Abstract
Ataxia telangiectasia (A-T) is a progressive childhood disorder characterized most notably by cerebellar degeneration and predisposition to cancer. A-T is caused by mutations in the kinase ATM, a master regulator of the DNA double-strand break response. In addition to DNA-damage signaling defects, A-T cells display mitochondrial dysfunction that is thought to contribute to A-T pathogenesis. However, the molecular mechanism leading to mitochondrial dysfunction in A-T remains unclear. Here we show that lack of ATM leads to reduced mitochondrial DNA (mtDNA) integrity and mitochondrial dysfunction, which are associated to defective mtDNA repair. While protein levels of mtDNA repair proteins are essentially normal, in the absence of ATM levels specifically of DNA ligase III (Lig3), the only DNA ligase working in mitochondria are reduced. The reduction of Lig3 is observed in different A-T patient cells, in brain and pre-B cells derived from ATM knockout mice as well as upon transient or stable knockdown of ATM. Furthermore, pharmacological inhibition of Lig3 in wild type cells phenocopies the mtDNA repair defects observed in A-T patient cells. As targeted deletion of LIG3 in the central nervous system causes debilitating ataxia in mice, reduced Lig3 protein levels and the consequent mtDNA repair defect may contribute to A-T neurodegeneration. A-T is thus the first disease characterized by diminished Lig3.
Keywords: ATM, mitochondria, mitochondrial DNA repair, DNA ligase 3, Ataxia Telangiectasia
1. Introduction
Ataxia Telangiectasia (A-T) is a neurodegenerative disorder that leads primarily to cerebellum degeneration and predisposes patients to cancer [1]. A-T patients present with a plethora of additional symptoms such as immunodeficiency, growth retardation, premature aging and insulin resistance [2]. While the molecular mechanisms associated with A-T neurodegeneration are unknown, mitochondrial dysfunction occurs in A-T patients [3,7] and, like in many neurodegenerative disorders [8], may play an important role in disease pathogenesis. A-T is caused by mutations in the gene encoding the DNA-damage response kinase ATM, absence of which is associated not only with nuclear genomic instability but also altered mitochondrial metabolism, including increased reactive oxygen species (ROS) production, decreased electron transport activity, and altered mitochondrial membrane polarization [3,7]. However, the molecular cause of mitochondrial dysfunction in A-T remains unknown and may represent a key feature for fully understanding A-T etiology and disease progression, and finding new therapeutic targets for the disease.
In trying to understand the role of mitochondria in A-T pathogenesis we turned to analysis of mtDNA integrity, a key feature required to maintain mitochondrial function but still overlooked in the context of A-T. Here we show that lack of ATM leads to decreased mtDNA integrity and impaired mitochondrial function in patient-derived cells and in tissues from an ATM knockout (KO) model. We also identified that in the absence of ATM mitochondrial base excision repair (BER) is impaired specifically due to decreased protein levels of DNA ligase III (Lig3), which is the only DNA ligase operating in mitochondria. Treatment of cells with MG132, a proteasome inhibitor, increased levels of Lig3 and pharmacological inhibition of Lig3 in ATM- proficient cells phenocopied the mtDNA repair defects detected in A-T. Our data confirms previous work showing that Lig3 is required to maintain mtDNA integrity and function, and highlight a new function of ATM in regulating DNA Lig3 stability and consequently mtDNA repair. Our results also indicate that targeting mitochondrial dysfunction, including mtDNA instability, may help alleviate some symptoms associated with A-T. Our findings can help explain the complex and variable phenotype of the disease, which is reminiscent of mitochondrial disorders, and support the notion that A-T may be a mitochondrial disease. Finally, A-T becomes the first example of a human disease characterized by diminished DNA Lig3 levels and function.
2. Materials and Methods
2.1. Cell culture and animal samples.
AG01522 (WT), AG04405 fibroblasts (A-T), GM07532 and GM02052 (A-T) were grown in MEM medium supplemented with 15% FBS and 1% Penicillin/streptomycin. These fibroblasts are available at the Coriell Repository (NJ). Nonimmortalized A-T fibroblasts were consistently used in lower passage (usually 2-4 from the passage obtained by Coriell) as compared to the WT counterpart (up to 10 passages) so as to avoid potential confounding effects associated to telomere shortening. We also routinely monitored changes in cell cycle regulation, cell morphology and doubling time as these are typically altered when cells are senescent or approaching senescence. Cerebrum and cerebellum from 3 ATM null mice and 3 WT littermates were analyzed (details on the animals can be found in D’Souza et al., 2012). Animals were maintained at the Yale Medical School vivarium and handled according to IACUC protocol. All experiments presented were performed at least 3 times using cultures grown at different times.
2.2. H2O2 treatment and mtDNA integrity analysis.
Protocols for H2O2 exposure and QPCR analysis of mtDNA integrity were followed as previously described [10,12]. Briefly, cells were exposed to 200 μM H2O2 for 60 min. Following treatment genomic DNA was isolated and a large (8.9 for human and 10kb for mouse) and a small fragment of the mtDNA were amplified using specific primers. Amplification of sample of interest was compared to that of control and these numbers were used to estimate the amount of lesions present on the mtDNA. Each experiment was independently reproduced at least 3 times; each DNA sample was submitted to 2 QPCR reactions; the average of the 2 reactions was then considered as N=1. For more details see references 11 and 12. Repair kinetics were performed both under normal growth conditions (MEM) and with supplemented medium (see doubling time below), and showed similar results ruling out confounding effects associated to cell cycle.
2.3. H2O2 exposure and kinetics of H2O2 decomposition.
The same number of WT and A-T- patient derived cells were plated and 16h later cells were exposed to 200 μM of H2O2 for 60 min. Immediately after the treatment cells were washed, harvested and the different mitochondrial parameters analyzed (see below). To measure kinetics of H2O2 decomposition (Fig. S3), amounts of H2O2 were followed over time using Amplex red and a H2O2 standard curve as reported earlier by us [10]. Each experiment was repeated at least 3 independent times.
2.4. Mitochondrial ROS, membrane potential and ATP measurements.
For mitochondrial superoxide measurements, after exposure to H2O2 (as above) cells were loaded with Mitosox fluorescent probe 10 μM (Invitrogen) for 10 min at 37°C. The data were collected by FACS. FCCP was employed to completely uncouple mitochondria and as such abolish the mitochondrial-generated superoxide signal. The signal from FCCP-treated samples was then subtracted from the samples and these final values were used for the comparisons. Mitochondrial membrane polarization was determined after incubation of cells for 10 min with JC-1 (10 μM) (Invitrogen, T3168) by FACS. Total ATP production was estimated using a Luciferase based assay (Invitrogen, A22060) and an ATP standard curve; the data were normalized to cell numbers.
2.5. Doubling time.
Cells were counted and 300,000 cells of each cell type were plated in either regular medium (MEM + 15% FBS) or supplemented medium (MEM + 15% FBS + pyruvate + 50 μg/mL uridine + 3.5 mM glucose) to support glycolytic growth. Upon reaching confluency, cells were trypsinized, counted and replated (300,000). Cell growth was followed using this protocol for 3 weeks; error bars represent SEM for each cell type from 2 independently grown cultures (stock frozen at independent times).
2.6. Mitochondrial content estimation using Mitotracker Green.
Cells were treated as above and immediately after the exposure to 200 μM H2O2 as well as 24h later, cells were incubated with 200 nm Mitotracker green (Invitrogen) for 30 min before submitting for FACS analysis. ImageStream was used to assure the signals were solely mitochondrial. Fluorescence values were compared between H2O2-treated samples and non- H2O2-treated control at both time points. Experiments were reproduced 3 independent times.
2.7. Isolation of mitochondrial from fibroblast and mouse tissue.
Mitochondrial isolations from human fibroblast were performed as recently reported by us [43]. The preparation of mitochondria from the different mouse tissues was performed following published protocols [44] with minor modifications. Briefly frozen cerebrum and cerebellum were washed free of blood with homogenization buffer (0.250 sucrose, 10 mM HEPES pH 7.4, 1 mM EGTA, 1mM DTT) supplemented with protease inhibitors (Sigma). The ratio of homogenization buffer to tissue was 1:10. The minced tissue was then transferred to a Teflon/glass homogenizer and homogenized at 2,000 rpm for 5 min. The homogenate was differentially centrifuged at 600Xg for 10 min and 10,000Xg for 10 min. The supernatant was saved as cytosolic fraction and the pellet was resuspended in mitochondria suspension buffer (0.250 M sucrose, 3 mM EGTA/Tris and 10 mM Tris/HCl, pH 7.4) and washed three times using the same buffer. The washed mitochondrial pellet was stored at −80°C for further experimental use. Protein concentrations were determined using the Lowry method, with BSA as standard.
