Abstract
Chromatin structure has emerged as a key contributor to spatial and temporal control over the initiation of DNA replication. However, despite genome-wide correlations between early replication of gene-rich, accessible euchromatin and late replication of gene-poor, inaccessible heterochromatin, a causal relationship between chromatin structure and replication initiation remains elusive. Here, we combined histone gene engineering and whole-genome sequencing in Drosophila to determine how perturbing chromatin structure affects replication initiation. We found that most pericentric heterochromatin remains late replicating in H3K9R mutants, even though H3K9R pericentric heterochromatin is depleted of HP1a, more accessible, and transcriptionally active. These data indicate that HP1a loss, increased chromatin accessibility, and elevated transcription do not result in early replication of heterochromatin. Nevertheless, a small amount of pericentric heterochromatin with increased accessibility replicates earlier in H3K9R mutants. Transcription is de-repressed in these regions of advanced replication but not in those regions of the H3K9R mutant genome that replicate later, suggesting that transcriptional repression may contribute to late replication. We also explored relationships among chromatin, transcription, and replication in euchromatin by analyzing H4K16R mutants. In Drosophila, the X Chromosome gene expression is up-regulated twofold and replicates earlier in XY males than it does in XX females. We found that H4K16R mutation prevents normal male development and abrogates hyperexpression and earlier replication of the male X, consistent with previously established genome-wide correlations between transcription and early replication. In contrast, H4K16R females are viable and fertile, indicating that H4K16 modification is dispensable for genome replication and gene expression.
Animal cells duplicate large, complex genomes by initiating replication at distinct locations within the genome at different times during S phase. An evolutionarily conserved feature of this regulatory paradigm is a temporal order of DNA replication initiation events that results in characteristically early and late replicating regions of the genome (Rhind and Gilbert 2013). Such “replication timing” (RT) programs appear at early stages of animal development and ensure genome integrity during cell proliferation (Shermoen et al. 2010; Mantiero et al. 2011; Collart et al. 2013; Hamperl and Cimprich 2016; Yuan and O'Farrell 2016; Almeida et al. 2018). Importantly, RT is associated with mutational burden and SNP density, as spontaneous mutations occur less frequently in early compared to late replicating regions of the genome (Stamatoyannopoulos et al. 2009; Donley and Thayer 2013). Furthermore, perturbed RT is thought to be an early epigenetic event that predisposes cancer and disease-associated genome rearrangement (Ryba et al. 2012; Donley and Thayer 2013). Notwithstanding their importance, mechanisms that control where and when DNA replication initiates within an animal genome remain poorly understood.
In contrast to replication initiation in single-celled eukaryotes such as budding yeast, replication of animal genomes does not initiate at well-defined sequence motifs (Bell and Stillman 1992; MacAlpine et al. 2010; Miotto et al. 2016). Rather, two levels of genome organization have emerged as putative regulators of replication initiation: three dimensional arrangement of DNA within the nucleus and local chromatin structure, characterized in part by differential DNA accessibility (i.e., differential nucleosome occupancy) (Hiratani et al. 2008; Pope et al. 2014; Heinz et al. 2018). Current models posit that these features of genome organization regulate replication by influencing trans-acting factor recruitment to sites of replication initiation (i.e., origins) (Mantiero et al. 2011; Collart et al. 2013; Pope et al. 2014; Das et al. 2015; Miotto et al. 2016; Rivera-Mulia and Gilbert 2016). In all metazoan organisms examined to date, transcriptionally active, accessible euchromatin generally replicates early during S phase, whereas transcriptionally repressive, inaccessible heterochromatin generally replicates late (Bell et al. 2010; Eaton et al. 2011; Lubelsky et al. 2014). Despite strong genome-wide correlations between replication and chromatin structure in animal cells, efforts to determine a causal relationship between the two have been hampered by imprecise methods for manipulating chromatin structure in vivo. We therefore developed an approach for altering the distribution of accessible chromatin throughout the genome and determined if and how these changes in chromatin structure affect genome replication.
Strategies to manipulate chromatin structure in animal cells often involve perturbation of factors that establish, interpret, or remove histone post-translational modifications (PTMs). Although informative, these studies cannot precisely determine functional roles for histone PTMs in DNA replication because most histone-modifying enzymes also have nonhistone substrates that may participate in DNA replication (Glozak et al. 2005; Huang and Berger 2008). Therefore, to reduce potential pleiotropic effects of mutating histone-modifying enzymes, we employed a strategy in Drosophila to more precisely manipulate chromatin structure by mutating the histone genes themselves, an approach that is not currently feasible in other animal models. This strategy involves deleting the endogenous wild-type histone genes and replacing them with transgenic copies encoding a single amino acid substitution that prevents PTMs of a particular histone residue (Günesdogan et al. 2010; McKay et al. 2015). Here, we determine how two different histone mutations that affect chromatin organization and transcription in heterochromatin (H3K9R) and euchromatin (H4K16R), respectively, affect DNA replication initiation throughout the genome.
Results
Profiling replication timing in a Drosophila tissue
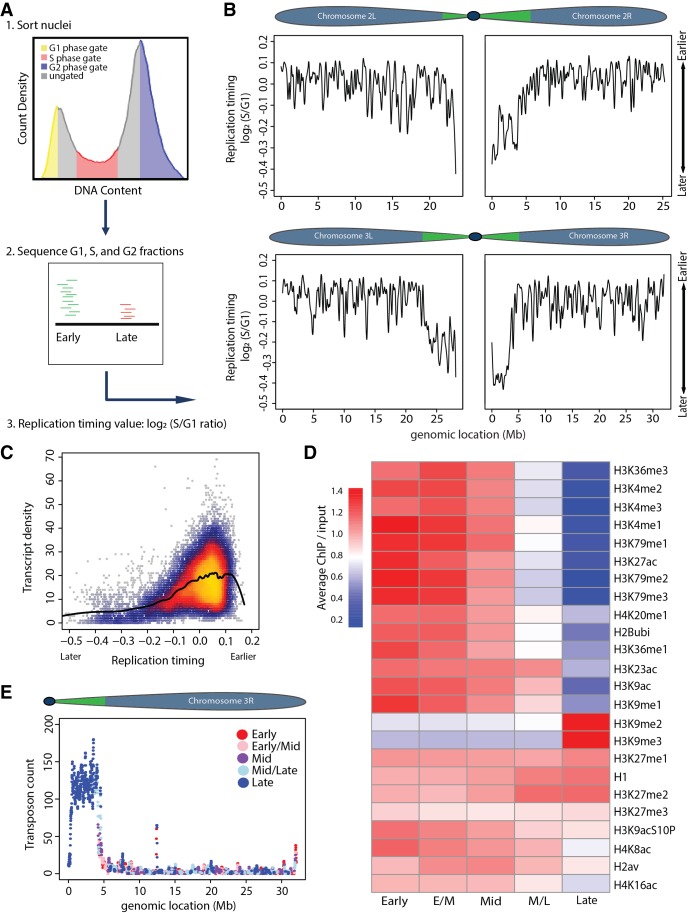
To probe the relationship between chromatin structure and replication in an intact animal, we adapted a genome-wide measure of RT for use in Drosophila wing imaginal discs, a relatively simple epithelium of proliferating diploid cells (Koren et al. 2014; Sasaki et al. 2017; Siefert et al. 2017). Our method is based on the premise that, in S phase cells, early replicating DNA sequences are overrepresented relative to late replicating ones, due to a higher probability of replication initiation (Rhind et al. 2010; Mantiero et al. 2011; Collart et al. 2013; Das et al. 2015). Consequently, RT data are a proxy for the propensity of replication initiation in a particular region of the genome. We performed whole-genome sequencing on DNA isolated from populations of G1 and S phase nuclei collected from wing discs by fluorescence-activated cell sorting (FACS) (Fig. 1A). Replication profiles were generated by determining the log2-transformed S/G1 read count at 100-kb intervals using a 10-kb slide across the genome (Methods; Fig. 1B; Supplemental Figs. S1, S2A), where larger values indicate earlier replication and smaller values indicate later replication. We chose 100-kb windows with a 10-kb slide because they produced the least amount of noise relative to smaller windows (Supplemental Fig. S1). RT values generated from independent S phase samples were highly reproducible (Supplemental Fig. S3).
