SUMMARY
The fusion oncoprotein CBFβ-SMMHC, expressed in leukemia cases with chromosome 16 inversion, drives leukemia development and maintenance by altering the activity of the transcription factor RUNX1. Here we demonstrate that CBFβ-SMMHC maintains cell viability by neutralizing RUNX1-mediated repression of MYC expression. Upon pharmacologic inhibition of the CBFβ-SMMHC/RUNX1 interaction, RUNX1 shows increased binding at three MYC distal enhancers. There it represses MYC expression by mediating the replacement of SWI/SNF complex component BRG1 with polycomb-repressive complex component RING1B, leading to apoptosis. Combining the CBFβ-SMMHC inhibitor with the BET- inhibitor JQ1 eliminates inv(16) leukemia in human cells and a mouse model. Enhancer-interaction analysis indicated that the three enhancers are physically connected with the MYC promoter, and genome-editing analysis demonstrated that they are functionally implicated in deregulation of MYC expression. This study reveals a mechanism whereby CBFβ-SMMHC drives leukemia maintenance, and suggests that inhibitors targeting chromatin activity may prove effective in inv(16) leukemia therapy.
INTRODUCTION
Cancer cells adapt to mutations that alter the function of transcription factors and chromatin-associated factors to survive. In leukemia, these factors often drive leukemia initiation and maintenance. The transcription factor complex core-binding factor is composed of a stabilizing subunit CBFβ and the DNA-binding subunit RUNX (encoded by three genes: RUNX1, RUNX2 and RUNX3). Together, RUNX1 and CBFβ regulate pathways associated with proliferation, survival and differentiation (Blyth et al., 2005). The genes encoding CBFβ and RUNX1 are frequent targets of mutations in hematologic malignancies. The chromosome inversion inv(16)(p13;q22), found in 8% of acute myeloid leukemia (AML) cases, fuses the CBFB and MYH11 genes to produce the leukemic oncoprotein CBFβ-SMMHC (Liu et al., 1993). This fusion protein has higher affinity and altered stoichiometry for RUNX1 relative to the native CBFβ (Cao et al., 1997; Lukasik et al., 2002). During development, CBFβ-SMMHC expression blocks definitive hematopoiesis and embryos die at mid-gestation (Castilla et al., 1996), a similar phenotype to that of Runx1- and Cbfb-knock out embryos (Wang et al., 1996a; Wang et al., 1996b), indicating that CBFβ-SMMHC has a dominant negative effect on RUNX function. In adult hematopoiesis, allelic CBFβ-SMMHC expression alters hematopoietic stem cell (HSC) differentiation, with clonal expansion of the short-term HSCs and pre-leukemic myeloid progenitor cells (Kuo et al., 2006; Xue et al., 2014). These myeloid pre-leukemic progenitors are leukemia precursors, which acquire cooperating mutations to induce myeloid leukemia in mice (Castilla et al., 1999; Xue et al., 2014).
RUNX1 associates with chromatin-modifying proteins, including histone deacetylases (Guo and Friedman, 2011), acetyltransferases (Kitabayashi et al., 2001; Kitabayashi et al., 1998) and methyltransferases (Reed-Inderbitzin et al., 2006; Vu et al., 2013; Zhao et al., 2008) in hematopoiesis. These interactions regulate RUNX1 affinity to DNA and its transcriptional activity, and modulate its association with activating and repressing chromatin complexes (Lichtinger et al., 2010). However, the role of RUNX1 in establishing chromatin-associated complexes that maintain the survival of AML cells and how they modulate leukemia maintenance remain poorly understood.
The SWItch/Sucrose Non-Fermentable (SWI/SNF) protein complex remodels histone-DNA interactions and is associated with active regulatory regions of the genome, and some of its subunits are mutated in cancer (Kadoch and Crabtree, 2015). In AML, chromatin-associated complexes, including SWI/SNF and BET family of bromodomain-proteins, promote enhancer activity to mediate the survival of the leukemia stem cells (Blobel et al., 2011; Zuber et al., 2011b). Conversely, the accumulation of polycomb-repressive complexes (PRC1 and PRC2) is associated with tri-methylation of lysine-27 in histone H3 (H3K27me3), local compaction of the chromatin structure around the enhancers and silencing of target gene expression (Di Croce and Helin, 2013). The homeostasis of hematopoiesis critically depends on PRC function as these proteins regulate self-renewal and differentiation of HSCs (Lessard and Sauvageau, 2003; van der Lugt et al., 1994), as well as inhibit differentiation and proliferation of myeloid progenitor cells (Cao et al., 2016).
The protooncogene MYC regulates the balance between self-renewal and differentiation of HSCs (Wilson et al., 2004), and is essential for lymphoid (de Alboran et al., 2001; Douglas et al., 2001) and megakaryocytic/erythroid development (Guo et al., 2009). Downregulation of MYC expression, however, is critical during myeloid differentiation, since MYC repression promotes granulopoiesis while ectopic expression blocks granulopoiesis (Gowda et al., 1986; Holt et al., 1988; Johansen et al., 2001). The epigenetic regulation at the MYC locus plays a critical role in myeloid differentiation and leukemia. The SWI/SNF and BRD4 complexes regulate MYC expression from the distal super-enhancer BDME (BRD4-dependent MYC enhancer), 1.7 megabases (Mb) downstream from its transcription start site (TSS, (Shi et al., 2013; Yashiro-Ohtani et al., 2014)). In leukemia, MYC regulates expression of its normal high-affinity targets and a new set of targets in a tumor type–specific manner (Kress et al., 2015). The SWI/SNF ATPase subunit BRG1 (Brahma related gene 1) associates at BDME to maintain MYC levels in mixed-lineage AML cells (Shi et al., 2013; Vradii et al., 2005). In addition, BDME function regulates leukemia maintenance in gamma secretase inhibitor-resistant T-cell acute lymphoblastic leukemia (Yashiro-Ohtani et al., 2014).
The mechanisms underlying oncogenic CBFβ-SMMHC function and the role of epigenetic complexes in the maintenance of inv(16) AML are poorly understood. We have developed a bivalent inhibitor, AI-10–49, that disrupts CBFβ-SMMHC binding to RUNX1, and specifically induces apoptosis of inv(16) AML cells (Illendula et al., 2015). In this study, we combined pharmacologic, genomic and genetic approaches to define the mechanism by which CBFβ-SMMHC inhibition drives apoptosis in inv(16) AML cells. This effort uncovered a RUNX1 mediated mechanism of MYC regulation at distal enhancers by the replacement of activating chromatin complexes for repressive complexes. This study also provides proof-of-principle evidence for the enhanced efficacy of the combination therapy in the treatment of inv(16) AML.
RESULTS
Inhibition of CBFβ-SMMHC activity by AI-10–49 represses MYC transcript expression
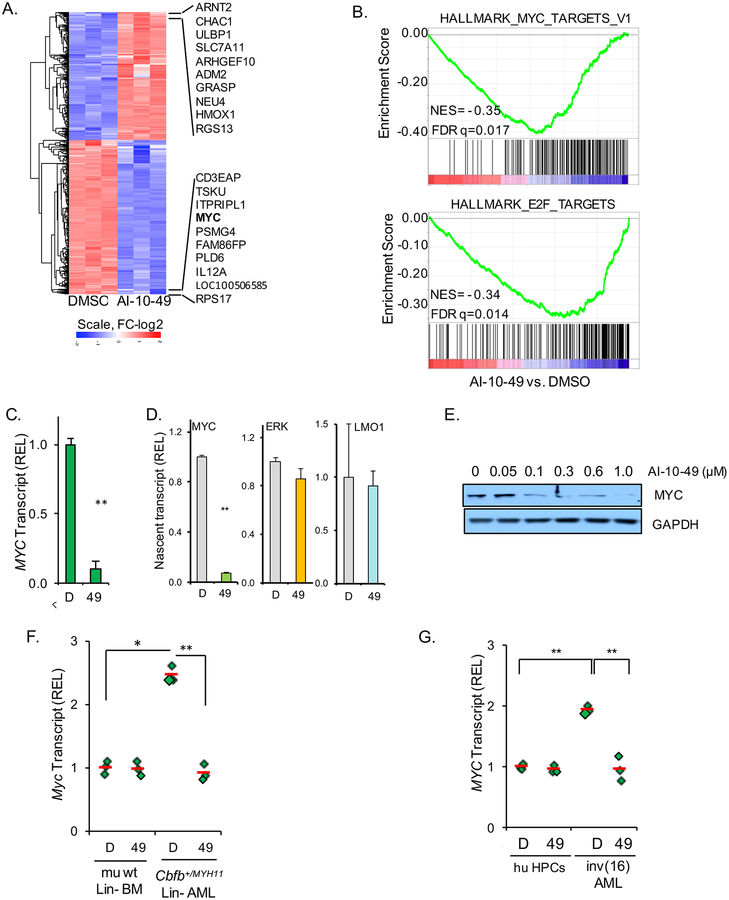
The expression of CBFβ-SMMHC is critical for inv(16) AML blast survival, and the small molecule inhibitor AI-10–49 selectively triggers apoptosis of human and mouse inv(16) AML cells (Illendula et al., 2015). In order to identify pro-apoptotic AI-10–49 targets, we performed RNA-sequencing analysis in AI-10–49 treated inv(16) AML cell line ME-1. Expression analysis identified 591 up- and 696 downregulated genes (>2 fold change, FDR<0.01; Figure 1A). Amongst the most repressed genes, there was an over 10-fold repression of MYC levels. Gene Set Enrichment Analysis (GSEA) revealed that AI-10–49 treatment was associated with MYC and E2F signatures (Figure 1B), and MYC-associated pathways, including cell cycle, amino acid metabolism and ribosome biogenesis (Figures S1A and S1B).
Figure 1. AI-10–49 inhibits MYC transcriptional program in inv(16) AML cells.
(A) Heat map representation of differentially expressed genes in RNA-seq analysis between DMSO and AI-10–49 treated ME-1 cells. A total of 591 genes (in red) are up- and 696 genes (in blue) down-regulated in AI-10–49 treated cells (>2fold change, FDR<0.01). (B) Gene set enrichment analysis showing biological processes and signaling pathways correlated with AI-10–49-treatment. (FDR) false discovery rate; (NES) normalized enrichment score. (C) MYC transcript levels in ME-1 cells treated with DMSO (D) or AI-10–49 (49), estimated by RT-PCR. (D) MYC, ERK and LMO1 nascent transcript levels in treated ME-1 cells, estimated by transient transcriptome qRT-PCR. (E) MYC protein levels in treated ME-1 cells, by immunoblotting. (F, G) MYC transcriptional levels in mouse lineage negative wild type bone marrow (mu wt Lin- BM) and lineage negative CBFβ+/MYH11 leukemic cells (F), and human CD34+ cord blood (HPCs) and human primary CD34+ inv(16) AML cells (G); each symbol represents the average of a triplicate experiment, and the average value of three samples is shown in red. Experiments in A, C and D were performed in triplicate treatments. Error bars represent the SD. Significance was calculated using unpaired t-test (C, D, F and G), *P < 0.05, or **P < 0.005. See also Figure S1.
We validated that AI-10–49 directs significant repression of MYC transcript levels (10-fold; p<0.005; Figure 1C) in ME-1 cells but not in non-inv(16) AML cell lines (Figure S1C). We further demonstrated that this repression is at the transcription step by evaluating nascent RNA levels for MYC and two unrelated control genes, ERK and LMO1, in ME-1 cells. To this end, we used “transient-transcription-RT-PCR”, a modification of the “transient-transcription-sequencing” protocol (Schwalb et al., 2016), to determined that expression of MYC nascent transcript was significantly reduced over 12 fold (p<0.005; Figure 1D). Accordingly, MYC protein levels were also inhibited at 0.1μM and greater concentrations in ME-1 cells (Figures 1E and S1D). Expression analysis in primary mouse and human cells indicated that MYC is expressed in inv(16) AML cells at higher levels than in normal bone marrow hematopoietic progenitor cells, and that its expression is repressed to basal levels by AI-10–49 (Figures 1F and 1G). These results demonstrate that AI-10–49 treatment represses CBFβ-SMMHC-induced MYC expression in inv(16) AML.
MYC is required for the survival of inv(16) AML leukemia initiating cells
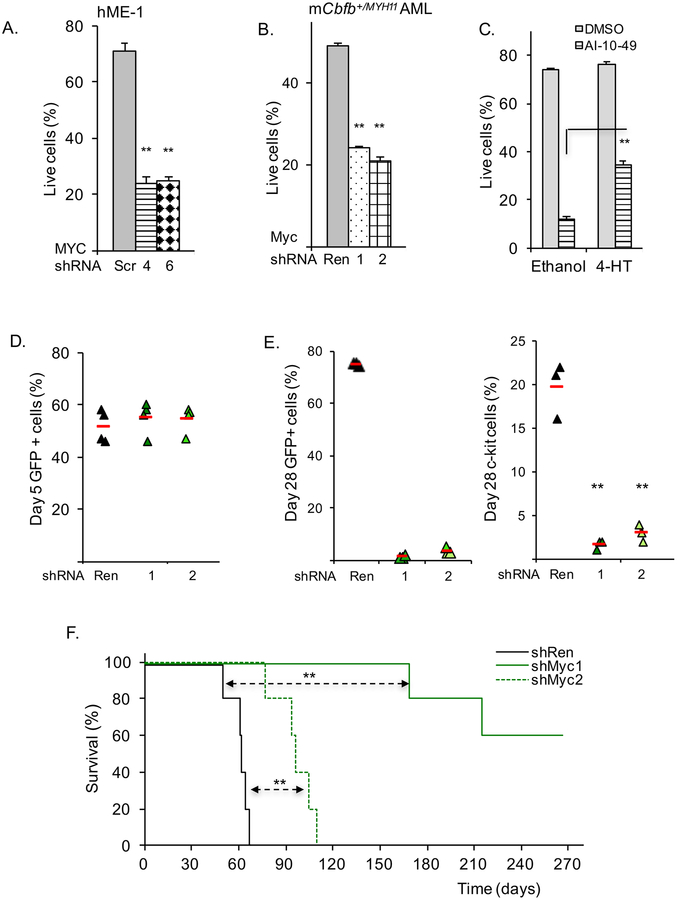
We analyzed whether MYC levels regulate survival in inv(16) AML cells. The knockdown of MYC, using MYC-shRNAs, reduced viability of ME-1 cells 66% and primary mouse CBFβ+/MYH11 leukemic cells 70% (Figures 2A, 2B and S2A). Furthermore, ectopic MYC expression resulted in a partial rescue of AI-10–49 mediated apoptosis (Figures 2C). These results suggest that reduced MYC levels mediate AI-10–49 induced apoptosis. In addition, MYC knockdown in six non-inv(16) AML cell lines revealed that, with the exception of NB4 cells, the viability of other AML cell types also depend on MYC levels (Figures S2C and S2D). These results indicate that although many AML types require MYC expression for survival, the inhibitor AI-10–49 specifically reduces MYC levels of inv(16) AML cells.
Figure 2. MYC is required for the survival of inv(16) cells.
(A-B) Viability analysis (Annexin V/7AAD assay) in ME-1 (A) and in primary mouse Cbfb+/MYH11 leukemic (B) cells transduced with control or MYC shRNAs. Each data point represents the mean of triplicate experiments. (C) Viability analysis (Annexin V/7AAD assay) in ME-1 cells expressing MYC-ER treated with DMSO or AI-10–49. Each data point represents the mean of triplicate experiments. (D, E, F) Percentage of Cbfb+/MYH11 leukemic cells (GFP+ or c-kit+ cells) in peripheral blood of recipient mice, 5 (D) and 28 (E and F) days after transplantation; average values shown as lines. (G) Kaplan-Meier survival curve of mice transplanted with control (shRen) or Myc shRNA transduced leukemic cells. Error bars represent the SD. Significance was calculated using Levene’s test (A-F), or log-rank test (G). *P < 0.05, or **P < 0.005. See also Figure S2.
Considering that MYC silencing is required for granulocytic differentiation of myeloid cells (Johansen et al., 2001), we investigated whether MYC knockdown promoted differentiation in ME-1 cells. The fraction of myeloid cells (CD15+ and CD11b+) 14 days after transduction was not significantly altered (Figure S2B), suggesting that MYC repression primarily directs apoptosis and not differentiation of inv(16) AML cells.