2.8. Preparation of nuclear and mitochondrial extracts for ligase assay.
All steps were performed at 4°C. The pelleted nuclei from cells were resuspended in the hypotonic nuclei extraction buffer containing 10 mM Tris-HCl pH 7.4, 10 mM MgCl2, 10 mM KCl 1 mM DTT and 350 mM NaCl and a cocktail of protease inhibitors. After 1h of incubation, the extracted nuclei were centrifuged at 50,000Xg for 1h in an Allegra 64X Beckman centrifuge. The cleared supernatant was collected and adjusted to 10% glycerol and stored at −80°C. The protein concentrations were determined using the Lowry assay. Mitochondrial extracts were prepared according to previous protocol with slight modification [26]. The intact pelleted mitochondria was given hypotonic treatment using chilled hypotonic buffer 10 mM Tris-HCl (pH 7.4), 1mM EDTA (pH 8.0) and was left for incubation at 4°C for 30 min. The lysed mitochondria pellet was sonicated for two pulses of 20 seconds at 40A using an ultrasonic homogenizer 4710 series (Cole Parmer Instrument Co). After sonication, lysed mitochondrial suspension was centrifuged at 50,000Xg for one hour to pellet out membrane protein. Further, the lysed mitochondrial protein suspension was concentrated using Amicon Ultra, centrifugal filters 3 kDa cut off as per manufacturer instructions. The obtained mitochondrial lysate was dialyzed against buffer (50 mM Tris, pH 7.5, 0.1 mM EDTA, 10 mM β-mercaptoethanol, 10 mM phenylmethylsulfonyl fluoride and 10% glycerol), then aliquoted and stored at −80°C for further use. Protein concentration was determined using the Lowry protein assay.
2.9. DNA ligase assays.
To assess re-ligation ability of extracts, cells were treated or not with H2O2 and the different fractions obtained by differential centrifugation. Re-ligation of cut pUC18 was followed as per previous protocols [26]. Briefly, pUC18 was cut with either PstI or Xbal and incubated overnight at 16°C with extracts from WT or A-T samples. Following this period the plasmid DNA was precipitated and purified, and run on agarose gels [26]. Reactions were supplemented with exogenous ATP and T4 DNA ligase was used as positive control. Data were reproduced multiple times using lysates that were obtained from independently grown cultures or animals. Each ligation reaction was also performed at least 3 independent times.
2.10. Proteasome Inhibition.
WT and A-T derived fibroblasts were treated with 25 μM of the proteasome inhibitor MG132 for 8h and inhibition of the proteasome was gauged based on a luminescence assay (Proteasome-Glo Chymotrypsin-Like based assay, G8662, Promega). The assay was performed as per manufacturer instructions in 3 independent samples. All analyses were done in triplicate.
2.11. SDS-PAGE and Western blotting.
Patient fibroblasts were washed in dishes and directly resuspended in 250 μl volume of lysis buffer containing (10 mM Tris-HCl pH 7.8, 200 mM KCl, 20% glycerol, 0.1 mM EDTA, 4% SDS, 1 mg/ml of each protease inhibitor (pepstatin, aprotinin, chymostatin and leupeptin), 1mM PMSF by vortexing and pipetting. Protein samples were then quantified using the Lowry assay and run on Novex NuPage 4-12% Bis-Tris gradient gel (Invitrogen). Protein extracts were prepared at least 2 independent times from each cell line, for which multiple biological replicates were grown for subsequent analyses. Western blot analyses of Lig3 in lymphoblastoid, HME-CC and pre-B cells were performed in total lysates kindly provided by Dr. Richard Paules (NIEHS). The EBV-transformed lymphoblastoid cell lines (AG14772, WT) and GM03332 (A-T) were obtained from the NIGMS Human Genetic Mutant Cell Repository (Camden, NJ). HME-CC cells stably expressing shRNA for either LacZ or ATM was previously described [29]. Lysates from (v)-Abl kinase-transformed pre-B cells were derived from an ATM KO mouse expressing the Eμ,-Bcl2 transgene (from Dr. Barry Sleckman, Washington University School of Medicine). The following primary antibodies were used: mouse anti-ATM (GeneTex, 1:1000), mouse anti-DNA ligase III (B. D Transduction Laboratories, 1:5000), goat anti-ND1 (Santa Cruz, 1:200), rabbit anti-Fen1 (Bethyl, 1:1000), anti-phospho AMPK (Thr 172) and total AMPK (Abcam 1:500), and pol gamma (Santa Cruz, 1:100). All secondary antibodies were from Li-Cor Biosciences. Blots were visualized using the Odyssey image analysis system (Li-Cor Biosciences, Cambridge, UK).
2.12. Real time PCR analysis of Lig3 RNA.
Total RNA from AG01522 and AG04405 was purified using the RNeasy kit (Qiagen) and reverse transcribed using the Reverse Superscriptase III (Invitrogen). The expression of DNA Lig3 was measured with a quantitative real time PCR assay. Expression levels were calculated as a ratio of the RNA level for Lig3 gene relative to the RNA level for β-actin in the same cDNA sample. Primer sequences for Lig3 gene are Forward primer: 5`GGCAGCAGGTACACCAAAGAA3` and reverse primer 5`TGGGTCTTCGTGTTGTAGCTA3. For each RNA sample, the real-time PCR was performed in triplicate.
2.13. Statistical Analysis.
Student’s unpaired t-test or ANOVA were used to evaluate statistical significance.
3. Results
3.1. Mitochondrial DNA integrity is decreased and mitochondrial function is compromised in the absence of ATM.
Proper mitochondrial function requires the maintenance of multiple copies of the ~16-kb mitochondrial genome (mtDNA) that encodes critical protein components of the electron transport chain (ETC) and ATP synthase [9]. Damage to mtDNA results in decreased oxidative phosphorylation (OXPHOS) activity and can also increase ROS generation that further damage this organelle leading to a vicious cycle of mitochondrial demise [9,10]. Thus mtDNA damage can initiate and contribute to a cascade of mitochondrial and cellular dysfunction. We hypothesized that persistent mtDNA damage may cause mitochondrial dysfunction in A-T patients. To test this hypothesis we analyzed mtDNA integrity in wild type (WT) and A-T samples using the gene-specific quantitative PCR (QPCR) assay that specifically measures replication blocking lesions in mtDNA [11,12]. Briefly, genomic DNA was isolated from independent cultures and a large (8.9Kb and 10 kb for human and mouse, respectively) and a small (221bp and 117 bp for human and mouse, respectively) fragment of the mitochondrial genome were quantitatively amplified. Amplification of the A-T sample was compared to amplification of the WT counterpart and these values were used to estimate the amount of lesions present in mtDNA. Data presented in Fig. 1 were normalized to mtDNA content (Fig. S1), and indicate a ~4-fold increase in mtDNA damage in the absence of ATM in two different non-immortalized, non-transformed fibroblasts from A-T patients (Fig. 1A) and in the cerebellum from ATM null mice (Fig. 1B).
Fig. 1. Basal levels of mtDNA damage are increased in the absence of ATM leading to mitochondrial dysfunction.
