Abstract
A growing collection of bacterial riboswitch classes is being discovered that sense central metabolites, coenzymes, and signaling molecules. Included among the various mechanisms of gene regulation exploited by these RNA regulatory elements are several that modulate messenger RNA (mRNA) translation. In this review, the mechanisms of riboswitch-mediated translation control are summarized to highlight both their diversity and potential ancient origins. These mechanisms include ligand-gated presentation or occlusion of ribosome-binding sites, control of alternative splicing of mRNAs, and the regulation of mRNA stability. Moreover, speculation on the potential for novel riboswitch discoveries is presented, including a discussion on the potential for the discovery of a greater diversity of mechanisms for translation control.
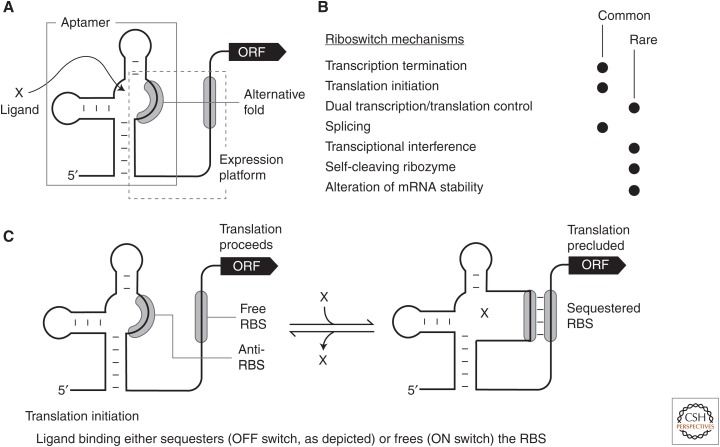
Riboswitches are RNA gene-control structures commonly found in the 5′ untranslated regions (UTRs) of messenger RNAs (mRNAs) where they sense and respond to small molecule or ion ligands (Breaker 2011; Serganov and Nudler 2013; Sherwood and Henkin 2016). In most instances, binding of a target ligand to the aptamer domain of the riboswitch triggers changes in the folding pattern of the expression platform (Fig. 1A), which alters the level of gene expression by one or more of many possible mechanisms. Several diverse mechanisms for riboswitch-mediated gene regulation have been established (Fig. 1B), and herein I will focus primarily on the mechanisms that more directly involve the regulation of mRNA translation.
Figure 1.
Schematic representations of common riboswitch expression platform arrangements. (A) Riboswitches typically carry a single ligand-binding aptamer (gray box) located upstream of (and slightly overlapping) the expression platform (dashed box). Folding changes in the aptamer, brought about by ligand binding, cause folding changes in the expression platform to regulate gene expression by various mechanisms. (B) List of experimentally validated or predicted riboswitch gene-control mechanisms. Processes by which the mechanisms highlighted in bold italic font regulate translation and are discussed in the text. (C) Schematic representation of a riboswitch that permits translation in the absence of ligand (left), but inhibits translation when bound to ligand (right). In the model depicted, ligand binding sequesters the ribosome-binding site (RBS) and prevents ribosome binding to the messenger RNA (mRNA). Alternatively, some riboswitch RNAs liberate the RBS on ligand binding to promote ribosome binding and translation.
It is important to note that several other long-studied gene-control systems in bacteria exploit some of the same regulatory mechanisms as do riboswitches. For example, the classic attenuation mechanisms first discovered in the early 1970s (Yanofsky 2000) require that the translation machinery report on the abundance of an aminoacylated transfer RNA (tRNA) of interest (e.g., for tryptophan) to regulate expression of the main open reading frame (ORF). This sensory process involves a change in translation speed through a region of repetitive tryptophan codons in an upstream ORF (uORF), which leads to alternative folding of the nascent mRNA transcript and premature transcription termination if the aminoacylated tRNA is abundant. Similarly, tryptophan can be directly sensed by TRAP, which is a protein factor that forms multimers and binds the 5′UTR of its target mRNA when tryptophan is abundant (Antson et al. 1999). These types of gene-control systems, which appear to be quite common, require complex-folded biopolymers (protein or RNA factors) other than the 5′UTR to serve as the sensor for the target ligand. In contrast, riboswitches serve both as the ligand sensor (aptamer) and as the device (expression platform) that communicates the requirement to regulate gene expression. As a result, these systems have been classified separately (Nahvi et al. 2002; Breaker 2011).
Additional examples of regulatory systems that involve 5′UTR structures exist, including T box RNAs, which directly sense non-amino-acylated tRNAs (Green et al. 2010), and RNA thermosensors, which respond to specific temperatures (Kortmann and Narberhaus 2012). These systems are also usually classified separately from riboswitches even though they too use many of the same mechanisms for regulating gene expression. Specifically, T box RNAs indirectly sense their target amino acids by binding to tRNAs that lack an aminoacyl modification. Although Watson–Crick base-pairing is involved in RNA-RNA (tRNA-T box) complex formation (Grigg et al. 2013), base-pairing appears to be less extensive than that observed for short RNA (sRNA) regulators that bind to 5′UTRs to trigger expression changes (Gottesman and Storz 2011). Interestingly, thermosensors do not need to form a binding site for a ligand at all, given that they respond to changes in temperature. Although all RNA structures can respond to heat as their thermal melting temperatures are approached, biologically relevant thermosensors need to form a metastable structure that is precisely tuned to change gene expression at a temperature useful to the cell. This appears to be a relatively easy demand to meet, as RNA thermosensor domains (Grosso-Becera et al. 2015) are typically far simpler than the complex structures formed by riboswitch aptamers (McCown et al. 2017).
