Abstract
Numerous large-scale genomic studies of matched tumor-normal samples have established the somatic landscapes of most cancer types. However, the downstream analysis of data from somatic mutations entails a number of computational and statistical approaches, requiring usage of independent software and numerous tools. Here, we describe an R Bioconductor package, Maftools, which offers a multitude of analysis and visualization modules that are commonly used in cancer genomic studies, including driver gene identification, pathway, signature, enrichment, and association analyses. Maftools only requires somatic variants in Mutation Annotation Format (MAF) and is independent of larger alignment files. With the implementation of well-established statistical and computational methods, Maftools facilitates data-driven research and comparative analysis to discover novel results from publicly available data sets. In the present study, using three of the well-annotated cohorts from The Cancer Genome Atlas (TCGA), we describe the application of Maftools to reproduce known results. More importantly, we show that Maftools can also be used to uncover novel findings through integrative analysis.
With advances in cancer genomics and reduction in costs, whole-genome sequencing (WGS) and whole-exome sequencing (WXS) of large cohorts of cancer samples have become the mainstream way of determining genetic abnormalities associated with cancer (Mardis and Wilson 2009; Vogelstein et al. 2013; Wheeler and Wang 2013). Along with the many ongoing studies, a plethora of published large-scale data sets offer opportunities for reanalysis to advance our understanding of cancer genome and biology. Such cohort-based large-scale characterizations often produce large amounts of data in the form of somatic variants containing single-nucleotide variants (SNV) and small insertion/deletions (indels). Somatic variants provide baseline data for many analyses such as driver gene detection, pathway analysis, mutational signatures, and estimation of tumor heterogeneity, to name a few (Alexandrov et al. 2013a; Lawrence et al. 2013b). However, these downstream analyses of mutational data often entail many computational/statistical approaches, which are laborious, time-consuming, and cumbersome. On the other hand, visualization of these complex and heterogeneous data plays key roles in genomic studies, with researchers finding it difficult to generate complicated publication-quality images, such as oncoplots and lollipop plots. Although a number of tools and software exist for each of these tasks, they all require specific input data format (Ding et al. 2014). Although tools such as Mutational Significance in Cancer (MuSiC) offer multiple analysis domains in a single software package, they require large alignment files and computational resources, hindering their usage in analyzing public data sets (Dees et al. 2012).
To address these issues, we developed a user-friendly, R Bioconductor package, which we termed “Maftools.” Maftools offers a multitude of analysis and visualization modules while only requiring a single input text file containing somatic variants in MAF format. MAF is a standard tab delimited text file format introduced by TCGA for storing and distributing somatic variants, containing complete somatic landscape of the cohort. MAF also has several advantages over variant call format (VCF) in storing annotations of hundreds of samples while maintaining readability and portability. With MAF files as a standard input, functions are implemented in Maftools to perform many commonly used statistical and computational analyses in cancer genome studies, including but not limited to driver gene detection and analysis of pathways, de novo signatures, and clinical parameter enrichment. Maftools also provides options to integrate and analyze copy number variation (CNV) data generated by programs such as genomic identification of significant targets in cancer (GISTIC) and circular binary segmentation (CBS) algorithms (Olshen et al. 2004; Mermel et al. 2011). In addition, Maftools can be used to perform variant annotations and other common tasks such as data format conversion and subset operations. Usage of Maftools is straightforward with self-explanatory functions and is implemented as an open source R package available through the Bioconductor project. Maftools is independent of alignment files, facilitating analysis of public data sets through data-mining approaches. Lastly, Maftools provides various plotting functions to help researchers generate intricate publication-quality images.
To demonstrate the application and performance of Maftools, we used three different TCGA cohorts: esophageal carcinoma (ESCA; WXS; N = 186), acute myeloid leukemia (AML; WXS; N = 192), and breast invasive carcinoma (BRCA; WGS; N = 96), which all contain both somatic variants as well as copy number variations (The Cancer Genome Atlas Network 2012b, 2013, 2017). ESCA is unique in its mutational pattern and its two molecularly distinct subtypes—esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC)—as we and others have characterized previously (Lin et al. 2014, 2018a, b; Hao et al. 2016). AML was selected because it has several well-known dysregulated pathways and clinical associations. WGS data from BRCA is ideal for the demonstration of rainfall plots and identification of hypermutated genomic regions (known as “Kataegis”) (Alexandrov et al. 2013b).