The requirement for Myc function in inv(16) leukemia was tested in mice transplanted with CBFβ-SMMHC-expressing leukemic cells (Nras+/LSL-G12D,CBFβ+/56M,Mx1Cre; Nras/CM; (Xue et al., 2014)) transduced with Renilla- or 2 Myc-shRNAs (Figures S2E and S2F). Engraftment efficiency of leukemic cells in bone marrow five days after transplantation was similar between groups (Figure 2D). However, the fraction of leukemic cells (estimated as GFP+ or c-kit+ (immature) cells) in peripheral blood 28 days after transplantation revealed a marked reduction in Myc shRNA groups, suggesting that Myc is required for the maintenance of inv(16) leukemic cells (Figure 2E). Furthermore, the median leukemia latency in mice with reduced Myc levels was significantly prolonged from 62 days (Renilla group) to incomplete penetrance (shMyc1) and 96 days (shMyc2; p<0.005, Figure 2F). Leukemic cells from either Myc-shRNA group expressed Myc at similar levels to Renilla group (Figure S2G), indicating that Myc knockdown was not maintained in these clones. Therefore, these in vitro and in vivo experiments in mice demonstrate that modulation of MYC levels is critical to the survival and expansion of inv(16) leukemia.
AI-10–49 cooperates with JQ1 to reduce inv(16) AML cell survival
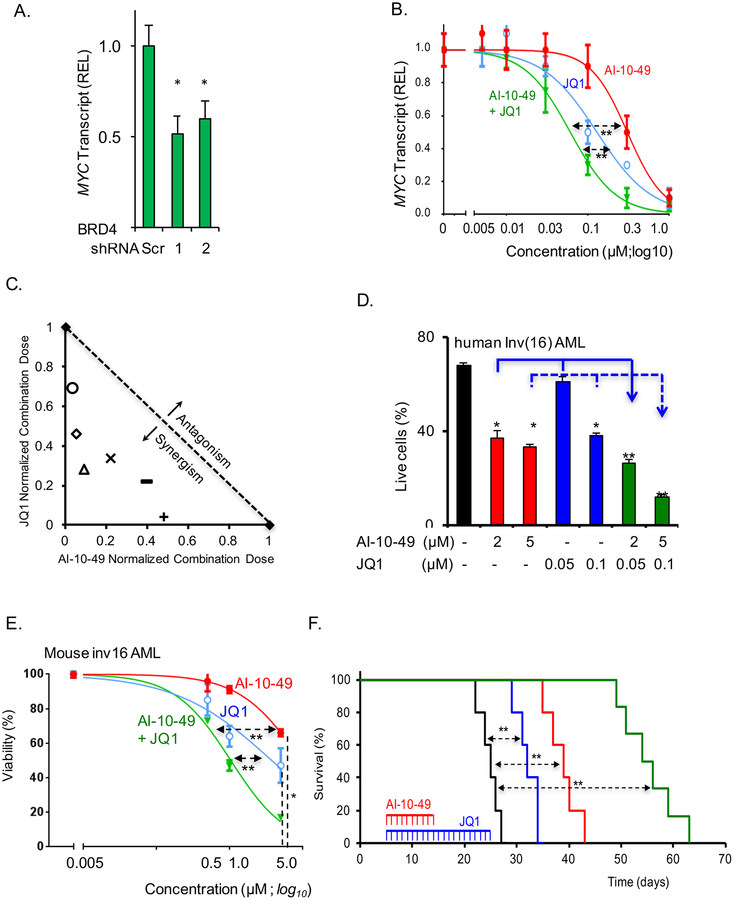
Bromodomain (BRD) proteins have been established as key drivers of oncogenic transcription factors such as MYC (Delmore et al., 2011). The BET-family of BRD inhibitors, such as JQ1, efficiently repress MYC levels (Delmore et al., 2011), and second generation BRD-inhibitors are being tested in the clinic. We confirmed that MYC expression depends on BRD4 in inv(16) AML by estimating MYC levels in ME-1 cells upon BRD4 knockdown (Figures 3A and S3A). We tested whether inv(16) AML cells are sensitive to combination treatment with AI-10–49 and JQ1. The MYC transcript and protein levels were readily reduced by AI-10–49 and JQ1 treatment of ME-1 cells, and combined treatment showed a cooperative effect in MYC transcript levels (Figures 3B and S3B). Remarkably, the combination index (CI) analysis (Chou, 2010) of dose response combined treatment consistently showed CI values below 1 (Figure 3C and S3C), indicative of a substantial synergy on ME-1 cell growth. The cooperativity between these inhibitors was further demonstrated in primary human inv(16) AML and mouse inv(16) leukemic cells (Figures 3D and 3E, respectively).
Figure 3. Inhibition of MYC by AI-10–49 and JQ1 leads to synergistic efficacy against inv(16) leukemia cell survival.
(A) MYC transcript level analysis in ME-1 cells transduced with BRD4 shRNAs (sh1 and sh2), by qRT-PCR. (B) MYC transcript level analysis after dose response treatment with AI-10–49 and JQ1. (C) Isobologram plot showing synergism between AI-10–49 and JQ1 in combined trement. (D-E) Viability analysis of primary inv(16) AML cells (D) and mouse leukemic cells (E) treated with AI-10–49 and JQ1. (F) Kaplan-Meier survival curve of mice (n = 5–6 per group) transplanted with mouse leukemic cells and treated with DMSO (black line), JQ1 (blue line), AI-10–49 (red line) or JQ1 and AI-10–49 (green line). Error bars represent the SD. Significance was calculated using unpaired t-test (A), Levene’s test (B-E), or log rank test (F). *P < 0.05, or **P < 0.005. See also Figure S3.
To test the efficacy of AI-10–49 and JQ1 in vivo, we analyzed the leukemia latency of mice transplanted with CBFβ-SMMHC-expressing (Nras+/LSL-G12D/CBFβ+/56M/Mx1Cre; Nras/CM) leukemia cells. Five days after transplantation, mice were treated with vehicle (dimethyl sulfoxide, DMSO), AI-10–49, JQ1, or combined treatment (Figure 3F). The median latency of leukemia was prolonged from 25 days (DMSO) to 32 (JQ1) and to 39 days (AI-10–49) when treated with one inhibitor (p=0.00184), and to 55 days with both inhibitors (p<0.005). Leukemia burden analysis at day 25 of transplantation (1 day after the last JQ1 session) revealed a significant reduction of leukemia cells (c-kit+ cells; p<0.005) in peripheral blood (Figure S3D). The combined treatment in human cord blood CD34+ hematopoietic progenitors (Figure S3E) and in wild type C57BL/6J mice did not reveal measurable toxicity (Figures S3F–S3J). Collectively, these findings provide in vitro and in vivo proof-of-principle for the potential use of combined therapy with AI-10–49 and BET inhibitors in inv(16) AML.
AI-10–49 enhances genome wide RUNX1 DNA binding
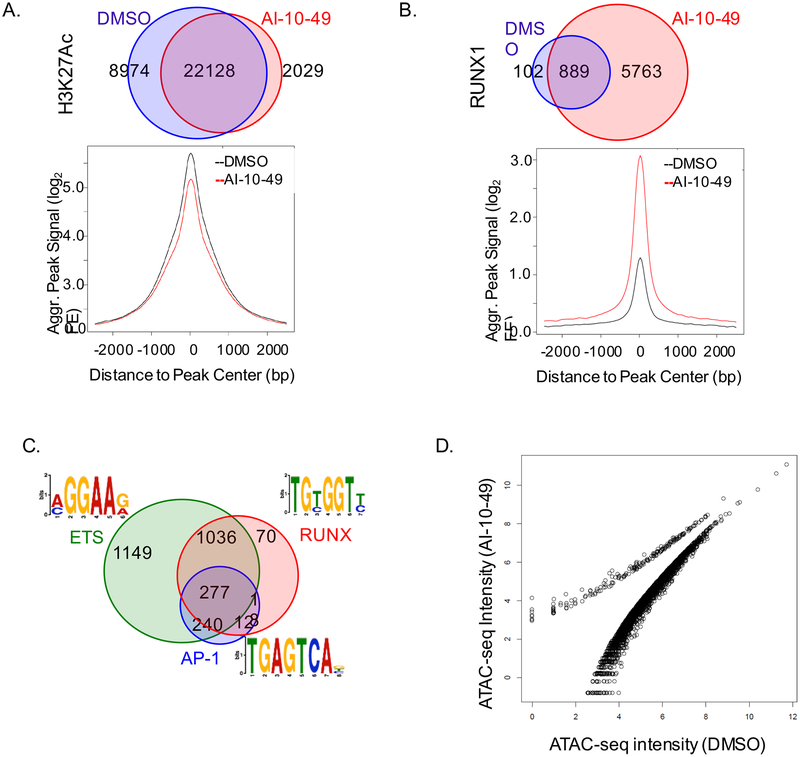
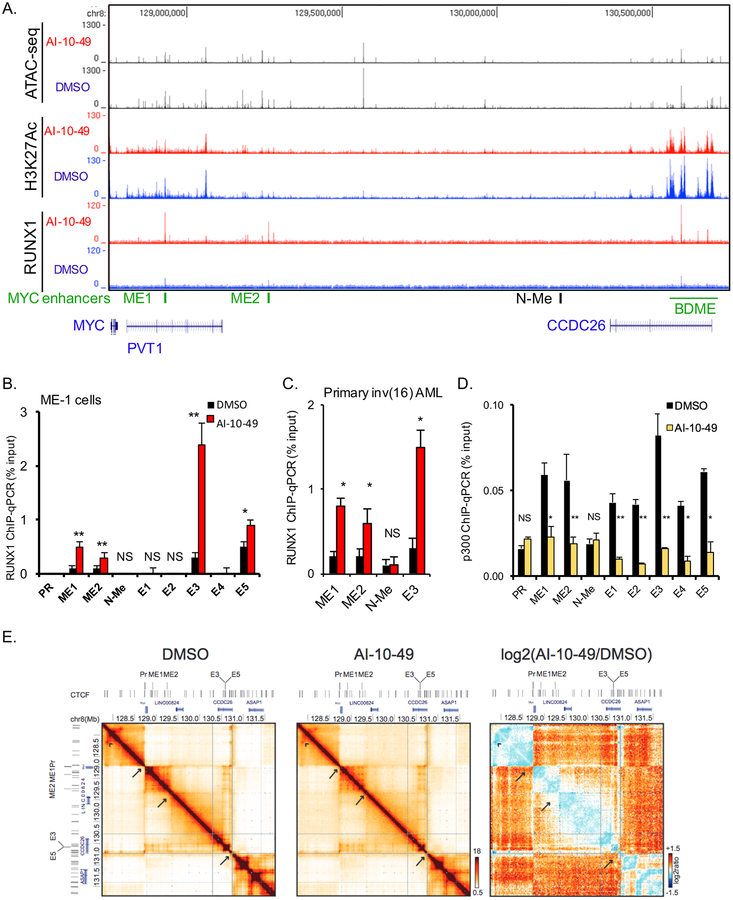
We have shown that AI-10–49 inhibits CBFβ-SMMHC / RUNX1 binding, and increases the occupancy of RUNX1 to selected RUNX target gene promoters (Illendula et al., 2015). To understand the impact of AI-10–49 in RUNX1 association with the chromatin, we conducted chromatin immune-precipitation followed by next generation sequencing (ChIP-seq) in AI-10–49 treated ME-1 cells. Analysis of Histone3-Lysine27 acetylation (H3K27ac) peaks, which mark transcriptionally active regions, indicated a global decrease in positive peaks (Figure 4A), suggesting that AI-10–49 triggers a global reduction in the activity of enhancers and promoters. ChIP-seq analysis for RUNX1 revealed that AI-10–49 treatment induces a 7-fold increase (991 sites for DMSO versus 6,652 for AI-10–49) in RUNX1 binding to target regulatory elements (Figure 4B). Examination of the frequency distribution of RUNX1 binding at the peak center and at the nearest TSS revealed a clear increase in RUNX1 signal intensity (Figures 4B and S4A). Analysis of peak distribution indicated a relative enrichment for promoter regions (Figure S4B). These results demonstrated that AI-10–49-mediated acute release of RUNX1 from CBFβ-SMMHC can trigger RUNX1 re-localization to regulatory regions in inv(16) AML cells. Motif analysis revealed that RUNX1 occupied elements were highly enriched for ETS and AP-1 binding motifs (Figure 4C). Association of RUNX1 with ETS factors has been reported in RUNX1-ETO positive leukemic cells (Ptasinska et al., 2012). AP-1 transcription factors were up-regulated by RUNX1 during megakaryocytic differentiation and recruited to RUNX1-occupied sites lacking AP-1 motifs (Pencovich et al., 2011). These data suggest that RUNX1 may cooperate with ETS factors to regulate gene expression during AI-10–49 treatment in inv(16) AML cells.
Figure 4. Global modification of RUNX1 binding to chromatin in inv(16) AML cells.
(A, B) Venn diagram of peak distribution (top) and aggregated peak signal from peak center (bottom) for H3K27ac (A) and RUNX1 (B) ChIP-seq peaks in ME-1 cells treated with DMSO (black) or AI-10–49 (red). (C) Motif analysis of RUNX1 associated peaks genome wide in AI-10–49 treated ME-1 cells. (D) Scattered plot representing open chromatin peaks by ATAC-seq analysis in DMSO and AI-10–49 treated cells. See also Figure S4.
RUNX1 can regulate chromatin remodeling during myeloid differentiation in mice (Hoogenkamp et al., 2009). Since we observed that AI-10–49 increases RUNX1 association with DNA, we evaluated whether AI-10–49 can modulate chromatin accessibility in ME-1 cells, using Assay for Transposase-Accessible Chromatin with high throughput sequencing (ATAC-seq; (Buenrostro et al., 2013)). Analysis of ATAC-seq data in cell treated with DMSO or AI-10–49 revealed that AI-10–49 induced a general reduction in chromatin accessibility (Figure 4D). These results suggest that AI-10–49-mediated RUNX1 acute release promotes a global reduction in chromatin accessibility.
RUNX1 represses MYC expression through direct binding at three downstream enhancer elements
Active enhancers regulate oncogene expression in cancer, including tumor-type specific distal enhancers that regulate oncogenic MYC expression in solid tumors (Hnisz et al., 2013; Loven et al., 2013; Pomerantz et al., 2009) and leukemia (Fulco et al., 2016; Herranz et al., 2014; Shi et al., 2013). We therefore hypothesized that AI-10–49-mediated RUNX1 function may repress MYC expression by perturbing active distal enhancers. To test this hypothesis, we analyzed a 4 Mb genomic region surrounding the MYC-TSS. Analysis of the 2Mb downstream region revealed no significantly change chromatin accessibility (by ATAC-seq) and a small reduction in H3K27ac mark (Figures 5A and S5A). Analysis of RUNX1 ChIP-seq peaks identified three elements with marked increased RUNX1 peaks in AI-10–49 treated cells. The primary RUNX1 peak was located within the BDME super-enhancer (BRD4-mediated MYC enhancer), 1.7 Mb downstream of the MYC TSS (Figure 5A). The BDME associates with the SWI/SNF proteins BRG1 and BRD4 to regulate MYC expression in myeloid cells and in mixed-lineage leukemia (Shi et al., 2013). The primary RUNX1 peak located at the BDME element 3 (E3) of its 5 conserved elements. The two other RUNX1 peaks, called MYC enhancer 1 and 2 (ME1 and ME2), were located at 0.18 Mb at 0.5 Mb downstream MYC-TSS, respectively. RUNX1 peaks were not detected in the MYC promoter or in the 2Mb region upstream of MYC (Figure S5B). RUNX consensus binding sites (TGYGGT) were identified at the MYC promoter, ME1, ME2, and BDME elements E3 and E5. Analysis of transcription factor binding sites in a 200bp at each side of the RUNX consensus binding site identified ETS sites at E2 and E3 elements (Table S1). We evaluated by ChIP-qPCR changes in RUNX1 peaks at eight sites, including MYC promoter, ME1 and ME2 enhancers and the five BDME elements. In addition, we tested the +1.4Mb N-ME (the T-cell leukemia associated NOTCH-dependent MYC enhancer, also referred as NDME; (Herranz et al., 2014; Yashiro-Ohtani et al., 2014)) as a non-myeloid control enhancer. RUNX1 peaks were significantly increased by AI-10–49 treatment at ME1 (5-fold, p<0.005), ME2 (3-fold, p<0.005), E3 (8-fold, p<0.005) and E5 (1.8-fold, p<0.05), but not at the MYC promoter, N-ME, E1, E2 and E4 (Figure 5B). Enhanced RUNX1 peaks at ME1, ME2 and E3, but not at N-ME, were also observed in human primary inv(16) AML samples (p<0.05; Figure 5C).
Figure 5. RUNX1 increases association with chromatin at three distal MYC enhancers in inv(16) cells.
