Abstract
Ammonium assimilation is linked to fundamental cellular processes that include the synthesis of non-essential amino acids like glutamate and glutamine. In Saccharomyces cerevisiae glutamate can be synthesized from α-ketoglutarate and ammonium through the action of NADP-dependent glutamate dehydrogenases Gdh1 and Gdh3. Gdh1 and Gdh3 are evolutionarily adapted isoforms and cover the anabolic role of the GDH-pathway. Here, we review the role and function of the GDH pathway in glutamate metabolism and we discuss the additional contributions of the pathway in chromatin regulation, nitrogen catabolite repression, ROS-mediated apoptosis, iron deficiency and sphingolipid-dependent actin cytoskeleton modulation in S.cerevisiae. The pleiotropic effects of GDH pathway in yeast biology highlight the importance of glutamate homeostasis in vital cellular processes and reveal new features for conserved enzymes that were primarily characterized for their metabolic capacity. These newly described features constitute insights that can be utilized for challenges regarding genetic engineering of glutamate homeostasis and maintenance of redox balances, biosynthesis of important metabolites and production of organic substrates. We also conclude that the discussed pleiotropic features intersect with basic metabolism and set a new background for further glutamate-dependent applied research of biotechnological interest.
Keywords: Glutamate dehydrogenase, GDH1, GDH2, GDH3, Ammonium assimilation, GABA shunt, ROS-mediated apoptosis, Chromatin regulation, Nitrogen catabolite repression, S. cerevisiae
Background
Ammonium assimilation into carbon chains follows specific biosynthetic routes that lead to the production of non-essential amino acids including glutamate. Initial observations in bacteria showed two major mechanisms that can be used for the production of glutamate. The glutamine synthetase (GS) and glutamate synthase (GOGAT) (GS–GOGAT) mechanism that occurs when cells grow in low ammonia concentrations and the glutamate dehydrogenase (GDH) pathway that has a lower energy cost and is used by the cells in excess of ammonium and phosphate [1].
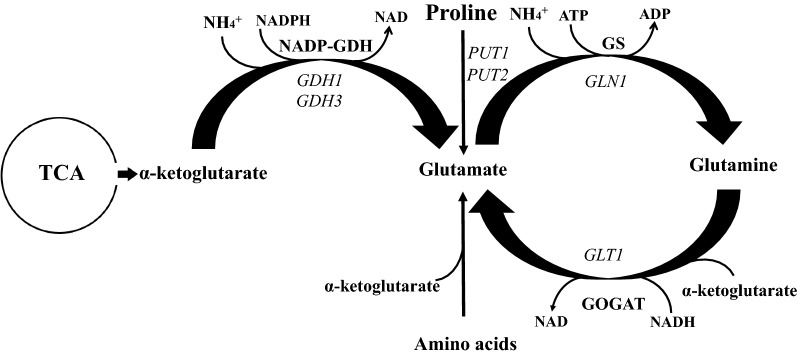
Central nitrogen metabolism in Saccharomyces cerevisiae hosts the same two conserved mechanisms for glutamate production [2–5]. GS–GOGAT pathway in yeast has a marginal contribution in glutamate synthesis both in fermentation and respiratory conditions, while the GDH pathway has the prominent role [5, 6]. Yeast strains lacking the GDH route, present a proline utilization pathway (PUT) that can contribute together with the GS–GOGAT in glutamate biosynthesis and nitrogen assimilation [7]. The relative contribution of PUT is being dictated by the nitrogen sources [7]. To our knowledge, S. cerevisiae is the only organism having three pathways for glutamate synthesis (Fig. 1) with the produced glutamate responsible for the 85% of the total cellular nitrogen and glutamine for the remaining 15% [8–10].
Fig. 1.
Glutamate production and nitrogen assimilation in S. cerevisiae. The figure was adapted from Fig. 1 [7]. GDH1 glutamate dehydrogenase 1, GDH3 glutamate dehydrogenase 3, PUT1 proline oxidase, PUT2 Δ1-pyrroline-5-carboxylate dehydrogenase, GLN1 glutamine synthetase (GS), GLT1 NAD(+)-dependent glutamate synthase (GOGAT), TCA tricarboxylic acid cycle
In this review, we summarize the current state of knowledge on the GDH pathway in S. cerevisiae and we aim to present the pleiotropic effects of the pathway in addition to its well-described role in glutamate metabolism. We discuss the findings presented in recent research and primary literature papers and we make a coherent argument about the topic.
The GDH pathway in S. cerevisiae
In yeast, glutamate can be synthesized through the activation of the GDH pathway when ammonium is present in abundant levels. This can be achieved through the activation of MEP permeases (Mep1, Mep2, Mep3) that facilitate the entrance of ammonium in the cell and its retention in the cytoplasm [9, 11]. Under sufficient ammonium levels, the GDH pathway catalyzes the synthesis of glutamate using α-ketoglutarate and ammonium through the NADP linked action of GDH (1):
| 1 |
The NADP-dependent GDH enzyme in yeast is encoded by GDH1 and GDH3 [12]. GDH1 and GDH3 are paralogous genes with GDH3 originating from an ancestral event of whole-genome duplication [6, 13] or interspecies hybridization [14]. Despite the high conservation of GDH1 and GDH3, Gdh1p exhibits higher utilization rates of α-ketoglutarate under glucose conditions compared to Gdh3p [15, 16]. This observation makes Gdh1 the primary (hyperbolic) NADP-GDH enzyme and Gdh3 the cooperative NADP-GDH isoform in the GDH pathway of S. cerevisiae. A recent study addressed the question whether the different utilization rates of α-ketoglutarate by Gdh1p and Gdh3p correlate with their evolutionary origin [6]. The authors compared the NADP-GDH activity of S. cerevisiae with that in closely related yeast species. The kinetic properties of NADP-GDH activity that derived from yeast species with either constitutively respiratory metabolism, or intermediate fermentative capacity were similar to the Gdh1 and Gdh3 isoforms of S. cerevisiae and complemented the total NADP-GDH activity. Based on this it was concluded that the different utilization rates of α-ketoglutarate by Gdh1p and Gdh3p were independent of their evolutionary origin [6].
In terms of localization, Gdh1p is found in the cytosol and the nucleus as opposed to Gdh3p that is localized in the mitochondria and the nucleus [17, 18]. This different localization seems to be evolutionarily retained due to the urge of a wise cellular exploitation of α-ketoglutarate that in many organisms acts as a signal and coordinates carbon and nitrogen metabolism [19]. Growing evidence shows that modulation of the intracellular α-ketoglutarate levels could constitute an important mechanism of metabolic control that can also interfere with many physiological processes [6, 20]. Enzyme purification experiments showed that Gdh1p and Gdh3p are hexamers (a6 50 kDa oligomeric structure) with the in vivo total NADP-GDH pool being a quite dynamic mixture of Gdh1p and Gdh3p monomers [15]. It was observed that under glucose fermentative growth the pool consisted mainly of Gdh1p monomers [12, 15]. GDH3 is a glucose-repressed gene and consequently the presence of Gdh3 protein in the pool was very low [15, 21–23].
The allosteric regulation of NADP-GDH activity is influenced by α-ketoglutarate and NADP, and not by small molecules (e.g. GTP, AMP) or amino acids as has been reported for other GDH proteins, including human GDH [24].
GDH1 transcriptional regulation and phase-specific degradation of Gdh1 protein
The regulation of GDH1 under glucose conditions is performed by nitrogen catabolite repressor (NCR)-sensitive activators, Leu3p and activators exclusive for respiratory growth such as the HAP complex that coordinates nuclear and mitochondrial gene expression [21, 25, 26]. Under ethanol conditions, GDH1 derepression is mediated by the Gcn4 and Hap4 transcriptional activators and is amplified by Gln3 [21, 27]. Experiments measuring the β-galactosidase activity of GDH1 promoted-lacZ, and nucleosome scanning assays in cells grown in glucose or ethanol with ammonia as nitrogen source, found that GDH1 transcription occurs throughout all growth phases in yeast [6, 15, 21, 23]. This is achieved through different members of the SAGA remodeling complex that modify the chromatin for GDH1 expression under different carbon sources [21]. The constant expression of GDH1 implied that its transcription proceeds normally during the different growth phases including the diauxic shift, when yeast cells reprogram their metabolism to enter the respiration phase. However, during the post-diauxic shift, the Gdh1p/Gdh3p ratio decreases and most of the NADP-GDH activity is attributed to Gdh3p [15]. The decrease of the NADP-GDH activity in ethanol growing cells was initially referred to be controlled through post-translational modifications [28] that could modulate the proportion of Gdh1p versus Gdh3p monomers that constitute the NADP-GDH pool [21]. Indeed, proteomic studies revealed that Gdh1p can be a potential target of ubiquitin attachment [29, 30]. Additional studies showed that Gdh1 protein is subjected to a “stationary phase-specific degradation” that occurred at the diauxic shift [23]. Through a series of point mutations and protein quantification experiments, it was concluded that Lys-426 (K426) in the C-terminal box is essential for the observed stationary phase-specific degradation of Gdh1p [23].