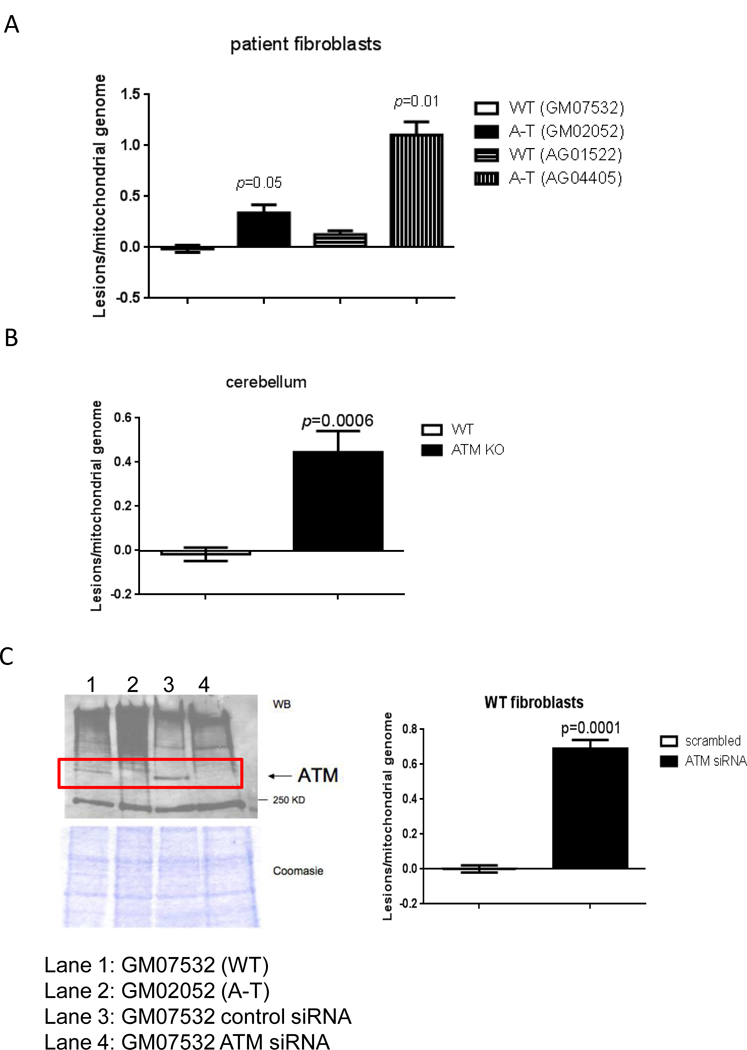
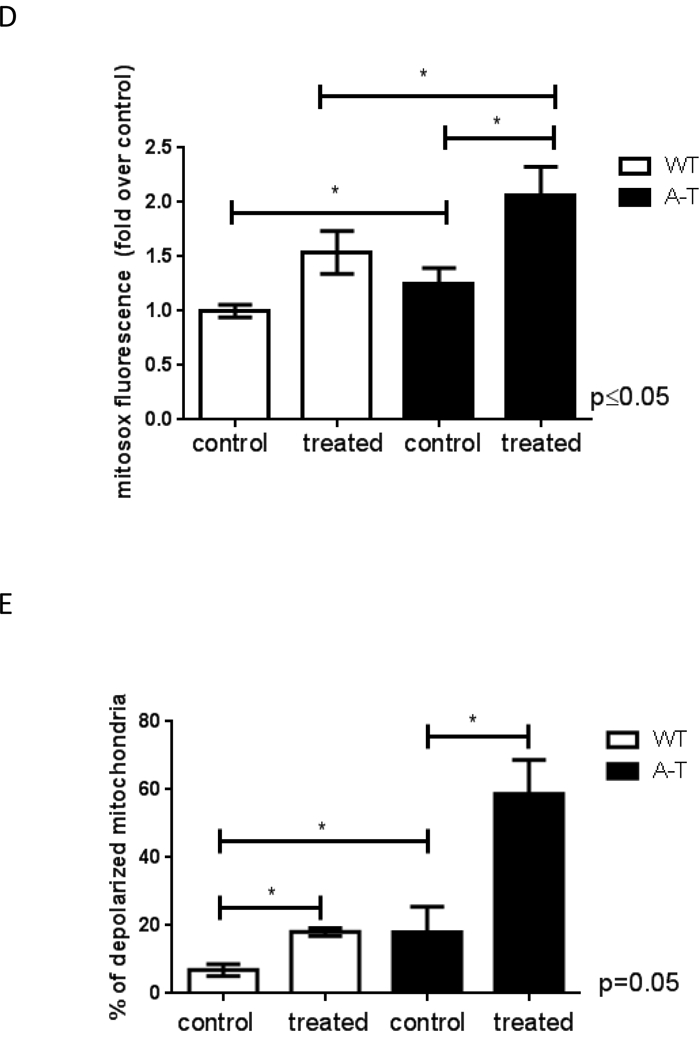
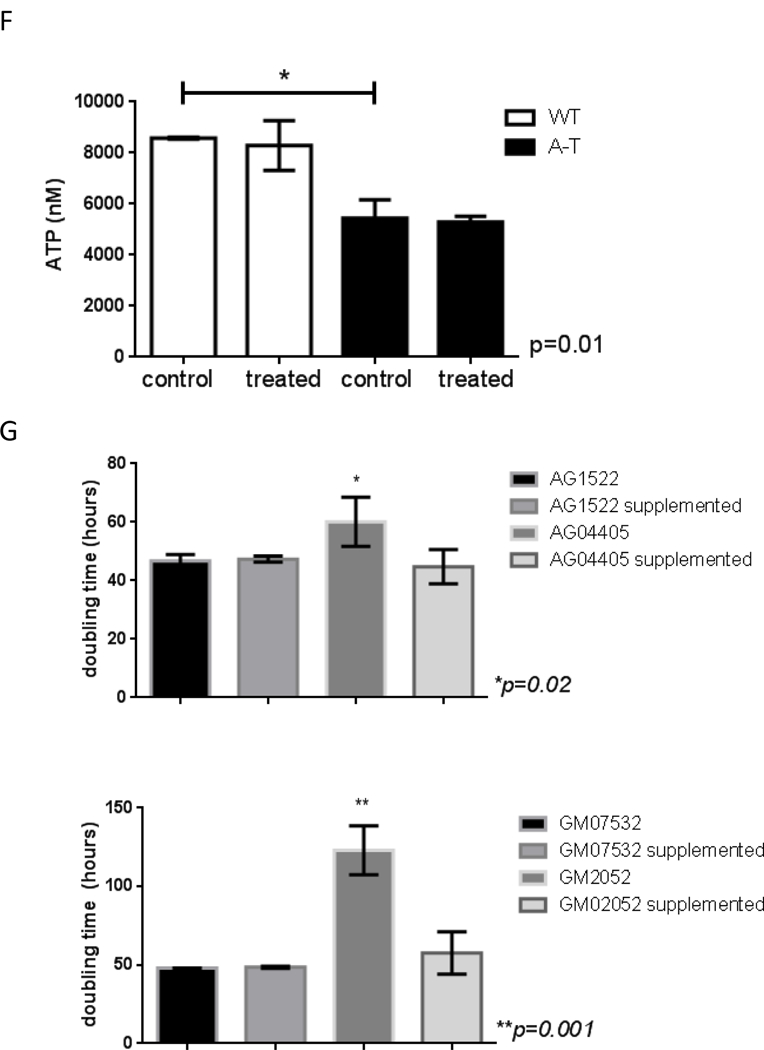
Genomic DNA was isolated from ATM-proficient and deficient samples and mtDNA integrity was analyzed using QPCR. Data were normalized to mtDNA content using amplification of the small mtDNA fragment (Santos et al., 2006). DNA was isolated 3 independent times and QPCR was performed two independent times with each DNA isolate, error bars represent ± SEM; (A) patient-derived cells and (B) mouse tissue (C) siRNA was transiently transfected in WT cells and levels of ATM analyzed by immunoblots 4 days after transfections (left). MtDNA damage was estimated in cells transfected with scrambled-control or siRNA targeted to ATM (right). QPCR was performed two independent times for each DNA sample from 3 independent experiments, error bars represent ± SEM. WT and A-T cells were submitted to 60 min treatment with 200 μM H2O2. Immediately after treatment cells were collected and: (D) mitochondrial ROS measured using Mitosox by FACS, N=3; * indicates significance between control WT and control A-T, and between A-T control and treated samples; (E) mitochondrial membrane polarization judged using JC-1 by FACS, N=3, * indicates significance using WT control as reference or between A-T control and treated cells. (F) Steady state levels of ATP were estimated using a luciferase-based assay. Experiments were reproduced at least 3 independent times; error bars represent ± SD. = p ≤0.05. * indicates significance between WT and A-T control. (G) Doubling time was calculated by following cell growth and counting cells each time they reached confluency. Results represent the mean doubling time calculated over a period of 3 weeks for two independently grown cultures for each cell type.
A limitation of our approach is that the WT and A-T fibroblasts were not derived from an isogenic background. Therefore, to rule out that basal damage reflected intrinsic differences in mitochondrial metabolism in the unrelated cell lines, we depleted ATM in WT GM07532 fibroblasts using siRNA and analyzed mtDNA integrity by QPCR. Increased mtDNA lesions were also observed in this context (Figure 1C) confirming that ATM deficiency is sufficient for accumulation of mtDNA damage. Consistent with decreased mtDNA integrity, analysis of mitochondrial function revealed that ATM-deficient cells produced increased mitochondrial ROS (Fig. 1D) had more depolarized mitochondria (Fig. 1E, representative dot plot on Fig. S2) and lower steady state levels of ATP (Fig. 1F), all of which are in agreement with previous reports [3,4,6,7]. ROS generation and membrane depolarization were further exacerbated upon exposure to exogenous hydrogen peroxide (H2O2, 200 μM for 60 min), suggesting that ATM protects mitochondria from oxidative damage. The sensitization of mitochondria of ATM- deficient cells to H2O2 exposure could not be simply explained by reduced antioxidant capacity because H2O2 was decomposed at a similar rate regardless of the presence of ATM (Fig. S3). The altered mitochondrial parameters observed in the absence of ATM were sufficient to impair cell growth as A-T cells had significantly longer doubling time compared to WT counterparts (Fig. 1G). This growth defect was rescued by supplementation of the medium with glucose, pyruvate and uridine (Fig. 1G), a classical means used to support glycolytic growth under conditions in which OXPHOS is impaired.
