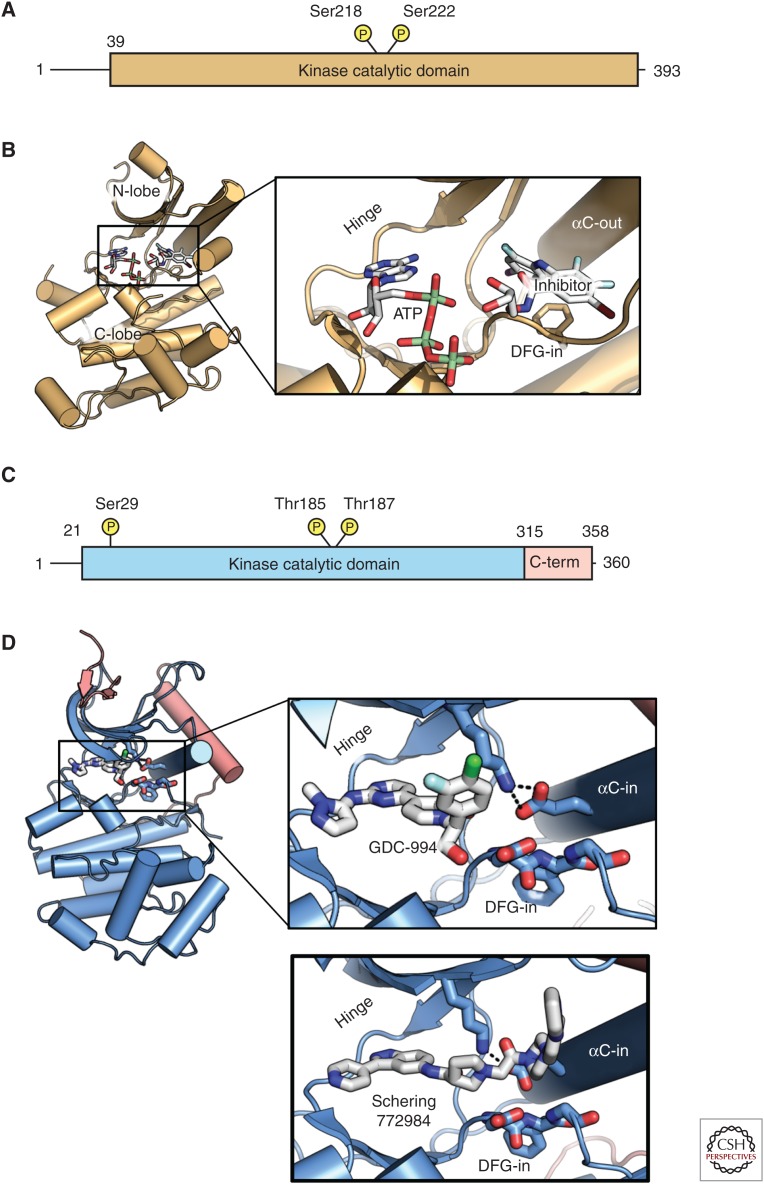
In addition to approaches to targeting RAF itself, MEK inhibitors have also been approved for BRAF-V600E/K melanomas. Phosphorylation by BRAF at two sites in the MEK activation loop (Ser218 and 222 in MEK1) correlates with release of MEK from BRAF, MEK activation, and downstream signaling to ERK. MEK1 and MEK2 are unusual dual-specificity kinases that phosphorylate ERK on both serine and tyrosine residues within the ERK activation loop. Unlike RAF, MEK does not possess extensive accessory domains analogous to the RAF NRR. In contrast, MEK is primarily comprised of a kinase domain with short flanking amino- and carboxy-terminal regions with sequences that modulate MEK activity by mechanisms, including facilitating interactions with binding partners and dictating subcellular localization (Fig. 4). MEK interacts with RAF by forming a face-to-face heterodimer leaving RAF free to dimerize with another RAF protomer, such that dimeric BRAF can bind two copies of MEK simultaneously (Haling et al. 2014). Unlike most kinases, MEK binds ATP with high affinity, which makes development of non-ATP competitive inhibitors the more preferred approach for this kinase. Accordingly, the majority of MEK inhibitors developed to date, including the two clinically approved inhibitors cobimetinib and trametinib, both bind to MEK in the presence of ATP within an allosteric binding pocket. The affinity of these inhibitors is increased in the presence of ATP, supporting a noncompetitive mechanism of action (Fig. 3) (Ohren et al. 2004; Caunt et al. 2015).