This phase specific degradation of Gdh1p and its substitution by Gdh3p in the NADP-GDH activity pool seems to be favorable under glucose deprivation. As discussed above, Gdh1p utilizes α-ketoglutarate at higher rates compared to Gdh3p, and contributes to glutamate production when yeast cells ferment, and thus are in the exponential phase. Glucose deprivation signifies the entrance of the yeast cells into a stationary-phase survival mode. The transition from fermentation to stationary phase conditions is accompanied by a dramatic growth reduction and a sharp drop in protein synthesis characteristic for stationary phase survival [31, 32]. In addition, transitioning to stationary phase, requires yeast cells to recruit different defense mechanisms that will protect them from ROS-mediated damage that influences lifespan [33]. Lee et al. [23] suggested that the consequence of significant decrease in protein synthesis will be also reflected as significant decrease in amino acid synthesis, since a variety of amino acids, including glutamate, become unnecessary in stationary phase cells. Therefore, it may be more beneficial for yeast to substitute Gdh1p with Gdh3p through the phase-specific expression of GDH3 and the simultaneous degradation of Gdh1p [23]. The authors observed that Gdh3p seems to be more suitable for the stationary phase survival in which glutamate is mainly required for ROS defense mechanisms (discussed below). The role of GDH1 gene in glutamate biosynthesis was also investigated in aerobic metabolism [16]. It was observed that yeast cells lacking GDH1 were unable to divide in acetate/raffinose media, containing ammonia as primary nitrogen source. Furthermore, 13C-enrichement experiments confirmed that incorporation of 13C into glutamate was nearly undetectable when gdh1Δ cells were incubated in [1, 2-13C]-acetate/raffinose. The NADP-GDH activity was measured to be less than 15% in gdh1Δ cells compared to wild type, confirming the important and primary contribution of Gdh1p in glutamate synthesis under aerobic conditions as well [16].
GDH3 transcriptional regulation and the role of the GDH path in ROS-mediated apoptosis
The transcription of GDH3 occurs extensively during the stationary phase [15, 23]. The activity of Gdh3p presents a 20 to 140-fold increment when cells are grown under aerobic conditions [12]. Under these conditions the majority of the total NADP-GDH activity is attributed to Gdh3p monomers that can contribute up to 70% to the pool, especially when cells enter or remain in aerobic metabolism for several days [15]. Under acetate/raffinose conditions with ammonia as the only nitrogen source, yeast cells lacking GDH3 gene had a significant impairment in glutamate synthesis [16]. The increase of the NADP-dependent GDH activity observed in gdh1Δ mutants was presumably due to Gdh3p that seems to play a prominent role in glutamate metabolism under aerobic conditions [12, 16]. However, glutamate synthesis under aerobic conditions was insufficient and required additionally the activity of Gdh1p [16]. The expression of both GDH3 and GDH1 is required to achieve wild-type growth in respiration [12, 16]. The transcriptional regulation of GDH3 is controlled by carbon sources and not by nitrogen catabolite repression as in the case of GDH1 [22, 34]. The glucose-repressed expression of GDH3 is attributed to the condensed chromatin organization of its promoter. Remodeling of the chromatin at the promoter region under non-fermentative carbon sources or under carbon limiting conditions is performed by the SWI/SNF and SAGA complexes [22]. In addition, microarray-based deacetylation maps in yeast revealed that GDH3 gene is located in a HAST domain (Hda1-Affected SubTelomeric region) where several metabolic genes are glucose-repressed through the action of Hda1 (histone deacetylase 1) [35]. Under diauxic shift, HDA1 gene is downregulated and through the counteracting activity of Gcn5p acetyltransferase (the catalytic subunit of the SAGA), SWI/SNF complex allows the expression of GDH3 under respiratory conditions [35]. Gdh3p is stable without being susceptible to post-translational modifications or to phase-specific degradation as Gdh1p. However, it has a lower catalytic capacity in vivo compared to Gdh1p and thus it cannot fulfill the extended needs or substitute the role of Gdh1p during the exponential growth phase in glucose. As such, impairment or lack of GDH3 does not affect either glutamate production or the survival and growth rates of glucose-grown cells [6, 15]. The low activity of GDH3 is not attributed to its promoter but to dissimilarities in the amino acid sequence between the two expressed Gdh isoforms. These dissimilarities allows Gdh3p to be more fit for processes that take place in specific cellular compartments like the mitochondria [23]. This is plausible in accelerated evolution which in many instances permits only one of two paralogues to diverge from ancestral functions and to acquire either new or complementary capabilities that favor new metabolic adaptations [13]. Based on this it was observed that the GDH pathway through GDH3, and not GDH1, is necessary for the resistance to stress-induced apoptosis in stationary-phase yeast cells [23]. Specifically, yeast cells lacking GDH3 exhibited sensitivity to thermal and oxidative stress as well as oxidative stress-dependent accumulation of ROS that led to apoptotic cell death [23].
Yeast is equipped with mitochondrial enzymes that scavenge free radicals. Among the ROS-scavenging mechanisms, glutathione system (GSH system) is probably the most important intracellular oxidative defense mechanism [23]. The GSH system consists of glutathione (GSH), glutathione reductase and glutathione peroxidase (GPx). Hydrogen peroxide (H2O2) detoxification is performed by GPx which requires glutathione as reducing power [36]. GSH biosynthesis requires glutamate as primary substrate [36, 37]. Glutamate concentration in stationary yeast cells lacking GDH3 was 20% less compared to wild type cells [23]. This caused glutamate deficiency subjecting the cells to GSH depletion and thus the observed ROS accumulation that led to apoptosis [23]. A similar phenomenon was observed when stationary GDH3 mutant yeast cells, were exposed to thermal stress [23]. In addition, it was observed that GDH3 deletion potentiated ROS generation when yeast cells were treated with ebselen, an antioxidant compound responsible for ROS-mediated cytotoxicity in excessive amounts [38]. Gdh3p was considered the molecular target of ebselen which ceased NADP-GDH activity through the formation of selenyl-sulfide bonds with cysteine residues. It was also observed that the NADP-GDH inactivation might had a critical function in the proteolysis of H3 histone in yeast [38–40], as discussed below.
The role of GDH path in ammonia production
The GDH pathway in S. cerevisiae is also responsible for the degradation of glutamate. Contrary to mammals and other organisms, the GDH pathway in yeast presents decoupled functions in terms of glutamate biosynthesis and glutamate catabolism. The NADP-GDH activity of Gdh1 and Gdh3 isoforms is unidirectional and specific for glutamate synthesis, while glutamate catabolism is performed via the action of the oxidizing form of GDH (NAD-GDH) that is also present in the GDH pathway. The NAD-GDH activity in yeast is encoded by GDH2 gene and catalyzes the oxidative deamination of glutamate to α-ketoglutarate and ammonium [4] (Fig. 2) (2):
| 2 |
Fig. 2.