3.2. MtDNA repair is deficient in the absence of ATM.
As comparable half-life of H2O2 (Fig. S3) rules out that H2O2 remains longer and thus induces more damage in A-T fibroblasts, we next tested whether the increased steady-state level of mtDNA lesions in ATM-deficient cells was due to inefficient mtDNA repair. We measured mtDNA repair kinetics by exposing WT and A-T fibroblasts to 200 μM H2O2 for 60 min (a non-cytotoxic dose), and allowing recovery for up to 48h before analyzing mtDNA integrity using QPCR. ATM had no effect on the amount of mtDNA lesions present immediately after H2O2 exposure (Fig. 2A-C, see time 0h). Strikingly A- T cells were defective in the removal of oxidative damage from mtDNA, only repairing 30%- 40% of lesions in the first 24 hours as compared to 60%−80% repair in the WT (Fig. 2A and B). Even 48 hours after H2O2 exposure, only 70% of mtDNA damage was repaired in the A-T cells, while no lesions were detectable in the WT sample (Fig. 2B). This 2- to 3-fold reduction in mtDNA repair efficiency was recapitulated when ATM was depleted using siRNA (Fig. 2C) but was not observed upon pharmacological inhibition of the ribonucleotide reductase (RR, Fig. 2D), an enzyme responsible for de novo dNTP synthesis required for mtDNA replication and repair in quiescent mammalian cells [13] which we showed previously is impaired in the absence of ATM [4].
Fig. 2. MtDNA repair is impaired in the absence of ATM.
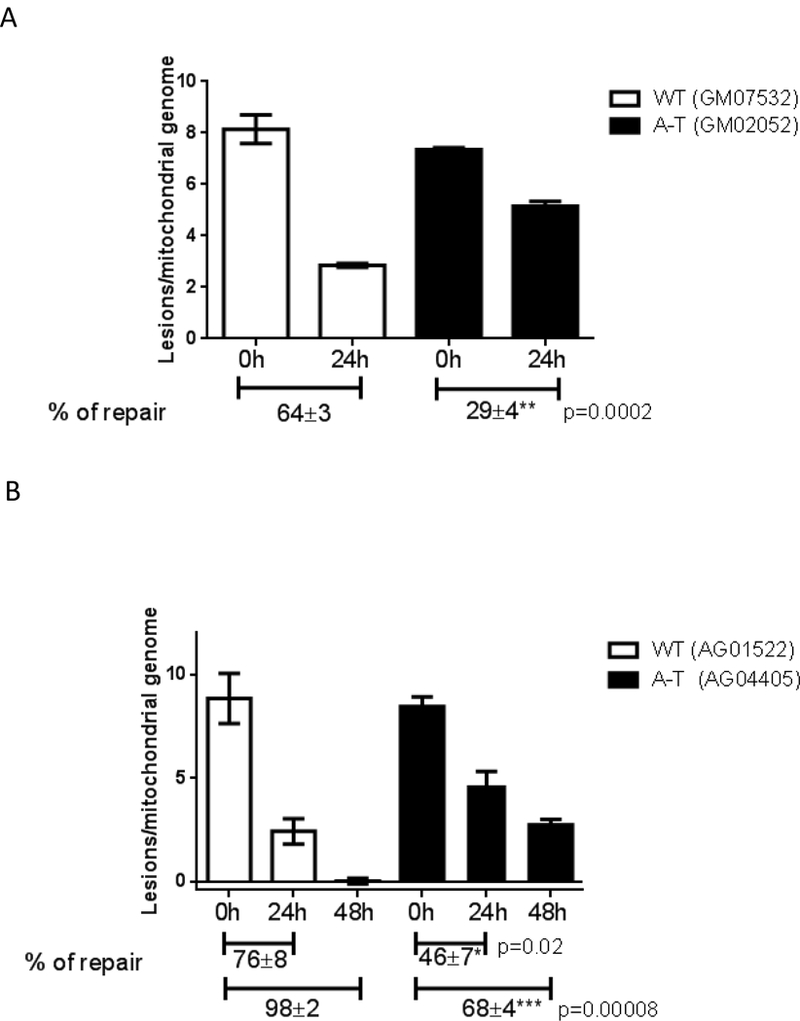
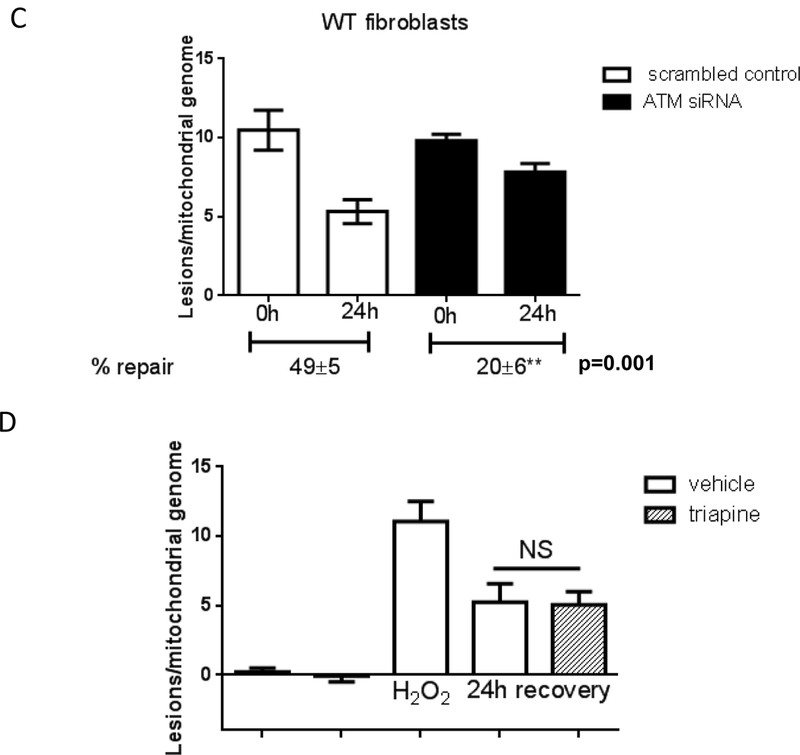
WT controls or A-T cells were submitted to treatment with 200 μM H2O2 for 60 min and allowed to recover for up to 48h. MtDNA integrity was analyzed using QPCR; data were normalized to mtDNA copy number. Experiments were reproduced at least 3 independent time; error bars reflect ± SEM. (A) Patient- derived fibroblasts GM07532 (WT) and GM02052 (A-T), (B) patient-derived cells AG01522 (WT) and AG04405 (A-T). (C) mtDNA repair kinetics was evaluated in GM07532 (WT) upon knock down of ATM using siRNA. (D) Kinetics of mtDNA repair was followed in GM07532 (WT) up to 24h after H2O2 exposure in the presence or absence of 1 μM triapine, a RR-inhibitor. NS = not-significant.
Defective clearance of mitochondria due to impaired autophagy or the specific degradation of mitochondria, mitophagy, could lead to accumulation of dysfunctional mitochondria and persistence of oxidative mtDNA damage. Accordingly, it has been reported that ATM modulates autophagy through an AMPK and mTOR-dependent pathway under oxidative stress [14] and that mitophagy is impaired in an ATM KO mouse model [6]. However, three independent results argue against the observed persistence of oxidative mtDNA damage being caused by defective autophagy/mitophagy. First, the H2O2 exposure used in this study failed to activate AMPK in A-T or WT cells, but was adequate to induce phosphorylation of AMPK in an unrelated fibroblast cell line (Fig. S4A), indicating that our experimental conditions can activate AMPK. Second, estimation of mitochondrial content using Mitotracker green did not reveal differences in accumulation of mitochondria after treatment with H2O2 (Fig. S4B, representative traces on Fig. S4C). Finally, the levels of the mitochondrial-localized matrix protein, mtHSP70, and the mtDNA-encoded protein, ND1, were similar prior to and after H2O2 treatment in each cell type (Fig. S4D), confirming that mitochondria are not accumulating in the absence of ATM.
Collectively these results indicate that the ability to remove damaged mitochondria is likely not different between WT and A-T cells under our experimental conditions, and support the hypothesis that increased mtDNA damage in ATM-deficient cells persists because of defective oxidative mtDNA repair.
3.3. DNA Lig3 levels and activity are reduced in A-T.
The only known DNA repair pathway operating in mitochondria for the removal of oxidative lesions is base excision repair (BER). Proteins that work in mitochondrial BER (mtBER) are nuclear-encoded, and some are splicing or translational variants of nuclear BER proteins [15,16]. Similar to nuclear BER, mtBER recognizes and removes the damaged base using a DNA glycosylase (such as OGG1 or NTH1) leading to the formation of abasic sites, also known as apurinic/apyrimidinic (AP) sites. AP sites are cleaved by the mitochondrial APE-1 endonuclease and lyase activity of DNA polymerase gamma (pol γ), and the resulting single-strand break is processed by either short-patch (where a single nucleotide is replaced) or long-patch BER (where 2-10 new nucleotides are synthesized). The latter may involve the flap-endonuclease 1 (FEN1), DNA2 or ExoG [16-18]. Filling of the mtDNA gap is performed by DNA pol γ, and the repair patch is sealed by DNA Lig3 [15,16]. Interestingly, total cell lysates from A-T cells exhibited efficient BER activity because nuclear BER is functional [19]. Therefore, even though nuclear and mtBER share similar enzymes and pathways, our data suggest that ATM-deficient cells are less efficient at one or more of the BER steps that are specific to mtDNA repair.