Results
Maftools package overview
As described above, the Maftools package was developed to bring the majority of standard analysis and visualization modules into a single channel through implementation of well-established statistical and computational approaches, while only requiring a single and unaltered input data format.
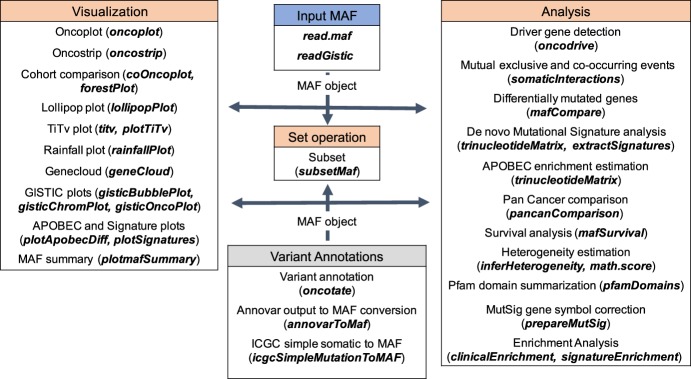
Functions of Maftools are divided into three main modules (Fig. 1): analysis, visualization, and annotation. Each of these modules and key functions are described below.
Figure 1.
Overview of Maftools package. Table headers describe available modules, namely, Visualization, Set operation, Variant annotations, and Analysis. A small description and the corresponding function (bold italics) in Maftools package are provided for every module. The typical workflow begins with MAF object creation either by reading an MAF file or by converting existing annotations to an MAF object, which is later passed to a desired function as an input (arrows). The visualization module includes functions to generate publication ready plots from the input MAF object, whereas the analysis module offers functions to perform commonly performed analyses in cancer genomics. The variant annotation module performs variant annotations using oncotator API and format conversion of annotations generated by programs such as ANNOVAR. The set operation includes function for subsetting MAF object based on user-defined queries.
Visualization
Clear and concise visualization of large-scale genomic data is a key step in displaying critical information in an effective, precise, and easy-to-comprehend manner. The visualization module in the Maftools package offers various plotting functions to generate customizable and feature-rich publication-quality plots from both somatic mutation and CNV data sets. Some of the visualizations offered by Maftools include oncoplots (also known as waterfall plots, coMut plots, and oncoprints) to display somatic landscape, lollipop plots (also known as stick plots, needle plots, and stem plots) to illustrate the distribution of variants on a linearized protein structure, summary plots to summarize mutation load, transition/transversions plot, and rainfall plot to visualize Kataegis phenotype.
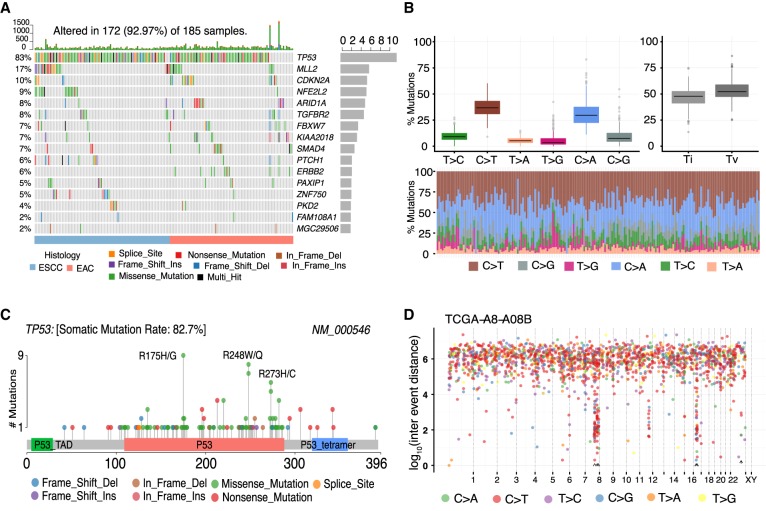
Using ESCA and BRCA cohorts, we show some of the key visualizations generated using Maftools. Figure 2A, known as oncoplots, displays significantly mutated genes (FDR < 0.1) identified by MutSigCV algorithm in ESCA cohort (Lawrence et al. 2013b). Genes are sorted by mutational frequency, and samples are sorted and ordered according to tumor histology, thereby differentiating the mutational spectrum between and within subtypes of ESCA. Options are available to include annotations as bottom annotation bars to display clinical parameters. The transition and transversion plot (Fig. 2B) summarizes SNVs into six categories. Figure 2C shows a lollipop plot for a highly mutated gene, TP53, in ESCA. To keep up with the consistency of the plot design, lollipop plots generated by Maftools follow the same visual aesthetics of the commonly used online tools available as a part of cBioPortal (Cerami et al. 2012). Supplemental Figure S1 lists the rest of the visualization options available for plotting mutational data.