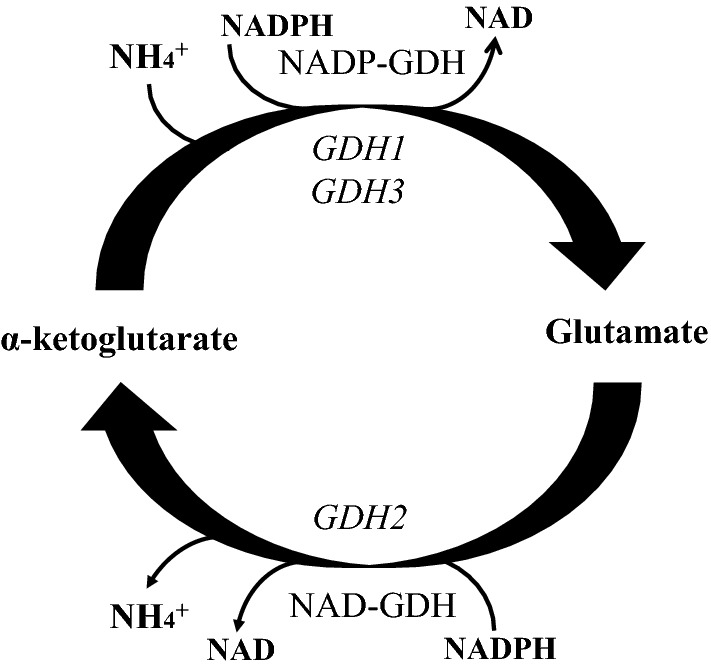
Schematic presentation of the GDH pathway. Synthesis of glutamate occurs through the action of NADP-GDH (encoded by GDH1 and GDH3 genes). NAD-GDH activity (encoded by GDH2) is responsible for glutamate degradation and release of ammonium and α-ketoglutarate. GDH2 glutamate dehydrogenase 2
The presence of the NAD-GDH enzyme was initially described with an unclear role in glutamate metabolism [2]. Years later, it was shown that GDH2 is encoded on chromosome IV of S. cerevisiae with a protein-coding sequence containing more than 1000 amino acids, and a strict mitochondrial function that is post-translationally regulated by phosphorylation [41, 42]. As shown, Gdh2p can be converted from an active NAD-dependent glutamate dehydrogenase to an inactive form by phosphorylation through cAMP-dependent and cAMP-independent protein kinases [42]. However, the functional phosphorylation sites have not yet been identified [43].
Initially, it was speculated that yeast cells lacking GDH1 could use GDH2 to promote glutamate biosynthesis using ammonia as sole nitrogen source [4]. Through experiments performed both in S. cerevisiae and in A. nidulans this speculation was gradually abandoned [4]. The NAD-GDH levels were much higher in cells grown in the presence of glutamate compared to those obtained from cells grown in ammonia. Furthermore, the concentration of the NAD co-factor in the cytoplasm was higher under glucose growth conditions, and thus, it was forcing the reaction towards glutamate degradation in order for the NAD–NADH equilibrium to be obtained [4].
Through the enzymatic activity of Gdh2p the breakdown of glutamate provides adequate levels of ammonia in yeast cells [4, 9, 44]. Indeed, the catabolism of glutamate via the NAD-GDH activity is the major pathway of ammonia generation in vivo. In experiments performed under glucose minimal conditions with glutamate as the only nitrogen source, GDH2 deletion caused limited growth and impaired utilization of the specific amino acid leading to low amounts of intracellular ammonia [4].
The role of GDH2 in glutamate homeostasis was further examined in cells grown under aerobic conditions [16]. Under acetate/raffinose conditions with ammonia as nitrogen source, the NAD dependent activity of GDH was 20-fold higher compared to that in cells grown in glucose. The disruption of GDH2 was not deleterious to glutamate homeostasis as expected [16]. In fact, gdh2Δ cells presented wild type growth and did not display any deficiencies due to glutamate homeostasis impairment neither under glucose nor under non-fermentable carbon sources [16, 44].
GDH2 transcriptional regulation and interaction with GDH3
GDH2 is subjected to transcriptional regulation by glucose [44]. Specifically, it is repressed in glucose and elevated under non-fermentable carbon sources and amino acids as nitrogen source [16, 44]. This strict control is expected due to the anapleurotic role of Gdh2p in Krebs cycle. It is known that Gdh2p replenishes the α-ketoglutarate levels under aerobic conditions [44]. It was considered that the transcriptional activation of GDH2 would require gene regulators necessary in respiratory growth. However, GDH2 transcription is independent of the HAP complex, but is regulated by the GATA-type transcriptional activators Gln3 and Ure2 [44, 45]. It was shown that GDH2 regulation requires two activation and four repression sites present on its promoter. One activation site promotes transcription under glucose starvation [44], while the other one (UASNTR) is implicated in nitrogen catabolite repression [46]. Under nitrogen limitation, Gln3 binds to the UASNTR and to an adaptor protein (Hfi1) responsible for the integrity of the SAGA transcription activator complex [47]. In the presence of preferred nitrogen sources, the expression of GDH2 is repressed by the transcriptional regulator Ure2 which sequesters Gln3 into the cytoplasm [4, 45]. It is also found that the expression of GDH2 is regulated by the concurrent action of Gcn4 and Gln3 [48]. These two regulatory networks have been thought to interact [49–51], putting forward the existence of a physiological relation between Gln3 and Gcn4 [52–54]. Under nitrogen derepressive conditions and amino acid deprivation, Gcn4 and Gln3 form part of a transcriptional complex that binds on GDH2 promoter and dictates its expression [48].
GDH2 genetically interacts with GDH3 and controls stress-induced apoptosis [23]. The role of GDH2 was investigated in stress-induced apoptosis in stationary phase cells that lacked GDH3 [23]. It was observed that deletion of GDH2 gene in a gdh3Δ background increased the resistance under thermal or oxidative stress by decreasing ROS accumulation. The apoptosis was suppressed by GDH2 deletion through the elevated levels of glutamate and glutathione present in the double mutant. Under the tested conditions, deletion of GDH2 compensated the depletion of intracellular glutamate and glutathione (GSH) followed by stress-induced apoptotic cell death and reinforced further the idea that Gdh2p is responsible only for glutamate catabolism and not its production [23].
Additional roles of the GDH pathway
Gdh1p and Gdh3p in nitrogen catabolite repression (NCR)
Saccharomyces cerevisiae can utilize a wide variety of nitrogen-based compounds [8]. Despite the broad nitrogen assimilation, not all nitrogen sources can support yeast growth equally well or trigger the same cellular responses [34]. Using these two assumptions, the nitrogen sources in yeast are empirically classified into “rich” or “preferred” and “poor” or “non-preferred” [55]. The “preferred” nitrogen sources are incorporated into glutamate through the GDH or GS–GOGAT pathways and the resulting carbon substrates are readily integrated in metabolism. The “poor” nitrogen sources, including branched-chain amino acids, aromatic amino acids, and methionine, are transferred to α-ketoglutarate by transaminases, forming glutamate [55, 56]. However, the resulting deaminated carbon compounds are converted through the Ehrlich pathway, to non-catabolizable and growth-inhibitory fuse oils [57]. In addition, under “poor” nitrogen sources, activation of the general control of amino acid biosynthesis (GAAC) mechanism is observed. GAAC activation is mediated by the transcription factor (TF) Gcn4 responsible for the expression of a large number of genes involved in amino acid biosynthesis [55, 58]. Although we recognize the importance of GAAC mechanism, we consider that a more detailed discussion on the regulation of GAAC under “poor nitrogen” sources has been reviewed elsewhere [55, 56] and exceeds the purposes of this review.
The preferential expression of genes involved in nitrogen metabolism is primarily controlled in S. cerevisiae by a transcriptional mechanism known as nitrogen catabolite repression (NCR). Through NCR, baker’s yeast can downregulate the expression of genes involved in the utilization of non-preferable nitrogen sources when preferable nitrogen compounds are available [11, 59]. NCR mechanism comprises four transcription factors (Gln3, Gat1, Dal80 and Gzf3) with a zinc-finger DNA binding domain that recognizes the GATA motif in the promoter of target genes [60]. NCR in yeast is additionally controlled by Ure2, a transcription factor responsible for the translocation of the four latter TFs into the nucleus [61]. According to different experimental studies, in the presence of ammonium, Ure2 binds Gln3 in the cytoplasm and prevents its translocation to the nucleus [60, 62]. This allows Dal80 and Gzf3 to repress the expression of NCR-sensitive genes involved in utilization of alternative nitrogen sources [60]. Ure2 releases Gln3 under nitrogen starvation or in the presence of non-preferable nitrogen sources [34], and upon release, Gln3 translocates into the nucleus. Gln3 and Gat1 act together, mediating the transcription of NCR-sensitive genes [63, 64].