Figure 1.
Measuring genome-wide replication timing in vivo. (A) Experimental paradigm: (1) Nuclei were FACS sorted into G1 (yellow), S (red), and G2 (blue) populations based on DNA content. (2) Sequenced DNA was mapped to the dm6 genome. More reads map to early than late replicating sequences. (3) Log2 S/G1 ratio generates RT profiles. Normalizing to G1 or G2 phase controls gave similar results. (B) LOESS regression line showing average yw (“yellow, white” control genetic background used for all fly lines) RT values (log2 S/G1) in 100-kb windows with 10-kb slide across Chr 2 and 3. Chromosome schematics show approximate locations of constitutive pericentric heterochromatin (green) and largely euchromatic arms (blue) (Riddle et al. 2011; Hoskins et al. 2015). (C) Heatscatter plot of yw log2 S/G1 (RT) versus gene density at all 10-kb windows across the genome with LOESS regression line (black). (D) Heat map of relative modENCODE histone PTM enrichment in bins of equally sized RT quintiles (early, early/mid, mid, mid/late, and late) generated using RT values (log2 S/G1) within 100-kb windows. modENCODE data are from third instar larvae (Celniker et al. 2009; see Supplemental Materials for accession numbers). Color indicates average enrichment of all windows within a quintile. Scale of heat map was capped at 1.4 to better represent distribution of values, as H3K9me2/me3 was greatly enriched in late replicating domains compared to other PTMs (see Supplemental Fig. S2E for noncapped H3K9me2/me3 heat map). (E) Plot of transposon number in 100-kb windows across Chr 3R with RT quintile (as determined in D) indicated by color.
Our wing disc replication profiles are similar to those previously generated from Drosophila cell lines and most closely correlate with RT data obtained from a cell line derived from the same developmental stage as wing discs (Supplemental Fig. S4; Lubelsky et al. 2014). Replication domain sizes ranged from 20–570 kb (Supplemental Fig. S2C), closely matching previous measurements (MacAlpine et al. 2004). Consistent with previous studies in zebrafish embryos and in fly and mammalian cultured cells (Bell et al. 2010; Eaton et al. 2011; Lubelsky et al. 2014; Petryk et al. 2016; Siefert et al. 2017), we found that earlier replication correlates with higher gene density (Fig. 1C), higher levels of transcription (Supplemental Fig. S2D), and the presence of activating histone PTMs such as H3K4me and H3K9ac (Fig. 1D). In contrast, later replication occurred in gene-poor regions (Fig. 1C) and was enriched in transposons (Fig. 1E; Supplemental Fig. S2G,H) and repressive histone PTMs, such as H3K9me2/me3 (Fig. 1D; Supplemental Fig. S2E,F). Our RT data revealed that, in wild-type wing discs, the pericentric heterochromatin replicates later than the mostly euchromatic chromosome arms (Fig. 1B; Supplemental Fig. S2B), consistent with prior cytological observations (Taylor 1960; Shermoen et al. 2010). Despite replicating at largely different times on average, both pericentric and euchromatic regions contained earlier and later replicating domains within them, such that the earliest replicating domains in pericentric heterochromatin exhibited similar values to the latest replicating domains on euchromatic chromosome arms. Thus, highly reproducible replication profiles from Drosophila tissue can be generated that match general features of replication found in other systems.
Replication timing is largely unchanged in H3K9R mutants
To determine how chromatin structure influences replication, we first tested if modification of H3K9 determines the difference in RT between heterochromatin and euchromatin. Defining features of heterochromatin are the presence of methylated H3K9 (H3K9me) and Heterochromatin Protein 1a (HP1a). HP1a binds H3K9me and facilitates heterochromatin formation through multimerization of HP1a molecules and recruitment of other factors (Canzio et al. 2011; Larson et al. 2017; Strom et al. 2017). Previously, we showed that H3K9R mutants are depleted of H3K9me and HP1a within pericentric heterochromatin (Penke et al. 2016). In addition, we found that loci within the pericentric heterochromatin of H3K9R mutants are nucleosome-depleted relative to controls, as measured by increased FAIRE-seq signal (Formaldehyde-Assisted Isolation of Regulatory Elements) (Penke et al. 2016).
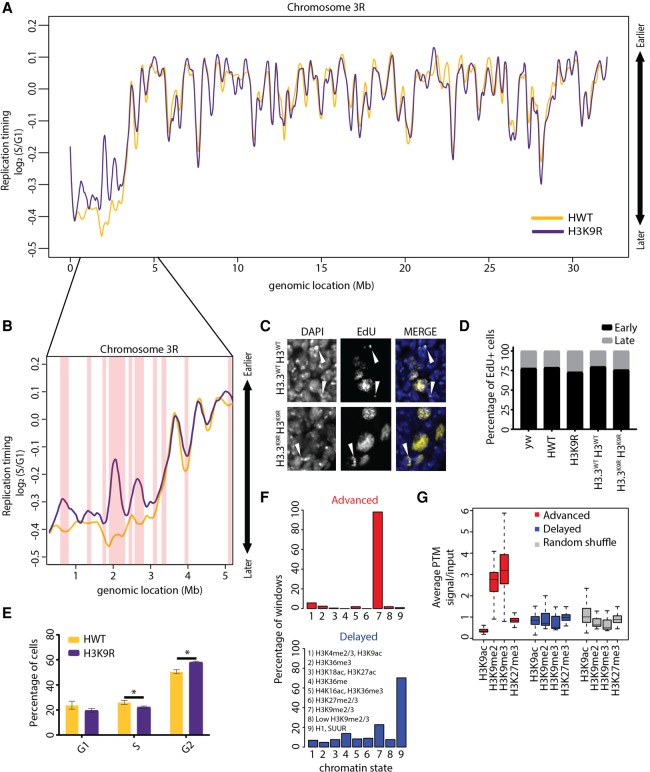
If increasing chromatin accessibility directly resulted in earlier replication initiation, we would expect large-scale advancement of RT at nucleosome-depleted H3K9R pericentromeres. We assigned RT values to 100-kb windows tiled 10-kb across the genome and used stringent significance thresholds (P < 0.01 [adjusted for multiple testing], absolute log2 fold change > 0.1; limma) to identify differential RT between H3K9R and control. Approximately 97% of the H3K9R genome has a similar replication profile compared to control, including much of the pericentric heterochromatin (Fig. 2A; Supplemental Fig. S5; Supplemental File S1). Consistent with these findings, cytological analysis of H3K9R imaginal cells revealed colocalization of the 359-bp pericentric satellite repeat on the X with late-patterned EdU incorporation at DAPI-bright chromocenters (Supplemental Fig. S6A), demonstrating that X pericentric heterochromatin remains late replicating. Late replicating H3K9R pericentric heterochromatin is not due to compensation by H3.3 variant histones because H3.3K9R; H3K9R mutants incapable of producing any H3K9me also contain late-patterned EdU incorporation at DAPI-bright chromocenters (Fig. 2C,D). FACS analysis revealed a small but statistically significant decrease in the number of S phase cells in H3K9R wing discs, indicating that cell cycle phasing is only slightly perturbed (Fig. 2E). These data show that H3K9 modification is dispensable for RT across most of the genome and that pericentric heterochromatin lacking HP1a and a closed chromatin configuration generally remains late replicating.
Figure 2.