(A) Analysis of ATAC-seq and K3K27ac and RUNX1 ChIP-seq profiles in a 2 Mb genomic region downstream of MYC. The three enhancer regions (ME1, ME2 and BDME) are depicted in green. (B-D) ChIP-qPCR analysis for RUNX1 in DMSO or AI-10–49 treated cells (B), in DMSO or AI-10–49 treated human primary CD34+ inv(16) AML cells (C), and for p300 in DMSO or AI-10–49 treated ME1 (D). (E) 5C interaction matrices for the MYC locus (−1Mb to +3Mb) in DMSO (left panel) and AI-10–49 (middle panel) treated ME-1 cells. The right panel shows the log2(AI-10–49/DMSO) ratio of the interaction matrices (blue color scheme: higher interaction frequencies in DMSO treated cells; orange color scheme: higher interaction frequencies in AI-10–49 treated cells). Arrows indicate TAD boundaries, arrowhead points to an example of a CTCF-CTCF looping interaction. Significance was calculated using unpaired t-test, *P < 0.05, or **P < 0.005. See also Figures S5 and S6.
In order to further investigate the activity of ME1, ME2 and E3 in inv(16) AML, we analyzed the transcription factor binding profile downstream of MYC, previously reported in ME-1 cells (Mandoli et al., 2014). We confirmed the presence of RUNX1 peaks at the three sites, and determined that RUNX1 co-factors p300, HDAC1, the E-box transcription factor HEB and the ETS transcription factor ERG were also found at ME1, ME2 and E3 (Figure S6). We evaluated if AI-10–49 treatment would alter binding of the transcription activator p300 in the MYC locus, using ChIP-PCR. As shown in Figure 5D, p300 was significantly reduced in ME1, ME2 and in the 5 elements of BDME (p<0.05) but not at N-ME. These data further support the enhancer activity of ME1, ME2 and E3 in inv(16) AML cells in association of RUNX1 complexes, and demonstrates that AI-10–49 treatment inhibits transcription activation factors at these regulatory sites.
The ME1, ME2 and E3 enhancers physically interact with the MYC promoter
A critical determinant of enhancer activity in the regulation of MYC expression is the identification of physical interactions between regulatory elements. Considering that distant MYC enhancers have been reported up and downstream of MYC in a variety of cancers, we analyzed the DNA interactions in a 4 Mb region around the MYC locus, including 1 Mb upstream and 3 Mb downstream of the MYC TSS, utilizing chromosome conformation capture carbon copy (5C; (Dostie et al., 2006)) in ME-1 cells. Chromatin interaction maps for this region showed the presence of a series of Topologically Associating Domains (TADs;(Dixon et al., 2012; Nora et al., 2012)). TADs are contiguous regions with generally elevated interaction frequencies separated by boundaries (Figure 5E, arrows) that often contain CTCF bound sites across which fewer interactions occur. The MYC gene is located near the left boundary of a large TAD that contains several subregions, one that contains ME1 and ME2, and another that contains the BDME encompassing CTCF sites and E3/E5. We analyzed the interactions of the MYC promoter in more detail by plotting the 5C interaction frequency of the promoter with the entire domain in a 4C-style interaction plot (Figure S5C). Interaction frequencies generally decrease as function of genomic distance, as expected. We observed several peaks superimposed on this general trend, representing specific looping interactions (Dekker et al., 2013). We identified a strong interaction of the MYC promoter with ME1, ME2, and BDME. Interestingly, within the superenhancer we also identified three peaks of elevated interactions, the two strongest of which contain CTCF binding sites (Figure S5C, arrowheads). These interactions may involve physical connections between the superenhancer and CTCF sites near the MYC promoter. In addition, a local peak of interactions is observed between the MYC promoter and the E3/E5 enhancer. These results were confirmed by plotting interactions between ME1, ME2 and E3 and the entire domain (Figure S5C), which showed that all these elements interact with each other. We also identified two loci located between ME1 and ME2 that interact with these elements (Figure S5C, arrows), which coincide with sites of open chromatin and H3K27Ac but not with RUNX1 peaks (Figure 5A), suggesting the presence of additional functional elements with local interactions.
In AI-10–49 treated cells the overall conformation of the region was not altered (Figure 5E, left and middle panels). However, we observed quantitative differences. First, when we plotted the ratio of the interactions maps obtained with AI-10–49 treated cells and control cells we observed elevated interactions between loci located in adjacent TADs (Figure 5E, right panel), indicating a weakening of the strength of TAD boundaries. This may be the result of a modest reduction in G1 cells in AI-10–49 treated cultures. Second, the interactions among the MYC promoter, ME1, ME2 and E3 appeared more prominent, while the interactions between the MYC promoter and the other elements of BDME containing sites bound by CTCF, were reduced. These experiments provide critical evidence that the MYC promoter physically interacts with RUNX1-associated enhancers ME1, ME2 and E3 in inv(16) AML cells, and that these enhancers also physically interact with each other.
AI-10–49 induces a switch of SWI/SNF active to PRC repressive complexes at the AML-associated MYC enhancers
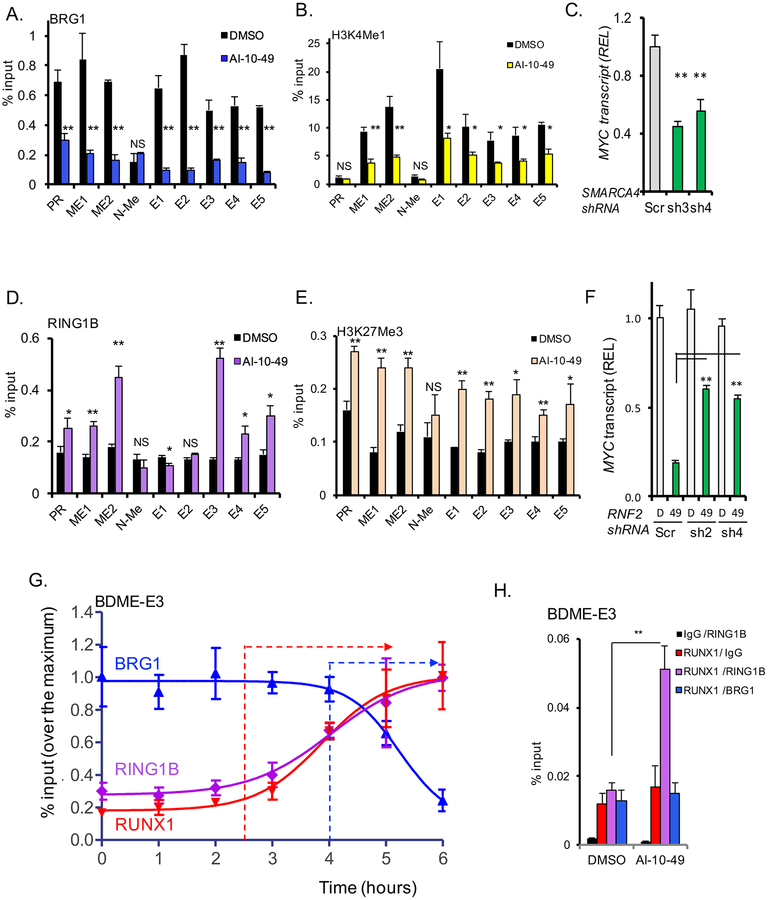
We reasoned that the increase in RUNX1 binding at the MYC enhancers could inhibit the active chromatin complexes. Therefore, we assessed AI-10–49-mediated changes in BRG1 occupancy, a component of the SWI/SNF complex that participates in BDME-mediated MYC expression (Shi et al., 2013). ChIP-qPCR analysis in ME-1 cells revealed that BRG1 is displaced from the MYC promoter, ME1, ME2, and BDME elements (Figure 6A) whereas total BRG1 levels were unchanged (Figure S7A). The active enhancer-specific histone mark histone H3 Lys 4 mono-methylation (H3K4me1) mark (Loven et al., 2013), was significantly reduced at ME1, ME2 and ME3 but not at N-ME and MYC promoter (p<0.05; Figure 6B). In addition, SMARCA4 knockdown (the BRG1 gene) reduced MYC levels (p<0.005; Figures 6C and S7B). These results suggest that inhibition of CBFβ-SMMHC activity leads to the removal of SWI/SNF activating complexes in the MYC distal enhancers.
Figure 6. AI-10–49 induces a switch of activation for repressive marks at RUNX1 associated MYC enhancers.
(A-B) ChIP-qPCR analysis of treated ME-1 cells at promoter (PR) and eight MYC enhancers (ME1, ME2, N-Me, and BDME elements E1 to E5) for BRG1 (A) and H3K4me1 (B). (C) MYC transcript level analysis in ME-1 cells transduced with scramble (Scr) or SMARCA4 shRNAs (sh3 and sh4), estimated by qRT-PCR. (D-E) ChIP-qPCR analysis of treated ME-1 cells at MYC promoter and MYC enhancers for RING1B (D), and H3K27Me3 mark (E). (F) MYC transcript level analysis in ME-1 cells transduced with scramble (Scr) or RNF2 shRNAs (sh2 and sh4) and treated with DMSO (D) or AI-10–49 (49), estimated by qRTPCR. (G) Time course ChIP-qPCR analysis of RUNX1, RING1B and BRG1 binding at E3 in treated ME-1. (H) Quantitative ChIP-re-ChIP of treated ME-1 ChIPed for RUNX1 or IgG, and re-ChIPed for IgG (red), RING1B (violet), or BRG1 (blue), at the E3 enhancer. Results from triplicate experiments are shown; error bars represent the SD. Significance was calculated using unpaired t-test; *P < 0.05, or **P < 0.005. See also Figure S7.
The combined activity of PRC repressive complexes at enhancers can instill gene silencing by tri-methylation of lysine 27 of histone H3 and ubiquitination of histone H2 lysine 119 (Blackledge et al., 2015). Considering that RUNX1 recruits the PRC1 subunit RING1B to the chromatin during hematopoietic differentiation (Yu et al., 2012), we hypothesized that RUNX1 may direct PRC1 binding to chromatin at the MYC locus in inv(16) AML. We evaluated if features associated with repressive chromatin, including RING1B and H3K27me3, were modified by AI-10–49. ChIP-qPCR analysis indicated that RING1B is specifically recruited to ME1, ME2 and E3 but not at N-ME, whereas total RING1B levels were not altered by AI-1049 treatment (Figures 6D and S7A). Similarly, AI-10–49 treatment significantly increased H3K27me3 marks at these enhancers (p<0.005; Figure 6E). In addition, MYC expression was partially rescued by RNF2 knockdown (the RING1B gene), or by pharmacologic inhibition of RING1B activity (PRT4165) in treated inv(16) AML cells (Figures 6F, S7C, and S7D).
To define the dynamics of chromatin complex replacement associated with AI-10–49 treatment in inv(16) AML, we performed a time-course ChIP-qPCR for RUNX1, RING1B and BRG1 at E3, in AI-10–49 treated ME-1 cells (Figure 6G). RUNX1 and RING1B binding showed a similar pattern, increasing at approximately 2.5 hours, and reaching 90% occupancy by 5 hours. Conversely, BRG1 binding was reduced between 4 and 6 hours of treatment. The observed delay between RUNX1/RING1B occupancy and the reduction in BRG1 at E3 supports a complex replacement model. In addition, we evaluated the interaction between RUNX1 and BRG1 or RING1B at E3, utilizing ChIP-re-ChIP technique. This analysis revealed that RUNX1 specifically interacts with RING1B but not with BRG1 at E3, and that this interaction is induced by AI-10–49 treatment (Figure 6H). Furthermore, co-immunoprecipitation analysis of ME-1 nuclear lysates confirmed that RUNX1 can bind to BRG1 and to RING1B (Figure S7E). These results indicate that AI-10–49 induces RUNX1-mediated repression of MYC expression by RUNX1-directed recruitment of PRC-repressive complexes to the MYC enhancers, thereby evicting the SWI/SNF activating complexes. Furthermore, these results suggest that RUNX1 interactions with chromatin complexes may be locus and enhancer specific.
MYC expression and viability of inv(16) AML cells depend on the activity of ME1, ME2 and E3 enhancers.
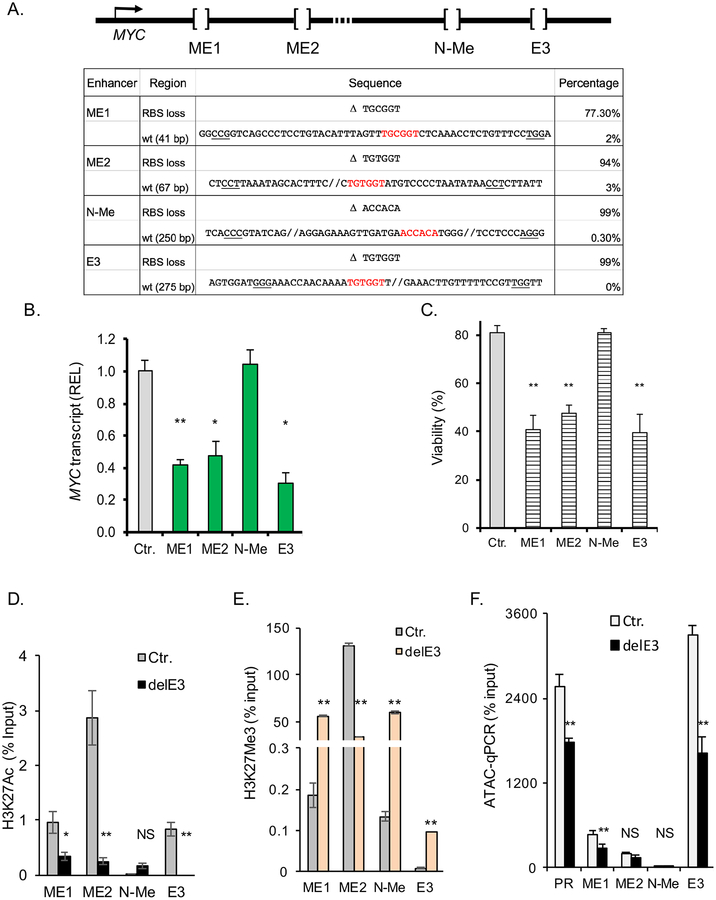
To establish the functional significance of the three enhancers identified in inv(16) AML cells, we evaluated whether single deletion of each enhancer, utilizing CRISPR/Cas9 technology, is sufficient to alter MYC expression. Sequence analysis of ME-1 cells transfected with Cas9 and 2 sgRNAs for each enhancer to produce small deletions surrounding the RUNX1 sites within the enhancers, showed frequent deletions of 41bp (ME1), 67bp (ME2) and 275 bp (within the approximately 2800bp of E3) and the overall frequency of deleted RUNX binding site (RBS) alleles ranged between 77% and 99% (Figures 7A and S8A). Further, these single deletions resulted in 40% to 50% reduction in MYC expression (Figure 7B), and in reduction of cell viability (60% to 70%; Figure 7C).
Figure 7. Deletion of three RUNX1-associated MYC enhancer elements impairs MYC expression and viability of inv(16) AML.
(A) Schematic of CRISPR/Cas9 mediated deletion of MYC enhancer elements (top), and frequency of deletions with RUNX1 binding site (RBS, bold) loss, and of wild type (wt) alleles at each element by sequencing analysis (bottom); PAM sequences used are underlined. (B, C) MYC expression analysis by qRT-PCR (B) and viability (Annexin V-/7AAD-) analysis (C) of ME-1 cells edited for single MYC enhancers, using CRISPR/Cas9. (D, E) Analysis of H3K27Ac (D) and H3K27me3 (E) marks in ME-1 cells edited for E3 enhancer (delE3), at ME1, ME2, N-Me, and E3 enhancers, using CUT&RUN. (F) ATAC-qPCR analysis of ME-1 cells transfected with empty vector/Cas9 (Ctr.) or E3 enhancer sgRNA/Cas9 (delE3) at MYC promoter (PR) and ME1, ME2, N-Me, and E3 enhancers. Results from triplicate experiments are shown; error bars represent the SD. Significance was calculated using unpaired t-test (B, D-F) or Levene’s test (C). *P < 0.05, or **P < 0.005. See also Figure S8.
Finally, to assess the impact of RUNX1-associated enhancer elements in the overall architecture of the locus, we used E3-deleted ME-1 cells, by CRISPR/Cas9, were used to test changes in H3K27Ac and H3K27me3 at ME1, ME2, N-ME, and E3 neighboring region, using CUT&RUN-qPCR. E3 deletion induced a significant reduction of H3K27Ac (p<0.05) and increase of H3K27me3 (P<0.005) marks at ME1 and E3 enhancers (Figures 7D and 7E, respectively). In addition, ATAC-qPCR analysis in the E3-deleted cells resulted in a significant reduction of open chromatin at the MYC promoter, ME1, and E3 neighboring region (p<0.05), although ME2 and N-ME showed no significant changes (Figure 7F).
Taken together, these data demonstrate that ME1, ME2 and E3 function as enhancers to maintain MYC expression levels and the viability of inv(16) AML cells. Furthermore, these results suggest that the architecture of the MYC locus is sensitive to E3 activity in inv(16) AML.