Oxidative stress causes the phosphorylation of DNA Lig3 by Cdk2, which is followed by the ATM-dependent dephosphorylation of Lig3 [20]. As the only known DNA ligase in mitochondria [21], Lig3 was previously shown to be required to maintain mtDNA integrity [22] and recently shown to be essential for mtDNA maintenance and organismal survival in addition to its important role in nuclear BER [23,24]. In the nucleus, deficiency in Lig3 can be compensated by Ligl [24,25]. Based on these considerations, we hypothesized that decreased mitochondrial Lig3 activity was a prime candidate to explain the mtDNA repair defect observed in cells lacking ATM. To address this question, WT and A-T cells were treated with H2O2 and mitochondria isolated based on differential centrifugation. Nuclear and mitochondrial lysates were then used to assay DNA ligase activity using a linearized plasmid as previously described [26]. Briefly, pUC18 was cut with either PstI or Xbal and incubated overnight at 16°C with extracts from WT or A-T samples. Following this period the plasmid DNA was precipitated and purified, and run on agarose gels [26]. Reactions were supplemented with exogenous ATP and T4 DNA ligase was used as positive control. Data presented in Fig. 3 is representative of different biological replicates in which lysates were obtained from independently grown cultures; ligation assay was also performed at least 3 independent times. Consistent with our hypothesis, mitochondrial extracts from A-T patient fibroblasts were indeed impaired in the ability to re- ligate the linearized plasmid DNA compared to extracts from WT cells (representative gel on Fig. 3 upper panels). Defective ligase activity was specific to isolated mitochondria as ATM- deficient nuclear extracts were capable of ligating the same linear DNA substrate (Fig. 3 lower left panel), likely reflecting the functional redundancy of DNA Ligl and Lig3 in the nucleus [24,25]. ATM-dependent reduction of mitochondrial ligase activity was recapitulated in extracts from cerebrum and cerebellum from the ATM KO mice although the signal obtained with tissue was low (Fig. S5A). Results were reproducible irrespective of the linearizing restriction endonuclease used (PstI or Xbal).
Fig. 3. Mitochondrial Lig3 activity is reduced in the absence of ATM.
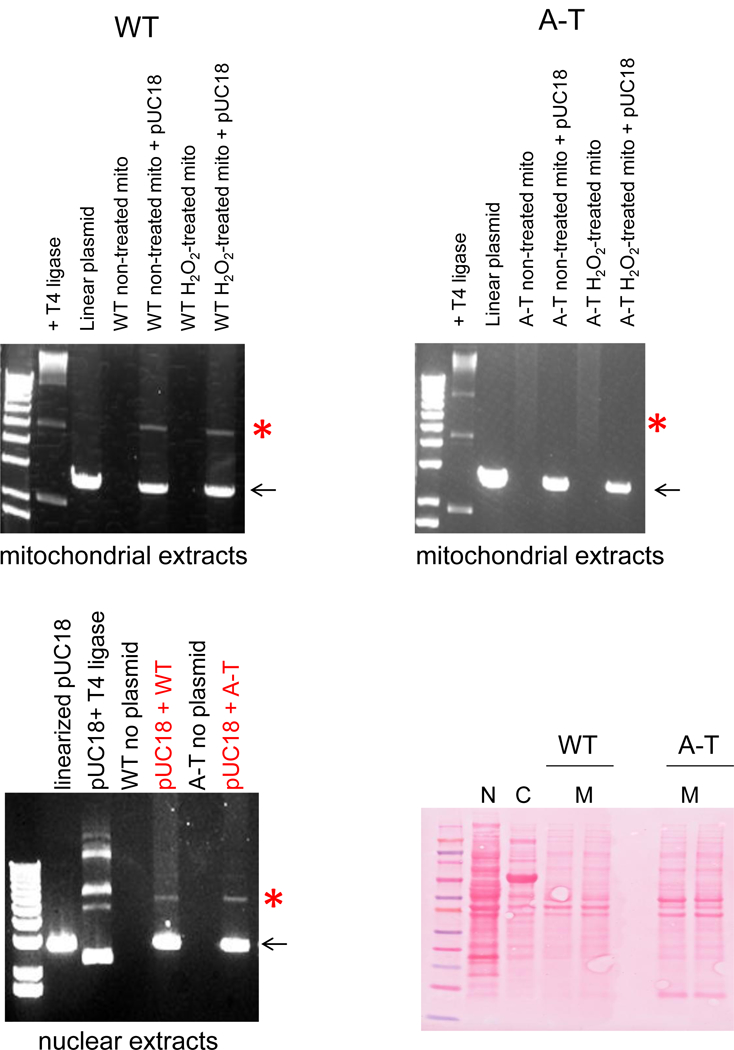
WT and A-T cells were submitted to 200 μM H2O2 for 60 min and mitochondria isolated. pUC18 was cut with PstI and incubated with 10 μg of mitochondrial extracts at 16°C overnight. The DNA was then purified and ran on agarose gels. (A) Data obtained with WT cells (left) and with the A-T counterpart (right). Arrow indicates linearized plasmid; red asterisk indicates expected ligated product. Lower left panel shows data using 10 μg of nuclear extracts; only non-H2O2 treated cells were assayed. Right lower panel depicts Ponceau staining of lysates to assure use of comparable protein concentration in the ligase assay. N: nuclear, C: cytosol and M: mitochondria.
3.4. ATM regulates Lig3 protein stability and mtDNA repair.
Phosphorylation of nuclear DNA Lig4 by DNAPK targets Lig4 for degradation [27]. As under oxidative stress Lig3 is dephosphorylated in an ATM-dependent manner [20], it is plausible that in A-T Lig3 is predominantly phosphorylated and, analogously to Lig4, is targeted for degradation. Alternatively, it is possible that mitochondria of A-T cells cannot efficiently import the nuclear- encoded Lig3 due to higher proportion of depolarized mitochondria (Fig. 1E) and maintenance of mitochondrial membrane polarization is a requirement for import of mitochondrial matrix proteins [28]. In either case, reduction in mitochondrial Lig3 amounts could limit mtDNA repair. As the mitochondrial amount of the nuclear-encoded mitochondrial-imported mtHSP70 were not decreased after H2O2 exposure (Fig. S4D), we conclude that mitochondrial protein import is not limiting under our experimental conditions. Therefore, we next used Western blotting to evaluate the levels of DNA Lig3 in crude (whole cell), purified nuclear and mitochondrial extracts.
Lig3 amounts were reduced to about 50% in A-T patient-derived samples in all cellular compartments analyzed (Fig. 4A and Fig. S6A, left panel; blots are representative, graphs depict average signal intensity of Lig3 normalized to actin or ND-1 in three independent lysates). Similar results were obtained in ATM null brain tissue (Fig. 4B and Fig. S6A, right panel). It is clear based on these findings that the decline in mitochondrial Lig3 levels simply mimics decreased nuclear amounts. Decrease in total levels of Lig3 was also reproduced in an unrelated A-T immortalized lymphoblastoid cell lines (Fig. S6B), in human mammary epithelial cell cultures (HME-CC) in which ATM was stably depleted using shRNA [29] (Fig. S6C) and in pre- B cells derived from an independent ATM KO animal (Fig. S6D).
Fig. 4. Protein levels of Lig3 specifically are regulated by ATM.