The methods used to discover novel classes of riboswitch candidates have had to be modified over the years to continue to yield new findings. This necessity can be understood by recognizing that data hinting that riboswitches indeed existed were derived from bacterial genetics experiments, and that the riboswitches first observed are members of the most common classes. For example, some of the earliest genetic data sets suggesting the existence of riboswitches for lysine (Boy et al. 1979), flavin mononucleotide (FMN) (Gusarov et al. 1997), and adenosylcobalamin (AdoCbl) (Lundrigan et al. 1991) riboswitches were generated and published years before definitive experimental proofs of these riboswitches were made. These three riboswitch classes are among the 10 most abundant classes known. Therefore, they were more likely than rare riboswitch classes to have been encountered by genetics-based mutational screening or other genetic studies.
As each newly found common riboswitch class has been revealed, it becomes increasingly more difficult to uncover evidence pointing to the remaining undiscovered classes. This is because the undiscovered classes tend to be rarer (fewer number of representatives) than those discovered previously (McCown et al. 2017). It became clear that genetic methods would not be effective in searching for additional riboswitch classes. Fortunately, new methods involving comparative sequence analyses that are performed by powerful computer algorithms have been used with much success to search for additional riboswitch classes (e.g., see Barrick et al. 2004; Weinberg et al. 2007, 2010). With improvements in these computation search methods, coupled with the expanding collection of genomic DNA sequence data, it seems certain that many additional riboswitch classes and representatives of many other types of RNA-based gene-control systems will be discovered in the coming years.
More than 38 distinct classes of ligand-binding riboswitches have been experimentally validated to date (Arachchilage et al. 2017; McCown et al. 2017), and it has been proposed (Ames and Breaker 2010; Breaker 2011; McCown et al. 2017) that many hundreds of additional classes remain to be discovered just among the bacterial genomes whose sequences have previously been determined. Thus, it also seems certain that a greater diversity of structures and mechanisms will be uncovered for riboswitches, and some of these might further blur the lines between the various types of RNA-based regulatory systems noted above. Currently, some of the most common mechanisms for riboswitch-mediated gene control involve the direct regulation of translation initiation, or the inhibition of protein production more indirectly by altering mRNA stability or by changing the primary sequence of mRNAs via alternative splicing. The details of some of these riboswitch mechanisms for translation control are discussed below.
DIRECT TRANSLATION CONTROL BY RIBOSWITCHES
After intrinsic transcription terminator control, the next-most-common gene-control mechanism used by many of the known riboswitch classes involves the occlusion or liberation of the ribosome-binding site ([RBS] or Shine-Dalgarno [SD] sequence) (Barrick and Breaker 2007). RBS sequences typically involve a purine-rich segment of approximately six nucleotides located a short distance upstream of the translation start codon, and these small genetic elements are present in ∼77% of the protein-coding mRNAs among thousands of bacterial species examined (Omotajo et al. 2015). The RBS sequence can form a Watson–Crick base-pairing interaction with 16S ribosomal RNA in the process of translation initiation (Laursen et al. 2005). Thus, ligand binding to a riboswitch aptamer can direct folding of an expression platform to either display the RBS for access by ribosomes, or block ribosomes from binding to the mRNA (Fig. 1C).
Numerous examples of riboswitch classes that use this direct control of translation are predicted to exist (Barrick and Breaker 2007). However, only a few examples have been experimentally tested by examining individual riboswitch representatives. By using comparative sequence analysis methods, the precise mechanisms for regulating RBS access were divided into three specific types. For the first type, the RBS could serve as an integral part of the aptamer structure (Fig. 1C), and therefore ligand binding precludes ribosome access to yield a riboswitch that functions as a genetic OFF switch in the presence of ligand. Riboswitches for AdoCbl or its close derivatives use this mechanism, as shown at atomic resolution from X-ray crystallography data (Johnson et al. 2012).
A second type has the RBS located at some distance downstream of the aptamer structure. In this case, the RBS does not directly participate in the formation of the aptamer. Therefore, controlling ribosome access must involve alternative folding of structures apart from the aptamer that either hide or openly display the RBS. This mechanism, which was originally proposed for some bacterial thiamin pyrophosphate (TPP) riboswitches (Miranda-Ríos et al. 2001; Rodionov et al. 2002; Winkler et al. 2002), typically involves the use of “anti-RBS” sequences that can base-pair to the RBS sequence in a ligand-regulated manner. As a result, this type of regulation can yield either genetic ON or OFF switches, depending on whether ligand binding to the riboswitch aptamer sequesters the anti-RBS sequence or deploys it. Experimental evidence for TPP riboswitches of this mechanistic type includes (1) ligand-triggered changes in the structural flexibility of the RBS (Winkler et al. 2002; Rentmeister et al. 2007), (2) effects on gene expression with constructs carrying mutations in the anti-RBS sequence (Winkler et al. 2002), (3) monitoring of structural equilibria between translation competent and inactive states (Lang et al. 2007), (4) ligand-dependent modulation of 30S ribosomal subunit binding to mRNA (Ontiveros-Palacios et al. 2008), and (5) bioinformatic discovery of conserved competitor sequences (anti-anti-RBS) for anti-RBS elements (Rodionov et al. 2002; Chauvier et al. 2017).
A third type has the RBS sequence located within an intrinsic terminator stem, such that formation of the terminator stem will both cease transcription before the ORF is made, and also sequester the RBS if the full-length mRNA has already been produced. This expression platform architecture is very common for TPP riboswitches, as observed via comparative sequence analysis of representatives from diverse bacterial species (Rodionov et al. 2002). Presumably, this dual mechanism is desirable because it offers exceptional regulatory and energy efficiency. For example, if plenty of TPP is available in the cell, ligand binding will terminate transcription before the region corresponding to the adjoining ORF has been fully transcribed. However, if TPP is low in concentration, full-length mRNAs can be made, but then can still permit RBS sequestration by the same TPP-binding interaction if this compound later attains sufficient concentration.