DISCUSSION
In the present study, we have investigated the mechanism underlying the dependency of inv(16) AML cells on the function of the fusion protein CBFβ-SMMHC. We demonstrate that acute release of the RUNX1 transcription factor from CBFβ-SMMHC directs the replacement of chromatin remodeling complexes at three MYC distal enhancer elements and induces apoptosis in inv(16) AML cells. This alteration in chromatin dynamics has therapeutic implications as it remarks that combined therapies using AI-10–49 and inhibitors of factors associated with chromatin complexes may result in excellent treatment in inv(16) AML.
Our study provides new insights on how transcription factor function can determine leukemia maintenance, as it demonstrates that one mechanism by which CBFβ-SMMHC maintains inv(16) AML cell viability is by impeding RUNX1 repression of MYC expression. Interestingly, RUNX1 has not been directly associated with MYC expression in hematopoiesis, suggesting that acute availability of RUNX1 in leukemia may set a different mechanistic scenario than that in hematopoietic differentiation.
The balance between SWI/SNF and PRC epigenetic complexes modulates enhancer activity. SWI/SNF has oncogenic function in AML (Shi et al., 2013) and rapidly evicts PRC1 from chromatin in a BRG1-dependent manner (Stanton et al., 2017). Therefore, the competition between BRG1 and RING1B activities seem to be critical for inv(16) AML survival. Our finding that CBFβ-SMMHC inhibition promotes the disassociation of the BRG1 and association of RING1B provides a mechanistic explanation for the observed phenotype. We determined that RUNX1 does not bind at the MYC promoter but that it binds at three distal enhancers downstream of MYC. Three lines of evidence strengthen the direct role of RUNX1 in MYC repression. First, the 5C assays indicate that the three RUNX1-bound enhancers physically interact with each other and with the MYC promoter in inv(16) AML cells. Second, the deletion of discrete DNA regions that include the RUNX1 binding sites in each of these enhancers is sufficient to reduce MYC expression, alter chromatin marks in the region, and induce apoptosis. Third, ChIP-re ChIP assays indicate that RUNX1 directly binds to RING1B at E3 after treatment. Therefore, acute RUNX1 activity, probably in consort with other co-factors, may direct the eviction of SWI/SNF complexes and binding of PRC repressive complexes.
Considering the enhancer specificity in the regulation of MYC expression among hematologic malignancies (Herranz et al., 2014; Ryan et al., 2015; Shi et al., 2013), our results suggest that different combination of enhancers could regulate MYC in different leukemias. The complexity in the interaction between enhancers and transcription factors was recently reported by the Kelliher group (Choi et al., 2017), showing that RUNX1 regulates MYC expression in T-ALL, at least in part, by modulating NOTCH1 occupancy at the N-ME enhancer. Such specificity may also occur among AML sybtypes. The recent study by the Spitz group (Bahr et al., 2018) extends the findings reported by the Vakoc group highlighting the role of the BDME enhancer in regulating Myc expression during hematopoietic differentiation and in a MLL-AF9 model of AML, using BDME enhancer knock out mice. We propose that RUNX1 may, in concert with other transcription factors, modulate the activity of “AML-subtype common” enhancers (such as BDME) in combination with “AML-subtype specific” enhancers, such as ME1 and ME2 in inv(16) AML. This specificity may be driven by the availability of specific transcription factors with binding sites at the primed enhancers in different leukemias, and their potential interaction with chromatin complexes.
Our ATAC-seq and 5C data indicate that RUNX1-mediated replacement of chromatin complexes and subsequent repression of MYC expression took place without altering the architecture of the locus. This suggests that disruption of enhancer-promoter looping may not be required to repress target gene expression. Instead, changes in transcription factor availability and subsequent association with chromatin complexes at pre-established loops may be sufficient to disrupt target gene expression.
Finally, the synergism found between AI-10–49 and JQ1 in inv(16) AML treatment has important translational implications as it suggests that combined treatment of leukemia-associated transcription factor inhibitors with bromo-domain (or with other components associated with chromatin remodeling complexes) inhibitors may provide effective therapies that bypass resistance and toxicity.
STAR *METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Lucio H. Castilla (Lucio.Castilla@UMassMed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal experiments were performed in accordance with a protocol reviewed and approved by the University of Massachusetts Institutional Animal Care and Use Committee. Mice carrying knock in Cbfb+/MYH11 and Nras+/G12D oncogenic alleles has previously described (Xue et al., 2014) and maintained at the animal facility at University of Massachusetts Medical School with all protocols approved by the University of Massachusetts Medical School Animal Care Committee (Certificate A1266). C57BL/6J mice for transplantation and toxicology experiments were obtained from Taconic Biosciences (Germantown).
Cell Lines
The inv(16) AML ME-1 cells (40 years old male) were cultured in RPMI 1640 with 20 % fetal bovine serum, 25 mM HEPES, 100 U/ml Penicillin, 100 mg/ml Streptomycin and 1μl/ml Plasmocin. The cell lines U937 (37 years old male), K562 (53 years old female), Jurkat (14 years old male), Kasumi-1 (7 years old male), THP-1 (1 year old male), NB4 (23 years old female) and MV4:11 (10 years old male) cells were cultured in RPMI 1640 with 10 % fetal bovine serum, 100 U/ml Penicillin, 100 mg/ml Streptomycin and 1μl/ml Plasmocin. 293T/17 cells were cultured in DMEM with 10 % fetal bovine serum, 100 U/ml Penicillin, 100 mg/ml Streptomycin and 1μl/ml Plasmocin.
Primary Hematopoietic Cell Cultures
Human cord blood samples were collected from the UMASS Memorial Medical Center, and CD34+ cells were isolated using the CD34 Microbead kit (Miltenyi Biotec). The use of the cord blood samples for research purposes was approved by the Ethics Committee of the University of Massachusetts Medical School. Human AML samples were received from AML banks by Martin Carroll (University of Pennsylvania) and Carsten Mueller-Tidow (University Hospital, Heidelberg). Hematopoietic CD34+ cells were isolated from human AML bone marrow samples using the CD34 Microbead kit (Miltenyi Biotec). All patients gave written consent for use of their samples. Personal information from AML and cord blood samples was unavailable as the samples were anonymized. Human cord blood CD34+ cells as well as human primary leukemic cells were cultured in StemSpan SFEM II (STEMCELL Technologies), 100 U/ml Penicillin and 100 mg/ml Streptomycin supplemented with 10 ng/mL human recombinant TPO, 10 ng/mL human recombinant FLT3L (10 ng/mL), 100 ng/mL human recombinant SCF, 10 ng/mL human recombinant IL3, and 20 ng/mL human recombinant IL-6.
Murine bone marrow cells were isolated by crushing femur and tibia of the hind legs of mice. Wild type mouse bone marrow cells were cultured in StemSpan Serum-Free Expansion Medium (SFEM, STEMCELL Technologies), 100 U/ml Penicillin and 100 mg/ml Streptomycin supplemented with 6 ng/ml murine recombinant IL3, 10 ng/ml murine recombinant IL6, and 50 ng/ml murine recombinant SCF. Cbfb+/MYH11 and Nras+/G12D leukemic bone marrow and spleen cells were cultured in StemSpan Serum-Free Expansion Medium (SFEM, STEMCELL Technologies), 100 U/ml Penicillin and 100 mg/ml Streptomycin supplemented with 10 ng/ml murine recombinant IL3 and 50 ng/ml murine recombinant SCF.
METHOD DETAILS
Generation of shRNA constructs
MYC lentiviral shRNA plasmids were obtained from UMMS RNAi Core Facility. The puromycin resistance cassette in pLKO plasmids was replaced by green fluorescent protein (GFP) using standard cloning techniques. RNF2, SMARCA4 and BRD4 shRNAs were generated as follows: first puromycin cassette was replaced from pLKO.1 - TRC cloning vector (Addgene #10878) with GFP using standard cloning techniques followed by cloning specific shRNA oligonucleotides at AgeI and EcoRI site. shRNA oligonucleotides sequences are provided in the Key Resource Table.
Retroviral Production and Transduction
For studying Myc silencing in inv(16) leukemic cells, 293T/17 packaging cells were transfected with 8 μg retroviral constructs co-expressing GFP (Myc (shMyc) or Renilla luciferase (shRen); ref. (Zuber et al., 2011a)) and 4 μg-Ecopac packaging plasmid with Fugene transfection reagent. Retroviral supernatants were collected at 40 and 64 hrs, pooled and concentrated using Retro-X Concentrator (Clonetech), following the manufacturer’s instructions.
Cbfb+/56M/Mx1Cre/Nras+/G12D conditional knock-in mice were treated with polyinosinicpolycytidylic acid (pIpC). Leukemic spleen cells from six to fourteen week-old (both sexes) mice were lineage (Lin) depleted using EasySep® Mouse Hematopoietic Progenitor Cell Enrichment Kit (Stem cell Technologies), following the manufacturer’s instructions. 1–2×106 Lin- cells were transduced twice, 24 hrs apart, by spin infection with shRNA retroviruses. Cells expressing GFP were sorted 24 hrs after the second infection. For the evaluation of knockdown efficiency by immunblotting, lysates were prepared 2 days after sorting of transfected cells.
Lentiviral Production and Transduction
293T/17 packaging cells were transfected with 6 μg of lentiviral constructs co-expressing GFP (pLKO scramble or pLKO MYC shRNAs), 6 μg of packaging plasmid psPAX2 (Addgene plasmid #12260) and 3 μg envelope plasmid pMD2.G (Addgene plasmid #12259) with Fugene transfection reagent. Lentiviral supernatants were collected at 40 and 64 hrs, pooled and concentrated using Lenti-X Concentrator (Clonetech), following the manufacturer’s instructions. 2–3×106 ME-1 cells were transduced twice by spin infection with lentiviruses with a time gap of 24 hrs. Cells expressing GFP were sorted 24 hrs after second infection. Expression of shRNA knockdown protein was tested 2 days after sorting by immunoblotting, or 4 to 5 days after sorting by qRT-PCR.
CRISPR/Cas9 – mediated deletion of the enhancer regions
The sgRNAs specific for 5’ to the region of interest were cloned in pLentiCRISPRv2 (Addgene #52961). sgRNAs corresponding to 3’ to the region of interest were cloned in pDecko-mCherry (Addgene #78534). The puromycin resistance cassette in pLentiCRISPRv2 was replaced by a GFP gene using standard cloning techniques. All sgRNA cloning was done in respective plasmids using standard guide RNA cloning method. Briefly, top and bottom strand guide RNA oligos were phosphorylated using T4 Polynucleotide Kinase (New England BioLabs), annealed and inserted into the vectors at BsmB1 site. Guide RNA’s cloned inside the pLentiCRISPRv2GFP were transfected into 293T/17 cells using the FuGENE® 6 method according to the manufactures instructions. 48 hrs after transfection, genomic DNA was isolated and PCR was carried out to amplify the region of interest. PCR product was reannealed and treated with T7 endonuclease according to the manufactures instruction. The reaction was later resolved on 2% agarose gel and product was analyzed. Oligonucleotide names and sequences (5’−3’) are listed in Table S2. 2–3×106 ME-1 cells were nucleofected with CRISPR/Cas9 plasmids (2μg each) using Nucleofector™ Technology (Lonza Biologics) with the program X-01 and Amaxa® Cell Line Nucleofector® Kit V. Samples were sorted by flow cytometry 24 hrs later. Cells were cultured overnight and dead cells were eliminated by dead cell removal kit (Miltenyi Biotec).
Mi-Seq Analysis
To identify deletion pattern with CRISPR/Cas9 plasmids, transfected ME-1 cells were sorted, and genomic DNA was isolated 48 hrs later. First round of PCR was carried out using the primers to introduce a portion of the Illumina adaptor sequences and products were gel purified. Sequences of primers used for first round are provided in Table S2. A second round of PCR was carried out to introduce the sample indices and resulting products (100–200 bp) were gel purified, combined in a library and sequenced by Illumina Mi-Seq (150bp-paired end).
Cell Viability Assay
Cell viability was estimated using the MTT kit, CellTiter 96® AQueous One Solution (Promega). 20,000 cells/well were seeded in triplicate into 96-well plates. After 24 hrs to 72 hrs 20 μl MTT reagent was added to wells with cells or medium (blank), absorption at 490 nm was measured using SpectraMax M5 plate reader (Molecular Devices).
Inducible expression of MYC using MYC-ER
2×106 ME-1 cells were nucleofected with Nucleofector™ Technology (Lonza Biologics) with 6 μg of ScaI-linearized MYC-ER plasmid (Addgene#19128; (Ricci et al., 2004)) and plated on 6-well plate containing phenol red-free RPMI/10% with charcoal-treated fetal bovine serum. Selection with 3 μg/ml puromycin began 48 hrs after transfection and lasted 48 hrs followed by dead cells removal by dead cell removal kit (Miltenyi Biotec). Cells expressing MYC-ER were treated with ethanol or 500 nM 4-HT for 9 hrs followed by treatment with DMSO or AI-10–49 (1 μM) for 24 hrs and assessed live cells (7AAD- Annexin V-) by Annexin V assay.
Leukemia transplantation studies in mice
In vivo Myc silencing by shRNAs:
For testing role of Myc silencing in inv(16) leukemia survival, Cbfb+/MYH11 ;Nras+/G12D leukemic cells from six to fourteen week-old (both sexes) mice were transduced with shRNAs, 50×103 GFP+ leukemic cells were transplanted into sub-lethally (650 rads) irradiated eight to ten week old wild type C57BL/6 female mice (n = 5 per group). Analyzed GFP+ cells in bone marrow 5 days after transplantation. Analyzed GFP+ cells and ckit+ cells in peripheral blood 28 days after transplantation. Mice were sacrificed after visible characteristics of AML, including reduced motility and grooming activity, hunched back, and pale paws (anemia). Leukemic cells were extracted from spleen and analyzed at time of euthanasia as previously described (Kuo et al., 2006).
In vivo drug efficacy analysis in mice:
For testing efficacy of AI-10–49 and JQ1 in leukemia survival, 1×103 Cbfb+/MYH11 ;Nras+/G12D leukemic cells from six to fourteen week-old (both sexes) mice were transplanted into sub-lethally (650 rads) irradiated eight to ten week old wild type C57BL/6 female mice (n = 5–6 per group). Treatment with vehicle (DMSO), AI-10–49, JQ1 or from day 5 post-transplantation, intraperitoneally. AI-10–49 was administered at 200 mg/kg/day from day 5 to day 14, and JQ1 at 50 mg/kg/day from day 5 to day 25. Mice were sacrificed after visible characteristics of AML, as mentioned above.
Dose response of AI-10–49 and JQ1
To find cooperation of AI-10–49 with JQ1 in inhibiting MYC transcript levels, ME-1 cells were treated with AI-10–49 and/or JQ1 at seven concentrations (0, 0.005, 0.01, 0.03, 0.1, 0.3, and 1μM) for each compound, and MYC transcript levels analyzed by qRT-PCR at 6 hrs. To find cooperation of AI-10–49 with JQ1 in inv(16) cell survival, ME-1 cells were treated with AI-10–49 and/or JQ1 at concentrations shown above, and cell viability analyzed by MTT assay at 72 hrs; human primary inv(16) CD34+ cells were treated with AI-10–49 and/or JQ1 at varying concentrations (JQ1: 0.5 and 01μM; AI-10–49: 2 and 5μM) and cell viability analyzed by Annexin V assay at 48 hrs; and mouse primary inv(16) lineage negative cells [Cbfb+/MYH11 ;Nras+/G12D leukemic cells from six to fourteen week-old (both sexes) mice] were treated with AI-10–49 and/or JQ1 at concentrations 0.5, 1 and 5μM, and cell viability analyzed by MTT assay at 72 hrs.
AI-10–49 and JQ1 Synergy Analysis
The IC50 was estimated using the drc package in R with lower bound set to 0 (Ritz et al., 2015). CIs were calculated using the formula a/A + b/B. Synergisms, additive effect and antagonism of combine treatment assays are defined as CI< 1, CI = 1 and CI > 1 respectively, utilizing the Chou-Talalay Method (Chou, 2010).
Toxicology Studies
Eight to ten week-old wild type C57BL/6 female mice were treated with vehicle (DMSO), AI-10–49, JQ1 or both began via intra peritoneal injection. AI-10–49 was administered at 200 mg/kg/day for 10 days. JQ1 was administered at 50 mg/kg/day for 21 days. Mice were evaluated for signs of toxicity, including grooming, motility, and body weight. 24 hrs after last injection, peripheral blood cells were analyzed by flow cytometry. Mice were then euthanized for spleen and bone marrow analysis.