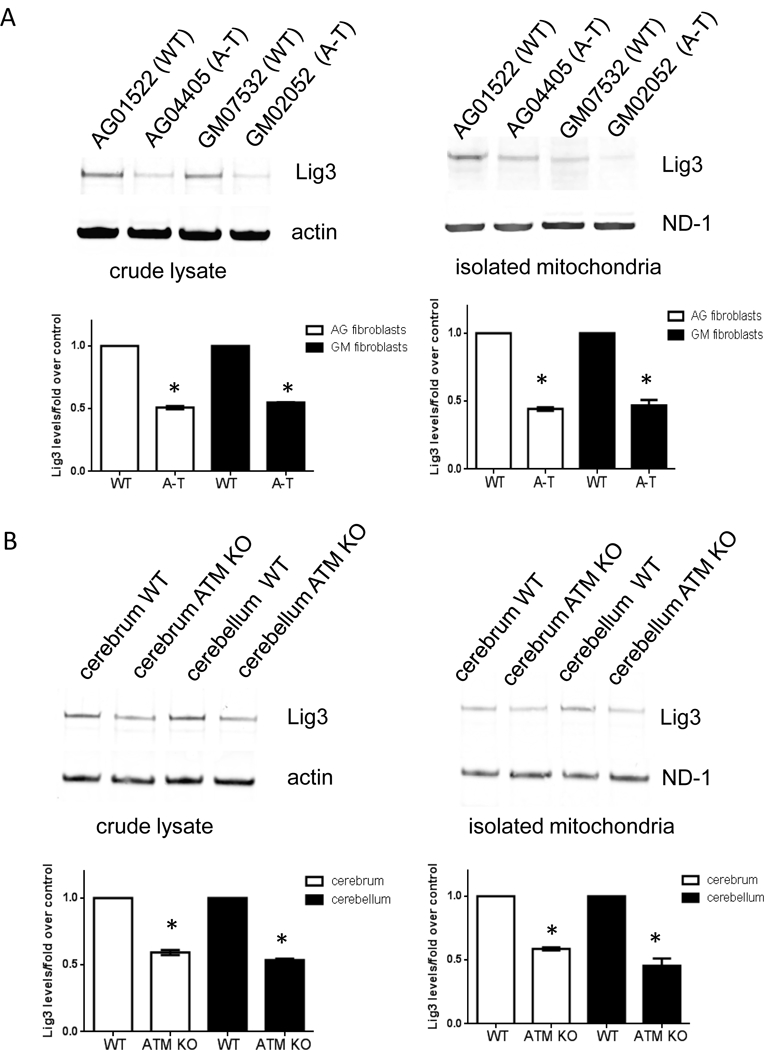
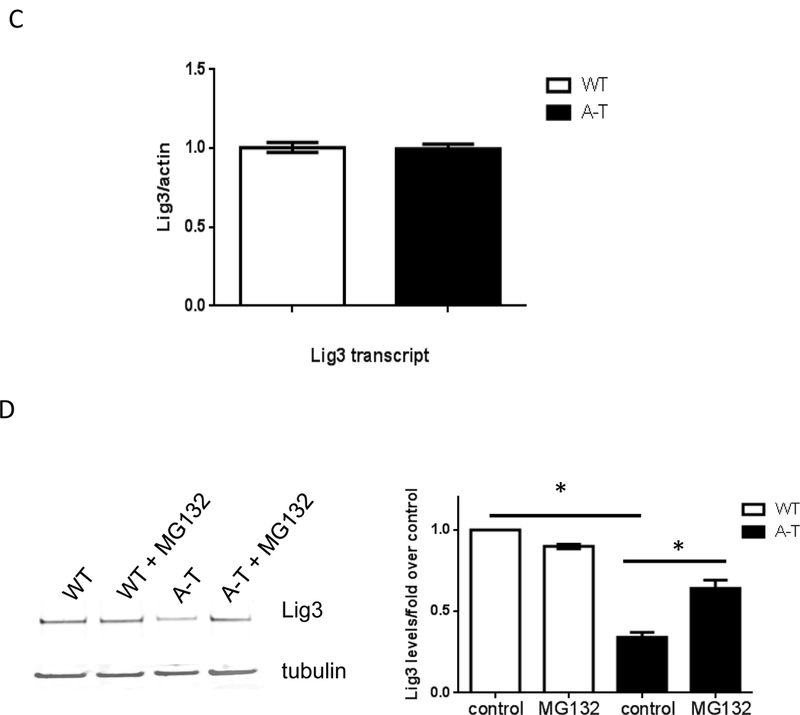
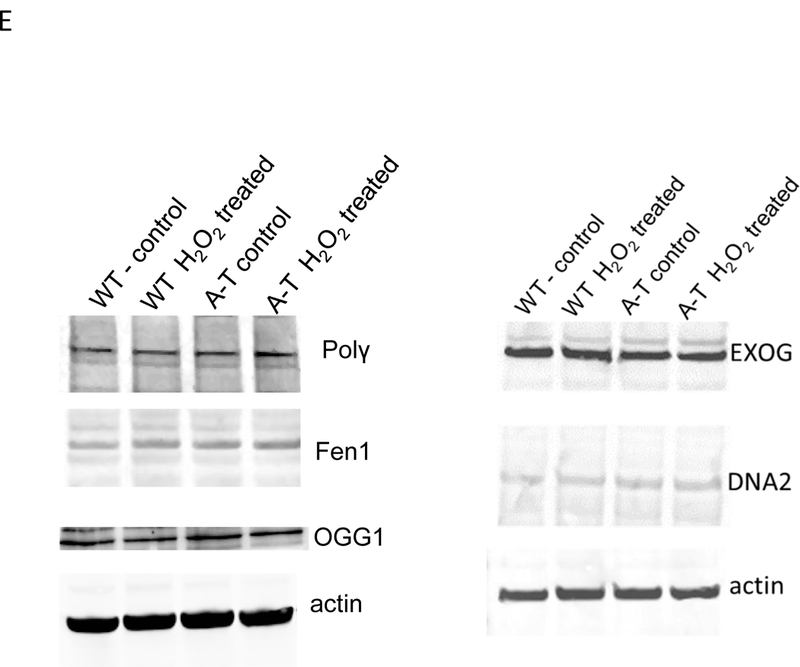
Lig3 levels were probed by Western blotting in crude (left) and isolated mitochondria (right) from (A) patient-derived fibroblasts and (B) tissue from WT and ATM KO animals. ND-1 is encoded by the mtDNA and was used as loading control for mitochondrial lysates. Graphs depict levels of Lig3 normalized to β-actin or ND-1. *=p ≤ 0.05. (C) Total RNA was isolated from WT and A-T fibroblasts and Lig3 transcript level was gauged by RT-PCR; data were normalized to actin. N=3 from independent RNA isolations; PCR was performed in triplicate. (D) WT and A-T cells were exposed to 25 μM of the proteasome inhibitor MG132 for 8h when cells were lysed and Lig3 levels were evaluated by Western blotting. Graph depicts levels of Lig3 normalized to tubulin. *=p ≤ 0.05. (E) Protein levels of the mtBER proteins Pol γ, OGG1, FEN1, EXOG and Dna2 were evaluated in WT and A-T cells prior to and after exposure to 200 μM H2O2 for 60 min. Blots are representative of 2-3 independent experiments.
Reduction in Lig3 was not manifested at the RNA level (Fig. 4C), suggesting that ATM regulates Lig3 stability via a post-transcriptional mechanism (e.g. post-translational modifications coupled to protein stability). Accordingly, pharmacological inhibition of the proteasome with MG132 increased levels of Lig3 in A-T patient fibroblasts (Fig. 4D) consistent with the notion that ATM regulates Lig3 stability.
To confirm that BER was not globally affected by the absence of ATM, we analyzed by Western blotting total levels of other mtBER proteins such as mitochondrial DNA Pol γ, OGG1, FEN1, EXOG and Dna2 in WT and A-T-derived fibroblasts prior to and after H2O2 exposure. We chose these proteins because they represent/act in different steps of the mtBER reaction. No changes in levels of any of these proteins were observed irrespective of ATM status or of H2O2 treatment (Fig. 4E). Taken together, results presented in Fig. 4 suggest that Lig3 turnover, specifically, is regulated by ATM.
That reduced Lig3 in ATM-deficient cells is sufficient to elicit mtDNA repair defect and mitochondrial dysfunction suggests that Lig3 activity is limiting for mtBER. If this is indeed the case, then increasing Lig3 amounts in A-T cells should reverse the defects in mtDNA repair. To test this premise, we compared mtDNA repair kinetics in A-T cells in the absence or presence of the proteasome inhibitor MG132. We took this approach as we reasoned that targeting exogenous Lig3 to mitochondria of A-T cells would still lead to its degradation due to the absence of ATM. While we were able to inhibit the proteasome within the time frame of our DNA repair experiments (Fig. 5A), the increase in Lig3 levels was consistently below the WT values under our experimental conditions (see example on Fig. 4D) and was insufficient for significantly impacting mtDNA repair kinetics. Longer exposure as well as higher doses of MG132 proved cytotoxic to the cells.
Fig. 5. Pharmacological inhibition of Lig3 recapitulates the mtDNA repair defects observed in A-T.