NCR has been described to respond to glutamine and glutamate deficiencies through the activation of Gln3 or Gat1 respectively, while ammonium is considered a distinct signal for NCR that acts independently of the glutamine and glutamate levels [34]. In addition, NCR is controlled by TOR signaling that mediates cell growth and metabolism under different nitrogen and carbon sources. Experimental studies showed that inhibition of TORC1 resulted in nuclear accumulation of Gln3 and Gat1 and transcriptional depression of NCR-sensitive genes [52, 54]. However, as discussed below, growing literature suggests that TORC1 is not the only regulator of these specific GATA-type transcription factors [34, 61, 65, 66]. Specifically, GDH1 was found to have a prominent role in nitrogen-responsive activities [34]. Under nitrogen repressive conditions the derepression of NCR-sensitive genes, like GAP1 (general amino acid permease 1), was due to the drastic effect that GDH1 deletion had on the localization and function of the essential GATA-type activator, Gat1. It was observed that lack of GDH1 mediated the accumulation of Gat1 into the nucleus and thus derepressed the transcription of GAP1 gene. This finding also questioned the role of GDH1 on the other GATA-type activator, Gln3. Upon GDH1 deletion, a highly derepressed expression of DAL5, a NCR-sensitive gene that requires both Gat1 and Gln3 for its expression, was observed. GDH1 deletion caused ammonium accumulation as expected, but surprisingly did not affect the subcellular distribution and the concentrations of glutamine as well as glutamate [34]. This suggested that the Gat1 and Gln3-mediated expression of DAL5 and GAP1 was independent of the glutamine/glutamate levels. Indeed, the GDH1-based repression of DAL5 and GAP1 was strongly correlated with the activity of Gdh1 enzyme per se [34]. As shown, GAP1 gene was depressed in yeast strains encoding a mutation responsible for the inactivation of the catalytic site of Gdh1 (Gdh1pK110L). On the contrary, the transcriptional repression of GAP1 was strong when the NADP-GDH activity was restored by expressing the GDH3 gene, and partially restored, when the bacterial GDHA was expressed in gdh1 mutant strains [34]. However, the signals for the GDH-dependent negative regulation of NCR-sensitive genes require further investigation.
Gdh1 and Gdh3 enzymes in chromatin regulation in yeast
The role of Gdh1p in transcriptional silencing was found to be crucial through the proteolysis of H3 histone in yeast (“H3-clipping” in the N-tail) [40]. This effect has been described previously in animal tissues [39, 67]. In yeast however, the proteolysis of H3 histone had been observed initially in sporulating and stationary phase cells, through the action of the vacuolar serine protease Prb1 [68]. Recent studies revealed that deletion of GDH1 gene increased “H3-clipping” in log phase cells revealing an inhibitory role of Gdh1p on the N-terminus cleavage of H3 [40]. Another observation was that Gdh1p mediated the silencing of sub-telomeric regions through the recruitment of SIR complex [40]. This highlighted the association of an enzyme primarily described in metabolism, with epigenetic processes. Specifically, GDH1 deletion led to decreased binding of Sir2 protein on the telomeres, causing elevated transcript levels of genes affected by the loss of the SIR complex [40]. GDH1 was found to regulate chromatin through its catalytic activity [40]. Specifically, upon GDH1 deletion, the elevated levels of α-ketoglutarate, and not those of NADH, resulted in the observed telomeric silencing defects [40]. The authors described physical association of Gdh1p on specific telomeric loci that were under the transcriptional control of SIR complex. Additional experiments showed that α-ketoglutarate levels changed when Gdh1p was depleted from the nucleus. This suggested that Gdh1p possibly affects the levels of α-ketoglutarate at the SIR-regulated telomeric loci.
The intracellular levels of α-ketoglutarate seems to have a “mechanistic” role also in other organisms. Experiments in mice and nematodes (Caenorhabditis elegans) showed that modulation of the intracellular levels of α-ketoglutarate interfere with gene transcription and longevity [52, 53, 69]. Moreover, modulation of α-ketoglutarate levels found to interfere with the epigenetic state and cellular fate of mouse embryonic stem cells [70]. GDH3 is also implicated in both metabolism and chromatin configurations [40]. Specifically, loss of telomeric silencing was observed in the double gdh1Δ gdh3Δ mutant grown both in ethanol and glucose. Under ethanol conditions deletion of either GDH gene led to mild silencing defects. Also, increased expression of GDH3 partially complemented the gdh1Δ phenotype. This demonstrated overlapping functions of the two isoforms that affect heterochromatin regulation [40].
The contradictory roles of GDH1 and GDH2 in cold-growth defects in yeast strains
The role of GDH1 and GDH2 found to be contradictory when investigated in yeast strains under cold-growth conditions [71]. Using recombinant strains of S. cerevisiae, it was observed that overexpression of GDH1 had detrimental effects on yeast growth at 15 °C creating a cold-sensitive yeast phenotype. On the contrary, overexpression of GDH2 was favoring yeast growth providing a growth advantage in the same conditions [71].
The GDH pathway interferes with the recycling of the essential coenzymes NADH–NAD by controlling the levels of α-ketoglutarate [72]. NADH–NAD homeostasis is crucial for proper cellular responses under environmental changes [73]. Ballester-Tomás et al. [71], suggested that growth temperatures below the optimal require a proper redox NADH–NAD balance that was probably disrupted through the overexpression of GDH1. However, under the examined conditions, the overexpression of GDH2 seemed to regulate this NADH–NAD imbalance, through the increased oxidation of NADH [71]. This was in accordance with previous studies reporting that increased NADH oxidation altered the distribution of metabolic fluxes and sustained yeast growth at suboptimal temperature conditions [74, 75]. Furthermore, an additional study showed the activity of NAD-related genes governs cold growth in yeast and that GDH2 is a cold-growth favoring gene [76].
Shifts to low growth temperatures create increased intracellular H2O2 levels and induced expression of antioxidant genes implicated in glutathione synthesis [77, 78]. Under cold-growth conditions, Ballester-Tomás et al. [71] showed that concurrent ectopic overexpression of GDH1 and GDH2 compensated the observed accumulation of ROS. The authors suggested that this is consistent with the role of GDH1 and GDH2 in glutamate synthesis and its possible implication to oxidation stress defense through the glutathione system. Specifically, glutamate can prevent cold-induced ROS accumulation through the synthesis of glutathione that requires glutamate as a precursor molecule and serves in ROS removal.
Implications of the GDH pathway in actin cytoskeleton, endocytosis and iron deficiency
Proper function of Gdh3p has a role in the sphingolipid-dependent suppression of reduced viability on starvation (RVS) defects that include inability to grow under nutrient starvation or osmotic stress [79]. RVS genes encode the calmodulin-binding, actin-associated, amphiphysin-like lipid raft proteins Rvs167 and Rvs161 which are not essential for yeast viability. However, the Rvs proteins contain a conserved BAR domain that appears to regulate endocytosis, actin cytoskeleton structure and nuclear events. As such, under nutrient depletion, recessive mutations or total loss of the RVS genes, cause growth abnormalities and cytoskeletal and endocytosis defects [80, 81]. Specifically, it was observed that the proper function of Gdh3p altered the growth defects provoked upon RVS and SUR4 deletions in yeast [79]. SUR4 in yeast encodes an elongase involved in fatty acid and sphingolipid biosynthesis. As it seems, Gdh3p participates in a sphingolipid-dependent manner to the restoration of growth under glucose starvation. In addition, Gdh3p was found to physically interact with three domains of the Rvs167p which forms with Rvs161p a complex, that regulates cell polarization, actin cytoskeleton, endocytosis and cell cycle [79]. The role of Gdh3p in that configuration is not known and it will be interesting to examine whether its catalytic activity is primarily involved.
Iron deficiency has a significant impact on amino acid biosynthesis in yeast [10]. It has been observed that many transcripts involved in amino acid metabolism are regulated during iron deficiency and that most of the amino acids affected include an iron-dependent step in their synthesis [82]. Transcriptomic analysis showed that upon iron deficiency, genes implicated in the GS–GOGAT pathway were downregulated while genes of the GDH pathway were upregulated [10]. Specifically, yeast cells exhibited a sixfold downregulation of GLT1 (encodes the NAD-dependent glutamate synthase–GOGAT) as opposed to GDH3 that was 4.5-fold upregulated under the same conditions [10]. This made GDH pathway an iron-independent pathway compared to GS-GOGAT that is iron-dependent. The reason for this differentiation lies on glutamate synthase that is a Fe–S requiring enzyme. Indeed, under iron deficiency the activity of glutamate synthase presents a 20-fold decrease [82]. As already mentioned, the vast majority of nitrogen-containing molecules in yeast acquire their nitrogen from glutamate and to a lesser extent from glutamine, so continuous synthesis of these two amino acids is critical and highly regulated under iron-limiting conditions.
Can the GDH pathway provide additional information on the role of GABA shunt in Saccharomyces cerevisiae?