Analysis of replication timing in H3K9R mutants. (A) Log2 S/G1 RT values at 100-kb windows with 10-kb slide for 12× HWT (histone wild type; yellow) and 12× H3K9R (purple) plotted across Chr 3R. See Supplemental Figure S5 for other chromosomes. (B) Approximately 5-Mb region of the pericentromeric heterochromatin of Chr 3R. Red vertical bars designate significant RT changes between H3K9R and HWT (P < 0.01, P value adjusted for multiple testing; absolute log2 fold change > 0.1; limma). (C) H3.3WT H3WT and H3.3K9R H3K9R (see Supplemental Materials for full genotype) first instar brains pulse-labeled for 1 h with EdU (yellow) and stained for DNA (blue; DAPI). White arrowheads designate late patterned EdU incorporation. (D) Percentage of EdU+ cells with early or late EdU incorporation patterns from ∼200 cells per genotype. There is no difference between genotypes (P > 0.05, χ2 test). (E) Cell cycle indices for HWT (yellow) and H3K9R (purple) wing disc cells acquired via FACS (calculated using the Dean-Jett-Fox model). Error bars indicate standard deviation of three experiments. (*) P < 0.05. (F) All advanced (red) or delayed (blue) 10-kb windows in H3K9R mutants were assigned to the nine chromatin states defined in flies (Kharchenko et al. 2011). Shown are the percentages of windows that overlap each chromatin state. (G) Average enrichment of modENCODE H3K9ac, me2, me3, and H3K27me3 signal from third instar larvae at 10-kb windows of advanced (red), delayed (blue), or randomized set of windows (Celniker et al. 2009).
Advanced replication occurs at newly accessible chromatin in H3K9R mutants
Despite largely unchanged RT in H3K9R mutants, 3% of the genome nevertheless exhibited altered RT (∼2% advanced and ∼1% delayed). Importantly, these changes do not result from pre-existing copy number differences between the G1 genomes of H3K9R and control (Supplemental Fig. S1). We used these changes to investigate the relationship between chromatin structure and replication initiation (Supplemental Fig. S6B). We found that the majority (82.1%) of earlier replicating 100-kb windows in H3K9R mutants are located in pericentric heterochromatin (Fig. 2B) or on the small fourth chromosome (Supplemental Fig. S5), which is primarily heterochromatic (Haynes et al. 2007). Importantly, these changes are unlikely to be caused by changes in the expression of genes encoding replication factors or other protein-coding genes, as the H3K9R mutation does not significantly affect their expression (Supplemental File S2; Penke et al. 2016). In contrast, 76.2% of later replicating 100-kb windows are located along euchromatic chromosome arms (Fig. 2A; Supplemental Fig. S5).
To compare our RT data to other genome features like histone PTMs, we assigned an RT value to nonoverlapping 10-kb windows across the entire genome (see Supplemental Materials). Notably, 10-kb windows with advanced replication in H3K9R mutants are enriched for H3K9me2/me3 in a wild-type genome and not for other histone PTMs such as H3K27me3, a marker of facultative heterochromatin (Fig. 2F,G; Supplemental Fig. S6C). This observation suggests that advanced replication is a direct effect of the H3K9R mutation, even though most regions enriched for H3K9me2/me3 do not change RT in H3K9R mutants. In contrast, delayed replication was not correlated with H3K9me2/me3 and instead occurred preferentially in chromatin environments relatively devoid of histone PTMs, referred to as “Black” chromatin (Fig. 2F,G; Supplemental Fig. S6C; Filion et al. 2010).
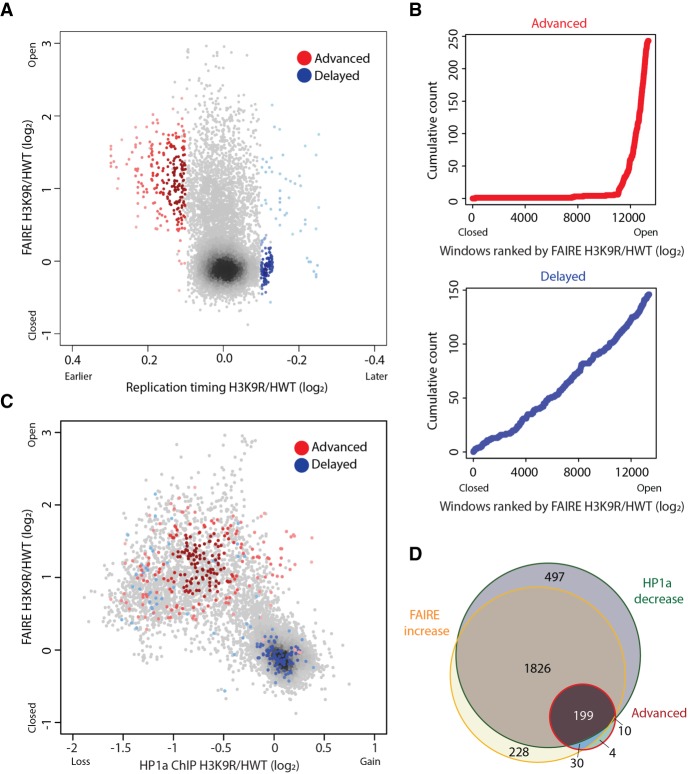
We hypothesized that if chromatin structure directly influences replication, then RT changes should occur at newly accessible chromatin in H3K9R mutants. To compare chromatin accessibility and RT in H3K9R mutants, we compared FAIRE-seq and RT values at 10-kb windows across the genome (Methods; Penke et al. 2016). While most pericentric regions included in the current genome assembly are more accessible in H3K9R mutants compared to control (Penke et al. 2016), we found that the vast majority (92.9%) of windows with increased FAIRE signal do not display altered RT (Fig. 3A). Thus, despite established correlations between accessible chromatin and early replication, increasing chromatin accessibility by H3K9R mutation does not invariably result in earlier replication.
Figure 3.
Open chromatin is permissive to advancement but not delay of replication timing. (A) Heatscatter plot of the H3K9R/HWT ratio of RT values (log2 S/G1) versus the H3K9R/HWT ratio of FAIRE signal at all 10-kb windows across the major chromosome scaffolds; 10-kb windows with significantly advanced (red) or delayed (blue) RT are indicated. Darker color indicates higher density of windows. (B) Cumulative count of advanced (red) or delayed (blue) 10-kb windows ordered by increasing FAIRE signal in H3K9R compared to HWT. (C) Heatscatter plot of the H3K9R/HWT ratio of HP1a ChIP signal versus the H3K9R/HWT ratio of FAIRE signal at all 10-kb windows across the major chromosome scaffolds. (D) Venn diagram of all 10-kb windows with significantly altered FAIRE or HP1a signal in H3K9R compared to HWT (P < 0.01; edgeR). For all panels, significantly different RT was determined as P < 0.05, log2 fold change > 0.1 using limma.
Importantly, this conclusion does not mean that high chromatin accessibility makes no contribution to early replication. Indeed, nearly all windows (230/243) that exhibit significantly advanced replication in H3K9R mutants also have increased FAIRE signal (Fig. 3A–D; Supplemental Fig. S7A,E). This result suggests that a more accessible chromatin environment may be necessary for earlier replication in H3K9R mutants. In contrast, most windows with delayed RT exhibit no change in FAIRE signal, suggesting that delayed replication occurs independently from chromatin accessibility changes in H3K9R mutants (Fig. 3A–D; Supplemental Fig. S7A,F).
We made similar observations when considering HP1a chromatin binding (Supplemental Fig. S7B), which we previously showed is depleted from regions of the H3K9R genome that largely overlap regions of increased chromatin accessibility (Supplemental Fig. S7C; Penke et al. 2016). HP1a is depleted at 217 of the 243 10-kb windows that advanced RT in H3K9R mutants (Fig. 3C,D; Supplemental Fig. S7B,D,F). However, the majority (94.7%) of windows that lose HP1a in H3K9R mutants do not have altered RT. These results indicate that HP1a loss does not invariably result in advanced replication in H3K9R mutants, although it may be necessary. Overall, our observations suggest that the hallmarks of heterochromatin—high levels of H3K9me and HP1a within a relatively inaccessible chromatin environment—are not necessary for maintaining late replication of most pericentric heterochromatin in animal cells.