Although the TPP class has been the primary model riboswitch for the study of RBS-sequestration mechanisms, many other riboswitch classes also use similar strategies to control translation. As novel riboswitch classes are discovered, new opportunities become available to assess the types of gene-control mechanisms used by these ligand-binding RNAs. Indeed, bioinformatic analyses of validated riboswitches and riboswitch candidates frequently reveal sequence signatures that indicate their gene-control mechanisms involve RBS availability (e.g., see Vitreschak et al. 2002; Rodionov et al. 2003; Weinberg et al. 2007, 2010, 2017). The basic regulatory framework for each of these riboswitches will likely fall into one of the three general types described above. However, it is clear that regulation of translation initiation can be just the beginning of a more complex gene-control cascade. The two systems described below show the potential for complexity with riboswitches that initially regulate ribosome access to an RBS.
The lysine riboswitch associated with the lysC gene of Escherichia coli exploits an anti-RBS sequence to preclude translation of the ORF in the presence of its target amino acid (Caron et al. 2012). Elsewhere in this same lysine-bound structure, two adjacent locations for the action of RNase E are presented as non-base-paired RNA. As a result, the mRNA is targeted for degradation while simultaneously translation is precluded. In this case, the two mechanisms used to reduce the amount of protein made from the mRNA are not interdependent. It is not essential that translation cease by RBS occlusion for mRNA degradation to proceed, and vice versa. Lysine binding by the riboswitch aptamer is needed only to trigger a difference in the folded state of the RNA that simultaneously triggers the two unrelated events that both independently reduce gene expression.
Another example of possible dual-acting riboswitch mechanisms helps explain a long-standing paradox that was evident when the first riboswitch validation studies began to be published. It was speculated that AdoCbl riboswitches from E. coli (Nou and Kadner 2000; Nahvi et al. 2002) and Salmonella typhimurium (Nahvi et al. 2004) regulate translation, presumably by an RBS occlusion mechanism like that shown for AdoCbl riboswitches from other species (Johnson et al. 2012). However, why would cells exploit a riboswitch that controls translation of only the first gene in an operon that contains multiple ORFs? This is particularly puzzling for the S. typhimurium mRNA that carries the large cob operon coding for 25 proteins that function in the long biosynthetic pathway for coenzyme B12. It would be exceedingly wasteful for the riboswitch to regulate the translation of only the first gene by occluding its RBS on AdoCbl binding, but still allow the cell to synthesize the entire mRNA and synthesize all the other 24 proteins encoded by this multi-ORF message.
It was speculated (Link and Breaker 2009; Peters et al. 2009; Roth and Breaker 2009; Breaker 2012) that perhaps the regulation of translation initiation triggers the action of the Rho termination factor (Kriner et al. 2016) that can terminate transcription before the rest of the mRNA can be synthesized. Specifically, inhibition of translation initiation prevents ribosomes from binding to the nascent mRNA, which otherwise would block the access of Rho to permit continued transcription and production of the complete mRNA. Intriguingly, a representative of an Mg2+-II riboswitch and also a representative of an FMN riboswitch have been reported to regulate Rho-dependent transcription termination (Hollands et al. 2012). However, these riboswitches do not necessarily achieve transcription termination by precluding translation initiation of the downstream ORF. Indeed, the FMN riboswitch example experiences Rho-mediated termination even before the start codon of the ORF is transcribed. Clearly, additional investigations will be needed to more fully explore the mechanisms by which riboswitches directly control translation, and to discover the associated regulatory processes that might ensue when riboswitches sense their cognate ligands.
RIBOZYME REGULATION OF TRANSLATION BY SMALL MOLECULES
Of course, ribosomes, the machines that catalyze the translation process, are directly involved in riboswitch-mediated gene regulation. Therefore, all bacterial riboswitch classes that use a mechanism that sequesters or reveals the ribosome-binding site of an mRNA are regulating ribozyme function by controlling the binding of 16S RNA to its mRNA target. So, in addition to the direct regulation of translation by riboswitches as described in the preceding section, there are several additional examples wherein riboswitches involve the action of ribozymes to indirectly regulate translation. These collaborations between riboswitches and likely ancient ribozyme classes provide intriguing evidence that some modern riboswitches might have their evolutionary origins in the RNA world (Vitreschak et al. 2004; Breaker 2012), a time before proteins emerged to take on many of these same roles.
The first such ribozyme-associated system to be described is the glmS riboswitch class (Barrick et al. 2004; McCown et al. 2012), whose members selectively bind the sugar derivative glucosamine-6-phosphate (GlcN6P) to promote RNA-catalyzed RNA strand scission (Winkler et al. 2004; Ferré-D'Amaré 2010). This self-cleaving ribozyme action promotes the rapid ribonuclease-mediated degradation of the adjoining ORF (Collins et al. 2007) coding for the protein enzyme GlmS (glutamine-fructose-6-phosphate amidotransferase). The regulatory logic established by this dual “ribozyme-riboswitch” is quite simple. GlmS proteins catalyze the biosynthesis of GlcN6P, which accumulates to levels that trigger the self-destructing glmS ribozyme to cleave the 5′ terminus from the full-length mRNA. However, the mRNA self-cleaves upstream of the coding region (Winkler et al. 2004), which left open the question of precisely how the action of the ribozyme ultimately prevents translation. It was eventually shown that, subsequent to ribozyme-mediated mRNA scission of the 5′UTR, RNase J1 recognizes the trimmed portion of the mRNA carrying the ORF and rapidly destroys this RNA to prevent further translation (Collins et al. 2007).
For the glmS ribozyme-riboswitch example, the riboswitch ligand (GlcN6P) is also the cofactor that directly promotes ribozyme-mediated RNA strand scission (Viladoms and Fedor 2012; Bingaman et al. 2017). However, riboswitches can control the action of other ribozymes by more remote mechanisms. In the next section on eukaryotic riboswitches, the regulation of alternative splicing by TPP riboswitches is presented in detail. Specifically, the spliceosomal apparatus that is so important for removing introns in many eukaryotic species is a ribozyme at its core (Fica et al. 2013), and there are several strategies used by fungal and plant riboswitches to regulate alternative splicing.