Flow Cytometry
For flow-cytometry, 2×106 cells were washed twice with 2% FBS in PBS and stained for 20–60 min at 4°C in the dark and analyzed with a BD LSRII flow cytometer. Bone marrow hematopoietic stem and multi-lineage progenitors were analyzed as LSK+: Lin(−) [Gr1(−), Mac1(−), Ter119(−), B220(−), CD3(−)], c-kit(+), Sca1(+); common myeloid progenitors, CMP: Lin(−)Sca1(−)kit(+)CD34(+)CD16/32(−); granulocyte/monocyte progenitor, GMP: Lin(−)Sca1(−)kit(+)CD34(+)CD16/32(+); and megakaryocyte/erythroid progenitors, MEP: Lin(−)Sca1(−)kit(+)CD34(−)CD16/32(−). Flow cytometry analysis was performed using FlowJo Software.
Annexin V Assay
For testing role of MYC silencing in inv(16) cell survival, ME-1 cells and and primary Cbfb+/MYH11 ;Nras+/G12D mouse leukemic cells were transduced with scramble or MYC shRNAs and assessed live cells (7AAD- Annexin V-) by Annexin V assay. Annexin assay was conducted 14 days after viral infection for ME-1 cells or 7 days for primary mouse leukemic cells.
For testing role of MYC enhancer deletion in inv(16) cell survival, ME-1 cells were transfected with empty vector/Cas9 (Ctr.) or MYC enhancer sgRNA/Cas9, sorted cells 24 hrs later and assessed live cells (7AAD- Annexin V-) by Annexin V assay 14 days later.
For detection of apoptotic cell death, the Annexin V Apoptosis Detection Kit I (BD Bioscience) was used as per manufacturer’s instructions. Briefly, cells were centrifuged 2000 rpm for 10 min, resuspended in 100 μl 1× Annexin V binding buffer, added 5 μl Annexin-PE and 10 μl 7AAD and incubated for 15 min at room temperature in the dark followed by adding 500 μl 1× Annexin binding buffer. Cell viability was determined as the percent of 7-AAD negative/Annexin V negative cells with a BD LSRII flow cytometer.
Quantitative RT-PCR Analysis
To evaluate MYC transcriptional regulation by AI-10–49 in inv(16) cells, ME-1 cells were treated with 1 μM AI-10–49 for 6 hrs. mRNA was isolated from three independent experiments and conducted qRT-PCR for MYC.
To evaluate MYC transcriptional regulation by AI-10–49 in primary cells, lineage negative mouse bone marrow cells from eight to ten week-old wild type C57BL/6 female mice, lineage negative mouse Cbfb+/MYH11 leukemic cells from sixteen to twenty four week-old wild type (both sexes) mice, human cord blood CD34+ cells and human primary inv(16) leukemic CD34+ cells were treated with 5 μM AI-10–49 for 24 hrs. mRNA was isolated and conducted qRT-PCR for MYC.
To find MYC transcriptional regulation during BRD4 silencing, ME-1 cells were transduced with scramble (Scr) or BRD4 shRNAs (sh1 and sh2), mRNA was isolated from three independent experiments 7 days after transduction and MYC expression was estimated by qRT-PCR analysis.
To find MYC transcriptional regulation during SMARCA4 silencing, ME-1 cells were transduced with scramble (Scr) or SMARCA4 shRNAs (Sh3 and sh4), mRNA was isolated from three independent experiments 7 days after transduction and MYC expression was estimated by qRT-PCR analysis.
To find rescue of AI-10–49 mediated inhibition of MYC transcription during RNF2 silencing, ME-1 cells were transduced with scramble (Scr) or RNF2 shRNAs (sh2 and sh4). 7 days after transduction cells were treated with DMSO (D) or with 0.6 μM AI-10–49 (49) for 6 hrs. mRNA was isolated from three independent experiments and MYC expression was estimated by qRTPCR analysis.
For testing role of MYC enhancer deletion in MYC transcriptional regulation in inv(16) cells, ME-1 cells were transfected with empty vector/Cas9 (Ctr.) or MYC enhancer sgRNA/Cas9, sorted cells 24 hrs later and isolated mRNA from three independent experiments 24 hrs later. MYC expression was estimated by qRT-PCR analysis.
mRNA was isolated with a PureLink® RNA Mini Kit (life technologies) and cDNA synthesis was performed with a SuperScript III kit (Life Technologies), per the manufacturers’ instructions. Quantitative PCR analysis was conducted on an Applied Biosystems StepOnePlus System with Power SYBR® Green PCR Master Mix (Applied Biosystems). Expression levels were determined with the ΔCt method and normalized to ACTB mRNA. Sequences of primers are provided in Table S2.
Transient transcriptome PCR
Transient transcriptome PCR was carried out using a modified transient transcriptome sequencing (TT-Seq) protocol (Schwalb et al., 2016). ME-1 cells were treated with DMSO/AI-10–49 (1 μM) for 6 hr and washed with 1× PBS. Cells were further incubated for 5 min at 37°C/5% CO2 with 500 μM 4 sU-containing media (0.5×106 cells/ml). Total RNA was isolated from PureLink® RNA Mini Kit and briefly sonicated with a bioruptor (Diagenode) for 30 secs Thiol-specific biotinylation of RNA samples were carried out in biotinylation buffer (10 mM Tris pH 7.4, 1 mM EDTA in water) at 37°C for 2 hrs with rotation (200 μl of EZ-Link™ HPDP-Biotin of 1mg/mL stock was used for 100 μg of RNA). RNA was further isolated using chloroform isolation method followed by NaCl, isopropanol precipitation and ethanol wash. Dynabeads™ MyOne™ Streptavidin C1 beads were processed according to the manufactures instruction with 0.1N NaOH, 50mM NaCl and re-suspended in binding buffer containing 10 mM Tris-Cl 7.4, 300 mM NaCl and 0.1% Triton X-100. Further, RNA samples were incubated with Dynabeads™ MyOne™ Streptavidin C1 beads for 20 min at room temperature. Samples were washed two times each with high salt buffer (50 mM Tris-Cl 7.4, 2M NaCl, 0.5% Triton X-100),10 mL H2O), binding buffer and low salt buffer (5 mM Tris-Cl 7.4, 0.1% Triton X-100). 4sU labelled RNA was eluted using 100 mM DTT at 65°C for 5 min and RNA was isolated using standard phenol-chloroform extraction method. cDNA was synthesized and qPCR was carried out as mentioned before. Expression levels were determined with the ΔCt method and normalized to GAPDH.
RNA Sequencing
ME-1 cells were treated with DMSO or AI-10–49 (1 μM) for 6 hrs. mRNA was isolated from three independent experiments with a PureLink® RNA Mini Kit. RNA concentration was quantified with a NanoDrop spectrophotometer (Thermo Scientific). RNA integrity was evaluated with a 2100 Bioanalyzer with an RNA 6000 kit (Agilent Technologies). Libraries were prepared with a TruSeq RNA library preparation kit (Illumina). Libraries were quantified by qPCR, normalized and pooled before sequencing with paired-end 90-bp reads on an Illumina HiSeq2000 in triplicate.
RNA-seq data analysis:
Raw reads from RNA-seq experiment were assessed for their quality using fastqc, followed by alignment to the reference human genome (hg19) using tophat v2.0.9, bowtie2/2.1.0 (Trapnell et al., 2009) with the default setting except the following parameters: --b2-very-sensitive --mate-inner-dist 160 --mate-std-dev 80 --no-coverage-search --transcriptome-index=hg19_knownGene_transcriptome_data. Differential gene expression analysis was performed by cufflinks v2.2.0 (Trapnell et al., 2012). Genes with a false discovery rate below 0.05 and a fold change greater than two were considered to be significantly differentially expressed and used for subsequent analysis.
Chromatin Immunoprecipitation (ChIP) qPCR
ME-1 cells were treated with DMSO or AI-10–49 (1 μM) for 6 hrs. Cross-linking of proteins to DNA was accomplished by the addition of 1 % formaldehyde for 10 min to cultured cells at room temperature. After neutralization with glycine, cells were lysed in lysis buffer with protease inhibitors and samples were sonicated to an average DNA length of 200–400 bp with a bioruptor (Diagenode). After sonication, the chromatin was immunoprecipitated with 5–10 μg of antibody of interest at 4 °C overnight. Antibody bound complexes were isolated with Dynabeads (Life Technologies). DNA was purified using phenol-chloroform isoamyl-alcohol method. Immunoprecipitated DNA was analyzed by qPCR on a StepOnePlus System (Applied Biosystems) with Power SYBR® Green PCR Master Mix and calculated as % of input. Details of the antibodies used and sequences of primers are provided in the Key Resource Table and Table S2.
For ChIP qPCR in human primary AML sample with inv(16), CD34+ cells were enriched using CD34 MicroBead Kit (Miltenyi Biotec) and cultured overnight followed by dead cell removal by dead cell removal kit (Miltenyi Biotec). Cells were treated with DMSO/ AI-10–49 (5μM) for 8 hrs followed by ChIP procedure mentioned above.
For ChIP-re-ChIP, 1×107 ME-1 cells were used per immunoprecipitation. ME-1 cells were treated with DMSO or AI-10–49 (1 μM) for 6 hrs. After first ChIP and washing as described above, the immune complexes were eluted in 100 μl (30 min at 37°C), elutes from five ChIPs were combined, and mix was diluted 10× using ChIP lysis buffer without SDS. After addition of the second-round ChIP antibody, the samples were incubated overnight at 4°C and treated as described above.
Chromatin immunoprecipitation followed by sequencing (ChIP-seq)
ME-1 cells were treated with DMSO or AI-10–49 (1 μM) for 6 hrs. Chromatin immunoprecipitation was conducted as mentioned above. DNA concentration was quantified with a NanoDrop spectrophotometer (Thermo Fisher Scientific). DNA integrity was evaluated with a 2100 Bioanalyzer (Agilent Technologies). Libraries were prepared with in house ChIP-Seq Library preparation kit. Libraries were quantified by qPCR, normalized and pooled before sequencing with single-end 50-bp reads on an Illumina HiSeq4000).
ChIP-seq data analysis:
ChIP-seq reads were aligned to the human genome (hg19) with Bowtie2 v2.1.0 (Langmead and Salzberg, 2012) with the standard default settings. Only the reads with mapping quality greater than 20 were kept, and the duplicated reads were removed using picard tools v1.96 (https://broadinstitute.github.io/picard/). Peak calling was performed with MACS2 v2.1.0 (Zhang et al., 2008) with default parameters. Input was used as a control for peak-calling. The narrowPeak files were generated by macs2 with a q-value threshold of 0.01, and the bigwig files were generated with the signal as fold enrichment by macs2 following the procedure at https://github.com/taoliu/MACS/wiki/Build-Signal-Track.
Cleavage Under Targets and Release Using Nuclease (CUT&RUN)
CUT&RUN was performed according to the protocol reported by the Henikoff laboratory [http://blocks.fhcrc.org/steveh/papers/CUT&RUN_protocol.htm] (Skene and Henikoff, 2015), with modifications. ME-1 cells were nucleofected with CRISPR/Cas9 plasmids, sorted by flow cytometry 24 hrs later and cultured overnight. 2×104 cells were washed in cold PBS and resuspended in 1 mL Nuclei isolation buffer, NE buffer (20 mM HEPES-KOH pH 7.9, 10 mM KCl, 0.5 mM Spermidine, 0.1% Triton X-100, 20% Glycerol, 1× EDTA-free protease inhibitor) and incubated on ice for 10 min. After spinning down the cells at 600g, cells were resuspended in 600 μl NE buffer. 100 μl Bio-Mag Plus Concanavalin A coated beads (Polysciences) were added to nuclei and rotated at room temperature for 10 min. Supernatant was removed by placing beads and nuclei on magnetic stand and resuspended in 1 mL blocking buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 0.1 % BSA, 2mM EDTA, 1× protease inhibitor cocktail). Nuclei were incubated in blocking buffer for 5 min and washed in wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, 0.1 % BSA, 1× EDTA free protease inhibitor cocktail). Wash buffer was removed on magnetic stand and nuclei were resuspended in 250 μl wash buffer. Primary antibody (H3K27Ac, H3K27Me3 or IgG) was added in 1:100 ratio and incubated overnight. Next morning nuclei were washed in wash buffer twice and resuspend in 250 μl wash buffer. Protein A-MNase (provided by Henikoff lab) (1:400 in 250 μl wash buffer) was added to nuclei and incubated for 1 hr at 4°C, washed twice in 1 ml wash buffer and resuspended in 150 μl wash buffer. 7.5 μl nuclei solution (5% of total volume) was taken as input and frozen. Rest of the nuclei solution was equilibrated to 0°C in metal block and added 3 μl of 100 mM CaCl2. The reaction was stopped after 30 min by adding 150 μl 2×STOP solution (200 mM NaCl, 20 mM EDTA, 4 mM EGTA, 50 μg/ml RNase A, 40 μg/ml glycogen).
Reaction tubes along with input samples were incubated at 37 °C water bath for 20 min to digest RNA and release CUT&RUN fragments from the insoluble nuclear chromatin. Nuclei were centrifuged at 16000g for 5 min at 4°C and supernatant was collected. 3 μl 10% SDS (to 0.1%), and 2.5 μl Proteinase K (20 mg/ml) were added to the supernatant and incubated at 70 °C for 10 min. 300 μl buffered phenol-chloroform-isoamyl solution (25:24:1) was added followed by vortexing and the aqueous phase was collected by centrifugation. 2 μl glycogen (2 mg/ml) and 750 μl 100% ethanol was added to the aqueous phase, chilled on ice and centrifuged at full speed for 5 min. The pellet was washed again in 100% ethanol and air dried. Finally, the pellet with genomic DNA was dissolved in 30 μl 0.1× TE8 (1 mM Tris, 0.1 mM EDTA pH 8.0).
DNA library preparation:
Sequencing libraries were prepared from the genomic DNA fragments as described (Henikoff et al., 2011; Kasinathan et al., 2014), using the KAPA DNA polymerase library preparation kit. The following steps were performed for library preparation:
-
(A)
End repair, phosphorylation, adenylation: Extracted genomic DNA fragments were end-repaired, phosphorylated adenylated using Taq DNA polymerase, T4 DNA polymerase and T4 PNK. Reaction conditions were as follows: 12 °C 15 min, 37 °C 15 min, 72 °C 20 min, 4 °C on hold.
-
(B)
Adapter ligation: In 50 μl of end repaired/adenylated DNA, 5 μl of 1.5 μM adapter were added. 55 μl of 2× rapid DNA ligase buffer along with 5 μl Enzymatics DNA ligase was added to the reactions, mixed and incubated at 20°C for 15 min.
-
(C)
Ligation cleanup: Ligation reaction was cleaned up using Agencourt AMPure beads (Beckman Coulter), by magnetic separation and purified DNA was eluted in Tris-HCl.
-
(D)
Library enrichment: Purified DNA was used for library enrichment. 30 μl of KAPA PCR mix was added to the 20 μl of purified DNA and mixed by pipetting followed by 14 cycles of PCR. Reaction conditions were as follows: 98°C 45 sec; 98°C 15 sec, 60°C 10 sec 13 Cycles; 72°C 1 min; 4°C hold.
-
(E)
PCR cleanup: PCR reaction was cleaned up using Agencourt AMPure beads (Beckman Coulter) (1.1× vol) and 80% ethanol. Adapter ligated DNA was eluted in 30 μL of 10 Mm Tris-HCl pH=8.
Quality control was performed for the libraries by DNA fragment analysis. Enriched libraries ran on the bioanalyzer at around 300 base pair. Immunoprecipitated DNA was analyzed by qPCR on a StepOnePlus System (Applied Biosystems) with Power SYBR® Green PCR Master Mix and calculated as % of input. Sequences of primers are provided in Table S2.
Assay for Transposase-Accessible Chromatin with sequencing (ATAC-seq)
To profile for accessible chromatin regions, we used ATAC-seq as described elsewhere (Buenrostro et al., 2015) with the following modifications: ME-1 cells (50,000) were treated with DMSO or AI-10–49 (1 μM) for 6 hrs followed by washing once with 1× PBS by centrifugation using 5 min at 500g and 4°C with low acceleration and brake settings. Cell pellets were re-suspended in 50μl of cold lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and nuclei were pelleted by centrifugation for 10 min at 500g, 4C. Supernatant was discarded and nuclei were re-suspended in 25 μl reaction buffer containing 2.5 μl of Tn5 transposase and 12.5 μl of TD buffer (Nextera Sample preparation kit, Illumina). The reaction was incubated at 37°C for 45 min. Immediately following transposition, tagmented DNA was purified using a Qiagen MiniElute PCR Purification Kit. For library amplification, two sequential PCRs were conducted with indexing primers included in the Nextera Index kit and NEBNext High-Fidelity 2× PCR Master Mix. After the first PCR, the libraries were enriched for fragments less than 600 by using Agencourt AMPure XP 5 mL Kit (Beckman Coulter). A second PCR was conducted with the similar settings followed by size enrichment by Agencourt AMPure XP 5 mL Kit. DNA was eluted and concentration was measured with a Qubit fluorometer (Life Technologies) and library quality evaluated using 2100 Bioanalyzer (Agilent Technologies). The libraries where sequences in 100 bp paired-end on an Illumina HiSeq2000.