Figure 4.
Domain organization and structure of mitogen-activated protein kinase (MEK) and extracellular signal-related kinase (ERK). (A) Domain organization of MEK1. (B) Structure of the MEK kinase domain (PDB 1S9J). Inset shows features of interest, including the adenosine triphosphate (ATP) site, allosteric inhibitor site, and the αC-helix that adopts an “out” conformation, including disruption of the catalytic salt bridge. (C) Domain structure of ERK. (D) Structure of ERK kinase bound to GDC-0994 (blue with additions to the canonical kinase domain in red; PDB 5K4I). C-term, Carboxy terminal.
In contrast to the clinically approved BRAF inhibitors, trametinib and cobimetinib do not induce paradoxical activation. However, similar to BRAF inhibitors, the mechanism of action of these MEK inhibitors modulates their ability to blunt MAPK signaling effectively in KRAS mutant tumors. Class I MEK inhibitors such as cobimetinib show efficacy in BRAF-V600E tumors, but have limited efficacy in KRAS mutant tumors (Hatzivassiliou et al. 2013). Studies have shown that this class of MEK inhibitor disables negative feedback in KRAS mutant tumors and, over time, results in pathway reactivation as observed with induction of phosphorylated MEK and phosphorylated ERK. Multiple studies have shown that this pathway reactivation is highly dependent on RAF kinase activation and the combination of RAF and MEK inhibition results in more robust efficacy in KRAS mutant tumors (Hatzivassiliou et al. 2013; Lito et al. 2014). In contrast, class II MEK inhibitors such as G-573 have shown efficacy in KRAS mutant tumors, demonstrating sustained inhibition of the pathway, despite disabled negative feedback (Hatzivassiliou et al. 2013; Lito et al. 2014). This activity has been attributed to the dual role of this class as RAF-MEK inhibitors through trapping and sequestering an inactive RAF-MEK complex. Therefore, these MEK inhibitors do not induce phosphorylated MEK and ERK over time and effectively inhibit MAPK signaling pathways in KRAS mutant tumors. These differences in behavior of class I and class II inhibitors likely derive from subtle differences of their interaction with the MEK A-loop (in particular the strength of the hydrogen bond to Ser212) (Hatzivassiliou et al. 2013). This effectively alters the stability of the inactive BRAF-MEK complex and contributes to the extent of MEK phosphorylation suppression.
ERK INHIBITORS
ERK inhibitors have also been investigated for use in both BRAF-V600E and KRAS mutant tumors. As ERK is the terminal kinase in the pathway, it has numerous substrates, and inhibition of its activity can have a broad downstream effect. As such, ERK has been targeted for small molecule drug discovery for inhibitors to work as single agents or in conjunction with upstream kinase inhibitors. Like MEK, the overall ERK structure is simpler than that of RAF, suggesting less complicated regulation. Unlike MEK, well-characterized ERK inhibitors to date are ATP competitive and bind to αC-“in” conformation. This includes the ERK inhibitor GDC-0994, which has been tested clinically (Fig. 4) (Blake et al. 2016). More recently, a novel binding mode with the ERK inhibitor SCH772984 has been reported. High-resolution crystal structures of SCH772984 bound to ERK1 and ERK2 reveal that the extended structure of this inhibitor resulted in discrete interactions with an induced binding pocket located between the αC-helix and the phosphate-binding loop (P-loop). This binding mode maintains the DFG and catalytic salt bridge in an active conformation while modestly tilting the C-helix and slightly distorting the P-loop. It remains to be seen whether these differential binding conformations result in differences in clinical responses.