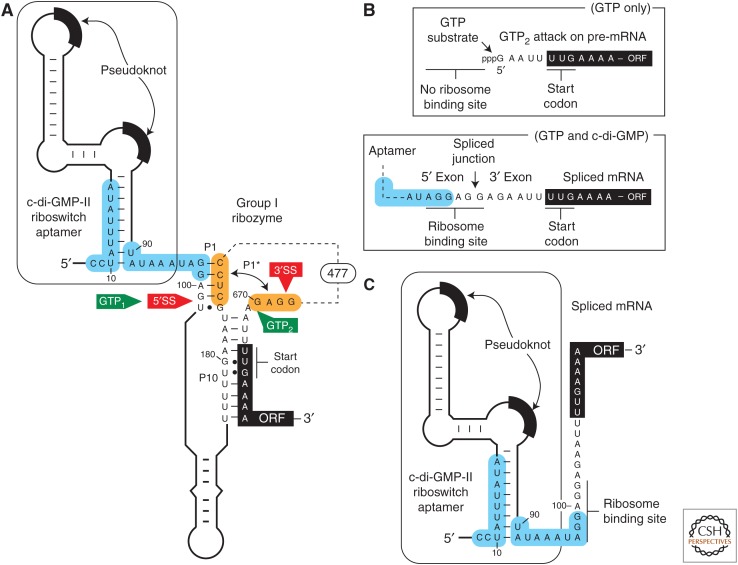
In addition, there are riboswitches associated with self-splicing ribozymes in bacteria that indirectly regulate translation. Two riboswitch classes are known to exist for the bacterial signaling molecule c-di-GMP (Sudarsan et al. 2008; Lee et al. 2010), the cyclic RNA dinucleotide made from two guanosine nucleotides (Römling et al. 2013). For example, there is a natural tandem arrangement in which a riboswitch aptamer for c-di-GMP was found (Lee et al. 2010) to reside immediately upstream of the 5′ splice site (5′SS) of a group I self-splicing ribozyme (Fig. 2) in the bacterial pathogen Clostridium difficile (recently renamed Clostridioides difficile) (Lawson et al. 2016). The riboswitch aptamer does not carry a typical expression platform like those that include either a transcription terminator or a structure to obscure an adjacent ribosome-binding site. Rather, ligand binding to the aptamer directly regulates ribozyme-mediated splicing by controlling the structural environment of the 5′SS.
Figure 2.
Regulation of self-splicing ribozyme function by a riboswitch aptamer. (A) Schematic representation of the 5′ untranslated region (5′UTR) of the CD3246 gene from Clostridium difficile, which carries a c-di-GMP-II riboswitch aptamer immediately upstream of a group I self-splicing ribozyme. The binding of c-di-GMP to the aptamer domain requires the structure depicted. This structure promotes ribozyme-catalyzed attack of GTP (called GTP1) at the 5′ splice site (5′SS) and subsequent attack of the 5′ exon’s 3′ terminus oxygen on the phosphorus of the 3′ splice site (3′SS) to yield a properly spliced messenger RNA (mRNA) product that can be translated. In the absence of c-di-GMP binding, portions of the aptamer and ribozyme shaded in blue reorganize to form an alternative base-paired structure. This disrupts the P1 stem that is required for proper splicing, and permits the formation of P1* to present an alternative site for GTP attack (called GTP2). GTP attack at this alternative site removes all but four nucleotides located upstream of the unusual UUG start codon for the ORF. (B) Two reaction products produced by the tandem aptamer-ribozyme construct depicted in A. (Top) Processed mRNA product generated by ribozyme action in the absence of c-di-GMP binding. Note that the ribosome-binding site (RBS) has been removed, and thus translation of the downstream open reading frame (ORF) is precluded. (Bottom) Processed mRNA product resulting from ribozyme splicing in the presence of c-di-GMP. Splicing creates a strong RBS that permits translation to occur. (C) Although the properly spliced mRNA (B, bottom) carries an RBS, the riboswitch aptamer presumably can still operate to occlude or reveal this sequence based on the availability of c-di-GMP. Specifically, the nucleotides shaded in blue that alternatively pair to regulate ribozyme splicing activity remain present in the properly spliced mRNA, and presumably can be exploited as an expression platform to continue to regulate gene expression as would a more common riboswitch.
This tandem arrangement between a riboswitch aptamer and a self-splicing ribozyme exploits the typical activities of each RNA device to create a far more sophisticated gene-control apparatus. As expected, the riboswitch aptamer can form two distinct structures that are important for indirectly regulating mRNA translation. When c-di-GMP is bound by the aptamer, the adjacent ribozyme can adopt a structure that presents the proper 5′SS in its active site (Fig. 2A). Thus, the presence of c-di-GMP promotes the fusion of the 5′ and 3′ exons, the complete removal of the ribozyme intron, and the generation of an RBS sequence that permits translation of the processed mRNA (Fig. 2B, bottom) (Chen et al. 2011). In contrast, the absence of c-di-GMP binding allows a reorganization of the aptamer sequence that prevents the use of the 5′SS, and yields a truncated RNA processing product that is missing an RBS (Fig. 2B, top). This truncated mRNA therefore cannot be readily translated.
This tandem aptamer-ribozyme system has two other notable features that increase the genetic complexity shown by such RNA devices. First, the properly spliced mRNA retains the c-di-GMP aptamer, and its ability to form alternative base-pairs based on the presence or absence of c-di-GMP remains intact (Fig. 2C). Therefore, it seems possible that translation of the processed mRNA could yet be regulated by a more conventional riboswitch mechanism of occluding or presenting the RBS, long after the self-splicing ribozyme has been self-extracted. Second, group I ribozymes initiate self-splicing by promoting the nucleophilic attack by a guanosine nucleotide (such as GTP) on the 5′SS, which means that sufficient quantities of both c-di-GMP and GTP are needed to yield translation-ready mRNA products. This means that the tandem system initially functions as a two-input Boolean logic gate, wherein both molecular inputs are needed to promote gene expression (Lee et al. 2010).