Figure 2.
Key plots generated by Maftools visualization module. (A) Oncoplot displaying the somatic landscape of ESCA cohort. Genes are ordered by their mutation frequency, and samples are ordered according to disease histology as indicated by the annotation bar (bottom). Side bar plot shows log10 transformed Q-values estimated by MutSigCV. (B) Transition and transversion plot displaying distribution of SNVs in ESCA classified into six transition and transversion events. Stacked bar plot (bottom) shows distribution of mutation spectra for every sample in the MAF file. (C) Lollipop plot displaying mutation distribution and protein domains for TP53 in ESCA with the labeled recurrent hotspots. Somatic mutation rate and transcript names are indicated by plot title and subtitle, respectively. (D) Rainfall plot for TCGA breast cancer sample TCGA-A8-A08B. Each point is a mutation color coded according to SNV class. Hypermutated genomic segments identified by the change-point method are highlighted by black arrowheads.
Multiple studies have reported hypermutated genomic regions in several cancer types (Nik-Zainal et al. 2012; Alexandrov et al. 2013a; D'Antonio et al. 2016). These genomic regions referred to as “Kataegis,” presumed to be the result of aberrant activity of apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) cytidine deaminases, tend to be enriched in C > T and C > G substitutions occurring within TpCpN trinucleotide contexts (Lada et al. 2012). The rainfallPlot function of Maftools visualizes the distribution of mutation spectrum with the simultaneous identification of Kataegis loci (Fig. 2D; Methods; Killick and Eckley 2014). From 96 WGS-derived TCGA BRCA samples, rainfallPlot identified 195 hypermutated genomic regions (Kataegis loci) that also contained 98 (of 132) previously identified Kataegis regions (Supplemental Table S1; D'Antonio et al. 2016).
Maftools can also be used to visualize and summarize copy number data generated by GISTIC or segmentation files generated by CBS algorithms. Copy number data can be easily integrated along with mutation data, further facilitating integrative analysis. Supplemental Figure S2 displays plotting options available to visualize CNV data.
Analysis
Mutational signatures and enrichment analysis
Every cancer, as it progresses, leaves behind a characteristic mutational pattern that can reveal its underlying mutagenic processes. Alexandrov et al. (2013a) have shown that such mutagenic processes can be identified by utilizing dimensional reduction techniques such as non-negative matrix factorization (NMF). Specifically, this method classifies SNVs into six different transition and transversion events, each of which is further classified into 16 subtypes based on immediate 5′ and 3′ bases surrounding the mutated base (Nik-Zainal et al. 2012; Alexandrov et al. 2013a). Typical de novo signature analysis includes frequency matrix generation and NMF decomposition. We implemented two functions, namely, trinucleotideMatrix and extractSignatures, to streamline the complete process. We further extended this analysis by signtaureEnrichment function to perform sample stratification, signature enrichment, and association analysis.