ATAC-seq data analysis:
The preprocessing of ATAC-seq data was followed as reported (Buenrostro et al., 2013). Briefly, the adaptors were removed using cutadapt program v 1.3, and reads were mapped onto the human genome hg19 assembly using Bowtie2 (Langmead and Salzberg, 2012). The standard default settings were modified to allow mapped paired-end fragments up to 2 kb. Only the reads with mapping quality greater than 20 were kept, and the duplicated reads were removed using picard tools v1.96 (https://broadinstitute.github.io/picard/), the reads from mitochondria were also removed. To visualize the mapped reads, the bigwig files were generated using deepTools2. Quality assessment of ATAC-seq data was performed using ATACseqQC (Ou et al., 2017). Reads enrichment were called by MACS2 v2.1.0 (Zhang et al., 2008) with default parameters using the reads with insert size less than 100 bp as nucleosome free regions.
ATAC-qPCR
For ATAC-qPCR, ME-1 cells were nucleofected with CRISPR/Cas9 plasmids with sgRNAs that delete a 275bp segment including the RUNX1 binding site, sorted by flow cytometry 24 hrs later and cultured overnight. ATAC libraries were prepared from three independent biological replicates as described above. 5% non-transposed genomic DNA was taken as input. Enrichment of open chromatin in the ATAC samples over input was performed by qPCR using primers for each regulatory element, and a region centromeric from the deleted E3 segment. Sequences of primers are provided in Table S2.
Co-immunoprecipitation
For co-immunoprecipitation assays, nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents. Antibodies were incubated with Dynabeads Protein A for three hours and crosslinked to beads by Dimethyl adipimidate. After washing beads with IP Buffer (50mM Tris, pH 7.4, 150mM NaCl, 1mM EDTA, 0.1% Tx-100, 1mM DTT and Protease inhibitor), resuspended beads in IP buffer, added 1mg nuclear extract and incubated in a rotator at 4C overnight. Beads were washed with IP buffer and protein complexes were eluted from beads by 50 mM glycine (pH 2.0) and analyzed by western blot. The membrane was probed with corresponding antibodies and detected using Clean-Blot IP Detection Reagent.
Immunoblotting
To evaluate MYC protein levels with AI-10–49 treatment in inv(16) cells, ME-1 cells were treated with 1 μM AI-10–49 for 6 hrs. ME-1 cells were lysed in modified RIPA buffer (50 mM Tris pH7.5, 150 mM NaCl, 1 % NP40, 0.25 % sodium deoxycholate and 1 mM EDTA) with phosphatase inhibitor (Sigma) and protease inhibitors (Millipore) for 15 min in ice followed by centrifugation. Protein concentrations were determined with the Biorad Proetin Assay (Biorad). Proteins were separated on precast 7.5–12% Mini Protean TGX gels at 60–80 V using the Mini Protean electrophoresis system and were blotted onto PVDF membrane at 100V for 90 min in a Mini Trans-Blot Cell. All antibodies were used as recommended by the manufacturer and mentioned in Key Resource Table. Relative band intensities were quantified using ImageJ software.
Carbon Copy Chromosome Conformation Capture (5C)
Experimental Design:
ME-1 cells were treated with DMSO or AI-10–49 (1 μM) for 6 hrs. The 3C libraries were generated as described before (Hnisz et al., 2016; Naumova et al., 2012), with the following modifications: 1) After HindIII digestion no SDS was added for restriction enzyme inactivation. 2) The ligation volume as 1.2 ml for 5×106 cells and a total of 3×107 ME-1 cells were used per 5C library preparation. 5C was carried out as previously described (Dostie et al., 2006; Ferraiuolo et al., 2012; Lajoie et al., 2009), with one modification: gel purification after adapter ligation was replaced by an Ampure step to remove unligated DNA. 5C primers were designed for a 3.98 Mb region (chr8: 127,753,661 – 131,737,521) around the MYC locus. 5C primers were designed at HindIII restriction sites using publicly available 5C primer design tools published previously (Lajoie et al., 2009). Primers were designed according to a double alternating scheme exactly as described before (Hnisz et al., 2016). We designed two primers for each HindIII fragment: one primer designed on the 5’ end of the fragment, and one on the 3’ end. For a fragment we either designed a right 5’ forward (FOR) and a left 3’ reverse (LREV) primer, or a right 5’ REV and a left 3’ LFOR primer. These two primer designs alternate along consecutive fragments throughout the entire region of interest. This design allows interrogation of all pairwise interactions among all fragments, which is not possible with a more simple alternating design used previously (Lajoie et al., 2009).
Primer settings:
U-BLAST, 3; S-BLAST, 50, 15-MER, 800, MIN_FSIZE, 100; MAX_FSIZE, 50,000; OPT_TM, 65; OPT_PSIZE, 40. The 5C primer tails were: FOR/LFOR: T7 sequence 5’-TAATACGACTCACTATAGCC-3’; REV/LREV: T3 sequence 5’-TCCCTTTAGTGAGGGTTAATA-3’. The full-length of all FOR/LFOR primers was 60 bases; the length of all REV/LREV was 61 bases. In total, we designed 359 forward (FOR), 367 left forward (LFOR), 367 reverse (REV) and 367 left reverse (LREV) primers that combined interrogate 532,158 long-range chromatin interactions.
Generation of 5C libraries:
A 5C multiplex primer annealing reaction was performed overnight at 50°C. Pairs of annealed 5C primers were ligated at the same temperature using Taq-DNA ligase for 1hr. Seven ligation reactions were performed to generate 5C libraries, except for the second biological replicate for AI-10–49-treated cells, where 14 ligation reactions were performed. Each ligation contained 6×105 genome copies, except for the second biological replicate for AI-10–49-treated cells, which contained 4 ×105 genome copies. Each primer was added to a final amount of 0.325 fmole. Ligated 5C primer pairs, which represent a specific ligation junction in the 3C library and thus a long-range interaction between the two corresponding loci, were then amplified using 20 cycles of PCR with T7 and T3R universal tail primers that recognize the common tails of the 5C forward and reverse primers. Four separate amplification reactions were carried out for each annealing reaction described above and all the PCR products of each library were pooled together. This pool constitutes the 5C library. The libraries were concentrated using Amicon Ultra Centrifugal filters - 0.5ml 30K (Millipore) and purified with Qiaquick PCR purification kit.
5C read mapping:
5C libraries were sequenced on an Illumina HiSeq 4000 instrument, reads were mapped (with Novoalign mapping algorithm V3.02.00) and 5C interactions assembled exactly as described before (Lajoie et al., 2009; Sanyal et al., 2012). Data from the two biological replicates were pooled, producing a single interaction map for DMSO treated, and AI-10–49 treated cells. The summary statistics and the read depth of each 5C libraries can be found in in Table S3.
5C filtering and analysis:
5C matrices were processed using previously described methods (Lajoie et al., 2009; Sanyal et al., 2012). Briefly, first we removed 5C interactions that represent self-ligated restriction fragments. Second, in 5C PCR can lead to over amplification of individual pair-wise interactions (outliers). To remove these, we first calculated the average interaction frequency and standard deviation of all pair-wise interactions as a function of their genomic distance using LOWESS smoothing, as described in Sanyal et al. (Sanyal et al., 2012). This average value represents the expected interaction frequency for a pair of loci. We then calculated the observed/expected ratio for each interaction and expressed this as a z-score ((observed – expected)/standard deviation; (Sanyal et al., 2012)). Outliers were then defined as those interactions with a z-score greater than 20 in each dataset. We then took the union of all outliers identified in the four 5C datasets, and removed these interactions from all four datasets. Third, some primers strongly over or underperform leading to strongly enriched or depleted rows of interactions. To identify these primers we calculated to sum of all interactions detected with each of the 5C primers. We then defined over- and underperforming primers as those with a sum that is outside the 1.5 times the interquartile range (of the distribution of all row/col sums). We then took the union of all flagged primers across the four 5C matrices, and removed these from all four datasets. Fourth, we scaled the four matrices to the same number of total reads (5 ×107). Fifth, the matrices were balanced according to the ICE method so that the sum of each row and each column is equal (Imakaev et al., 2012). Sixth, data were binned at 20Kb (median) with a sliding window with 2.5 Kb steps, or at 15Kb (median) with a sliding window with 2.5 Kb steps when data were plotted as interaction profiles of single loci (4C-style plots). Seventh, matrices were balanced again after binning.
4C-style plots:
To display the interaction profiles (4C-style plots) of selected loci we extract rows for corresponding bins that overlap the Myc Promoter, ME1- ME2- and E3 enhancers from the 15 Kb binned 5C interaction matrix. We also calculated and plotted the LOWESS smoothed average plus and minus 1 standard deviation of 5C signal as a function of genomic distance (representing the expected 5C signal).
Gene Set Enrichment Analysis (GSEA)
Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) was used to determine the statistically significant molecular signatures with the AI-10–49 treatment. The input data for the GSEA were a complete table of genes ranked by the test statistics from the cuffdiff results and a catalog of functional gene sets from Molecular Signature Database (Molecular Signatures Database v6.0, www.broad.mit.edu/gsea/msigdb/msigdb_index.html). Default parameters were used. Gene sets with false discovery rate less than 0.25 were included.
Transcription Factor Binding Site Analysis
The search for transcription factor binding sites at enhancers was performed using 200bp of genomic sequence at each side of the RUNX1 binding site within ME1, ME2 and E3 (Table S1). The analysis was performed using Software Package for Transcription Factor Binding Site (TFBS) Analysis, from TFBSTools, an R/Bioconductor package (http://bioconductor.org/packages/release/bioc/html/TFBSTools.html).
Integrative analysis of high-throughput data
All downstream analyses were carried out using R v3.4.1, a system for statistical computation and graphics (Ihaka and Gentleman, 1996). Exploratory analyses and differential gene expression analysis were carried out with CummeRbund package v2.7.4 (http://compbio.mit.edu/cummeRbund/). The enriched regions/peaks of CHIP-seq and ATAC-seq were annotated to the hg19 genomic features with ChIPpeakAnno v3.9 (Zhu, 2013; Zhu et al., 2010). The heatmaps and density plots were also generated with ChIPpeakAnno. The Bioconductor DiffBind package v2.4.8 (Ross-Innes et al., 2012) was used to quantitate and identify genomic regions with differential binding by ATAC-seq or ChIP-seq reads between control and AI-10–49 treated samples. To identify motifs associated with RUNX1 binding sites, the weight matrices of different consensus binding sites for various transcription factors were obtained from Jaspar database (http://jaspar.genereg.net/). The overlapping of peaks with RUNX1 binding motif and those with other binding motifs were analyzed by ChIPpeakAnno. To visualize all genomic data, narrowPeak or bigwig files were uploaded to the UCSC Genome Browser. All genomic data are accessible at the Gene Expression Omnibus, and the accession numbers can be found in the Key Resource Table.
Quantification and Statistical Analysis
The statistical analyses were performed using R. For mouse leukemia survival analysis, the leukemia latency and P values were estimated using Survival package in R. P-value between groups was calculated using log-rank test. Relative cell viability, measured as the proportion of viable cells, was first arcsine-transformed to homogenize the variance. Levene’s test shows that the assumption of homogeneity of variances is met. One-way ANOVA was performed followed by predetermined contrasts for Figures 2A–2E, 3E, 7C, and S2D. Two-way ANOVA was performed followed by Tukey’s honest significance test for Figure 3D. Two-way ANOVA was performed to test the main effect and the interaction of two drug treatment (JQ1 and AI-10–49) on c-kit+ cells (Figure S3D). To compare the response curves (Figures 3B and 3C), an indicator variable was created to distinguish the two-drug treatment and one of the single-drug treatments. Two-way ANOVA was performed to compare a single-drug treatment and the two-drug treatment.