Although this particular tandem aptamer-ribozyme architecture is very rare, and is only found in various strains of C. difficile, there are reasons to believe that other tandem arrangements will be discovered. Indeed, we have identified several examples wherein riboswitch aptamers appear in close proximity to group II self-splicing ribozymes. Moreover, there are large numbers of self-cleaving ribozymes that are present in many bacterial and eukaryotic species (Jimenez et al. 2015). This creates many opportunities for riboswitch aptamers to regulate mRNA processing by controlling the action of ribozymes that cut RNA. Indeed, there exists some evidence that GlcN6P naturally affects the activity of a self-cleaving hepatitis delta virus (HDV)-class ribozyme from bacteria (Passalacqua et al. 2017). This regulation is different than that seen with glmS ribozymes that respond to the same compound (Winkler et al. 2004), as the effect is more subtle and appears to be via an allosteric mechanism rather than via the function of GlcN6P as a cofactor for the ribozyme. Moreover, it is not yet clear how this allosteric HDV ribozyme might ultimately regulate translation of the adjacent ORF.
EUKARYOTIC RIBOSWITCHES AND TRANSLATION REGULATION BY SPLICING
Confirmation that certain eukaryotic species exploit riboswitches was obtained by using comparative sequence analysis to uncover homologs of bacterial TPP riboswitches in fungi and plants (Sudarsan et al. 2003). Soon thereafter (Sudarsan et al. 2005), it was recognized that previously observed thiamin-dependent regulation of thiA pre-mRNA splicing in fungi (Kubodera et al. 2003) involves a representative of this riboswitch class, and that mutations in the TPP riboswitch were the source of resistance to the antimicrobial compound pyrithiamine in the fungus Aspergillus oryzae (Kubodera et al. 2000). These observations provided the first opportunities to examine the mechanisms by which eukaryotic riboswitches regulate gene expression outside of the bacterial and archaeal domains of life.
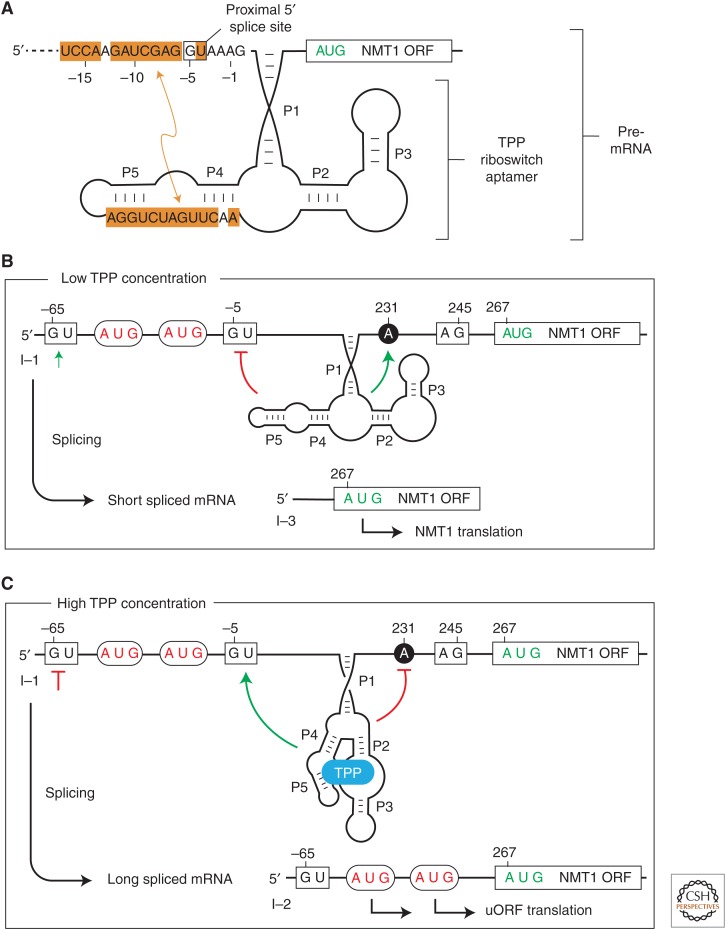
A detailed mechanism for eukaryotic TPP riboswitch function was first established by examining thiamin-mediated alternative splicing in the fungus Neurospora crassa (Cheah et al. 2007). This organism has three TPP riboswitches, each residing in an intron located in the 5′UTR of the NMT1 mRNA, or disrupting the coding regions of the mRNAs for THI4 and NCU01977. Extensive mutational analysis of riboswitch constructs in vivo revealed that the NMT1 riboswitch regulates alternative splicing in a TPP-dependent manner by forming alterative RNA structures to sequester or display one of two possible 5′SS.
In the absence of TPP, a portion of the riboswitch aptamer sequence can form Watson–Crick base-pairs with nucleotides adjacent to and including a 5′SS located proximal to the 3′SS (Fig. 3A). Thus, the spliceosomal machinery must choose a distal 5′SS, which yields a processed mRNA that can be efficiently translated to produce the desired NMT1 protein (Fig. 3B). However, in the presence of TPP, the aptamer nucleotides are sequestered, which liberates the proximal 5′SS and yields a processed mRNA with three ORFs, the main coding region for NMT1 plus two uORFs that serve as translational decoys (Fig. 3C). Thus, in the presence of TPP, the alternatively spliced mRNA attracts ribosomes to the uORFs, rather than permitting translation of the main ORF (Cheah et al. 2007).
Figure 3.
Translation regulation by a fungal thiamin pyrophosphate (TPP) riboswitch that controls alternative splicing. (A) Schematic representation of the region of the Neurospora crassa NMT1 pre–messenger RNA (mRNA) region that includes an intron and TPP riboswitch. The TPP aptamer (including stems P1 through P5) is stabilized on the binding ligand as depicted, or nucleotides from the P4 and P5 region can alternatively base-pair to a region encompassing a 5′SS. (B) Results of mRNA splicing when the TPP concentration is low. The alternative base-pairing depicted in A will sequester the proximal 5′SS to promote the use of a distal 5′SS. This produces a short mRNA splicing product wherein the main NMT1 open reading frame (ORF) can be translated. (C) TPP binding to the aptamer prevents occlusion of the proximal 5′SS, which results in a longer mRNA splicing product. This alternatively spliced RNA carries upstream ORFs (uORFs) that are translated instead of the main NMT1 ORF.