Elevated transposon expression accompanies advanced replication in H3K9R mutants
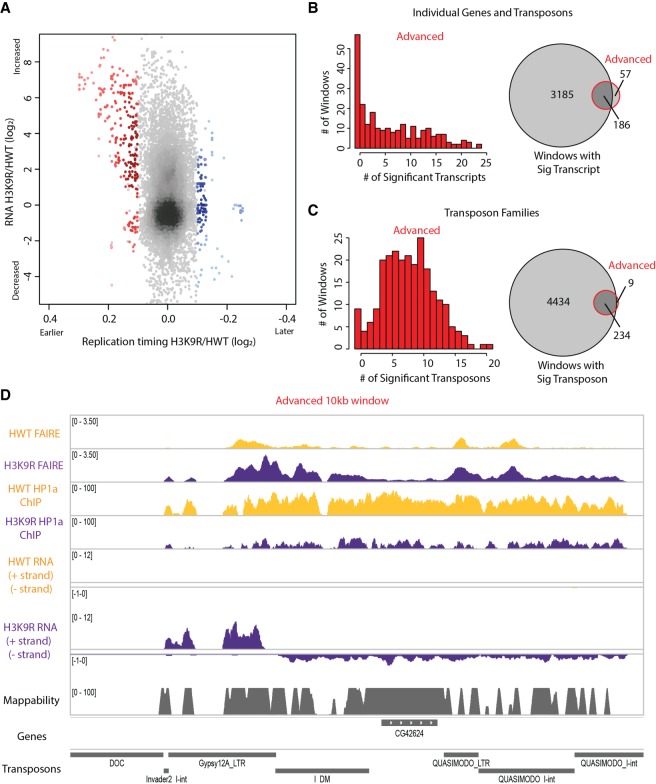
We next considered the transcriptional activity of domains of altered replication. We compared our newly generated RT profiles with our previously generated wing disc transcriptome profiles from H3K9R and control (Penke et al. 2016). We focused on transcripts (genes or transposons) most likely to drive RT changes by identifying the transcript that was most significantly different within each 10-kb window between H3K9R and control (i.e., the transcript with the lowest P-value in differential expression analysis; edgeR). We then compared the fold change of this transcript to the RT value assigned to the same 10-kb window. We found that only a small fraction (6.8%) of the 3371 10-kb windows containing a transcript with a significant expression change also exhibited an RT change (Fig. 4A,B; Supplemental Fig. S8A). This observation indicates that, despite strong correlations between active transcription and early replication (MacAlpine et al. 2004; Liu et al. 2012; Lubelsky et al. 2014; Rivera-Mulia and Gilbert 2016), transcriptional activity and RT are separable. Conversely, we found that the majority (76.5%) of windows with advanced replication in H3K9R mutants exhibited a change in gene expression (Fig. 4A,B; Supplemental Fig. S8B). Because most (97.3%) changes were increases in expression, we speculate that transcription might promote early replication initiation in pericentric heterochromatin. Similar results were obtained by using the average expression change of all transcripts that overlap each window with advanced RT, rather than the transcript with the most significant change in expression across the window (Supplemental Fig. S8G).
Figure 4.
Altered transposon expression occurs at advanced replication domains in H3K9R mutants. (A) Heatscatter plot of the H3K9R/HWT ratio of RT values (log2 S/G1) plotted versus the H3K9R/HWT ratio of RNA-seq signal at all 10-kb windows across major chromosome scaffolds. RNA-seq differences were determined based on the transcript with the lowest P-value across the 10-kb window; 10-kb windows with significantly advanced (red) and delayed (blue) RT are indicated (P < 0.05, log2 fold change > 0.1; limma). (B) Histogram of the number of differentially expressed transcripts in 10-kb windows of advanced replication (red; left). Venn diagram comparing the number of windows with differentially expressed transcripts and number of windows with advanced replication (right). (C) Histogram of the number of transposons belonging to a differentially expressed transposon family in 10-kb windows of advanced replication (red; left). Venn diagram comparing the number of windows with a transposon belonging to a differentially expressed transposon family to the number of windows with advanced replication (right). (D) Browser shot of a 10-kb window (Chr 3R: 2,130,000–2,140,000) with advanced replication. HWT (yellow) and H3K9R (purple) FAIRE-seq, HP1a ChIP-seq, and RNA-seq data plotted in the context of mappability, genes, and transposons.
Windows with advanced replication in H3K9R mutants have a high transposon density, unlike delayed windows which are gene-rich (Fig. 4D; Supplemental Fig. S9A). Low sequence mappability of most transposons likely inhibited our ability to detect all transcriptional changes within advanced replication domains (Supplemental Fig. S8C). Therefore, we also identified transposons belonging to families that were differentially expressed between H3K9R and control (Methods; Fig. 4C; Supplemental Fig. S8C–F). All 243 windows of advanced replication in H3K9R mutants contain either a transposon belonging to a family that was differentially expressed in H3K9R compared to control (96.4%) or that neighbored a window containing multiple differentially expressed transcripts (Fig. 4C,D). Although we cannot determine whether individual transposons within all 243 advanced windows changed expression, these data suggest that altered transcription may promote advancement of replication in H3K9R mutants.
Along with transposon enrichment (Supplemental Fig. S9A,B), advanced replication domains in H3K9R mutants are normally enriched for H3K9me2/me3 (Supplemental Fig. S9C) and exhibited a lower GC content (Supplemental Fig. S9D) compared to domains of increased chromatin accessibility or increased RNA expression with unaltered replication (FAIRE only and RNA only, respectively). Although transposon density distinguished advanced domains, the majority of domains with altered transposon expression have no change in RT (Fig. 4C; Supplemental Fig. S8D). Therefore, we surmise that altered transposon expression is necessary, but additional events must occur within accessible chromatin to advance replication.
Our data thus far indicate that increased chromatin accessibility and gene expression act upstream of advanced replication within pericentric heterochromatin in H3K9R mutants (Fig. 4D; Supplemental File S3). To further understand the relationship between transcription and DNA replication within transcriptionally active euchromatin, we investigated the Drosophila male X Chromosome, which replicates earlier in males than in females (Schwaiger et al. 2009; Bell et al. 2010; Lubelsky et al. 2014). In addition, a twofold hyperactivation of gene expression from the male X results in matched X-linked gene expression between XY males and XX females (Kuroda et al. 2016). We therefore generated a replication-dependent (RD) histone genotype (H4K16R) predicted to disrupt dosage compensation and determined the effect on gene expression and RT and whether these two processes could be uncoupled.
H4K16 is necessary for hyperexpression of the Drosophila male X Chromosome
The Drosophila dosage compensation mechanism is mediated by the Male-Specific Lethal (MSL) complex, which specifically localizes to and promotes higher gene expression from the male X. The MSL complex includes MOF, a histone acetyltransferase that acetylates lysine 16 of histone H4, resulting in higher levels of H4K16ac on male X Chromosomes relative to autosomes or the female X (Hilfiker et al. 1997; Smith et al. 2000; Gelbart et al. 2009). Furthermore, hyperacetylation of H4K16 correlates with increased chromatin accessibility of the male X (Bell et al. 2010). These data suggest that H4K16ac is required for dosage compensation in flies. In accordance with these findings, MOF mutations cause a male-specific lethal phenotype; however, MOF performs both H4K16-dependent and -independent functions (Hilfiker et al. 1997; Buscaino et al. 2003; Sykes et al. 2006). A requirement for H4K16 in dosage compensation, therefore, has not been directly tested.