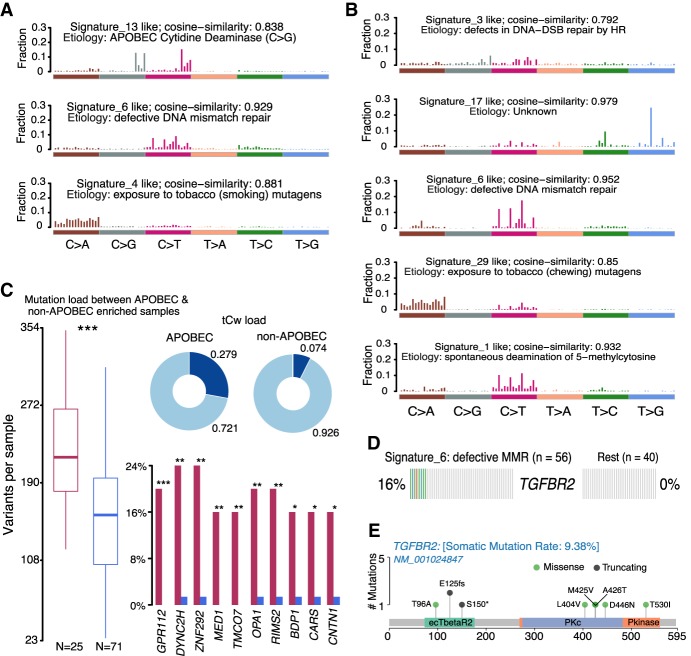
Applying these functions on EAC and ESCC cohorts, we robustly reproduced previous findings by Lin et al. (WXS), TCGA (WXS), and ICGC consortiums (WGS) (Secrier et al. 2016; The Cancer Genome Atlas Network 2017; Lin et al. 2018a). Based on cophenetic correlation metric, we identified three and five signatures in ESCC and EAC, respectively (false positive rate <0.01% in ESCC and <0.122% in EAC) (Fig. 3A,B; Supplemental Fig. S3A–D). De novo signatures identified in ESCC were enriched in APOBEC-related signature (COSMIC Signature 13; cosine similarity: 0.838) and tobacco mutagen signature (COSMIC Signature 4; cosine similarity: 0.881). In contrast, EAC samples had exclusively COSMIC Signature 17, which is associated with gastric acid reflux (cosine similarity: 0.979) (Dulak et al. 2013). In addition, DNA mismatch repair (MMR) signature (COSMIC Signature 6) was noted in both EACs and ESCCs (cosine similarity: 0.952 and 0.929, respectively), in agreement with previous reports (Lin et al. 2018a).
Figure 3.
Signature and APOBEC enrichment analysis. (A,B) Mutational signatures identified in ESCC and EAC, respectively. The y-axis indicates exposure of 96 trinucleotide motifs to overall signature. The plot title indicates best match against validated COSMIC signatures and cosine similarity value along with the proposed etiology. (C) Results from the APOBEC enrichment analysis in ESCC. The box plot shows differences in mutation load between APOBEC-enriched and nonenriched samples: (***) Wilcoxon rank-sum test; P < 0.001. Donut plots display the proportion of mutations in tCw context. Bar plots show the top 10 differentially mutated genes between APOBEC-enriched and non-APOBEC-enriched samples: (***) P < 0.001; (**) P < 0.01; (*) P < 0.05, Fisher's exact test. (D) Enriched TGFBR2 mutations in ESCC among samples associated with defects in DNA mismatch repair signature (COSMIC Signature 6). (E) Lollipop plot showing the distribution of mutations in TGFBR2 and truncating mutations (frameshift deletions and splice site) in the BETR2 domain.
Along with the signature analysis, we integrated a method described by Roberts et al. (2013) to estimate APOBEC enrichment in individual tumor samples. Consistent with the signature analysis, EACs showed no APOBEC enrichment, whereas 26% (25 of 96 samples) of ESCCs were enriched for APOBEC-associated mutations (APOBEC enrichment score >2) (Fig. 3C; Supplemental Table S2). In line with previous findings, mutation burdens among APOBEC-enriched samples were significantly higher than APOBEC-negative ones (median: 196 versus 136; Wilcoxon rank-sum test; P < 0.001) (Fig. 3C; Harris et al. 2002; Taylor et al. 2013). Furthermore, increased mutation rates within MED1, ZFP292, and GPCR genes were detected in APOBEC-enriched samples (Fisher's exact test P < 0.01). Of interest, these proteins play a role in maintaining genome integrity, and their mutational enrichment among APOBEC-high tumors suggests increased DNA damage and APOBEC-mediated genome instability (Fig. 3C; Parsons 2003; Swanton et al. 2015).