Data and Software Availability
The accession numbers for all genomic data for RNA-seq, ChIP-seq, ATAC-seq, and 5C reported in this paper are GEO: GSE101788, GSE101789, GSE101790, and GSE109764. These files are also accessible under the SuperSeries accession number GEO: GSE101791.”.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Antibodies | ||
| CD117 APC | BD Biosciences | Cat#553356 |
| Ly-6A/E APC-Cy7 | BD Biosciences | Cat#560654 |
| CD34 FITC | BD Biosciences | Cat#553733 |
| CD16/CD32 PE-Cy | eBiosciences | Cat#25-0161 |
| Ly-6G and Ly-6C Biotin | BD Biosciences | Cat#553124 |
| Cd11b Biotin | BD Biosciences | Cat#553309 |
| CD45R/B220 Biotin | BD Biosciences | Cat#553086 |
| CD3e Biotin | eBiosciences | Cat#13-0037-82 |
| Ter119 Biotin | BD Biosciences | Cat#553672 |
| Streptavidin eFluor® 450 | eBiosciences | Cat#48-4317-82 |
| CD11b PE | BD Biosciences | Cat#555388 |
| CD15 APC | BD Biosciences | Cat#561716 |
| CD117 APC | BD Biosciences | Cat#553356 |
| RUNX1 polyclonal | Abcam | Cat#ab23980 |
| H3K27ac polyclonal | Abcam | Cat#ab4729 |
| H3K4me1 polyclonal | Abcam | Cat#ab8895 |
| H3k27me3 monoclonal | Abcam | Cat#ab6002 |
| BRG1 monoclonal | EPITOMICS | Cat#2822-1 |
| RING1B polyclonal | Abcam | Cat#ab3832 |
| MYC polyclonal | Santa Cruz | Cat#N-262 |
| p300 polyclonal | BETHYL | Cat#A300-358A |
| GAPDH monoclonal | Cell Signaling | Cat#3683 |
| Biological Samples | ||
| human AML samples | UPENN | N/A |
| human AML samples | University Hospital Heidelberg | N/A |
| human cord blood | UMASS | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AI-10-49 | John Bushweller | N/A |
| JQ1 | James Bradner | N/A |
| JQ1 | ApexBio | Cat#A1910 |
| PRT 4165 | Tocris Bioscience | Cat#5047 |
| pIpC | GE healthcare | Cat#27-4732-01 |
| 4-Hydroxytamoxifen | Sigma | Cat#H7904 |
| Recombinant Human IL-3 | Peprotech | Cat#200-03 |
| Recombinant Human IL-6 | Peprotech | Cat#200-06 |
| Recombinant Human FLT3L | Peprotech | Cat#300-19 |
| Recombinant Human SCF | Peprotech | Cat#300-07 |
| Recombinant Human TPO | Peprotech | Cat#300-18 |
| Recombinant Murine IL-3 | Peprotech | Cat#213-13 |
| Recombinant Murine IL-6 | Peprotech | Cat#216-16 |
| Recombinant Murine SCF | Peprotech | Cat#250-03 |
| Critical Commercial Assays | ||
| PureLink® RNA Mini Kit | Life Technologies | Cat#12183018A |
| SUPERSCRIPT III | Thermo Fisher Scientific | Cat#18080-044 |
| Power SYBR® Green PCR Master Mix | Applied Biosystems | Cat#4367659 |
| Annexin V Apoptosis Detection Kit | BD Biosciences | Cat#559763 |
| NE-PER™ Nuclear and Cytoplasmic Extraction Reagents | Thermo Fisher Scientific | Cat#78833 |
| CellTiter 96® AQueous One Solution Cell Proliferation Assay | Promega | Cat#G3580 |
| True seq Nano DNA LT kit | Illumina | Cat#15041757 |
| Nextera DNA Sample Preparation Kit | Illumina | Cat#FC-121-1030 |
| KAPA Genotyping kit | Kapabiosystems | Cat#KK7352 |
| TruSeq RNA library kit | Illumina | Cat#RS-122-2001 |
| CD34 MicroBead Kit | Milteny Biotec | Cat#130-046-702 |
| EasySep® Mouse Hematopoietic Progenitor Cell Enrichment Kit | Stemcell Technologies | Cat#19756 |
| Amaxa® Cell Line Nucleofector® Kit V | Lonza | Cat#VCA-1003 |
| Deposited Data | ||
| SuperSeries of RNA-seq, ChIP-seq, ATAC-seq and 5C | This paper | GEO: GSE101791 |
| RNA-seq in ME-1 cells | This paper | GEO: GSE101788 |
| ChIP-seq in ME-1 cells | This paper | GEO: GSE101789 |
| ATAC-seq in ME-1 cells | This paper | GEO: GSE101790 |
| 5C in ME-1 cells | This paper | GEO: GSE109764 |
| ChIP-seq in ME-1 cells | Mandolli et al., 2014 | GEO: GSE46044 |
| ChIP-seq in K562 cells | Pugacheva et al., 2015 | GEO: GSE70764 |
| Experimental Models: Cell Lines | ||
| Human: ME-1 cells | DSMZ | Cat#ACC 537 |
| Human: U937 cells | ATCC | Cat#CRL-1593.2 |
| Human: K562 cells | ATCC | Cat#CCL-243 |
| Human: Jurkat cells | DSMZ | Cat#ACC-282 |
| Human: Kasumi-1 cells | ATCC | Cat#CRL-2724 |
| Human: THP-1 cells | ATCC | Cat#TIB-202 |
| Human: NB4 cells | DSMZ | Cat#ACC-207 |
| Human: MV4:11 cells | ATCC | Cat#CRL-9591 |
| Human: 293T/17 cells | ATCC | Cat#CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | Taconic Biosciences | N/A |
| Oligonucleotides | ||
| ChIP-qPCR Primers (See Table S2) | This paper | N/A |
| RT-PCR Primer (See Table S2) | This paper | N/A |
| sgRNA (See Table S2) | This paper | N/A |
| CUT&RUN Primer (See Table S2) | This paper | N/A |
| ATAC-qPCR Primer (See Table S2) | This paper | N/A |
| shRNA (See Table S2) | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: shRNA MYC4 | UMMS RNAi Core Facility | Cat#TRCN0000174055 |
| Plasmid: shRNA MYC6 | UMMS RNAi Core Facility | Cat#TRCN0000010390 |
| Plasmid: pLKO.1 puro | Sigma | Cat#SHC002 |
| Plasmid: shRNA Myc1 (Myc1891) CTCGAGAAGGTATATTGCTGTTGACA GTGAGCGCACGACGAGAACAGTTG AAACATAGTGAAGCCACAGATGTAT GTTTCAACTGTTCTCGTCGTTT GCCTACTGCCTCGGAATTC |
Zuber et al., 2011b | N/A |
| Plasmid: shRNA-Myc2 (Myc2105) CTCGAGAAGGTATATTGCTG TTGACAGTGAGCGCCTGCC TCAAACTTAAATAGTATAGTGAAGCCA CAGATGTATACTATTTAAGTTTGA GGCAGTTGCCTACTGCCTCGGAATTC |
Zuber et al., 2011b | N/A |
| Plasmid: shRNARen (Ren713) TGCTGTTGACAGTGAGCGCAGGAA TTATAATGCTTATCTATAG TGAAGCCACAGATGTATAGATA AGCATTATAATTCCTAT GCCTACTGCCTCGGA |
Zuber et al., 2011b | N/A |
| Plasmid: pLKO. 1 - TRC cloning vector | Moffat et al., 2006 | Addgene#10878 |
| Plasmid: psPAX2 | Trono Lab Packaging and Envelope Plasmids (unpublished) | Addgene#12260 |
| Plasmid: pMD2.G | Trono Lab Packaging and Envelope Plasmids (unpublished) | Addgene#12259 |
| Plasmid: pLentiCRISPRv2 | Sanjana et al., 2014 | Addgene#52961 |
| Plasmid: pDecko-mCherry | Pulido-Quetglas et al., 2017 | Addgene#78534 |
| Plasmid: MYC-ER | Ricci et al., 2004 | Addgene#19128 |
| Software and Algorithms | ||
| Bioconductor v3.4 | http://www.bioconductor.org | |
| ChIPpeakAnno v3.9 | Zhu, 2013; Zhu et al., 2010 | https://bioconductor.org/packages/release/bioc/html/ChIPpeakAnno.html |
| ATACseqQC v1.0.3 | Bioconductor package | https://bioconductor.org/packages/release/bioc/html/ATACseqQC.html |
| Bowtie2 v2.1.0 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS2 v2.1.0 | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| FastQC v0.10.1 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Tophat v2.0.9 | Trapnell et al., 2009 | https://ccb.jhu.edu/software/tophat/index.shtml |
| Cufflinks v2.2.0 | Trapnell et al., 2012 | http://cole-trapnell-lab.github.io/cufflinks/ |
| Diffbind v2.4.8 | Ross-Innes et al., 2012 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Picard tools 1.96 | Broad Institute | https://broadinstitute.github.io/picard/ |
| Bedtools v1.25.0 | Quinland et al., 2010 | http://bedtools.readthedocs.io/en/latest/ |
| Fastx-toolkit v.0.0.18 | Hannon Laboratory | http://hannonlab.cshl.edu/fastxtoolkit/ |
| GSEA | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| Molecular Signatures Database v6.0 | Broad Institute | http://software.broadinstitute.org/gsea/msigdb/index.jsp |
| GraphPad Prism 6.0 | GraphPad | https://www.graphpad.com |
| FlowJo Software | FlowJo LLC | https://www.flowjo.com |
| Other | ||
| RPMI 1640 Medium | Thermo Fisher Scientific | Cat#A10491-01 |
| Fetal Bovine Serum, Charcoal Stripped | Sigma | Cat#F6765 |
| StemSpan SFEM II | STEMCELL Technologies | Cat#09605 |
| StemSpan SFEM | STEMCELL Technologies | Cat#09650 |
| RBC Lysis Solution | 5 Prime | Cat#2301310 |
| MethoCult | STEMCELL Technologies | Cat#M3534 |
| Effectene | Qiagen | Cat#301425 |
| Fugene 6 Transfection Reagent | Promega | Cat#E2691 |
| Dynabeads® Protein G | Thermo Fisher Scientific | Cat#10004D |
| Dynabeads® Protein A | Thermo Fisher Scientific | Cat#10002D |
| Dynabeads™ MyOne™ Streptavidin C1 | Thermo Fisher Scientific | Cat#65001 |
| Agencourt AMPure XP 5 mL Kit | Beckman Coulter | Cat#A63880 |
| Bio-Mag Plus Concanavalin A coated beads | Polysciences | Cat#86057 |
| Protein A-MNase | Skene et al., 2015 | N/A |
| Plamsmocin | Invivogen | Cat#ant-mpt |
| Retro-X Concentrator | Clontech | Cat#631455 |
| Lenti-X Concentrator | Clontech | Cat#631231 |
| Halt™ Protease Inhibitor Cocktail | Thermo Fisher Scientific | Cat#778429 |
| Proteinase K (Fungal) | Thermo Fisher Scientific | Cat#25530-031 |
| T4 polynucleotide kinase | New England Biolabs | Cat#M0201 |
| Q5® High-Fidelity DNA Polymerase | New England Biolabs | Cat#M0491L |
| HindIII | New England Biolabs | Cat#R0104S |
| Dounce homogenizer, pestle A | Kimble Chase | Cat#885301-0002 |
| Amicon® Ultra - 0.5ml-30K | Millipore | Cat#UFC5030BK |
| T4 DNA ligase | Thermo Fisher Scientific | Cat#15224 |
| Taq DNA ligase and buffer | New England Biolabs | Cat#M0208S |
| Salmon testes DNA | Sigma | Cat#D7656 |
| QIAquick gel extraction kit | Qiagen | Cat#28704 |
| 4-Thiouridine | Carbosynth | Cat#NT06186 |
| EZ-Link™ HPDP-Biotin | Thermo Fisher Scientific | Cat#21341 |
| T4 PNK (10 U/μL) | New England Biolabs | Cat#M0201S |
| T4 DNA polymerase | Thermo Fisher Scientific | Cat#18005025 |
| Taq DNA polymerase | Thermo Fisher Scientific | Cat#EP0401 |
| 2× Rapid DNA ligase buffer | Enzymatics | Cat#B101L |
| Enzymatics DNA ligase | Enzymatics | Cat#L6030-HC-L |
| 5× KAPA buffer | Kapabiosystems | Cat#KK2502 |
| KAPA HS HIFI polymerase | Kapabiosystems | Cat#KK2502 |
| Clean-Blot IP Detection Reagent | Thermo Fisher Scientific | Cat#21230 |
Supplementary Material
Related to Figure 5. Transcription Factor consensus binding sites at ME1, Me2 and E3 Enhancers.
(Related to STAR Methods Key Resource Table). List of Oligonucleotide Sequences.
Related to Figure 5E: 5C Statistics.
Figure S1. AI-10–49 mediated MYC transcriptional changes is specific to inv(16) cells. Related to Figure 1. (A, B) Gene Set Enrichment Analysis of RNA-seq data depicting pyrimidine metabolism, cell cycle and ribosome biogenesis (A), and pathway signatures (B) positively correlated with AI-10–49 treatment in ME-1 cells. (FDR) false discovery rate; (NES) normalized enrichment score. (C) MYC transcript levels in non-inv(16) AML cells (U937, K562, Jurkat, Kasumi-1 and THP-1) treated with DMSO or 1μM AI-10–49 for 6 hrs. (D) Immunoblot depicting MYC and GAPDH protein levels in ME-1 cells treated with 1 μM AI-10–49 for 0 hrs, 2 hrs, 4 hrs, 6 hrs and 8 hrs.
Figure S2. Effect of MYC silencing in inv(16) AML cells. Related to Figure 2. (A) Time course analysis of cell viability (7AAD- Annexin V-) in ME-1 cells transduced with scramble (Scr) or two MYC shRNAs. (B) Flow cytometry analysis of granulocytic differentiation in ME-1 cells transduced with MYC shRNAs at day 14. (C, D) Analysis of MYC protein levels assessed by western blot analysis (C) and cell viability (7AAD- Annexin V-; D) of AML cell lines Kasumi-1, NB4, ME-1, THP1, MV4:11 and K562, 14 days after transduction with MYC shRNAs; each data point represents the mean of triplicate experiments; error bars represent the SD. (E) Immunoblot analysis of Myc and Gapdh protein levels mouse Cbfb+/MYH11 leukemic cells transduced with Renila (Ren) or Myc shRNAs 1 and 2. (F) Schematic representation of experimental design for in vivo evaluation of Myc shRNA knockdown experiments. (G) Immunoblot analysis of Myc and Gapdh protein levels in Cbfb+/MYH11 leukemic cells of leukemic mice (Ren, shMyc1 and shMyc2 groups) from secondary transplant assays shown in Figure 2G. Each band represents Myc total protein levels of leukemic cells isolated from a single mouse. Significance was calculated using Levene’s test (D). *P < 0.05, or **P < 0.005.
Figure S3. AI-10–49 cooperates with JQ1 in inv(16) AML. Related to Figure 3. (A) qRT-PCR analysis of BRD4 transcript levels in ME-1 cells transduced with scramble (Scr) or two BRD4 shRNAs (sh1 and sh2). (B) Immunoblot analysis of MYC and GAPDH protein levels in ME-1 cells treated with BET inhibitor JQ1 for 6 hrs. (C) Dose response viability analysis (MTT assay) of ME-1 cells treated with AI-10–49 and/or JQ1 for 72 hrs; the LD50 for each compound is: AI-10–49-LD50=0.468 μM, range=0.398–0.537 μM; JQ1-LD50= 0.344μM, range=0.228–0.460 μM; both at 95% confidence intervals. (D) Percentage of c-kit+ (leukemic) cells in peripheral blood 25 days after transplantation in respective groups, assessed by flow cytometry. (E) Viability analysis (MTT assay) of JQ1 and AI-10–49 in human cord blood CD34+ cells 48 hrs after treatment with AI-10–49 and/or JQ1 at the indicated concentrations. (F-J) Toxicology analysis of wild type mice treated with a daily dose of DMSO (D, black) or 200 mg/kg/day AI-10–49 (10 days) and 50 mg/kg/day JQ1 (21 days) (49+JQ1, green). Mice were analyzed 1 day after last treatment dose; body weight (F), spleen weight (G), bone marrow cellularity (H), percentage of stem and early progenitor cells [LSK+: Lin(−) Sca1(+) c-kit(+)] in bone marrow (I), percentage of progenitor cell compartments common myeloid progenitors [CMP: LSK-,CD34(+)CD16/32(−)], megakaryocyte/erythroid progenitors [MEP: LSK-, CD34(−)CD16/32(−)], and granulocyte/monocyte progenitors [GMP: LSK-, CD34(+)CD16/32(+)], in LSK- cells (J). Each symbol represents the mean of values from three animals; error bars represent the S.D. Significance was calculated using unpaired t-test (A) or Levene’s test (D). *P < 0.05, or **P < 0.005.
Figure S4. AI-10–49 leads to increased genome wide RUNX1 binding in ME-1 cells. Related to Figure 4. (A) genomewide (left) and transcription start site (TSS, right) centered RUNX1 aggregated peak signal in ChIP-seq dataset from AI-10–49 or DMSO treated ME-1 cells, and respective heat maps (bottom). (B) Gene distribution of H3K27Ac (top) and RUNX1 (bottom) peaks in ME-1 cells treated with DMSO (left) or AI-10–49 (right).
Figure S5. RUNX1 mediated chromatin changes at MYC enhancer elements with AI-10–49. Related to Figure 5. (A) ATAC-seq and ChIP-seq profiles for K3K27ac and RUNX1 at the +1.7 Mb BDME superenhancer. Five previously reported enhancer regions (E1 to E5) are depicted below the profile. (B) ChIP-seq profiles for K3K27ac and RUNX1 peaks in ME-1 cells treated with DMSO (blue) or AI-10–49 (red) in the 2Mb genomic region upstream of MYC-TSS. (C) 4C-style plots for 15 Kb bins (anchor bins) containing the MYC promoter (Myc-Pr), ME1, ME2, and E3/E5 enhancers for DMSO and AI-10–49 treated cells. Anchor bins are shown in orange, solid black lines represent the LOWESS mean (the expected interaction frequency as a function of genomic distance) and the dotted black lines are the LOWESS plus and minus 1 standard deviation. Red lines are the observed 5C interaction frequencies. Green dots and vertical dotted lines highlight the positions and interactions between Myc-Pr, ME1, ME2, and E3. Arrow-heads indicate interactions with CTCF sites around the BDME superenhancer. Arrows indicate peaks of interactions pointing to loci interacting with Myc-Pr, ME1, ME2, and E3. The CTCF binding data were used from ChIP-seq data previously reported in K562 leukemia cells (GSE70764; ref. (Pugacheva et al., 2015)).
Figure S6. Analysis of transcription factor ChIP-seq at MYC locus. Related to Figure 5. Transcription factor ChIP-seq analysis from (GSE46044; ref. (Mandoli et al., 2014) at the 2Mb downstream of the MYC TSS. Peak location for MYC promoter (blue) and ME1, ME2 and E3 (black) are shown as dotted line windows.
Figure S7. Related to Figure 6 (A) Immunoblot analysis for BRG1, RING1B and GAPDH in lysates of ME-1 cells treated with 1μM AI-10–49 at 2 to 8 hrs. (B, C) qRT-PCR analysis of SMARCA4 (B) and RNF2 (C) transcript levels in ME-1 cells transduced with scramble (Scr) or two gene specific shRNAs. Results from triplicate experiments shown; error bars represent the SD. (D) Evaluation of MYC transcript levels in ME-1 cells treated with RING1B inhibitor PRT 4165 for 8 days followed by treatment with DMSO/ AI-10–49 (0.6 μM) for 6 hrs, and MYC relative expression levels (REL) were estimated using qRT-PCR. Results from triplicate experiments shown; error bars represent the SD. (E) Co-immunoprecipitation analysis of RUNX1 binding to BRG1 and RING1B in nuclear extracts from ME-1 cells treated with DMSO/ AI-10–49 for 6 hrs. Significance was calculated as unpaired t-test (B-D).
Figure S8, related to Figure 7. Sequence analysis of deletions (A) by size and (B) by location, in inv(16) AML ME-1 cells treated with CRISPR-Cas9 and sgRNAs for ME1, ME2, N-Me and E3. Analysis performed utilizing CRISPR Genome Analyzer.
ACKNOWLEDGEMENTS
We thank James Bradner for providing JQ1, Steven Henikoff for providing the pA-MNase protein, and Dale Grainer and Michael Brehm for providing CD34+ cord blood cells. This work was supported by the NIH R01 CA204979 and Leukemia & Lymphoma Society - #6463–15 grants to L.H.C, and by the NIH R01 CA140398 and ASH Bridge grants to J.H.B.. J.A.P. was funded by the UMASS Cancer Center Postdoctoral Fellowship, the Alex’s Lemonade Stand Foundation-Young Investigator Award, and the Lauri Strauss Leukemia Foundation-Discovery Grant.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interests. J.H.B. and A.I. are co-inventors on patent US8748618B2, which includes AI-10–49.