Similar alternative base-pairing interactions are important for regulation of alternative splicing by another series of fungal TPP riboswitches (Li and Breaker 2013). However, several features of these systems are distinct from that described for the NMT1 gene. First, these riboswitches reside in introns that interrupt the coding region of the pre-mRNA, and so complete removal of the intron is required for translation of the full ORF to occur, as shown for the N. crassa gene called NCU01977. When TPP is bound by the riboswitch, numerous alternative splicing products result from the apparent random use of numerous possible 3′SS. All these splicing products still carry portions of the intron, which causes premature translation termination. In contrast, the entire intron is removed when TPP concentration in cells is low.
The switching mechanism for riboswitches of the type found in N. crassa NCU01977 involves alternative pairing between nucleotides (called α) that help form stem P1 when TPP is bound and a distal complementary region (called α′). Although these two complementary segments are separated by ∼500 nucleotides, formation of the α-α′ base-paired structure brings the proper 5′SS and 3′SS in proximity to promote complete removal of the intron, and translation of the resulting ORF to produce the full protein product (Li and Breaker 2013).
Regulation of alternative splicing by riboswitches also appears to be a very common mechanism used by eukaryotic riboswitches from diverse species. For example, it has been proposed (Croft et al. 2007) that TPP binding by a riboswitch in the THIC gene of the green alga Chlamydomonas reinhardtii promotes either of two alternative splicing events that lead to incomplete removal of an intron separating coding exons of the pre-mRNA. Translation machinery operating on the resulting processed mRNA yields a truncated protein that presumably is nonfunctional. In the absence of TPP, the intron structure rearranges to promote the use of splice sites resulting in removal of the entire intron, proper fusion of the ORF, and subsequent production of the full-length protein product.
An exceedingly common mechanism for TPP riboswitch-mediated control of gene expression in multicellular plants involves alternative splicing of an intron that resides downstream of the ORF. In this configuration, TPP binding to the riboswitch aptamer of thiamin-related transcripts promotes splicing of an intron in the 3′UTR of the pre-mRNA such that the polyadenylation signal sequence is removed (Bocobza et al. 2007; Wachter et al. 2007). Thus, TPP causes the destabilization of the transcript and reduces mRNA translation. In the absence of TPP, a portion of the riboswitch aptamer can base-pair with the 5′SS to prevent the removal of the intron and the polyadenylation signal. This base-pairing potential is conserved among numerous plant species (Wachter et al. 2007), suggesting that this mechanism of indirect regulation of translation by TPP riboswitch alteration of transcript polyadenylation and mRNA stability is widespread. To date, there are no known eukaryotic riboswitches that directly control translation without first making changes to the mRNA sequence by regulating RNA processing events. However, given the intimate roles played by tRNAs, ribosomal RNAs and various regulatory RNAs, there exist many opportunities for ligand-binding RNAs in eukaryotes to exploit translation control mechanisms other that those described above.
SPECULATION ON REGULATORY MECHANISMS AND CONTRIBUTIONS OF RIBOSWITCHES
It seems likely that there are thousands of distinct bacterial riboswitch classes that have yet to be discovered (Ames and Breaker 2010; Breaker 2011; McCown et al. 2017). Most of these classes will be present among the innumerable bacterial species whose genomes have yet to be sequenced. It has been estimated that ∼85% of the riboswitches encoded by the currently available genomic databases have already been identified (McCown et al. 2017), because members of the more than 38 riboswitch classes that have been discovered to date are so abundant. In other words, the many undiscovered riboswitch classes are so sparsely represented that they form only a small fraction of the total number of individual riboswitches.
If the assumptions that give rise to the numbers above are generally true, we can be confident in estimating the extent to which riboswitches directly regulate translation processes. Updated quantitation of the frequency of various riboswitch gene-control mechanisms is not available, but there are no indications from recent riboswitch discovery efforts to suggest that past tabulations (Barrick and Breaker 2007) are in error. Thus, it is likely that the two most common mechanisms by which riboswitches control gene expression in modern cells will remain (1) transcription termination via the regulated formation of intrinsic transcription terminator stems, followed closely in abundance by (2) translation initiation via the regulated display or occlusion of the ribosome-binding site. All other mechanisms are far rarer, although they each showcase the diversity of possible gene-control mechanisms exploited by regulatory domains made entirely of RNA.
After 15 years of intensive efforts to discover novel riboswitch classes, it is noteworthy that only one riboswitch class (TPP) has been found to be common in some eukaryotic lineages. Furthermore, it is not certain that additional riboswitch classes exist in eukaryotes (Bocobza and Aharoni 2014), particularly given that proteins undoubtedly serve as strong competition for RNAs as organisms evolve new sensors for various targets. However, the massive amount of noncoding RNA in eukaryotes, transcribed either as introns of protein-coding genes or as separately expressed transcripts such as long noncoding RNAs (e.g., Ulitsky and Bartel 2013), provides tremendous opportunities for ligand-sensing riboswitches to emerge and operate.
To date, no examples of metabolite- or ion-binding riboswitches have been discovered in humans or indeed any metazoans. If researchers wanted to initiate a concerted campaign to discover more eukaryotic riboswitches, how should they focus their searches? The genomic locations and biochemical functions of eukaryotic TPP riboswitches so far provide a strikingly uniform picture of what other eukaryotic riboswitches might look like if they indeed exist. All known TPP riboswitches in fungi and plants reside in introns, regardless of whether these introns are present in the 5′UTR, in intervening sequences disrupting the ORF, or in the 3′UTR. In each case, they appear to regulate alternative splicing, which ultimately regulates translation of the primary protein product either directly (determine integrity of main ORF) or indirectly (uORF regulation; determine mRNA stability). Eukaryotic TPP riboswitches have yet to be observed in RNA transcripts apart from the mRNAs for thiamin biosynthesis or transport proteins. Therefore, it seems prudent to focus future searches for eukaryotic riboswitches primarily on the alternatively spliced introns of metabolic genes, or genes that are regulated by signaling molecules. The introns of such genes would make excellent locations for metabolite-binding aptamers that selectively respond to their targets, and in the process can be removed by splicing so as to avoid interfering with the protein coding portion of the mRNA.