Furthermore, we implemented a method to perform sample classification and analysis of signature enrichment, wherein samples were assigned to identified signatures using k-means clustering of signature exposures, followed by Fisher's exact tests (Supplemental Fig. S4A,B). To test its performance, we compared APOBEC enrichment scores (measured by trinucleotideMatrix method) and APOBEC signature weight (measured by signatureEnrichment function). Notably, this comparison revealed a high correlation coefficient (Spearman's correlation coefficient: 0.9438; P-value < 2.2 × 10−6) (Supplemental Fig. S4C), highlighting the concordance between these two methods. We next performed an enrichment analysis to identify genes preferentially mutated along with a particular signature. Applying this method on ESCCs revealed exclusive mutations of the TGFBR2 gene in samples associated with COSMIC Signature 6 (Fisher's exact text; P < 0.001) (Fig. 3D; Supplemental Fig. S4D; Supplemental Table S3). Signature 6 has been associated with MMR deficiency and is characterized by C·G→T·A transitions at a NpCpG sequence context and C·G→A·T transversions at CpCpC. Several solid tumors associated with Signature 6—such as breast, colon, and ovarian cancers—are shown to be enriched for small indels, a characteristic feature associated with Microsatellite Instability (MSI) (Helleday et al. 2014). Also, TGFBR2 mutations have been previously shown to occur among MSI tumors in colon cancer (The Cancer Genome Atlas Network 2012a). Specifically, truncating mutations in TGFBR2 affecting BAT-RII domain, cause deficiency in mismatch repair (MMR) pathways (Biswas et al. 2008). Similar to observations in colon cancer, mutations in ecTBetaR2 domain of TGFBR2 were mostly loss of function (frame shift, indels, and splice site), further highlighting the role of TBGBR2-associated MMR-deficient pathways in ESCC (Fig. 3E).
Combined observations from signature and enrichment analyses suggest that the use of therapies such as methotrexate or poly(ADP-ribose) polymerase (PARP) inhibitors targeting MMR-deficient pathways may have therapeutic activity in ESCC (Martin et al. 2010).
Cohort comparison and Pfam domain summarization
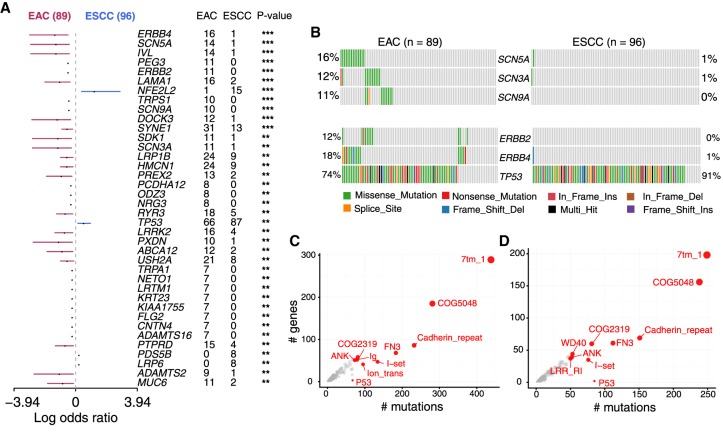
Different forms of cancers differ in their mutational burden and overall mutational landscape depending on tissue lineage and underlying mutagenic processes (Lawrence et al. 2013b). On the other hand, clinical parameters and histopathology contribute to tumor heterogeneity within a single cancer type. We implemented mafCompare function to identify such differentially mutated genes (DMGs) and pathways between two cohorts, wherein mutation load for each gene is compared by Fisher's exact tests. Comparison of ESCC and EAC cohorts revealed 38 genes to be differentially mutated (P < 0.01) (Fig. 4A; Supplemental Table S4). Among them, only four genes (NFE2L2, TP53, LRP6, and PDS5B) were significantly enriched in ESCC, and the other 34 genes were significantly enriched in EAC, largely validating our recent reports using different cohorts (Lin et al. 2018a). These 34 EAC-specific genes were enriched in ERBB signaling and pathways associated with sodium channel signaling (Fig. 4B). ERBB signaling pathway has been known to be dysregulated in EAC more frequently than ESCC (Fichter et al. 2014; The Cancer Genome Atlas Research Network 2017; Lin et al. 2018a). Alterations of genes involved in sodium signaling pathways have not been documented in EAC. Mutant genes of this pathway included SCN3A, SCN5A, and SCN9A, which belong to the family of voltage gated sodium channels (VGSCs) involved in action potential initiation and conduction in excitable cells such as cardiac and neuronal cells (Catterall 2012). VGSC factors are aberrantly expressed in several types of cancers and contribute to cell migration and metastasis (Schönherr 2005; Fiske et al. 2006; House et al. 2015). In addition, VGSC mutations in glioblastoma are associated with shorter survival rates of the patients (Joshi et al. 2011). However, their roles in EAC are unknown. Notably, uniformly distributed activating mutations in a mutually exclusive manner (Fig. 4B; Supplemental Fig. S5) strongly suggest that these may be gain-of-function variants.