Although the purpose of this review focuses on the known pleiotropic effects of the GDH pathway, we consider that it will be beneficial for the reader to briefly describe the GABA shunt in S. cerevisiae and suggest possible interactions with the GDH pathway. The GABA shunt is a pathway utilized in many organisms for the conversion of glutamate to succinate, via the formation of γ-aminobutyric acid (GABA) and succinic semialdehyde [83, 84]. The pathway involves the decarboxylation of glutamate by Gad1p and the sequential action of GABA aminotransferase (Uga1p) and succinic semialdehyde dehydrogenase (Uga2p) for the production of succinate. In S. cerevisiae, the expression of UGA1 and UGA2 is upregulated by GABA, whereas the expression of GAD1 has been linked with calcium levels since Gad1p is a calmodulin-binding protein [85, 86]. In addition, the expression of GAD1 is proposed to be regulated by TOR signaling [87]. Experimental studies showed that yeast utilizes and stores GABA as nitrogen source and can transport it using the inducible GABA-specific transport protein (encoded by UGA4), general amino acid permease Gap1 and proline-specific permease Put4 [88–90].
GABA shunt is still poorly understood in yeast, and has received less attention compared to other studied nitrogen pathways in S. cerevisiae [84, 87]. However, it is suggested that it has a role in cellular oxidative stress defense and is upregulated during the stationary phase, under nitrogen starvation [91]. Studies performed in yeast cells have shown that expression of GAD1 provided increased tolerance to oxidative agents compared to GAD1 mutant strains, while deletion of UGA1 and UGA2 rendered the cells hypersensitive compared to wild type strains [86]. The authors observed that the role of GABA shunt in oxidative stress was strictly dependent on the presence of the intact GABA catabolic pathway, and suggested the importance of GABA as a signal to stress responses, as described in other organisms [92]. A recent study attributed the strong growth inhibition of UGA2 yeast mutants, to the accumulation of succinate semialdehyde that is considered a potential toxic intermediate of GABA catabolism [93]. Finally, mutations in the GABA shunt genes resulted in yeast strains with reduced heat-stress tolerance compared to wild type [84]. The heat susceptibility observed in the mutant strains was correlated with the high intracellular ROS concentrations produced under the same growth conditions.
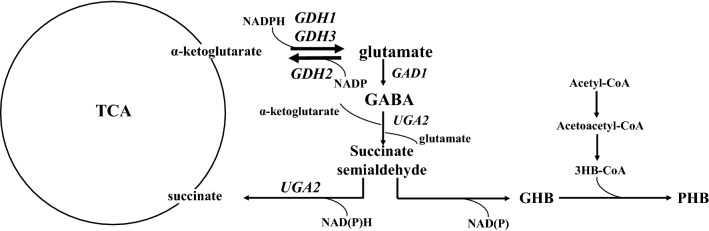
As discussed above, the GDH pathway has a role in cellular ROS defense in stationary phase cells and is linked with NADPH availability and NADH oxidation during yeast growth under suboptimal conditions. Although to our knowledge, there is no direct evidence of crosstalk between the GDH pathway and the GABA shunt, we speculate that the GDH pathway may interfere with the decarboxylating step of the GABA shunt by controlling the available glutamate via the action of Gdh1, Gdh2 and Gdh3 proteins under specific conditions. We also speculate that the GDH pathway may also control the fate of the last reaction of the GABA shunt, which involves the degradation of GABA to γ-aminobutaric acid, instead of succinate, as previously described in S. cerevisiae (Fig. 3). γ-aminobutaric acid is a substrate produced from GABA degradation through the action of γ-aminobutaric acid dehydrogenase. γ-aminobutaric acid can be utilized for the production of polyhydroxybutyrates (PHB), a type of complex macromolecules that accumulate as discrete granules in the cytosol of many microorganisms and serve as energy-storage molecules under imbalanced nutrient conditions [94]. During the last decades the properties of PHBs have been extensively investigated in bacteria [95–97]. Besides their role as carbon storage molecules, they were described as biopolymers with similar properties to common plastics with the unique ability to biodegrade [94]. Efforts to utilize bacteria for the production of PHBs in an industrial scale resulted in low yields of PHBs, leading to the search of other biotechnological platforms [87]. As discussed above, GDH pathway in yeast has an anapleurotic role and replenishes α-ketoglutarate for the production of succinate in Krebs cycle. GABA shunt also facilitates Krebs cycle by providing succinate via GABA catabolism. If the fate of GABA catabolism, favoring the production of γ-aminobutaric acid over succinate, is partially controlled by GDH pathway, then formation of PHBs via GABA under specific conditions, opens new opportunities to use S. cerevisiae for PHB-related compounds.
Fig. 3.
GABA catabolism and production of polyhydroxybutyrates (PHBs) in S. cerevisiae. The figure was adapted from Fig. 1 [87]. PHBs production starts with the reduction of succinate semialdehyde by γ-hydroxybutyric acid dehydrogenase to produce γ-hydroxybutyric acid. γ-hydroxybutyric acid and 3-hydroxybutyrate are polymerized by poly(3-hydroxybutyrate-co-4-hydroxybutyrate) synthase. 3HB formation from acetyl-CoA involves acetoacetyl-CoA thiolase and 3HB-CoA dehydrogenase. GDH1 glutamate dehydrogenase 1, GDH2 glutamate dehydrogenase 2, GDH3 glutamate dehydrogenase 3, GAD1 glutamate decarboxylase, UGA2 succinate semialdehyde dehydrogenase, GHB γ-hydroxybutyric acid, 3HB 3-hydroxybutyrate, P(3HB-co-4HB) poly(3-hydroxybutyrate-co-4-hydroxybutyrate)
Conclusions
The GDH pathway has a key role in glutamate homeostasis and ammonium assimilation in yeast cells. Synthesis of adequate levels of glutamate utilizing two GDH enzymes with different metabolic activities reflects a unique evolutionary advantage of S. cerevisiae. This ability can fulfill the requirements of both fermentative and respiratory metabolic growth under ammonium excess. This leads to an efficient exploitation of the carbon sources available at each given growth phase. The other essential adaptation in the GDH pathway of S. cerevisiae is the expression of a third GDH enzyme localized in the mitochondria that is primarily responsible for the degradation of glutamate and the anapleurosis of Krebs cycle by providing α-ketoglutarate. The presence of three GDH enzymes in yeast assures rapid nitrogen assimilation and glutamate biosynthesis during fermentation and promotes the wise use of α-ketoglutarate without disturbing the proper function of Krebs cycle under respiratory conditions.
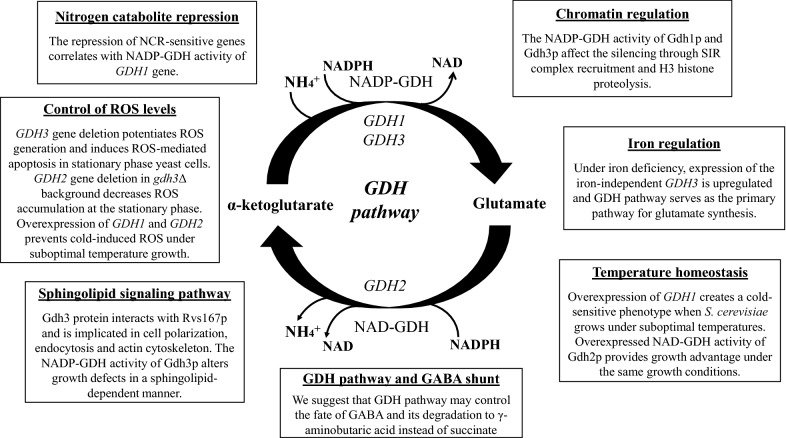
During the past years, cumulative evidence showed that the GDH pathway affects a broader range of cellular activities (Fig. 4). This is an established example showing that cellular metabolic status coordinates the correct function of different cellular compartments. Indeed, the GDH pathway through the action of Gdh1p and Gdh3p showed a strong positive effect on epigenetic processes that promote telomeric silencing. Additionally, the metabolic capacity of Gdh1p and Gdh3p isoforms is essential in the regulation of α-ketoglutarate levels that seems to be the signal also for the NCR-regulated gene expression. These findings demonstrate new roles for the two conserved NADP-GDH enzymes that were primarily described for their role in glutamate production. Additionally, the increased metabolic activity of NADP-GDH through GDH1 overexpression seemed to alter the NAD–NADH levels and thus yeast growth under suboptimal temperatures.
Fig. 4.