As noted above, such riboswitches might retain the ancient mechanism of working with RNA-based splicing machinery now present in spliceosomal RNAs to control the information flow from RNA to proteins. Eukaryotes do exploit some RNA-based signaling molecules, such as cAMP, cGMP, and certain cyclic dinucleotides, which have been proposed to reflect ancient signaling compounds from the RNA world (Nelson and Breaker 2017). Perhaps if riboswitches for additional signaling molecules derived from RNA nucleotides are discovered in bacteria, homologs will be uncovered in eukaryotes that respond to these same molecules. However, the ligands for eukaryotic riboswitches do not necessarily need to be of ancient origin. It is already apparent that eukaryotes largely lack examples of the most common and likely oldest classes of riboswitches being discovered in bacteria. If more complex eukaryotes do exploit riboswitches beyond just those for TPP, they might have more recently acquired the ability to respond to distinctly eukaryotic ligands such as hormones or other signaling molecules important only for multicellular life.
CONCLUDING REMARKS
The abundant information available on riboswitches shows that this mode of gene regulation relies heavily on mechanisms that directly regulate translation, primarily via controlling mRNA binding to ribosomes. However, a diversity of other mechanisms is used by riboswitches to regulate translation even after the full-length mRNA has been synthesized. Most of these processes involve intricate folding of the riboswitch aptamer and its associated expression platform, which must be achieved on a timescale that is relevant to the translation events that come later. These rapid RNA folding and ligand-binding events cannot easily be studied by traditional biochemical or genetic studies that involve examining purified RNAs in bulk or examining mutants in cell culture. Rather, some questions on the details of translation control will remain to be answered until biophysical analyses involving such techniques as fluorescent labeling, single-molecule analyses, and atomic-resolution modeling are fully used (Perez-Gonzalez et al. 2016).
Structural biologists have made truly remarkable progress in revealing the ligand-bound states of nearly all known riboswitch classes (Garst et al. 2011; Peselis and Serganov 2014; McCown et al. 2017). However, riboswitches have evolved to function with highly dynamic structures that can either change their shapes in real time based on ligand-binding events, or perhaps more commonly they make folding pathway decisions based on ligand binding while being actively synthesized by RNA polymerase. To fully understand the structural and kinetic properties of riboswitches, studies that can monitor individual RNAs in real time as they progress through various structural states or forks in their folding pathways will be necessary. Methods that use optical tweezers to evaluate RNA folding during the act of transcription (e.g., Frieda and Block 2012) appear to be particularly promising. However, monitoring similar kinetic processes that involve ribosome docking to a riboswitch-controlled RBS, or spliceosomal particles choosing splice sites whose availabilities are controlled by riboswitch folding, seem daunting.
Regardless of the challenges ahead, it is very important to continue to discover novel riboswitch classes and to examine the mechanistic details of their gene-control actions. This work inevitably expands our understanding of a common mode of gene control in bacteria, and reveals how small ligands can regulate complex processes like alternative splicing in eukaryotes. Furthermore, as the importance of synthetic biology grows in the future, simple and effective engineered genetic switches will be needed and riboswitches might be made to serve roles as designer sensors and switches. In particular, their ability to directly regulate protein synthesis could be exploited to yield long-lived mRNAs that quickly activate or deactivate translation by binding to externally supplied trigger molecules. Finally, each newly discovered natural riboswitch potentially provides a window into our evolutionary past. Even if modern riboswitches do not directly descend from ancient riboswitches, they yield clues that explain how RNAs can fold to sense diverse ligands and how these binding events can interface sometimes directly with modern ribozymes to regulate complex biochemical processes.
ACKNOWLEDGMENTS
R.R.B. thanks all members of the laboratory for many helpful discussions. Riboswitch research in the Breaker laboratory has been supported by the National Institutes of Health (NIH) (GM022778 and DE22340), Yale University, and by investigator support to R.R.B. from the Howard Hughes Medical Institute.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
- Ames TD, Breaker RR. 2010. Bacterial riboswitch diversity and analysis. In The chemical biology of nucleic acids (ed. Mayer G), Chap. 20 Wiley, Chichester, UK. [Google Scholar]
- Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401: 235–242. [DOI] [PubMed] [Google Scholar]
- Arachchilage GM, Sherlock ME, Weinberg Z, Breaker RR. 2017. SAM-VI RNAs selectively bind S-adenosylmethionine and exhibit similarities to SAM-III riboswitches. RNA Biol 10.1080/15476286.2017.1399232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingaman JL, Zhang S, Stevens DR, Yennawar NH, Hammes-Schiffer S, Bevilacqua PC. 2017. The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme. Nat Chem Biol 13: 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocobza SE, Aharoni A. 2014. Small molecules that interact with RNA: Riboswitch-based gene control and its involvement in metabolic regulation in plants and algae. Plant J 79: 693–703. [DOI] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. 2007. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev 21: 2874–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy E, Borne F, Patte JC. 1979. Isolation and identification of mutants constitutive for aspartokinase III synthesis in Escherichia coli K12. Biochimie 61: 1151–1160. [DOI] [PubMed] [Google Scholar]
- Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 43: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2012. Riboswitches and the RNA World. Cold Spring Harb Perspect Biol 4: a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Massé E, Lafontaine DA. 2012. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci 109: E3444–E3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvier A, Picard-Jean F, Berger-Dancause JC, Bastet L, Naghdi MR, Dubé A, Turcotte P, Perreault J, Lafontaine DA. 2017. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat Commun 8: 13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. 2007. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447: 497–501. [DOI] [PubMed] [Google Scholar]