Figure 4.
Cohort comparison and domain enrichment analysis. (A) Differentially mutated genes between EAC and ESCC displayed as a forest plot. Bars indicate 95% confidence interval of odds ratio. The adjacent table includes the number of samples in EAC and ESCC with the mutations in the highlighted gene. P-value indicates significance threshold: (***) P < 0.001; (**) P < 0.01; Fisher's exact test. (B) Mutated pathways involving genes associated with VGSC and ERBB signaling in EAC. Genes associated with these pathways are preferentially enriched in EAC, mutated in a mutually exclusive manner. (C,D) Frequently mutated pfam protein domains in EAC and ESCC, respectively. The top ten domains are highlighted. Bubble sizes are proportional to the number of genes containing the highlighted domain. Ion_trans domain is largely mutated in EAC.
The pfamDomains function in Maftools performs domain enrichment analysis. Grouping protein domains helps to identify most deregulated pathways and protein families involved in similar functions. In a separate analysis, EAC and ESCC showed similar pattern of mutated protein domains, particularly those within the top 10 (Fig. 4C,D; Supplemental Table S5). Interestingly, we observed that a specific domain, pfam00520 (Ion_trans), was affected more frequently in EAC (mutated 95 times across 41 genes) (Fig. 4C) compared to ESCC (mutated 35 times across 25 genes) (Fig. 4D). The Ion_trans domain is a family of six transmembrane helicases responsible for ion transportation, primarily involved in ion (Na+/Ca2+) signaling and L1CAM interaction pathways (Jiang et al. 2003). Importantly, these observations in the Ion_trans domain strongly support our earlier results of mutated pathways involving VGSCs (Fig. 4B) and the potential role of dysregulated sodium channels in EAC.
Somatic interactions, cancer genes, and clinical enrichment analysis
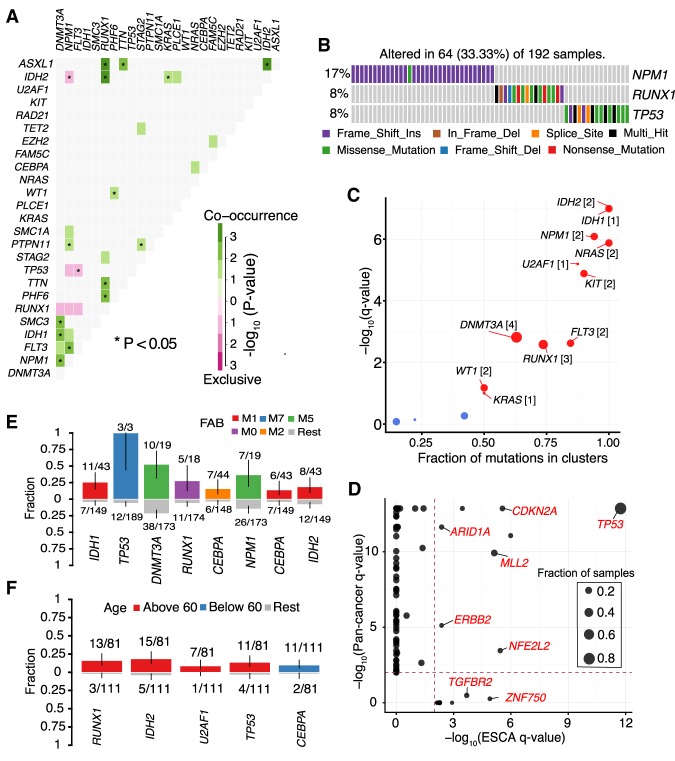
Recent evidence from cancer genomic studies demonstrates that key genes in dysregulated pathways are often mutated in a mutually exclusive manner (Yeang et al. 2008). Identification of such gene sets can reveal de novo pathways and underlying mechanisms of tumorigenesis. Here, we developed a somaticInteractions function, which facilitates the identification of gene sets mutated in either mutually exclusive or a co-occurring manner. To demonstrate its performance, we used the TCGA AML cohort, which has well-documented gene sets mutated in either a mutually exclusive or co-occurring manner (The Cancer Genome Atlas Research Network 2013). Importantly, our method accurately reproduced a number of these observations, such as TP53/FLT3 (mutually exclusive; P-value = 0.012) and NPM1/FLT3 (co-occurring; P-value = 0.00104) (Fig. 5A; Supplemental Table S6). Extending this method further to gene sets of larger size (N = 3) identified several pathways mutated in an exclusive manner (exact test P < 0.001), including well-known transcription factor fusion genes, NPM1, TP53, and RUNX1 (Fig. 5B; Supplemental Table S7; The Cancer Genome Atlas Research Network 2013).