The pleiotropic functions of GDH pathway in S. cerevisiae. The enzymatic activity of Gdh1p, Gdh3p and Gdh2p regulates glutamate homeostasis and influences other essential cellular processes
The GDH pathway was also implicated in the regulation of ROS under different conditions. The lack of GDH3 provoked yeast cells sensitivity to thermal and oxidative stress leading to apoptotic cell death due to ROS accumulation. Furthermore, treatment of gdh3Δ yeast strains with the drug ebselen, potentiated ROS generation and possibly affected the function of NADH-GDH as a histone H3 protease, as it was also observed in chicken [67]. GDH2 was found to genetically interact with GDH3 and GDH1 and regulate ROS levels. Finally, the GDH pathway was found to be the iron-independent pathway that regulates glutamate production in yeast cells grown in iron limiting conditions. The importance of the GDH pathway in glutamate homeostasis, nitrogen assimilation and its role to various cellular functions is not restricted only to yeast cells. It has been observed that glutamate production through the appropriate function of GDH pathway has a role in the mitochondrial retrograde signaling that affects changes in nuclear gene expression related to nutrient sensing and TOR signaling [26, 98], aging [99, 100] metabolism [20] and as recently shown, in different types of cancer [101–108]. Furthermore, malfunction of the GDH pathway has been implicated in several other human diseases including diabetes [109–111], neurodegenerative disorders [112], as well as congenital syndromes that affect mainly children [113].
Saccharomyces cerevisiae is a model organism frequently used for biotechnological purposes [114–117]. Being a successful biotechnological tool for many decades, yeast still remains a cell factory that can be used for further challenges regarding genetic engineering and maintenance of redox balances, biosynthesis of basic metabolites and control of biosynthetic pathways [114]. Hosting three different and independent GDH enzymes makes S. cerevisiae an excellent system for better understanding the GDH-derived glutamate and its fate on post translational modifications, epigenetics, chromatin regulation, signaling, oxidative stress defense mechanisms and efflux processes essential for redox homeostasis. Finally, the decoupled functions of the GDH pathway in S. cerevisiae, in terms of glutamate biosynthesis and catabolism can be used as a model to address important biotechnologically related questions on the role and regulation of organic substrates like α-ketoglutarate. This seems to be important due to the significance of α-ketoglutarate in stress responses, lifespan extension, cellular senescence, tumor suppressing conditions and human diseases.
Authors’ contributions
PM wrote the initial draft and designed the figures. GSF, FG, and DA revised and formatted the manuscript and the figures. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank the coordinator of the THALIS project Prof. A. Plaitakis and colleagues Prof. K. Tokatlidis and Dr. I. Zaganas. PM would also like to thank Ms. Maria Trypaki for her useful suggestions and comments on the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
The writing of the manuscript was supported by the European Union (European Social Fund – ESF) and by the Greek National Strategic Reference Framework (NSRF) THALIS: – UOC “Mitochondrial dysfunction in neurodegenerative diseases” (Grant Code 377226).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
P. Mara, Email: pmara@whoi.edu
G. S. Fragiadakis, Email: fragiad@imbb.forth.gr
F. Gkountromichos, Email: f.gkountromichos@campus.lmu.de
D. Alexandraki, Email: alexandr@imbb.forth.gr
References
- 1.Santero E, Hervás AB, Canosa I, Govantes F. Glutamate dehydrogenases: enzymology, physiological role and biotechnological relevance. In: Dehydrogenases. InTech; 2012.
- 2.Holzer H, Schneider S. Anreicherung und Trennung einer DPN-spezifischen und einer TPN-spezifischen Glutaminosaure Dehydrogenase aus Hefe. Biochem Z. 1957;5:361–369. [PubMed] [Google Scholar]
- 3.Tempest DW, Meers JL, Brown CM. Synthesis of glutamate in Aerobacter aerogenes by a hitherto unknown route. Biochem J. 1970;117:405–407. doi: 10.1042/bj1170405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller SM, Magasanik B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol. 1990;172:4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guillamon JM, Van Riel NAW, Giuseppin MLF, Verrips CT. The glutamate synthase (GOGAT) of Saccharomyces cerevisiae plays an important role in central nitrogen metabolism. FEMS Yeast Res. 2001;1:169–175. doi: 10.1016/S1567-1356(01)00034-4. [DOI] [PubMed] [Google Scholar]
- 6.Campero-Basaldua C, Quezada H, Riego-Ruíz L, Márquez D, Rojas E, González J, et al. Diversification of the kinetic properties of yeast NADP-glutamate-dehydrogenase isozymes proceeds independently of their evolutionary origin. Microbiologyopen. 2016;6:1–18. doi: 10.1002/mbo3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sieg AG, Trotter PJ. Differential contribution of the proline and glutamine pathways to glutamate biosynthesis and nitrogen assimilation in yeast lacking glutamate dehydrogenase. Microbiol Res. 2014;169:709–716. doi: 10.1016/j.micres.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper TG. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern JN, Jones EW, Broach JR, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. New York: Cold Spring Harbor Laboratory Press; 1982. pp. 39–99. [Google Scholar]
- 9.Magasanik B. Ammonia assimilation by Saccharomyces cerevisiae. Eukaryot Cell. 2003;2:827–829. doi: 10.1128/EC.2.5.827-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philpott CC, Leidgens S, Frey AG. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta Mol Cell Res. 2012;1823:1509–1520. doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/S0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- 12.Avendano A, Deluna A, Olivera H, Valenzuela L, Gonzalez A. GDH3 encodes a glutamate dehydrogenase isozyme, a previously unrecognized route for glutamate biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:5594–5597. doi: 10.1128/jb.179.17.5594-5597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 14.Marcet-Houben M, Gabaldón T. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker’s yeast lineage. PLoS Biol. 2015;13:1–26. doi: 10.1371/journal.pbio.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLuna A, Avendaño A, Riego L, González A. NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae: purification, kinetic properties, and physiological roles. J Biol Chem. 2001;276:43775–43783. doi: 10.1074/jbc.M107986200. [DOI] [PubMed] [Google Scholar]
- 16.Tang Y, Sieg A, Trotter PJ. 13C-metabolic enrichment of glutamate in glutamate dehydrogenase mutants of Saccharomyces cerevisiae. Microbiol Res. 2011;166:521–530. doi: 10.1016/j.micres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh W, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 18.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ninfa AJ, Jiang P. PII signal transduction proteins: sensors of α-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol. 2005;8:168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Wu N, Yang M, Gaur U, Xu H, Yao Y, Li D. Alpha-ketoglutarate: physiological functions and applications. Biomol Ther. 2016;24:1–8. doi: 10.4062/biomolther.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riego L, Avendaño A, DeLuna A, Rodríguez E, González A. GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem Biophys Res Commun. 2002;293:79–85. doi: 10.1016/S0006-291X(02)00174-2. [DOI] [PubMed] [Google Scholar]
- 22.Avendaño A, Riego L, DeLuna A, Aranda C, Romero G, Ishida C, et al. Swi/SNF-GCN5-dependent chromatin remodelling determines induced expression of GDH3, one of the paralogous genes responsible for ammonium assimilation and glutamate biosynthesis in Saccharomyces cerevisiae. Mol Microbiol. 2005;57:291–305. doi: 10.1111/j.1365-2958.2005.04689.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee YJ, Kim KJ, Kang HY, Kim HR, Maeng PJ. Involvement of GDH3-encoded NADP+-dependent glutamate dehydrogenase in yeast cell resistance to stress-induced apoptosis in stationary phase cells. J Biol Chem. 2012;287:44221–44233. doi: 10.1074/jbc.M112.375360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plaitakis A, Spanaki C, Mastorodemos V, Zaganas I. Study of structure–function relationships in human glutamate dehydrogenases reveals novel molecular mechanisms for the regulation of the nerve tissue-specific (GLUD2) isoenzyme. Neurochem Int. 2003;43:401–410. doi: 10.1016/S0197-0186(03)00028-7. [DOI] [PubMed] [Google Scholar]