KSR INHIBITORS
In addition to catalytic components, the RAF pathway is modulated by the pseudokinase KSR (KSR1/2). KSR possesses an overall domain structure analogous to that of BRAF but lacks the critical catalytic lysine residues thereby rendering it a pseudokinase. KSR has been described as having a scaffolding function as it can associate with many of the components of the MAPK pathway to potentiate signaling (Raabe and Rapp 2002). In particular, the kinase domain of KSR can homodimerize and interact with MEK (Brennan et al. 2011) or can heterodimerize and hence activate BRAF (and CRAF) (Rajakulendran et al. 2009). Although it is still unclear whether residual KSR enzymatic activity is essential for its biological function (Brennan et al. 2011), the ability of KSR to activate the pathway appears to depend in part on the conformation of the pseudokinase domain, suggesting that small molecules, which bind to KSR in specific conformational states, might be able to regulate its function. For instance, blocking the KSR active site by the mutation A587F stabilizes interactions with RAF and MEK, decreases pathway activity, and has been shown to prevent RAS transformation (Hu et al. 2011). This was originally ascribed to a latent catalytic activity of KSR, but could also be attributable to a sequestration of MEK by a KSR/RAF heterodimer. More recently, ATP-competitive inhibitors binding to KSR have been described and cocrystal structures reported (Dhawan et al. 2016). These compounds, exemplified by APS-2-79, bind to the KSR within the ATP-binding pocket and stabilize an inactive conformation such that heterodimerization with BRAF is blocked. Although the kinase selectivity of these tool compounds is not yet optimal and the target conformational state not clearly defined, they serve as good tool compounds to probe the effect of KSR conformation states and better understand how KSR regulates MAPK signaling downstream from KRAS.
CONCLUDING REMARKS
Identifying approaches to target RAS mutant tumors remains a critical unmet need in cancer therapy. As described in this review, there is strong rationale to target the MAPK signaling pathway downstream from KRAS; however, inhibition of any individual target in the pathway has had little single activity in KRAS mutant tumors to date. Some of this is dependent on the nature of the inhibitor used as described for the various classes of RAF and MEK inhibitors. Depending on the genetic context, molecules binding to the same target can either inhibit or activate the pathway in a drug-induced, conformation-specific manner. In other cases, efficacy has been limited by dose-limiting toxicities in the clinic. It remains to be seen whether combinations of agents targeting the MAPK pathway will show efficacy in RAS mutant tumors; many of those trials are currently underway.
However, given the profound differences in efficacy observed with subtle differences in binding modes of various MAPK inhibitors, the possibility exists that other classes of allosteric or conformation-specific inhibitors will show efficacy in this context. Discovery of such classes of inhibitors may require novel approaches such as phenotypic screens and novel fragment screening approaches directly targeting allosteric sites rather than the ATP-competitive binding sites. Allosteric binding sites for other kinases have been reported in the literature, including for oncogenic kinases such as EGFR and BCR-ABL (Zhang et al. 2010; Jia et al. 2016). Further, novel biological, biochemical, and structural data characterizing scaffolding proteins that regulate this pathway may provide new directions for either single agent or combination therapies to treat KRAS mutant cancers. Although much as been learned about the RAF pathway and its regulation in recent years, much remains to be discovered especially with regard to the development of therapeutics with transformative tolerability and durability targeting MAPK pathway-driven cancers.
Footnotes
Editors: Linda Van Aelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Anforth R, Tembe V, Blumetti T, Fernandez-Penas P. 2012a. Mutational analysis of cutaneous squamous cell carcinomas and verrucal keratosis in patients taking BRAF inhibitors. Pigment Cell Melanoma Res 25: 569–572. [DOI] [PubMed] [Google Scholar]
- Anforth RM, Blumetti TC, Kefford RF, Sharma R, Scolyer RA, Kossard S, Long GV, Fernandez-Penas P. 2012b. Cutaneous manifestations of dabrafenib (GSK2118436): A selective inhibitor of mutant BRAF in patients with metastatic melanoma. Br J Dermatol 167: 1153–1160. [DOI] [PubMed] [Google Scholar]
- Blake JF, Burkard M, Chan J, Chen H, Chou KJ, Diaz D, Dudley DA, Gaudino JJ, Gould SE, Grina J, et al. 2016. Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an extracellular signal-regulated kinase 1/2 (ERK1/2) inhibitor in early clinical development. J Med Chem 59: 5650–5660. [DOI] [PubMed] [Google Scholar]
- Blasco RB, Francoz S, Santamaria D, Canamero M, Dubus P, Charron J, Baccarini M, Barbacid M. 2011. C-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19: 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. 2012. Vemurafenib: The first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov 11: 873–886. [DOI] [PubMed] [Google Scholar]
- Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, Barford D. 2011. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472: 366–369. [DOI] [PubMed] [Google Scholar]
- Callahan MK, Rampal R, Harding JJ, Klimek VM, Chung YR, Merghoub T, Wolchok JD, Solit DB, Rosen N, Abdel-Wahab O, et al. 2012. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med 367: 2316–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caunt CJ, Sale MJ, Smith PD, Cook SJ. 2015. MEK1 and MEK2 inhibitors and cancer therapy: The long and winding road. Nat Rev Cancer 15: 577–592. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan NS, Scopton AP, Dar AC. 2016. Small molecule stabilization of the KSR inactive state antagonizes oncogenic Ras signalling. Nature 537: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. 2005. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell 17: 215–224. [DOI] [PubMed] [Google Scholar]
- Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, Bastian BC, Springer C, Marais R. 2006. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF disrupted cyclic AMP signaling. Cancer Res 66: 9483–9491. [DOI] [PubMed] [Google Scholar]
- Emuss V, Garnett M, Mason C, Marais R. 2005. Mutations of C-RAF are rare in human cancer because C-RAF has a low basal kinase activity compared with B-RAF. Cancer Res 65: 9719–9726. [DOI] [PubMed] [Google Scholar]
- Erijman A, Shifman JM. 2016. RAS/effector interactions from structural and biophysical perspective. Mini Rev Med Chem 16: 370–375. [DOI] [PubMed] [Google Scholar]
- Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H. 2016. Twenty years on: The impact of fragments on drug discovery. Nat Rev Drug Discov 15: 605–619. [DOI] [PubMed] [Google Scholar]
- Farrar MA, Alberol-Ila J, Perlmutter RM. 1996. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature 383: 178–181. [DOI] [PubMed] [Google Scholar]
- Foster SA, Klijn C, Malek S. 2016a. Tissue specific mutations in BRAF and EGFR necessitate unique therapeutic approaches. Trends Cancer 2: 699–701. [DOI] [PubMed] [Google Scholar]
- Foster SA, Whalen DM, Ozen A, Wongchenko MJ, Yin J, Yen I, Schaefer G, Mayfield JD, Chmielecki J, Stephens PJ, et al. 2016b. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell 29: 477–493. [DOI] [PubMed] [Google Scholar]
- Freeman AK, Ritt DA, Morrison DK. 2013. The importance of Raf dimerization in cell signaling. Small GTPases 4: 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Rana S, Paterson H, Barford D, Marais R. 2005. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell 20: 963–969. [DOI] [PubMed] [Google Scholar]
- Haling JR, Sudhamsu J, Yen I, Sideris S, Sandoval W, Phung W, Bravo BJ, Giannetti AM, Peck A, Masselot A, et al. 2014. Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling. Cancer Cell 26: 402–413. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, et al. 2010. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464: 431–435. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Haling JR, Chen H, Song K, Price S, Heald R, Hewitt JF, Zak M, Peck A, Orr C, et al. 2013. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature 501: 232–236. [DOI] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, et al. 2010. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Yu H, Kornev AP, Zhao J, Filbert EL, Taylor SS, Shaw AS. 2011. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc Natl Acad Sci 108: 6067–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Stites EC, Yu H, Germino EA, Meharena HS, Stork PJ, Kornev AP, Taylor SS, Shaw AS. 2013. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell 154: 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Yun CH, Park E, Ercan D, Manuia M, Juarez J, Xu C, Rhee K, Chen T, Zhang H, et al. 2016. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 534: 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, et al. 2010. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci 107: 14903–14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoulia Z, Wu Y, Ahmed TA, Xin Q., Bollard J, Krepler C, Wu X, Zhang C, Bollag G, Herlyn M, et al. 2016. An integrated model of RAF inhibitor action predicts inhibitor activity against oncogenic BRAF signaling. Cancer Cell 30: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. 2011. C-Raf is required for the initiation of lung cancer by K-RasG12D. Cancer Discov 1: 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, McKee AE, Ning YM, Hazarika M, Theoret M, Johnson JR, Xu QC, Tang S, Sridhara R, Jiang X, et al. 2014. FDA approval summary: Vemurafenib for treatment of unresectable or metastatic melanoma with the BRAFV600E mutation. Clin Cancer Res 20: 4994–5000. [DOI] [PubMed] [Google Scholar]
- Kornfeld K, Hom DB, Horvitz HR. 1995. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell 83: 903–913. [DOI] [PubMed] [Google Scholar]