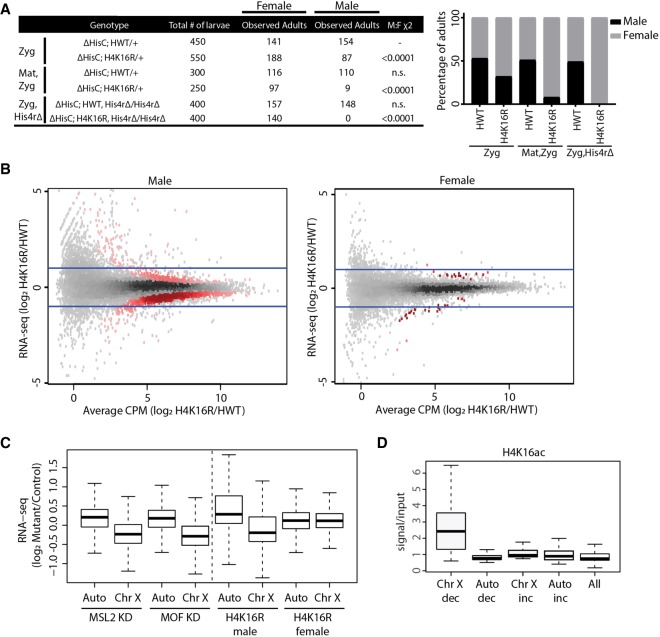
Similar to observations made using mutations in MOF and other MSL complex members (Lucchesi 1998), we found that male viability is significantly reduced in zygotic, RD H4K16R mutants (Fig. 5A). However, unlike mutation of MSL complex members, which causes fully penetrant male lethality (Lucchesi 1998), these H4K16R males can develop to adulthood (Fig. 5A), although they eclose later than their female siblings. A further reduction in male viability occurred when both maternal and zygotic sources of histones were RD H4K16R mutant (Fig. 5A). The Drosophila genome also contains a single-copy, replication-independent His4r gene, which is not located in the RD gene cluster but encodes an identical H4 protein. Combining the RD H4K16R zygotic genotype with a CRISPR-derived homozygous deletion of His4r resulted in complete male lethality (Fig. 5A). We therefore conclude that H4K16 function is required for male development. In contrast, females of all these H4K16R genotypes are viable, indicating that H4K16 modification is not generally required for organismal viability.
Figure 5.
H4K16 promotes hyperexpression of the Drosophila male X Chromosome. (A) Table showing the number of adult females and males resulting from homogeneous vial cultures of 50 first instar larvae of the indicated genotypes (see Supplemental Materials for crosses and complete genotypes). Either only the zygotic (rows 1 and 2) or the maternal and zygotic histones (rows 3 and 4) were of the replication-dependent HWT and H4K16R genotypes. In rows 5 and 6, homozygous deletion of the His4r gene was combined with zygotic, replication-dependent HWT and H4K16R genotypes. The χ2 comparisons were performed using the zygotic HWT male to female results (row 1) as the expected classes. The right panel shows the percentage of viable male (black) and female (gray) adults for H4K16R and HWT. (B) Heatscatter plot of the H4K16R/HWT ratio of RNA-seq signal of individual genes from third instar wing imaginal discs. Statistically different transcripts between H4K16R and HWT males (left panel) or females (right panel) are indicated in red (P < 0.05, edgeR). Blue lines indicate a twofold change. (C) Box plot of RNA-seq signal from autosomes and Chr X after MSL2 or MOF knockdown in male S2 cells (Zhang et al. 2010) and in H4K16R/HWT male and female wing discs on autosomes (Auto) and Chr X. (D) Average enrichment of modENCODE H4K16ac signal from male third instar larvae at 10-kb windows of significantly (P < 0.05) decreased (dec) or increased (inc) transcript expression between H4K16R and HWT males on Chr X and autosomes (Auto) or at all 10-kb windows (GSE49497) (Celniker et al. 2009).
We next performed gender-specific total RNA-seq from replication-dependent H4K16R and control wing discs, generated transcriptomes (Cufflinks), and identified differentially expressed transcripts between H4K16R males and females and their respective controls (Trapnell et al. 2012). We observed 1789 differentially expressed transcripts (608 increased and 1181 decreased) in H4K16R males relative to control males and 105 differentially expressed transcripts in H4K16R females relative to control females (39 increased and 66 decreased), indicating that the H4K16R effect on gene expression is greater in males than in females (P < 0.05, edgeR) (Fig. 5B). Of the 1181 genes with decreased expression in H4K16R males, 72% are located on the X. In addition, the majority (92%) of the down-regulated, X-linked genes in H4K16R males have a log2 fold change less than 1, which would be expected for a disruption in X Chromosome dosage compensation. In contrast, only 3.6% of genes with increased expression in H4K16R males are on the X.
We further examined chromosome-specific differential gene expression by assessing transcript abundance for the X separately from autosomes. We compared our H4K16R wing disc RNA-seq data to previously published data from the male Drosophila S2 cell line in which MSL2 or MOF had been depleted by RNAi (Zhang et al. 2010). Similar to MSL2 or MOF knockdown, global transcript abundance is decreased for genes on the X in H4K16R male wing disc cells compared to control but not for genes located on the autosomes or the female X (P < 0.05) (Fig. 5C). Importantly, 10-kb windows containing a significantly decreased transcript from the H4K16R male X are enriched in a wild-type genome for H4K16ac (P < 0.05) (Fig. 5D). In contrast, windows in H4K16R males from autosomes containing a decreased transcript, those with an increased transcript (either from the X or the autosomes), or all windows with a transcriptional change were not normally enriched for H4K16ac (Fig. 5D). These data directly demonstrate that H4K16 is a critical component of the Drosophila dosage compensation machinery. Moreover, this residue is not required for basal genome function, as female gene expression, viability, and fertility are unaffected.
H4K16 promotes early replication of the Drosophila male X Chromosome
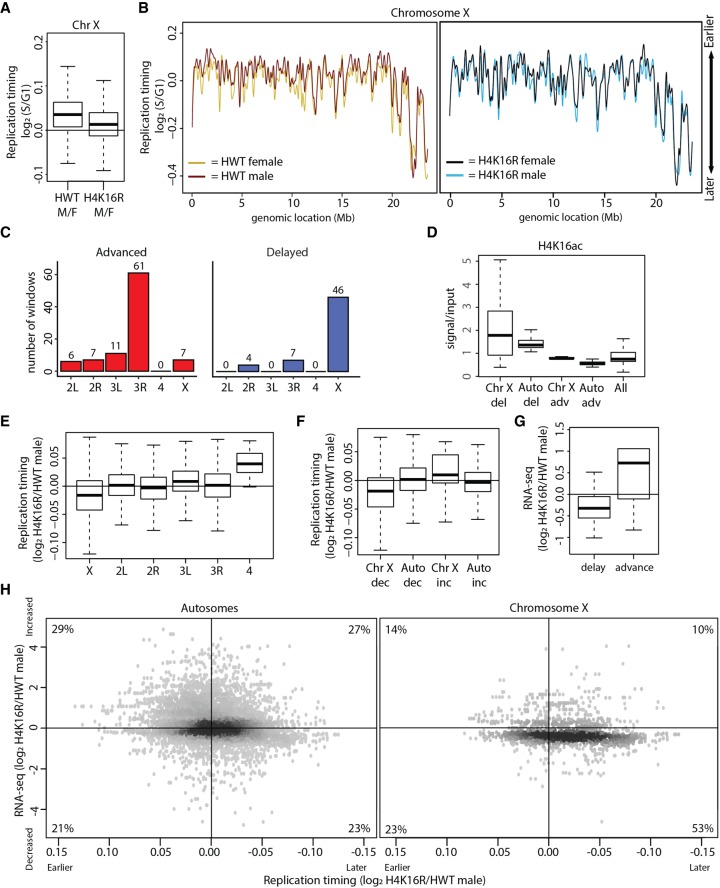
We next profiled RT in replication-dependent H4K16R and control male and female wing discs. When considering the major chromosome scaffolds using overlapping 100-kb windows, we observed very few significant replication changes between H4K16R and control in either females or males (0.04% and 1%, respectively) (Supplemental Figs. S10, S11). These data indicate that H4K16 is not globally required for maintenance of RT in flies. However, when we considered 100-kb windows only from the X Chromosome, we observed that the normally earlier replication of the male X relative to the female X was largely abrogated in H4K16R mutants (P < 0.05) (Fig. 6A,B), suggesting that H4K16ac promotes early replication of the male X Chromosome.