REFERENCES
- Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, et al. (2018). A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, and Klose RJ (2015). Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nature reviews Molecular cell biology 16, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Kalota A, Sanchez PV, and Carroll M (2011). Short hairpin RNA screen reveals bromodomain proteins as novel targets in acute myeloid leukemia. Cancer Cell 20, 287–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, and Neil JC (2005). The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 5, 376–387. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Gearhart MD, Gery S, Shojaee S, Yang H, Sun H, Lin DC, Bai JW, Mead M, Zhao Z, et al. (2016). BCOR regulates myeloid cell proliferation and differentiation. Leukemia 30, 1155–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Britos-Bray M, Claxton DF, Kelley CA, Speck NA, Liu PP, and Friedman AD (1997). CBF beta-SMMHC, expressed in M4Eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene 15, 1315–1327. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, Owens J, Eckhaus M, Bodine D, and Liu PP (1999). The fusion gene CBFβ-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nature Genetics 23, 144–146. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, Marin-Padilla M, Collins FS, Wynshaw-Boris A, and Liu PP (1996). Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell 87, 687–696. [DOI] [PubMed] [Google Scholar]
- Choi A, Illendula A, Pulikkan JA, Roderick JE, Tesell J, Yu J, Hermance N, Zhu LJ, Castilla LH, Bushweller JH, et al. (2017). RUNX1 is required for oncogenic Myb and Myc enhancer activity in T-cell acute lymphoblastic leukemia. Blood 130, 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC (2010). Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70, 440–446. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, and Alt FW (2001). Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55. [DOI] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, and Mirny LA (2013). Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 14, 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. (2011). BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, and Helin K (2013). Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 20, 1147–1155. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. (2006). Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome research 16, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas NC, Jacobs H, Bothwell AL, and Hayday AC (2001). Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol 2, 307–315. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Sanyal A, Naumova N, Dekker J, and Dostie J (2012). From cells to chromatin: capturing snapshots of genome organization with 5C technology. Methods 58, 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, and Engreitz JM (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda SD, Koler RD, and Bagby GC Jr. (1986). Regulation of C-myc expression during growth and differentiation of normal and leukemic human myeloid progenitor cells. J Clin Invest 77, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, and Friedman AD (2011). Phosphorylation of RUNX1 by cyclin-dependent kinase reduces direct interaction with HDAC1 and HDAC3. J Biol Chem 286, 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Niu C, Breslin P, Tang M, Zhang S, Wei W, Kini AR, Paner GP, Alkan S, Morris SW, et al. (2009). c-Myc-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Blood 114, 2097–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, and Henikoff S (2011). Epigenome characterization at single base-pair resolution. Proceedings of the National Academy of Sciences of the United States of America 108, 18318–18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, Xu L, Castillo-Martin M, Llobet-Navas D, Cordon-Cardo C, et al. (2014). A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nature medicine 20, 1130–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, and Young RA (2013). Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JT, Redner RL, and Nienhuis AW (1988). An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol 8, 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, et al. (2009). Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, and Gentleman R (1996). R: A language for data analysis and graphics. J Comput Graph Stat 5, 5. [Google Scholar]
- Illendula A, Pulikkan JA, Zong H, Grembecka J, Xue L, Sen S, Zhou Y, Boulton A, Kuntimaddi A, Gao Y, et al. (2015). Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFβeta-SMMHC delays leukemia in mice. Science 347, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, and Mirny LA (2012). Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature methods 9, 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR, and Tenen DG (2001). c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol 21, 3789–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, and Crabtree GR (2015). Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv 1, e1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinathan S, Orsi GA, Zentner GE, Ahmad K, and Henikoff S (2014). High-resolution mapping of transcription factor binding sites on native chromatin. Nature methods 11, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I, Aikawa Y, Nguyen LA, Yokoyama A, and Ohki M (2001). Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J 20, 7184–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I, Yokoyama A, Shimizu K, and Ohki M (1998). Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J 17, 2994–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Sabo A, and Amati B (2015). MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 15, 593–607. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, Le Beau MM, Kogan SC, and Castilla LH (2006). Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell 9, 57–68. [DOI] [PubMed] [Google Scholar]
- Lajoie BR, van Berkum NL, Sanyal A, and Dekker J (2009). My5C: web tools for chromosome conformation capture studies. Nature methods 6, 690–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, and Sauvageau G (2003). Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–260. [DOI] [PubMed] [Google Scholar]
- Lichtinger M, Hoogenkamp M, Krysinska H, Ingram R, and Bonifer C (2010). Chromatin regulation by RUNX1. Blood Cells Mol Dis 44, 287–290. [DOI] [PubMed] [Google Scholar]
- Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, and Collins FS (1993). Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 261, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, and Young RA (2013). Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik SM, Zhang L, Corpora T, Tomanicek S, Li Y, Kundu M, Hartman K, Liu PP, Laue TM, Biltonen RL, et al. (2002). Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat Struct Biol 9, 674–679. [DOI] [PubMed] [Google Scholar]
- Mandoli A, Singh AA, Jansen PW, Wierenga AT, Riahi H, Franci G, Prange K, Saeed S, Vellenga E, Vermeulen M, et al. (2014). CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia 28, 770–778. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298. [DOI] [PubMed] [Google Scholar]
- Naumova N, Smith EM, Zhan Y, and Dekker J (2012). Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods 58, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Yu J, Kelliher M, Castilla L, Lawson N, and Zhu LJ (2017). ATAC-seq Quality Control. Bioconductor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencovich N, Jaschek R, Tanay A, and Groner Y (2011). Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood 117, e1–14. [DOI] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. (2009). The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41, 882–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J, Hoogenkamp M, Wu M, Care M, McNeill H, et al. (2012). Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia 26, 1829–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EM, Rivero-Hinojosa S, Espinoza CA, Mendez-Catala CF, Kang S, Suzuki T, Kosaka-Suzuki N, Robinson S, Nagarajan V, Ye Z, et al. (2015). Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome biology 16, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Quetglas C, Aparicio-Prat E, Arnan C, Polidori T, Hermoso T, Palumbo E, Ponomarenko J, Guigo R, and Johnson R (2017). Scalable Design of Paired CRISPR Guide RNAs for Genomic Deletion. PLoS Comput Biol 13, e1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed-Inderbitzin E, Moreno-Miralles I, Vanden-Eynden SK, Xie J, Lutterbach B, Durst-Goodwin KL, Luce KS, Irvin BJ, Cleary ML, Brandt SJ, et al. (2006). RUNX1 associates with histone deacetylases and SUV39H1 to repress transcription. Oncogene 25, 5777–5786. [DOI] [PubMed] [Google Scholar]
- Ritz C, Baty F, Streibig JC, and Gerhard D (2015). Dose-Response Analysis Using R. PloS one 10, e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RJ, Drier Y, Whitton H, Cotton MJ, Kaur J, Issner R, Gillespie S, Epstein CB, Nardi V, Sohani AR, et al. (2015). Detection of Enhancer-Associated Rearrangements Reveals Mechanisms of Oncogene Dysregulation in B-cell Lymphoma. Cancer Discov 5, 1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nature methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, and Dekker J (2012). The long-range interaction landscape of gene promoters. Nature 489, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Fruhauf K, Demel C, Tresch A, Gagneur J, and Cramer P (2016). TT-seq maps the human transient transcriptome. Science 352, 1225–1228. [DOI] [PubMed] [Google Scholar]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, et al. (2013). Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 27, 2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, and Henikoff S (2015). A simple method for generating high-resolution maps of genome-wide protein binding. Elife 4, e09225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BZ, Hodges C, Calarco JP, Braun SM, Ku WL, Kadoch C, Zhao K, and Crabtree GR (2017). Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet 49, 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. (1994). Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 8, 757–769. [DOI] [PubMed] [Google Scholar]
- Vradii D, Zaidi SK, Lian JB, van Wijnen AJ, Stein JL, and Stein GS (2005). Point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and effects a transformation-like phenotype. Proceedings of the National Academy of Sciences of the United States of America 102, 7174–7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LP, Perna F, Wang L, Voza F, Figueroa ME, Tempst P, Erdjument-Bromage H, Gao R, Chen S, Paietta E, et al. (2013). PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep 5, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, and Speck NA (1996a). Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America 93, 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, et al. (1996b). The CBFβeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87, 697–708. [DOI] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, and Trumpp A (2004). c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev 18, 2747–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Pulikkan JA, Valk PJ, and Castilla LH (2014). NrasG12D oncoprotein inhibits apoptosis of preleukemic cells expressing CBFβeta-SMMHC via activation of MEK/ERK axis. Blood 124, 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, Knoechel B, Lanauze C, Louis L, Forsyth KS, et al. (2014). Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proceedings of the National Academy of Sciences of the United States of America 111, E4946–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, Chouinard CR, Labadorf A, Akie TE, Moran TB, et al. (2012). Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Molecular cell 45, 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, Zhang J, Dunne R, Xiao A, Erdjument-Bromage H, et al. (2008). Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev 22, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ (2013). Integrative analysis of ChIP-chip and ChIP-seq dataset. Methods Mol Biol 1067, 105–124. [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, and Green MR (2010). ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC bioinformatics 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, et al. (2011a). An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev 25, 1628–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. (2011b). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 5. Transcription Factor consensus binding sites at ME1, Me2 and E3 Enhancers.
(Related to STAR Methods Key Resource Table). List of Oligonucleotide Sequences.
Related to Figure 5E: 5C Statistics.
Figure S1. AI-10–49 mediated MYC transcriptional changes is specific to inv(16) cells. Related to Figure 1. (A, B) Gene Set Enrichment Analysis of RNA-seq data depicting pyrimidine metabolism, cell cycle and ribosome biogenesis (A), and pathway signatures (B) positively correlated with AI-10–49 treatment in ME-1 cells. (FDR) false discovery rate; (NES) normalized enrichment score. (C) MYC transcript levels in non-inv(16) AML cells (U937, K562, Jurkat, Kasumi-1 and THP-1) treated with DMSO or 1μM AI-10–49 for 6 hrs. (D) Immunoblot depicting MYC and GAPDH protein levels in ME-1 cells treated with 1 μM AI-10–49 for 0 hrs, 2 hrs, 4 hrs, 6 hrs and 8 hrs.
Figure S2. Effect of MYC silencing in inv(16) AML cells. Related to Figure 2. (A) Time course analysis of cell viability (7AAD- Annexin V-) in ME-1 cells transduced with scramble (Scr) or two MYC shRNAs. (B) Flow cytometry analysis of granulocytic differentiation in ME-1 cells transduced with MYC shRNAs at day 14. (C, D) Analysis of MYC protein levels assessed by western blot analysis (C) and cell viability (7AAD- Annexin V-; D) of AML cell lines Kasumi-1, NB4, ME-1, THP1, MV4:11 and K562, 14 days after transduction with MYC shRNAs; each data point represents the mean of triplicate experiments; error bars represent the SD. (E) Immunoblot analysis of Myc and Gapdh protein levels mouse Cbfb+/MYH11 leukemic cells transduced with Renila (Ren) or Myc shRNAs 1 and 2. (F) Schematic representation of experimental design for in vivo evaluation of Myc shRNA knockdown experiments. (G) Immunoblot analysis of Myc and Gapdh protein levels in Cbfb+/MYH11 leukemic cells of leukemic mice (Ren, shMyc1 and shMyc2 groups) from secondary transplant assays shown in Figure 2G. Each band represents Myc total protein levels of leukemic cells isolated from a single mouse. Significance was calculated using Levene’s test (D). *P < 0.05, or **P < 0.005.
Figure S3. AI-10–49 cooperates with JQ1 in inv(16) AML. Related to Figure 3. (A) qRT-PCR analysis of BRD4 transcript levels in ME-1 cells transduced with scramble (Scr) or two BRD4 shRNAs (sh1 and sh2). (B) Immunoblot analysis of MYC and GAPDH protein levels in ME-1 cells treated with BET inhibitor JQ1 for 6 hrs. (C) Dose response viability analysis (MTT assay) of ME-1 cells treated with AI-10–49 and/or JQ1 for 72 hrs; the LD50 for each compound is: AI-10–49-LD50=0.468 μM, range=0.398–0.537 μM; JQ1-LD50= 0.344μM, range=0.228–0.460 μM; both at 95% confidence intervals. (D) Percentage of c-kit+ (leukemic) cells in peripheral blood 25 days after transplantation in respective groups, assessed by flow cytometry. (E) Viability analysis (MTT assay) of JQ1 and AI-10–49 in human cord blood CD34+ cells 48 hrs after treatment with AI-10–49 and/or JQ1 at the indicated concentrations. (F-J) Toxicology analysis of wild type mice treated with a daily dose of DMSO (D, black) or 200 mg/kg/day AI-10–49 (10 days) and 50 mg/kg/day JQ1 (21 days) (49+JQ1, green). Mice were analyzed 1 day after last treatment dose; body weight (F), spleen weight (G), bone marrow cellularity (H), percentage of stem and early progenitor cells [LSK+: Lin(−) Sca1(+) c-kit(+)] in bone marrow (I), percentage of progenitor cell compartments common myeloid progenitors [CMP: LSK-,CD34(+)CD16/32(−)], megakaryocyte/erythroid progenitors [MEP: LSK-, CD34(−)CD16/32(−)], and granulocyte/monocyte progenitors [GMP: LSK-, CD34(+)CD16/32(+)], in LSK- cells (J). Each symbol represents the mean of values from three animals; error bars represent the S.D. Significance was calculated using unpaired t-test (A) or Levene’s test (D). *P < 0.05, or **P < 0.005.
Figure S4. AI-10–49 leads to increased genome wide RUNX1 binding in ME-1 cells. Related to Figure 4. (A) genomewide (left) and transcription start site (TSS, right) centered RUNX1 aggregated peak signal in ChIP-seq dataset from AI-10–49 or DMSO treated ME-1 cells, and respective heat maps (bottom). (B) Gene distribution of H3K27Ac (top) and RUNX1 (bottom) peaks in ME-1 cells treated with DMSO (left) or AI-10–49 (right).
Figure S5. RUNX1 mediated chromatin changes at MYC enhancer elements with AI-10–49. Related to Figure 5. (A) ATAC-seq and ChIP-seq profiles for K3K27ac and RUNX1 at the +1.7 Mb BDME superenhancer. Five previously reported enhancer regions (E1 to E5) are depicted below the profile. (B) ChIP-seq profiles for K3K27ac and RUNX1 peaks in ME-1 cells treated with DMSO (blue) or AI-10–49 (red) in the 2Mb genomic region upstream of MYC-TSS. (C) 4C-style plots for 15 Kb bins (anchor bins) containing the MYC promoter (Myc-Pr), ME1, ME2, and E3/E5 enhancers for DMSO and AI-10–49 treated cells. Anchor bins are shown in orange, solid black lines represent the LOWESS mean (the expected interaction frequency as a function of genomic distance) and the dotted black lines are the LOWESS plus and minus 1 standard deviation. Red lines are the observed 5C interaction frequencies. Green dots and vertical dotted lines highlight the positions and interactions between Myc-Pr, ME1, ME2, and E3. Arrow-heads indicate interactions with CTCF sites around the BDME superenhancer. Arrows indicate peaks of interactions pointing to loci interacting with Myc-Pr, ME1, ME2, and E3. The CTCF binding data were used from ChIP-seq data previously reported in K562 leukemia cells (GSE70764; ref. (Pugacheva et al., 2015)).
Figure S6. Analysis of transcription factor ChIP-seq at MYC locus. Related to Figure 5. Transcription factor ChIP-seq analysis from (GSE46044; ref. (Mandoli et al., 2014) at the 2Mb downstream of the MYC TSS. Peak location for MYC promoter (blue) and ME1, ME2 and E3 (black) are shown as dotted line windows.
Figure S7. Related to Figure 6 (A) Immunoblot analysis for BRG1, RING1B and GAPDH in lysates of ME-1 cells treated with 1μM AI-10–49 at 2 to 8 hrs. (B, C) qRT-PCR analysis of SMARCA4 (B) and RNF2 (C) transcript levels in ME-1 cells transduced with scramble (Scr) or two gene specific shRNAs. Results from triplicate experiments shown; error bars represent the SD. (D) Evaluation of MYC transcript levels in ME-1 cells treated with RING1B inhibitor PRT 4165 for 8 days followed by treatment with DMSO/ AI-10–49 (0.6 μM) for 6 hrs, and MYC relative expression levels (REL) were estimated using qRT-PCR. Results from triplicate experiments shown; error bars represent the SD. (E) Co-immunoprecipitation analysis of RUNX1 binding to BRG1 and RING1B in nuclear extracts from ME-1 cells treated with DMSO/ AI-10–49 for 6 hrs. Significance was calculated as unpaired t-test (B-D).
Figure S8, related to Figure 7. Sequence analysis of deletions (A) by size and (B) by location, in inv(16) AML ME-1 cells treated with CRISPR-Cas9 and sgRNAs for ME1, ME2, N-Me and E3. Analysis performed utilizing CRISPR Genome Analyzer.