- Kortum RL, Lewis RE. 2004. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol 24: 4407–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba S, Russo M, Sun C, Lazzari L, Cancelliere C, Grernrum W, Lieftink C, Bernards R, Di Nicolantonio F, Bardelli A. 2014. RAF suppression synergizes with MEK inhibition in KRAS mutant cancer cells. Cell Rep 8: 1475–1483. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Therrien M. 2015. Regulation of RAF protein kinases in ERK signalling. Nature Rev Mol Cell Biol 16: 281–298. [DOI] [PubMed] [Google Scholar]
- Lin WC, Iversen L, Tu HL, Rhodes C, Christensen SM, Iwig JS, Hansen SD, Huang WY, Groves JT. 2014. H-Ras forms dimers on membrane surfaces via a protein–protein interface. Proc Natl Acad Sci 111: 2996–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, Saborowski M, Kastenhuber E, Fellmann C, Ohara K, et al. 2014. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Tzivion G, Belshaw PJ, Vavvas D, Marshall M, Avruch J. 1996. Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature 383: 181–185. [DOI] [PubMed] [Google Scholar]
- Mizutani S, Inouye K, Koide H, Kaziro Y. 2001. Involvement of B-Raf in Ras-induced Raf-1 activation. FEBS Lett 507: 295–298. [DOI] [PubMed] [Google Scholar]
- Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, Dyba MA, Stefanisko K, Tarasov SG, Gursoy A, et al. 2015. GTP-dependent K-Ras dimerization. Structure 23: 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Arita T, Tsuchiya S, Donelan J, Chouitar J, Carideo E, Galvin K, Okaniwa M, Ishikawa T, Yoshida S. 2013. Antitumor activity of the selective pan-RAF inhibitor TAK-632 in BRAF inhibitor-resistant melanoma. Cancer Res 73: 7043–7055. [DOI] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. 1995. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature 375: 554–560. [DOI] [PubMed] [Google Scholar]
- Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, Roden C, Chalk CJ, Ardlie K, Palescandolo E, et al. 2012. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol 30: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, et al. 2004. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol 11: 1192–1197. [DOI] [PubMed] [Google Scholar]
- Owens DM, Keyse SM. 2007. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26: 3203–3213. [DOI] [PubMed] [Google Scholar]
- Peng SB, Henry JR, Kaufman MD, Lu WP, Smith BD, Vogeti S, Rutkoski TJ, Wise S, Chun L, Zhang Y, et al. 2015. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 28: 384–398. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. 2010. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480: 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. 2009. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci 106: 4519–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe T, Rapp UR. 2002. KSR—A regulator and scaffold protein of the MAPK pathway. Science STKE 2002: pe28. [DOI] [PubMed] [Google Scholar]
- Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. 2009. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461: 542–545. [DOI] [PubMed] [Google Scholar]
- Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M. 2002. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev 16: 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth LK, Hindley AD, O’Neill E, Kolch W. 2006. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol 26: 2262–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, et al. 2017. Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol 13: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. 2012. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med 366: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Han M. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83: 889–901. [DOI] [PubMed] [Google Scholar]
- Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. 1995. KSR, a novel protein kinase required for RAS signal transduction. Cell 83: 879–888. [DOI] [PubMed] [Google Scholar]
- Therrien M, Michaud NR, Rubin GM, Morrison DK. 1996. KSR modulates signal propagation within the MAPK cascade. Genes Dev 10: 2684–2695. [DOI] [PubMed] [Google Scholar]
- Udell CM, Rajakulendran T, Sicheri F, Therrien M. 2011. Mechanistic principles of RAF kinase signaling. Cell Mol Life Sci 68: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. 2004. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116: 855–867. [DOI] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. 2017. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell 168: 890–903 e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CK, Slupsky JR, Kalmes HA, Rapp UR. 2001. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res 61: 3595–3598. [PubMed] [Google Scholar]
- Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li Q, de Stanchina E, Abdel-Wahab O, Solit DB, Poulikakos PI, et al. 2015. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 28: 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, et al. 2010. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Spevak W, Zhang Y, Burton EA, Ma Y, Habets G, Zhang J, Lin J, Ewing T, Matusow B, et al. 2015. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature 526: 583–586. [DOI] [PubMed] [Google Scholar]