Figure 6.
H4K16R mutation reduces gene expression and delays replication of the male X Chromosome. (A) Box plot of HWT male/female and H4K16R male/female ratios of RT values (log2 S/G1) on Chr X. (B) LOESS regression line applied to log2 S/G1 averaged replicates from HWT female (yellow) and HWT male (maroon) and H4K16R female (black) and HWT male (blue) plotted across Chr X (100-kb windows, 10-kb slide). Note that the male X Chromosome generally replicates earlier in HWT, but not in H4K16R mutants. (C) Histogram of 10-kb windows with advanced (red) or delayed (blue) RT between H4K16R and HWT males on major chromosome scaffolds (P < 0.05; absolute log2 fold change > 0.1; limma). (D) Average enrichment of modENCODE H4K16ac signal from male third instar larvae at 10-kb windows of delayed (del) or advanced (adv) replication between H4K16R and HWT males on Chr X and autosomes (Auto) or at all 10-kb windows (GSE49497) (Celniker et al. 2009). (E) Box plot of the H4K16R/HWT ratio of male RT values (log2 S/G1) on all major chromosome scaffolds. (F) Box plot of the H4K16R/HWT ratio of male RT values (log2 S/G1) at 10-kb windows of decreased or increased RNA-seq signal on Chr X or autosomes (Auto) (P < 0.05). (G) Box plot of the H4K16R/HWT ratio of male RNA-seq signal at 10-kb windows of delayed or advanced RT (P < 0.05). (H) Heatscatter plot of the H4K16R/HWT ratio of male RT values (log2 S/G1) plotted versus the H4K16R/HWT ratio of male RNA-seq signal at all 10-kb windows across the autosomes (left) and Chr X (right). RNA-seq differences were determined based on the transcript with the lowest P-value across the 10-kb window. The percentage of 10-kb windows present in each quadrant is indicated.
To evaluate this effect more thoroughly, we assigned RT values to 10-kb windows across the genome using significance thresholds as for the H3K9R RT data. We identified 57 individual 10-kb windows in H4K16R males with delayed RT, and most (78%) of these were located on the X (Fig. 6C; Supplemental Fig. S12A). We identified 92 10-kb windows in H4K16R males with advanced RT, with most (94%) located on the autosomes (including 61 windows on Chr 3R) (Fig. 6C; Supplemental Fig. S12A). Windows from the H4K16R male X with delayed replication are enriched for H4K16ac in a wild-type genome, whereas those advanced windows (on the X or autosomes) or delayed windows on autosomes are not (P < 0.05) (Fig. 6D; Supplemental Fig. S12B). These data suggest that delayed replication in H4K16R males is a direct result of the H4K16R mutation, while regions of advanced replication may occur indirectly.
We were concerned that the small number of windows with an RT change identified using our significance cutoffs was masking a more general effect, as many X Chromosome replication delays in H4K16R males might be less than a log2 fold change of 0.1 and therefore not scored as significant. Indeed, chromosome-wide RT of the X in control males is advanced less than a log2 fold change of 0.1 relative to control females (Fig. 6A,B), in accordance with previous analyses (e.g., a change of ∼0.1 as described by Schwaiger et al. 2009; Lubelsky et al. 2014). Therefore, we analyzed replication in H4K16R males by assessing, as a group, all 10-kb windows located on an individual chromosome (X and 4) or an individual chromosome arm (2L, 2R, 3L, and 3R). Using this approach, replication of the X was found to be significantly delayed in H4K16R males relative to control males (Fig. 6E). No such effect was observed for the autosomes, consistent with a specific role for H4K16ac in promoting early replication of the male X. The small, heterochromatic fourth chromosome replicated earlier in both H4K16R males (Fig. 6E) and females (Supplemental Fig. S12C), which may result from an H4K16 function that is independent of X Chromosome dosage compensation. We conclude that H4K16 promotes early replication of the male X in Drosophila.
H4K16R mutation concurrently reduces gene expression and delays replication of the male X Chromosome
To explore the relationship between gene expression and RT of individual windows on the male X, we identified 10-kb windows containing a differentially expressed transcript in H4K16R males relative to controls and determined whether these windows also displayed altered replication. For the X, we found a significant correlation between decreased gene expression and later replication in H4K16R males, as well as a correlation between increased gene expression and earlier replication (P < 0.05) (Fig. 6F). No such correlation exists for windows on the autosomes (Fig. 6F). These correlations hold when we consider the converse relationship: 10-kb windows in H4K16R males with significantly delayed replication have decreased gene expression, whereas 10-kb windows with significantly advanced replication have increased gene expression (P < 0.05) (Fig. 6G). These data indicate that changes in RT correlate with changes in gene expression for the male X in H4K16R males.
Furthermore, scatterplots comparing transcription and replication between H4K16R males and controls resulted in different distributions. We found that 53% of all 10-kb windows on Chromosome X had both lower gene expression and later replication in H4K16R males compared to 23% on autosomes (Fig. 6H). A two-dimensional Peacock test (https://rdrr.io/cran/Peacock.test/; Peacock 1983) revealed that the distributions resulting from simultaneous comparison of the log2 fold change of the H4K16R/control ratio of transcript abundance and RT for all 10-kb windows on the X and the autosomes are statistically different (P < 5.9 × 10−317). These data indicate that H4K16 promotes the strong correlation between elevated gene expression and early replication of the Drosophila male X Chromosome.
Discussion
Studies of animal cells have revealed strong, genome-wide correlations between early replication of highly transcribed, accessible chromatin and late replication of lowly transcribed, inaccessible chromatin (Bell et al. 2010; Eaton et al. 2011; Lubelsky et al. 2014). Here, we explored potential causal relationships underlying these correlations by combining perturbation of chromatin structure using two different histone mutations (H3K9R and H4K16R) with genome-wide RT data.
We found that the male X Chromosome of H4K16R mutants experiences both reduced transcription and delayed RT, consistent with previous studies showing a strong correlation between transcriptional activity and RT (Aggarwal and Calvi 2004; MacAlpine et al. 2004; Schwaiger et al. 2009; Liu et al. 2012; Lubelsky et al. 2014). However, we also show that correlations between transcription and RT can be uncoupled: Active transposon expression in H3K9R mutants was not accompanied by earlier replication of most pericentric heterochromatin. Thus, activation of transcription does not always result in earlier replication. In addition, our analysis of H4K16R mutants show for the first time that H4K16 is required for proper dosage-compensated expression of the Drosophila male X Chromosome, as predicted by previous studies of factors that acetylate H4K16 (Hilfiker et al. 1997; Smith et al. 2000; Kuroda et al. 2016). The changes in autosomal gene expression we observed in H4K16R males are likely a secondary consequence of wholesale changes in gene expression on the X, as transcription factors are encoded on the X Chromosome.
HP1a binding to H3K9me is a defining feature of constitutive heterochromatin and is thought to be critical for most, if not all, aspects of heterochromatin function (Canzio et al. 2011; Larson et al. 2017). Therefore, we were surprised to find that pericentric heterochromatin generally remained late replicating relative to the euchromatic chromosome arms in H3K9R mutants. Indeed, despite decreased nucleosome density and loss of HP1a, replication at ∼97% of the genome remained unchanged in H3K9R mutants. Studies of the onset of late replication in the early fly embryo show that chromatin condensation and late replication of pericentric heterochromatin occur prior to H3K9me and HP1a recruitment, indicating that these two features of heterochromatin are not always required for late replication (Shermoen et al. 2010; Yuan and O'Farrell 2016). In fact, we observe a DAPI-bright heterochromatic chromocenter in H3K9R diploid nuclei, similar to that of wild-type nuclei, that colocalizes with late-patterned replication foci. Thus, pericentric heterochromatin retains many of its hallmarks despite loss of H3K9me and HP1a, suggesting additional features define heterochromatin function.