- 25.Hu Y, Cooper TG, Kohlhaw GB. The Saccharomyces cerevisiae Leu3 protein activates expression of GDH1, a key gene in nitrogen assimilation. Mol Cell Biol. 1995;15:52–57. doi: 10.1128/MCB.15.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 27.Yang R, Wek SA, Wek RC. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol Cell Biol. 2000;20:2706–2717. doi: 10.1128/MCB.20.8.2706-2717.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazon MJ, Hemmings BA. Regulation of Saccharomyces cerevisiae nicotinamide adenine dinucleotide phosphate-dependent glutamate dehydrogenase by proteolysis during carbon starvation. J Bacteriol. 1979;139:686–689. doi: 10.1128/jb.139.2.686-689.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starita LM, Lo RS, Eng JK, von Haller PD, Fields S. Sites of ubiquitin attachment in Saccharomyces cerevisiae. Proteomics. 2012;12:236–240. doi: 10.1002/pmic.201100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, et al. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paz I, Choder M. Eukaryotic translation initiation factor 4E-dependent translation is not essential for survival of starved yeast cells. J Bacteriol. 2001;183:4477–4483. doi: 10.1128/JB.183.15.4477-4483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herman PK. Stationary phase in yeast. Curr Opin Microbiol. 2002;5:602–607. doi: 10.1016/S1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- 33.MacLean M, Harris N, Piper PW. Chronological lifespan of stationary phase yeast cells; a model for investigating the factors that might influence the ageing of postmitotic tissues in higher organisms. Yeast. 2001;18:499–509. doi: 10.1002/yea.701. [DOI] [PubMed] [Google Scholar]
- 34.Fayyad-Kazan M, Feller A, Bodo E, Boeckstaens M, Marini AM, Dubois E, et al. Yeast nitrogen catabolite repression is sustained by signals distinct from glutamine and glutamate reservoirs. Mol Microbiol. 2016;99:360–379. doi: 10.1111/mmi.13236. [DOI] [PubMed] [Google Scholar]
- 35.Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/S0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- 36.Inoue Y, Matsuda T, Sugiyama KI, Izawa S, Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J Biol Chem. 1999;274:27002–27009. doi: 10.1074/jbc.274.38.27002. [DOI] [PubMed] [Google Scholar]
- 37.Kiriyama K, Hara KY, Kondo A. Oxidized glutathione fermentation using Saccharomyces cerevisiae engineered for glutathione metabolism. Appl Microbiol Biotechnol. 2013;97:7399–7404. doi: 10.1007/s00253-013-5074-8. [DOI] [PubMed] [Google Scholar]
- 38.Azad GK, Singh V, Mandal P, Singh P, Golla U, Baranwal S, et al. Ebselen induces reactive oxygen species (ROS)-mediated cytotoxicity in Saccharomyces cerevisiae with inhibition of glutamate dehydrogenase being a target. FEBS Open Bio. 2014;4:77–89. doi: 10.1016/j.fob.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandal P, Verma N, Chauhan S, Tomar RS. Unexpected histone H3 tail-clipping activity of glutamate dehydrogenase. J Biol Chem. 2013;288:18743–18757. doi: 10.1074/jbc.M113.462531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su XB, Pillus L. Functions for diverse metabolic activities in heterochromatin. Proc Natl Acad Sci USA. 2016;113:E1526–E1535. doi: 10.1073/pnas.1518707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemmings BA. Phosphorylation and proteolysis regulate the NAD-dependent glutamate dehydrogenase from Saccharomyces cerevisiae. FEBS Lett. 1980;122:297–302. doi: 10.1016/0014-5793(80)80460-1. [DOI] [PubMed] [Google Scholar]
- 42.Uno I, Matsumoto K, Adachi K. Regulation of NAD-dependent glutamate dehydrogenase by protein kinases in regulation of NAD-dependent glutamate dehydrogenase by protein kinases in Saccharomyces cerevisiae. J Biol Chem. 1984;259:1288–1293. [PubMed] [Google Scholar]
- 43.Oliveira AP, Ludwig C, Picotti P, Kogadeeva M, Aebersold R, Sauer U. Regulation of yeast central metabolism by enzyme phosphorylation. Mol Syst Biol. 2012;8:623–636. doi: 10.1038/msb.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coschigano PW, Miller SM, Magasanik B. Physiological and genetic analysis of the carbon regulation of the NAD-dependent glutamate dehydrogenase of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4455–4465. doi: 10.1128/MCB.11.9.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magasanik B. The transduction of the nitrogen regulation signal in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2005;102:16537–16538. doi: 10.1073/pnas.0507116102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nuñez de castro I, Ugarte M, Cano A, Mayor F. Effect of glucose, galactose, and different nitrogen-sources on the activity of yeast glutamate dehydrogenase (NAD and NADP-linked) from normal strain and impaired respiration mutant. Eur J Biochem. 1970;16:567–570. doi: 10.1111/j.1432-1033.1970.tb01118.x. [DOI] [PubMed] [Google Scholar]
- 47.Soussi-Boudekou S, André B. A co-activator of nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol. 1999;31:753–762. doi: 10.1046/j.1365-2958.1999.01187.x. [DOI] [PubMed] [Google Scholar]
- 48.Hernández H, Aranda C, Riego L, González A. Gln3–Gcn4 hybrid transcriptional activator determines catabolic and biosynthetic gene expression in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2011;404:859–864. doi: 10.1016/j.bbrc.2010.12.075. [DOI] [PubMed] [Google Scholar]
- 49.Ishida C, Aranda C, Valenzuela L, Riego L, Deluna A, Recillas-Targa F, et al. The UGA3-GLT1 intergenic region constitutes a promoter whose bidirectional nature is determined by chromatin organization in Saccharomyces cerevisiae. Mol Microbiol. 2006;59:1790–1806. doi: 10.1111/j.1365-2958.2006.05055.x. [DOI] [PubMed] [Google Scholar]
- 50.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, et al. Integration of general amino acid control and Target of Rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem. 2010;285:16893–16911. doi: 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell AP, Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2758–2766. doi: 10.1128/MCB.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 53.Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL, Hardwick JS, et al. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics. 2012;190:885–929. doi: 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, van Helden J, et al. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3065–3086. doi: 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazelwood LA, Daran J-M, van Maris AJA, Pronk JT, Dickinson JR. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol. 2008;74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hinnebusch AG. General and cross-pathway controls of amino acid biosynthesis. In: Brambl R, Marzluf GA, editors. Biochemistry and molecular biology. Berlin: Springer; 1992. pp. 319–414. [Google Scholar]
- 59.Ter Schure EG, Van Riel NAW, Verrips CT. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 60.Cajueiro DBB, Parente DC, Leite FCB, de Morais Junior MA, de Barros Pita W. Glutamine: a major player in nitrogen catabolite repression in the yeast Dekkera bruxellensis. Antonie van Leeuwenhoek Int J Gen Mol Microbiol. 2017;110:1157–1168. doi: 10.1007/s10482-017-0888-5. [DOI] [PubMed] [Google Scholar]
- 61.Georis I, Tate JJ, Cooper TG, Dubois E. Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J Biol Chem. 2011;286:44897–44912. doi: 10.1074/jbc.M111.290577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cox K, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J Biol Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minehart PL, Magasanik B. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1991;11:6216–6228. doi: 10.1128/MCB.11.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Georis I, Feller A, Vierendeels F, Dubois E. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol Cell Biol. 2009;29:3803–3815. doi: 10.1128/MCB.00399-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rai R, Tate JJ, Nelson DR, Cooper TG. Gln3 mutations dissociate responses to nitrogen limitation (nitrogen catabolite repression) and rapamycin inhibition of TorC1. J Biol Chem. 2013;288:2789–2804. doi: 10.1074/jbc.M112.421826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fayyad-Kazan M, Tate JJ, Vierendeels F, Cooper TG, Dubois E, Georis I. Components of Golgi-to-vacuole trafficking are required for nitrogen- and TORC1-responsive regulation of the yeast GATA factors. Microbiologyopen. 2014;3:271–287. doi: 10.1002/mbo3.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mandal P, Azad GK, Tomar RS. Identification of a novel histone H3 specific protease activity in nuclei of chicken liver. Biochem Biophys Res Commun. 2012;421:261–267. doi: 10.1016/j.bbrc.2012.03.149. [DOI] [PubMed] [Google Scholar]
- 68.Xue Y, Vashisht AA, Tan Y, Su T, Wohlschlegel JA. PRB1 is required for clipping of the histone H3 N terminal tail in Saccharomyces cerevisiae. PLoS ONE. 2014;9:1–7. doi: 10.1371/journal.pone.0090496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chin RM, Fu X, Pai MY, Vergnes L, Hwang H, Deng G, et al. The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carey BW, Finley LWS, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ballester-Tomás L, Randez-Gil F, Pérez-Torrado R, Prieto JA. Redox engineering by ectopic expression of glutamate dehydrogenase genes links NADPH availability and NADH oxidation with cold growth in Saccharomyces cerevisiae. Microb Cell Fact. 2015;14:1–11. doi: 10.1186/s12934-015-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nissen TL, Schulze U, Nielsen J, Villadsen J. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology. 1997;143:203–218. doi: 10.1099/00221287-143-1-203. [DOI] [PubMed] [Google Scholar]