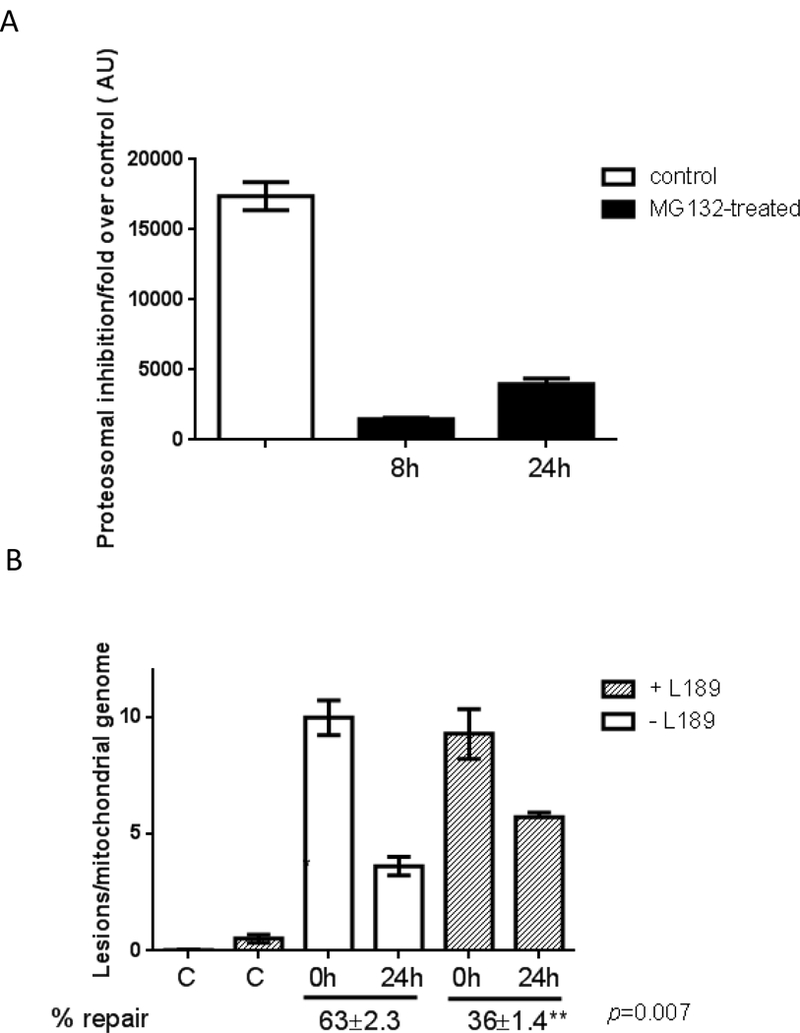
(A) AG04405 (A-T) cells were treated with 25 μM MG132 for 8h and the degree of proteasome inhibition was gauged up to 24h following MG132 exposure using a fluorescence assay. Results represent mean of 3 biological replicates, each of which was measured in duplicate. (B) AG01522 (WT for ATM) cells were exposed to 200 μM H2O2 for 60 min and mtDNA repair kinetics was followed using QPCR over 24h in the presence or absence of vehicle or 10 μM L189. Data were normalized to mtDNA copy number. Experiments were reproduced at least 3 independent time; error bars reflect ± SEM.
To circumvent this issue we took a complementary approach. A DNA ligase inhibitor, L189, was recently described and shown to inhibit DNA ligases by competitively binding to nicked DNA [30]. If Lig3 is indeed limiting for mtDNA repair, than pharmacological inhibition of Lig3 in WT cells should recapitulate the mtDNA repair defects observed in ATM-deficient cells (Fig. 2). This approach has the advantage of maintaining ATM-dependent regulation of Lig3 but the disadvantage of non-specific inhibition of DNA ligases. However, as only Lig3 works in mitochondria any effect of L189 on mtDNA integrity would reflect inhibited Lig3 activity. We treated AG01522 (WT for ATM) cells with 200 μM H2O2 for 60 min and allowed them to recover for 24h in the presence or absence of 10 μM L189 when mtDNA repair kinetics was analyzed. Treatment with L189 for 24h was not cytotoxic to cells, in agreement with a previous report [30] but phenocopied the deficiency in mtDNA repair detected in A-T cells at 24h (compare Fig. 5B to Fig. 2B). It is noteworthy that inhibition of Lig3 per se led to increased mtDNA damage (Fig. 5B second bar from left) consistent with enhanced basal mtDNA damage in ATM-deficient samples (Fig. 1) and with decreased mtDNA integrity observed upon knockdown of Lig3 [22].
Taken together, our data indicate that lower steady-state amounts of Lig3 in the absence of ATM become insufficient for mtDNA repair, which is associated to mitochondrial dysfunction and, along with the nuclear defects, likely play a role in A-T pathogenesis.
4. Discussion
In trying to understand mitochondrial dysfunction in A-T pathogenesis, we made the key observation that lack of ATM decreases mtDNA integrity and impairs removal of oxidative lesions from mtDNA, indicative of defective mtDNA repair (Fig. 1-2). Detailed scrutiny of the mechanisms associated with this phenotype identified that Lig3 levels are reduced in the absence of ATM, resulting in decreased Lig3 activity in mitochondria (Fig. 3-5). As Lig3 is the only mitochondrial DNA ligase [21], it follows that decreased Lig3 activity in mitochondria can lead to persistent mtDNA damage, mitochondrial dysfunction and perhaps be causative in the pathogenesis of A-T. Our data together with previous results showing that knockdown of Lig3 leads to reduced mtDNA integrity and mitochondrial respiration [22] and that targeted deletion of mitochondrial Lig3 in the central nervous systems results in severe ataxia in mice [23] support this hypothesis.
Our findings indicate that, like Lig4 [27], the stability of Lig3 may depend on its phosphorylation state. While it was previously shown that the E3 ubiquitin ligase CHIP can control the turnover of BER proteins, including Lig3, it was proposed that this regulation involved protein-protein (in this case Lig3 and XRCC1) and/or protein-DNA interactions in order to maintain adequate protein stability. However, the mechanism that allowed CHIP to differentiate between free and to-be degraded BER proteins remained unknown [31]. Given the fact that XRCC1 is not present in mitochondria and that our results show that Lig3 levels are reduced in all ATM-deficient samples analyzed, irrespective of genotoxic stress, it seems clear that stability of Lig3 must be accomplished through additional means. Our data not only indicate that ATM is a main regulator of Lig3 stability but also suggest involvement of phosphorylation events.
The observation that Lig3 is constitutively expressed and its protein levels are tightly regulated suggests that Lig3 is continuously required to maintain mtDNA integrity. Modulation of its stability by phosphorylation (less stable) and dephosphorylation (more stable) offers a rapid mode of regulation allowing Lig3 to accumulate in times of genotoxic stress. This mode of regulation is known to stabilize other important genome guardians such as p53 [32] and, relevant to mtDNA metabolism, the mitochondrial transcription factor TFAM [33]. It is not yet known how ATM regulates Lig3 levels, whether it needs to be activated through the classical double strand break (DSB) node or, perhaps, whether the newly recognized redox modifications on ATM [34] are required. Considering that under basal state no nuclear DNA damage was detected (data not shown) while increased mitochondrial ROS and reduced Lig3 levels were observed (Figs. 1 and 2), it is tempting to speculate that the redox sensor ability of ATM is involved. The ATM variants that cannot be activated by H2O2 [35] will be useful in addressing this question.
An essential role for Lig3 in mtDNA homeostasis was previously described although it was not firmly established whether its function was most relevant for mtDNA replication or repair [23,24]. Our data indicate that Lig3 function is absolutely required for mtDNA repair both at basal levels and under genotoxic stress although, as previously noted, mitochondrial ligase activity can be artificially compensated by other DNA ligases [24]. More recently, it was shown that levels of mitochondrial Lig3 are in large excess and thus dispensable for the viability of cultured cells while essential to maintain mtDNA [36]. In opposition to our results, the authors show that cells with severely reduced expression of Lig3 maintain normal mtDNA copy number and respiration, and show reduced viability in the face of oxidative damage. They also show increased mtDNA degradation in response to oxidative damage [36] which was not observed in our study. It is worth noting that this particular study used HeLa cells and it is possible that the observed phenotypes are associated to the tumorigenic potential of this particular cell line as compared to our non-transformed and primary fibroblasts. Of relevance to A-T, targeted deletion of Lig3 in the central nervous system leads to incapacitating ataxia in mice [23], a hallmark of A-T pathology. While the mechanisms underlying the cerebellar degeneration in A-T are currently unknown, it has been hypothesized that mitochondrial dysfunction and oxidative stress may be playing a role [3-7,37,38]. Based on our findings, we propose that the mitochondrial dysfunction caused by diminished levels of Lig3, the resulting decrease in DNA Lig3 activity and the consequential defect in mtDNA repair contribute to the cerebellar degeneration. Such effects can result in increased steady state levels of mitochondrial ROS production (as shown in Fig. 1) that in this scenario could not only perpetuate the cycle of mtDNA damage and mitochondria dysfunction, but would also put the nuclear genome at risk for oxidative damage and increased DSB. This effect compounding with the ATM-dependent DSB and redox signaling defect could lead to loss of sensitive neurons in the brain and eventually affect other tissues, a phenotype reminiscent of classical mitochondrial diseases [39]. Furthermore, this could also explain why AT carriers are at increased risk of developing cancers [40-43].