Figure 5.
Pathway and clinical enrichment analysis. (A) Mutually exclusive and co-occurring gene pairs in AML displayed as a triangular matrix. Green indicates tendency toward co-occurrence, whereas pink indicates tendency toward exclusiveness. (B) Significantly altered pathway identified in AML by CoMEt exact test involving NPM1, RUNX1, and TP53 genes mutated in mutually exclusive manner (P < 0.001). (C) Disease-associated driver genes identified by oncodrive in TCGA AML cohort (FDR < 0.1). Number of closely spaced mutational clusters are highlighted within brackets. (D) Pan cancer comparison of significantly mutated genes in ESCA identified by MutSigCV against Pan cancer driver genes. TGFBR2 and ZNF750 are exclusively mutated in esophageal cancer, whereas other drivers, such as TP53, and CDKN2A, are mutated in the global pan cancer cohort. (E,F) Bar plots displaying the association between genes and clinical features, French–American–British (FAB) classification and age group, respectively (P < 0.05, Fisher's exact test). Bars are annotated with the ratio of mutated samples to total samples. Error bars display 95% CI of binomial ratios. The y-axis denotes the fraction of samples associated with the phenotype.
Cancer driver genes are defined by their capability of providing selective growth advantage to cancer cells when genetically altered. Several mathematical approaches have been developed to identify such driver genes, based on mutational frequencies, functional impact, or by clonality modeling (Dees et al. 2012; Gonzalez-Perez and Lopez-Bigas 2012; Lawrence et al. 2013b; Zapata et al. 2017). To facilitate such detection, we built a function oncodrive based on OncodriveCLUST algorithm, which leverages the observation that a majority of the activating mutations within oncogenes are clustered around mutational hotspots (Tamborero et al. 2013). Applying oncodrive on the TCGA AML cohort identified 11 well-known oncogenes as significantly mutated (FDR < 0.1) (Fig. 5C). However, as noted in the original study, oncodrive is biased toward oncogenes with mutational hotspots and has less optimum performance in identifying potential tumor suppressors, such as TP53, with randomly distributed mutations across the gene body. Despite this, the majority of oncodrive results overlapped with driver genes identified by the widely used program MutSigCV (9 of 11) (Supplemental Fig. S6A; Lawrence et al. 2013b). The two drivers identified exclusively by oncodrive are known leukemic oncogenes, KIT and KRAS (Supplemental Fig. S6B,C). These results underscore the performance of oncodrive and suggest that combined approaches of multiple programs can increase the sensitivity of identifying driver genes.
Via the pancanComparison function, users can analyze MutSigCV results and compare them to pan cancer driver genes identified by analyzing more than 4000 samples across 21 different cancer types (Lawrence et al. 2014). Such comparison identifies common driver genes as well as tumor-specific ones. Applying pancanComparison on ESCA cohort revealed several common driver genes, including TP53 and CDKN2A. Importantly, tumor-specific driver genes, such as TGFBR2 and ZNF750, were uniquely mutated in ESCC, validating the findings from us and others (Fig. 5D; Lin et al. 2014).
The clinicalEnrichment in Maftools uses Fisher's exact test to perform both pairwise and groupwise comparisons to determine associations between mutated genes and clinical-pathological characteristics (categorical variables). Clinical-pathological data often includes histopathological classifications, race, gender, treatment, and smoking/drinking history, among others. In AML, testing for enrichments according to French–American–British (FAB) classifications reproduced well-known association patterns. For example, IDH1, IDH2 and CEBPA mutations were enriched within M1 subtype (AML with minimal maturation), and RUNX1 mutations were enriched among M0 subtype (undifferentiated AML; Fisher's exact test, P < 0.05) (Taketani et al. 2003; Patel et al. 2011). Similarly, DNMT3A mutations were frequent in M5 subtype (acute monocytic leukemia), in line with previous results (Fig. 5E; Yan et al. 2011). Furthermore, RUNX1, IDH2, U2AF1, and TP53 mutations were enriched among elder patients (age group >60; median = 68; N = 81; Fisher's exact test, P < 0.05), whereas CEBPA mutations were enriched among younger individuals (age group <60; median = 47; N = 111; Fisher's exact test, P < 0.05) (Fig. 5F).