Our data support a model in which compartmentalization of euchromatin and heterochromatin into different nuclear compartments is not disrupted by the loss of H3K9 modification. Furthermore, the arms and pericentric regions of Drosophila chromosomes may correspond, respectively, to the largely euchromatic compartment “A” and heterochromatic compartment “B” previously identified in human cells (Lieberman-Aiden et al. 2009). Accordingly, factors other than HP1a may remain associated with compartment “B” in H3K9R mutants, preventing large-scale advancement of RT at the pericentromere. One such factor could be Rif1, which is required for the onset of late replication of heterochromatin during early fly embryogenesis as well as for late replication in other species (Peace et al. 2014; Foti et al. 2016; Seller and O'Farrell 2018).
Nevertheless, we found reproducible and significant RT changes in H3K9R pericentric heterochromatin. By carefully analyzing features of these altered RT domains, we conclude that accessible chromatin does not invariably result in early replication, although early replication may require accessible chromatin. We propose that H3K9R mutation alters RT by disrupting local chromatin accessibility without affecting overall compartmentalization of heterochromatin (Larson et al. 2017; Strom et al. 2017). The events that function within accessible chromatin to dictate RT could include origin specification or origin activation. Origins of replication are licensed during G1 phase by the activity of origin specification factors (e.g., the origin recognition complex; ORC), and during S phase DNA replication initiates at a subset of licensed origins (Bell and Stillman 1992). Certain models describing temporal programs of replication initiation posit a stochastic process in which a higher density of licensed origins in accessible euchromatin increases the probability of replication initiation compared to inaccessible heterochromatin (Rhind et al. 2010; Das et al. 2015; Miotto et al. 2016). These models are consistent with observations that ORC complexes are most abundant where chromatin accessibility is also high (MacAlpine et al. 2010; Lubelsky et al. 2014; Miotto et al. 2016). However, other factors function within the licensed origin landscape to either promote or inhibit origin activation (Foti et al. 2016). Thus, changes in either origin licensing or activation could demarcate domains that advance replication within the permissive open chromatin environment created by the H3K9R mutation.
We also found that delayed replication domains are largely independent of altered chromatin accessibility or transcriptional changes in H3K9R mutants. We hypothesize that elevated accessibility of pericentric heterochromatin in H3K9R mutants functions as a “sink” for limiting replication factors, resulting in delayed replication of domains along chromatin arms as proposed for other replication factors (Foti et al. 2016). Another possible explanation for delayed replication in H3K9R mutants is the relocalization of HP1a to chromosome arms (Penke et al. 2016). Previous work has shown that tethering HP1a to a euchromatic domain delays RT (Pokholkova et al. 2014). However, we did not observe a strong correlation between 10-kb windows that gain HP1a and those that delay replication, perhaps because the amount of HP1a relocalization in H3K9R mutants was below a threshold necessary to affect replication. We note that domains of altered RT in H3K9R mutants do not match those previously identified after HP1a knockdown in Drosophila cultured cells (Supplemental Fig. S13; Schwaiger et al. 2010), potentially due to H3K9-independent functions of HP1a or to the exclusion of repetitive DNA from the microarray-based assay used in the previous study.
In summary, our study shows that correlations among chromatin configuration, transcription, and RT in animal cells can be mechanistically separated by mutation of specific histone residues, indicating modes of control for replication initiation that are independent of these features of animal genome structure and activity.
Methods
FACS and genomic DNA sequencing
12× HWT (Histone Wild Type), H3K9R, and H4K16R wing imaginal disc nuclei were sorted into G1, S, and G2 populations by a FACSAria II or III based on DAPI intensity and subsequently pelleted, flash-frozen, and stored at −80°C prior to DNA isolation and library preparation. Libraries were prepared with the Rubicon ThruPLEX DNA-seq kit and subjected to Illumina HiSeq 2500 single-end 50-bp sequencing.
RT characterization
Reads from G1, S, and G2 samples were aligned to the dm6 reference genome (Release 6.04) using Bowtie 2 (v2.3.2) default parameters (Langmead et al. 2009). Reads with a MAPQ score >10 were retained using SAMtools (v1.6) (Li et al. 2009). Replication timing values were obtained by averaging the S/G1 ratio of reads per million (RPM) value from each S phase replicate for a particular window size. Profiles were generated by plotting the RT value at each window versus genomic location. The limma statistical package was used to identify windows with significantly altered RT values between HWT and either H3K9R or H4K16R samples (lmFit, P value adjusted for multiple testing [P < 0.01]; absolute log2 fold change > 0.1) (Newville et al. 2014). Coordinates of chromatin states were obtained from Kharchenko et al. (2011) and converted to dm6 coordinates using the UCSC liftOver tool (Karolchik et al. 2004).
The genomic coordinates halfway between each peak and valley of an RT profile were used to determine replication domain sizes. Third instar larvae modENCODE ChIP-seq data were used to calculate histone PTM enrichment at 100-kb windows (accession numbers found in Supplemental Materials; ftp://data.modencode.org/D.melanogaster/Histone-Modification/ChIP-seq/raw-seqfile_ fastq/; Celniker et al. 2009). For each PTM, raw reads for two ChIP replicates and two input replicates were aligned to the genome using Bowtie 2 (v2.3.2) (Langmead et al. 2009). BEDTools coverage (v2.25.0) was used to quantify the number of reads mapping to each 100-kb window, with results normalized to read depth (Quinlan and Hall 2010). PTM enrichment was calculated by averaging the ChIP/input read counts for each replicate. Average PTM enrichment in five equally sized categories (early, early/mid, mid, mid/late, and late) of 100-kb windows ordered by RT values were represented as a heat map using the R package pheatmap (https://cran.r-project.org/web/packages/pheatmap/). The sum of the number of transcripts and the sum of RPMs of each transcript overlapping a 100-kb window was determined using our previously published RNA-seq data from third instar imaginal wing discs (Penke et al. 2016).
FAIRE, HP1a, and RNA analyses
H3K9R FAIRE-seq from third instar imaginal wing discs and H3K9R HP1a ChIP-seq from whole third instar larvae were obtained from GSE85374 (Penke et al. 2016). FAIRE and HP1a reads from three H3K9R and two HWT replicates were aligned to the genome using Bowtie 2 (v2.25.0) default parameters (Langmead et al. 2009). The number of reads overlapping 10-kb windows were normalized to read depth (FAIRE) or the number of uniquely mapping Drosophila virilis spike-in reads (HP1a). edgeR (v3.16.5) was used to calculate windows with significantly altered FAIRE or HP1a fold change (P < 0.01) (Robinson et al. 2010; McCarthy et al. 2012).
TopHat default parameters (v2.1.1) (Trapnell et al. 2012) were used to align single-end 50-bp RNA reads to dm6 from triplicate samples of HWT, H3K9R (obtained from GSE85374 [Penke et al. 2016]), HWT female, HWT male, H4K16R female, and H4K16R male third instar wing imaginal discs. Transcriptomes were generated using Cufflinks (v2.2.1) (see Supplemental Materials for parameters). For H3K9R experiments, we combined the Cufflinks-generated transcriptome with transposons annotated by RepeatMasker (Smit et al. 2013–2015). Differentially expressed transcripts were determined via edgeR statistical analysis (P value < 0.01) (Robinson et al. 2010; McCarthy et al. 2012). We identified transcripts overlapping each 10-kb window and selected the transcript with the lowest P value. For determination of RT values at 10-kb windows, refer to Supplemental Materials.