5. Conclusions
The main conclusions of our study are that in the absence of ATM DNA Lig3 levels are significantly dimished, negatively impacting mtDNA repair. The impact of ATM on mtDNA repair seems to be specific to Lig3 as no other mtBER protein was altered. Collectively, these results bring to light a new and previously unidentified impact of ATM on mtDNA repair. The importance of Lig3 regulation by ATM reported here in addition to previous work from the Tomkinson’s group [20] prompts more studies about the exact mechanism by which ATM maintains Lig3 stability and whether modulating the levels of Lig3 would be an effective treatment for A-T. In addition, reduced Lig3 protein amounts may provide an easy and effective biomarker of ATM dysfunction that can be used as a diagnostic tool in pre-symptomatic patients with familial history of A-T.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Ronald Mason for hosting Drs. Sharma and Santos at NIEHS. We would like to acknowledge the technical help with flow cytometry and real time PCR from Drs. Carl Bortner and Dr. Artiom Gruzdev, respectively, at NIEHS, and Drs. Douglas Ganini da Silva and Stela S. Palii (NIEHS) for critical review of the manuscript. We would also like to thank Dr. Alan Tomkinson (University of New Mexico) and Richard Paules for kindly sharing reagents, and Ms. Sharen Mckay and Dr. Brooke Christian, members from the Shadel laboratory, for providing the mouse tissues.
This work was partially funded by the Department of Defense, Army Research Office, grant number 56027LS to JHS.
Footnotes
Conflict of Interest
The authors declare no conflict of interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shiloh Y, and Ziv Y, The ATM protein kinase: regulating the cellular response to genotoxic stress, and more, Nat. Rev. Mol. Cell. Biol. 4, (2013) 197–210. [PubMed] [Google Scholar]
- 2.Ambrose M, Gatti RA, Pathogenesis of ataxia-telangiectasia: the next generation of ATM functions, Blood 121, (2013) 4036–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrose M, Goldstine JV, Gatti RA, Intrinsic mitochondrial dysfunction in ATM- deficient lymphoblastoid cells, Human. Mol. Gen. 16, (2007) 2154–2164. [DOI] [PubMed] [Google Scholar]
- 4.Eaton J, Lin ZP, Sartorelli AC, Bonawitz NC, Shadel GS, Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis, Journal of Clin. Inv. 177, (2007) 2723–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel AY, McDonald TM, Spears LD, Ching JK, Fisher JS, Ataxia telangiectasia mutated influences cytochrome c oxidase activity, Biochem. Biophys. Res. Commun. 2405, (2011) 599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A Valentin-Vega Y, H Maclean K, Tait-Mulder J, Milasta S, Steeves M, C Dorsey F, L Cleveland J, R Green D, B Kastan M, Mitochondrial dysfunction in ataxia-telangiectasia, Blood 119(6), (2012) 1490–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza AD, Parish IA, S Krause D, Kaech SM, Shadel GS, Reducing Mitochondrial ROS Improves Disease-related Pathology in a Mouse Model of Ataxia-telangiectasia, Mol. Ther. 21, (2012) 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johri A, Beal MF, Mitochondrial dysfunction in neurodegenerative diseases, J. Pharmacol. Exp. Ther. 342, (2012) 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Houten B, Woshner V, Santos JH, Role of mitochondrial DNA in toxic responses to oxidative stress, DNA Repair (Amst) 5, (2006) 145–152. [DOI] [PubMed] [Google Scholar]
- 10.Santos JH, Hunakova L, Chen Y, Bortner C, Van Houten B, Cell sorting experiments link persistent mitochondrial DNA damage with loss of mitochondrial membrane potential and apoptotic cell death, J. Biol. Chem. 278, (2003) 1728–1734. [DOI] [PubMed] [Google Scholar]
- 11.Santos JH, Meyer JN, Mandavilli BS, Van Houten B, Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells, Methods Mol. Biol. 314, (2006) 183–199. [DOI] [PubMed] [Google Scholar]
- 12.Kovalenko OA, Santos JH, Analysis of oxidative damage by gene-specific quantitative PCR. Curr. Protoc, Hum. Genet. 19, (2009) 1.1–19.1.13. [DOI] [PubMed] [Google Scholar]
- 13.Pontarin G, Ferraro P, Bee L, Reichard P, Bianchi V, Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells, Proc. Natl. Acad. Sci. U S A. 109, (2012) 13302–13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander A et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. U S A. 107, (2010) 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandavilli BS, Santos JH, Van Houten B, Mitochondrial DNA repair and aging, Mutat. Res. 509, (2002) 127–151. [DOI] [PubMed] [Google Scholar]
- 16.Sykora P, Wilson DM 3rd, Bohr VA, Repair of persistent strand breaks in the mitochondrial genome, Mech. Ageing Dev. 133, (2012) 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng L, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates, Mol. Cell. 32, (2008) 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tann AW, Boldogh I,, Meiss G, Qian W,, Van Houten B, Mitra S, Szczesny B, Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5’-EXO/endonuclease) in their repair, J. Biol. Chem. 286, (2011) 31975–31983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cappelli E, Rossi O, Chessa L, Frosina G, Efficient DNA base excision repair in ataxia telangiectasia cells, Eur. J. Biochem. 267, (2000) 6883–6887. [DOI] [PubMed] [Google Scholar]
- 20.Dong Z, Tomkinson AE, ATM mediates oxidative stress-induced dephosphorylation of DNA ligase IIIalpha, Nucleic Acids Res. 34, (2006) 5721–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellenberger T, Tomkinson AE, Eukaryotic DNA ligases: structural and functional insights, Annu. Rev. Biochem. 77, (2008) 313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakshmipathy U, Campbell C, Anti-sense mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity, Nucleic Acids Res. 29, (2001) 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair, Nature 471, (2011) 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simsek D, et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1- dependent repair, Nature 471, (2011) 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arakawa H, et al. Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells, Nucleic Acids Res. 40, (2012) 2599–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakshmipathy U, Campbell C, Double strand break rejoining by mammalian mitochondrial extracts, Nucleic Acids Res. 27, (1999) 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YG, Nnakwe C, Lane WS, Modesti M, Frank KM, Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase, J. Biol. Chem. 279, (2004) 37282–37290. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt O, Pfanner N, Meisinger C, Mitochondrial protein import: from proteomics to functional mechanisms, Nat. Rev. Mol. Cell. Biol. 11, (2010) 655–667. [DOI] [PubMed] [Google Scholar]
- 29.Palii SS, Cui Y, Innes CL, Paules RS, Dissecting cellular responses to irradiation via targeted disruptions of the ATM-CHK1-PP2A circuit, Cell Cycle 12, (2013) 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair, Cancer Res. 68, (2008) 3169–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons JL, Tait PS, Finch D, Dianova II, Allinson SL, and Dianov GL (2008) CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 29, 477–487. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins LM, Durell SR, Mazur SJ, Appella E, p53 N-terminal phosphorylation: a defining layer of complex regulation, Carcinogenesis 33, (2012) 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu B, et al. Phosphorylation of Human TFAM in Mitochondria Impairs DNA Binding and Promotes Degradation by the AAA(+) Lon Protease, Mol. Cell 49, (2012) 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT, ATM activation by oxidative stress, Science 330, (2007) 517–521. [DOI] [PubMed] [Google Scholar]
- 35.Guo Z, Deshpande R, Paull TT, ATM activation in the presence of oxidative stress, Cell Cycle 24, (2010) 4805–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shokolenko IN, Fayzulin RZ, Katyal S, McKinnon PJ, Wilson GL, Alexeyev MF. Mitochondrial DNA Ligase is dispensable for the viability of cultured cells, but essential for mtDNA maintenance. J Biol Chem. 2013. July 24 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamsler A, et al. Increased oxidative stress in ataxia telangiectasia evidenced by alterations in redox state of brains from Atm-deficient mice, Cancer Res. 61, (2001) 1849–1854. [PubMed] [Google Scholar]
- 38.Cosentino C, Grieco D, Costanzo V, ATM activates the pentose phosphate pathway promoting anti-oxidant defense and DNA repair, EMBO J. 30, (2011) 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vafai SB, Mootha VK, Mitochondrial disorders as windows into an ancient organelle, Nature 491, (2012) 374–383. [DOI] [PubMed] [Google Scholar]
- 40.Fletcher O et al. Missense variants in ATM in 26,101 breast cancer cases and 29,842 controls, Breast Cancer Association Consortium Cancer Epidemiol Biomarkers Prev. 19, (2010)2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliveira S et al. Genetic polymorphisms and cervical cancer development: ATM G5557A and p53bp1 C1236G, Oncol. Rep. 4, (2012) 1188–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen L et al. Association between ATM polymorphisms and cancer risk: a meta-analysis, Mol. Biol. Rep. 39, (2012) 5719–5725. [DOI] [PubMed] [Google Scholar]
- 43.Skowronska A et al. ATM germline heterozygosity does not play a role in chronic lymphocytic leukemia initiation but influences rapid disease progression through loss of the remaining ATM allele, Haematologica 97, (2012) 142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma NK et al. Human telomerase acts as a hTR-independent reverse transcriptase in mitochondria, Nucleic Acids Res. 40, (2012) 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frezza C, Cipolat S, Scorrano L, Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts, Nat. Protoc. 2, (2007) 287–295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