- Chen AGY, Sudarsan N, Breaker RR. 2011. Mechanism for gene control by a natural allosteric group I ribozyme. RNA 17: 1967–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC. 2007. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev 21: 3356–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. 2007. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci 104: 20770–20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR. 2010. The glmS ribozyme: Use of a small molecule coenzyme by a gene-regulatory RNA. Q Rev Biophys 43: 423–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li N, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. 2013. RNA catalyses nuclear pre-pRNA splicing. Nature 503: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda KL, Block SM. 2012. Direct observation of cotranscriptional folding in an adenine riboswitch. Science 338: 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT. 2011. Riboswitches: Structures and mechanisms. Cold Spring Harb Perspect Biol 3: a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Storz G. 2011. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3: a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NJ, Grundy FJ, Henkin TM. 2010. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 584: 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg JC, Chen Y, Grundy FJ, Henkin TM, Pollack L, Ke A. 2013. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci 110: 7240–7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso-Becera MV, Servín-González L, Soberon-Chávez G. 2015. RNA structures are involved in the thermoregulation of bacterial virulence-associated traits. Trends Microbiol 23: 509–518. [DOI] [PubMed] [Google Scholar]
- Gusarov II, Kreneva RA, Podcharniaev DA, Iomantas IV, Abalakina EG, Stoinova NV, Perumov DA, Kozlov II. 1997. Riboflavin biosynthetic genes in Bacillus amyloliquefaciens: Primary structure, organization and regulation of activity. Mol Biol (Mosk) 31: 446–453. [PubMed] [Google Scholar]
- Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. 2012. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci 109: 5376–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez RM, Polanco JA, Lupták A. 2015. Chemistry and biology of self-cleaving ribozymes. Trends Biochem Sci 40: 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE Jr, Reyes FE, Polaski JT, Batey RT. 2012. B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: Molecular zippers and switches. Nat Rev Microbiol 10: 255–265. [DOI] [PubMed] [Google Scholar]
- Kriner MA, Sevostyanova A, Groisman EA. 2016. Learning from the leaders: Gene regulation by the transcription termination factor Rho. Trends Biochem Sci 41: 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubodera T, Yamashita N, Nishimura A. 2000. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: Cloning, characterization and application as a dominant selectable marker for transformation. Biosci Biotechnol Biochem 64: 1416–1421. [DOI] [PubMed] [Google Scholar]
- Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. 2003. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett 555: 516–520. [DOI] [PubMed] [Google Scholar]
- Lang K, Rieder R, Micura R. 2007. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res 35: 5370–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. 2005. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson PA, Citron DM, Tyrrell KL, Finegold SM. 2016. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 40: 95–99. [DOI] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329: 845–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Breaker RR. 2013. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res 41: 3022–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link KH, Breaker RR. 2009. Engineering ligand-responsive gene-control elements: Lessons learned from natural riboswitches. Gene Ther 16: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundrigan MD, Koster W, Kadner RJ. 1991. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc Natl Acad Sci 88: 1479–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Winkler WC, Breaker RR. 2012. Mechanism and distribution of glmS ribozymes. Methods Mol Biol 848: 113–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23: 995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Ríos J, Navarro M, Soberón M. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci 98: 9736–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. 2002. Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043–1049. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Barrick JE, Breaker RR. 2004. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32: 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Breaker RR. 2017. The lost language of the RNA World. Sci Signal 10: eaam8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. 2000. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci 97: 7190–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotajo D, Tate T, Cho H, Choudhary M. 2015. Distribution and diversity of ribosome binding sites in prokaryotic genomes. BMC Genomics 16: 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros-Palacios N, Smith Am, Grundy FJ, Soberon M, Henkin TM, Miranda-Ríos J. 2008. Molecular basis of gene regulation by the THI-box riboswitch. Mol Microbiol 67: 793–803. [DOI] [PubMed] [Google Scholar]
- Passalacqua LFM, Jimenez RM, Fong JY, Lupták A. 2017. Allosteric modulation of the Faecalibacterium prausnitzii hepatitis delta virus-like ribozyme by glucosamine 6-phosphate: The substrate of the adjacent gene product. Biochemistry 56: 6006–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez C, Grondin JP, Lafontaine DA, Carlos Penedo J. 2016. Biophysical approaches to bacterial gene regulation by riboswitches. Adv Exp Med Biol 915: 157–191. [DOI] [PubMed] [Google Scholar]
- Peselis A, Serganov A. 2014. Themes and variations in riboswitch structure and function. Biochim Biophys Acta 1839: 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Kuan PF, Rowland JL, Keleş S, Landick R. 2009. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci 106: 15406–15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentmeister A, Mayer G, Kuhn N, Famulok M. 2007. Conformational changes in the expression of the Escherichia coli thiM riboswitch. Nucleic Acids Res 35: 3713–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2002. Comparative genomics of thiamin biosynthesis in procaryotes: New genes and regulatory mechanisms. J Biol Chem 277: 48949–48959. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2003. Regulation of lysine biosynthesis and transport genes in bacteria: Yet another RNA riboswitch? Nucleic Acids Res 31: 6748–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77: 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Breaker RR. 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78: 305–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood AV, Henkin TM. 2016. Riboswitch-mediated gene regulation: Novel RNA architectures dictate gene expression responses. Annu Rev Microbiol 70: 361–374. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. 2005. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol 12: 1325–1335. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321: 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: Genomics, evolution, and mechanisms. Cell 154: 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladoms J, Fedor MJ. 2012. The glmS ribozyme cofactor is a general acid-base catalyst. J Am Chem Soc 134: 19043–19049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30: 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2004. Riboswitches: The oldest mechanism for the regulation of gene expression? Trends Genet 20: 44–50. [DOI] [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK. 2007. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. 19: 3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35: 4809–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR. 2017. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res 45: 10811–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. 2000. Transcription attenuation: Once viewed as a novel regulatory strategy. J Bacteriol 182: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]