To identify differentially expressed transposon families, raw counts of all individual transposons within a family were summed and edgeR was used to determine significant (P < 0.05) differences between samples. In addition to transposon families, all transcripts identified in Cufflinks were included in this edgeR analysis to facilitate modeling of variability. Data were visualized using the Integrative Genomics Viewer (Robinson et al. 2011).
Data access
The data generated as a part of this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE114925.
Supplementary Material
Acknowledgments
This work was supported by NIH (National Institute of General Medical Sciences) Grants R01-GM124201 to R.J.D.; F31-GM115194 to T.J.R.P; R01-GM104097 to D.M.M.; R35-GM128851 to D.J.M.; and R01-GM129132 to A.G.M. In addition, R.L.A. and T.J.R.P. were supported in part by an NIH predoctoral training grant T32-GM007092. We thank the UNC Flow Cytometry and High Throughput Sequencing Core Facilities, supported in part by the Center for Strategic Scientific Initiatives, National Cancer Institute, P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. FACS results reported in this publication were supported in part by the North Carolina Biotechnology Center Institutional Support Grant 2012-IDG-1006. We thank Janet Dow and Corbin Jones for data acquisition and analysis advice. We thank Spencer Nystrom for critical reading of the manuscript and Agnes Ezekwesili and Esther Kwon for assistance with larval dissection.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.239913.118.
References
- Aggarwal BD, Calvi BR. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376. [DOI] [PubMed] [Google Scholar]
- Almeida R, Fernández-Justel JM, Santa-María C, Cadoret J-C, Cano-Aroca L, Lombraña R, Herranz G, Agresti A, Gómez M. 2018. Chromatin conformation regulates the coordination between DNA replication and transcription. Nat Commun 9: 1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128. [DOI] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D. 2010. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 17: 894–900. [DOI] [PubMed] [Google Scholar]
- Buscaino A, Kocher T, Kind JH, Holz H, Taipale M, Wagner K, Wilm M, Akhtar A. 2003. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol Cell 11: 1265–1277. [DOI] [PubMed] [Google Scholar]
- Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. 2011. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell 41: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. 2013. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SP, Borrman T, Liu VWT, Yang SC-H, Bechhoefer J, Rhind N. 2015. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res 25: 1886–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donley N, Thayer MJ. 2013. DNA replication timing, genome stability and cancer: Late and/or delayed DNA replication timing is associated with increased genomic instability. Semin Cancer Biol 23: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti R, Gnan S, Cornacchia D, Dileep V, Bulut-Karslioglu A, Diehl S, Buness A, Klein Felix A, Huber W, Johnstone E, et al. 2016. Nuclear architecture organized by Rif1 underpins the replication-timing program. Mol Cell 61: 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. 2009. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol 16: 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23. [DOI] [PubMed] [Google Scholar]
- Günesdogan U, Jäckle H, Herzig A. 2010. A genetic system to asses in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep 11: 772–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamperl S, Cimprich KA. 2016. Conflict resolution in the genome: how transcription and replication make it work. Cell 167: 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes KA, Gracheva E, Elgin SCR. 2007. A distinct type of heterochromatin within Drosophila melanogaster chromosome 4. Genetics 175: 1539–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz KS, Casas-Delucchi CS, Török T, Cmarko D, Rapp A, Raska I, Cardoso MC. 2018. Peripheral re-localization of constitutive heterochromatin advances its replication timing and impairs maintenance of silencing marks. Nucleic Acids Res 46: 6112–6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. 1997. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J 16: 2054–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang C-W, Lyou Y, Townes TM, Schübeler D, Gilbert DM. 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, et al. 2015. The release 6 reference sequence of the Drosophila melanogaster genome. Genome Res 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Berger SL. 2008. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18: 152–158. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Aleksyenko AA, Schwartz YB, Minoda A, Riddle BC, Ernst J, Sobo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila. Nature 471: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Handsaker RE, Kamitaki N, Karlić R, Ghosh S, Polak P, Eggan K, McCarroll SA. 2014. Genetic variation in human DNA replication timing. Cell 159: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda MI, Hilfiker A, Lucchesi JC. 2016. Dosage compensation in Drosophila—a model for the coordinate regulation of transcription. Genetics 204: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, McConnell K, Dixon M, Calvi BR. 2012. Analysis of model replication origins in Drosophila reveals new aspects of the chromatin landscape and its relationship to origin activity and the prereplicative complex. Mol Biol Cell 23: 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, MacAlpine DM. 2014. DNA replication and transcription programs respond to the same chromatin cues. Genome Res 24: 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC. 1998. Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr Opin Genet Dev 8: 179–184. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP. 2004. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, MacAlpine DM. 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero D, Mackenzie A, Donaldson A, Zegerman P. 2011. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J 30: 4805–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ, Klusza S, Penke TJR, Meers MP, Curry KP, McDaniel SL, Malek PY, Cooper SW, Tatomer DC, Lieb JD, et al. 2015. Interrogating the function of metazoan histones using engineered gene clusters. Dev Cell 32: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Ji Z, Struhl K. 2016. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci 113: E4810–E4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newville M, Stensitzki T, Allen DB, Ingargiola A. 2014. LMFIT: Non-linear least-square minimization and curve-fitting for Python. Zenodo 10.5281/zenodo.11813. [DOI] [Google Scholar]
- Peace JM, Ter-Zakarian A, Aparicio OM. 2014. Rif1 regulates initiation timing of late replication origins throughout the S. cerevisiae genome. PLoS One 9: e98501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock JA. 1983. Two-dimensional goodness-of-fit testing in astronomy. Mon Not R Astron Soc 202: 615–627. [Google Scholar]
- Penke TJR, McKay DJ, Strahl BD, Matera AG, Duronio RJ. 2016. Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev 30: 1866–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk N, Kahli M, d'Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen C-L, Hyrien O. 2016. Replication landscape of the human genome. Nat Commun 7: 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholkova GV, Koryakov DE, Pindyurin AV, Kozhevnikova EN, Belyakin SN, Andreyenkov OV, Belyaeva ES, Zhimulev IF. 2014. Tethering of SUUR and HP1 proteins results in delayed replication of euchromatic regions in Drosophila melanogaster polytene chromosomes. Chromosoma 124: 209–220. [DOI] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Gilbert DM. 2013. DNA replication timing. Cold Spring Harb Perspect Biol 5: a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Yang SC-H, Bechhoefer J. 2010. Reconciling stochastic origin firing with defined replication timing. Chromosome Res 18: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Jaffe JD, Kennedy C, Linder-Basso D, et al. 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res 21: 147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Mulia JC, Gilbert DM. 2016. Replication timing and transcriptional control: beyond cause and effect—part III. Curr Opin Cell Biol 40: 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Battaglia D, Chang BH, Shirley JW, Buckley Q, Pope BD, Devidas M, Druker BJ, Gilbert DM. 2012. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res 22: 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Rivera-Mulia JC, Vera D, Zimmerman J, Das S, Padget M, Nakamichi N, Chang BH, Tyner J, Druker BJ, et al. 2017. Stability of patient-specific features of altered DNA replication timing in xenografts of primary human acute lymphoblastic leukemia. Exp Hematol 51: 71–82.e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schübeler D. 2009. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes Dev 23: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Kohler H, Oakeley EJ, Stadler MB, Schübeler D. 2010. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res 20: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seller CA, O'Farrell PH. 2018. Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLoS Biol 16: e2005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O'Farrell PH. 2010. Developmental control of late replication and S phase length. Curr Biol 20: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefert JC, Georgescu C, Wren JD, Koren A, Sansam CL. 2017. DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res 27: 1406–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol 20: 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. 2009. Human mutation rate associated with DNA replication timing. Nat Genet 41: 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JH. 1960. Asynchronous duplication of chromosomes in cultured cells of Chinese hamster. J Biophys Biochem Cytol 7: 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, O'Farrell PH. 2016. TALE-light imaging reveals maternally guided, H3K9me2/3-independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev 30: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, MacAlpine DM, Oliver B. 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol 8: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.