- 73.Kato M, Lin SJ. Regulation of NAD+ metabolism, signaling and compartmentalization in the yeast Saccharomyces cerevisiae. DNA Repair. 2014;23:49–58. doi: 10.1016/j.dnarep.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heux S, Cachon R, Dequin S. Cofactor engineering in Saccharomyces cerevisiae: expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab Eng. 2006;8:303–314. doi: 10.1016/j.ymben.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 75.Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2007;104:2402–2407. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paget CM, Schwartz JM, Delneri D. Environmental systems biology of cold-tolerant phenotype in Saccharomyces species adapted to grow at different temperatures. Mol Ecol. 2014;23:5241–5257. doi: 10.1111/mec.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L, Onda K, Imai R, Fukuda R, Horiuchi H, Ohta A. Growth temperature downshift induces antioxidant response in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2003;307:308–314. doi: 10.1016/S0006-291X(03)01168-9. [DOI] [PubMed] [Google Scholar]
- 78.García-Ríos E, Ramos-Alonso L, Guillamón JM. Correlation between low temperature adaptation and oxidative stress in Saccharomyces cerevisiae. Front Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Germann M, Swain E, Bergman L, Nickels JT. Characterizing the sphingolipid signaling pathway that remediates defects associated with loss of the yeast amphiphysin-like orthologs, Rvs161p and Rvs167p. J Biol Chem. 2005;280:4270–4278. doi: 10.1074/jbc.M412454200. [DOI] [PubMed] [Google Scholar]
- 80.Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sivadon P, Bauer F, Aigle M, Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. MGG Mol Gen Genet. 1995;246:485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- 82.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, et al. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:1233–1243. doi: 10.1091/mbc.e03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shelp BJ, Mullen RT, Waller JC. Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci. 2012;17:57–59. doi: 10.1016/j.tplants.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 84.Cao J, Barbosa JM, Singh N, Locy RD. GABA transaminases from Saccharomyces cerevisiae and Arabidopsis thaliana complement function in cytosol and mitochondria. Yeast. 2013;30:279–289. doi: 10.1002/yea.2962. [DOI] [PubMed] [Google Scholar]
- 85.Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H. A plant glutamate decarboxylase containing a calmodulin binding domain Cloning, sequence, and functional analysis. J Biol Chem. 1993;268:19610–19617. [PubMed] [Google Scholar]
- 86.Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J Biol Chem. 2001;276:244–250. doi: 10.1074/jbc.M007103200. [DOI] [PubMed] [Google Scholar]
- 87.Bach B, Meudec E, Lepoutre JP, Rossignol T, Blondin B, Dequin S, et al. New insights into γ-aminobutyric acid catabolism: evidence for γ-hydroxybutyric acid and polyhydroxybutyrate synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:4231–4239. doi: 10.1128/AEM.00051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grenson M, Hou C, Crabeel M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J Bacteriol. 1970;103:770–777. doi: 10.1128/jb.103.3.770-777.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jauniaux J-C, Vandenbol M, Vissers S, Broman K, Ggrenson M. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae. Cloning of the PUT4 gene and study of PUT4 RNA levels in wild-type and mutant strains. Eur J Biochem. 1987;164:601–606. doi: 10.1111/j.1432-1033.1987.tb11169.x. [DOI] [PubMed] [Google Scholar]
- 90.André B, Hein C, Grenson M, Jauniaux J-C. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol Gen Genet MGG. 1993;237:17–25. doi: 10.1007/BF00282779. [DOI] [PubMed] [Google Scholar]
- 91.Rossignol T, Dulau L, Julien A, Blondin B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast. 2003;20:1369–1385. doi: 10.1002/yea.1046. [DOI] [PubMed] [Google Scholar]
- 92.Ramesh SA, Tyerman SD, Xu B, Bose J, Kaur S, Conn V, et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat Commun. 2015;6:1–9. doi: 10.1038/ncomms8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mekonnen DW, Ludewig F. Phenotypic and chemotypic studies using Arabidopsis and yeast reveal that GHB converts to SSA and induce toxicity. Plant Mol Biol. 2016;91:429–440. doi: 10.1007/s11103-016-0475-6. [DOI] [PubMed] [Google Scholar]
- 94.Madison LL, Huisman GW. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Steinbüchel A, Füchtenbusch B. Bacterial and other biological systems for polyester production. Trends Biotechnol. 1998;16:419–427. doi: 10.1016/S0167-7799(98)01194-9. [DOI] [PubMed] [Google Scholar]
- 96.Liu G, Zhu J, Yu M, Cai C, Zhou Y, Yu M, et al. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J Transl Med. 2015;13:144–154. doi: 10.1186/s12967-015-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen G, Jiang X, Guo Y. Synthetic biology of microbes synthesizing polyhydroxyalkanoates. Synth Syst Biotechnol. 2016;1:236–242. doi: 10.1016/j.synbio.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.González A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017;36:397–408. doi: 10.15252/embj.201696010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Segovia G, Porras A, Del Arco A, Mora F. Glutamatergic neurotransmission in aging: a critical perspective. Mech Ageing Dev. 2001;122:1–29. doi: 10.1016/S0047-6374(00)00225-6. [DOI] [PubMed] [Google Scholar]
- 100.Wang X, Patel ND, Hui D, Pal R, Hafez MM, Sayed-Ahmed MM, et al. Gene expression patterns in the hippocampus during the development and aging of Glud1 (Glutamate Dehydrogenase 1) transgenic and wild type mice. BMC Neurosci. 2014;15:37–54. doi: 10.1186/1471-2202-15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jeong SM, Xiao C, Finley LWS, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to dna damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jeong SM, Lee A, Lee J, Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. 2014;289:4135–4144. doi: 10.1074/jbc.M113.525949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang L, Moss T, Mangala LS, Marini J, Zhao H, Wahlig S, et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol Syst Biol. 2014;10:728. doi: 10.1002/msb.20134892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu G, Zhu J, Yu M, Cai C, Zhou Y, Yu M, et al. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J Transl Med. 2015;13:1–10. doi: 10.1186/s12967-014-0365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J, Wang G, Mao Q, Li S, Xiong W, Lin Y, et al. Glutamate dehydrogenase (GDH) regulates bioenergetics and redox homeostasis in human glioma. Oncotarget. 2016;5:1–12. [Google Scholar]
- 108.Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer. 2017;3:169–180. doi: 10.1016/j.trecan.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 110.Fahien LA, MacDonald MJ. The complex mechanism of glutamate dehydrogenase in insulin secretion. Diabetes. 2011;60:2450–2454. doi: 10.2337/db10-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li M, Li C, Allen A, Stanley CA, Smith TJ. Glutamate dehydrogenase: structure, allosteric regulation, and role in insulin homeostasis. Neurochem Res. 2014;39:433–445. doi: 10.1007/s11064-013-1173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Plaitakis A, Kalef-Ezra E, Kotzamani D, Zaganas I, Spanaki C. The glutamate dehydrogenase pathway and its roles in cell and tissue biology in health and disease. Biology. 2017;6:11. doi: 10.3390/biology6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stanley CA, Lieu YK, Hsu BYL, Burlina AB, Greenberg CR, Hopwood NJ, et al. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N Engl J Med. 1998;338:1352–1357. doi: 10.1056/NEJM199805073381904. [DOI] [PubMed] [Google Scholar]
- 114.Kavšček M, Stražar M, Curk T, Natter K, Petrovič U. Yeast as a cell factory: current state and perspectives. Microb Cell Fact. 2015;14:1–10. doi: 10.1186/s12934-015-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mattanovich D, Sauer M, Gasser B. Yeast biotechnology: teaching the old dog new tricks. Microb Cell Fact. 2014;13:34. doi: 10.1186/1475-2859-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hitzeman RA, Hagie FE, Levine HL, Goeddel DV, Ammerer G, Hall BD. Expression of a human gene for interferon in yeast. Nature. 1981;293:717–722. doi: 10.1038/293717a0. [DOI] [PubMed] [Google Scholar]
- 117.Ferrer-Miralles N, Domingo-Espín J, Corchero J, Vázquez E, Villaverde A. Microbial factories for recombinant pharmaceuticals. Microb Cell Fact. 2009;8:1–8. doi: 10.1186/1475-2859-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.