Variant annotations, format conversions, and subset operations
Maftools also includes functions to perform quick variant annotations and format conversions. Oncotate function takes raw variants stored in a simple tabular format and annotates them using oncotator's REST web API. However, often this process is relatively slower and time-consuming for larger inputs (most time is needed for connecting to API and retrieving annotations). Another widely used tool for variant annotations is Annotate Variation (ANNOVAR), capable of annotating a putative variant with several gene, region, and filter-based annotations. Tabular output files generated from ANNOVAR can be converted to MAF with function annovarToMaf, which parses and maps values from gene-based annotations into MAF-specific values. For converting VCF files to MAF, we recommend vcf2maf utility (https://github.com/mskcc/vcf2maf), which processes variant annotation (with Variant Effect Predictor) and transcript prioritization.
Similar to TCGA, ICGC is an international consortium providing large-scale cancer genome data. However, somatic variants from ICGC are in “simple somatic mutation format,” a standard tab delimited text file introduced by the ICGC consortium to store and distribute somatic variants. We implemented icgcSimpleMutationToMAF function, which converts ICGC mutation format files into MAF, thereby streamlining ICGC data processing. subsetMaf is another function in Maftools that allows subset operations of MAF files based on genes of interest, samples, or user-specified queries.
Discussion
With the rapid increase in genomic analysis, exponential growth has occurred in the availability of software to reanalyze and understand these data (Ding et al. 2014). MuSiC is one of the most widely used tools that offers several analysis modules to perform tasks such as identification of significantly mutated genes, pathway analysis, and clinical correlation tests (Dees et al. 2012). However, its application is rather limited by its dependency on alignment files, platform specificity, and large computational requirements. This further limits its application on public data sets, which are often restricted by the availability of raw alignment files. Several tools such as EmU and deconstructSigs offer statistical frameworks to identify de novo signatures and enrichment of known signatures (Fischer et al. 2013; Rosenthal et al. 2016). Other tools such as Dendrix, PathScan, and HotNet2 can be used for pathway and network analysis (Wendl et al. 2011; Vandin et al. 2012). However, these tools have different requirements of data formats and much time is required in preparing data sets and file format conversions, significantly hindering the efficiency of cancer researchers.
Here, we introduce an R Bioconductor package, Maftools, which integrates standard analyses modules that are frequently performed in cancer genomics and provides a plethora of visualization options to generate publication-quality images. Along with the analysis of somatic variants, Maftools allows easy integration of copy number data generated from either GISTIC or segmentation algorithms. Maftools utilizes well-established dimensional reduction and statistical methods, enabling reproducible data-driven research with the requirement of few lines of code. In addition, inclusion of clinical data can identify novel clustering and enrichment patterns. Moreover, Maftools is easy to use, platform independent, and does not rely on large alignment files, greatly facilitating the exploration of genomic data sets such as those from TCGA and ICGC.
Here, we reproduced many known results utilizing only MAF files from the published TCGA data sets. More importantly, we showed that Maftools can also be used to uncover novel findings through integrative analysis. Via implementation of well-established computational and statistical methods, Maftools provides a wide range of functions for cancer genomic analyses. In future updates, we will include gene expression as well as DNA methylation data for integrative multiomic analysis.
Methods
Data sets
TCGA MAF files (ESCA: http://dx.doi.org/10.7908/C1BV7FZC; LAML: http://dx.doi.org/10.7908/C1D21X2X) along with the clinical data, MutSigCV (v1.41), and GISTIC2 results for ESCA and AML cohort were obtained from Broad Firehose using firehose_get utility (analysis stamp: 2016_01_28). Somatic mutations for BRCA WGS samples were obtained from a published study (D'Antonio et al. 2016) and were later converted to MAF using vcf2maf utility. All raw input data and reproducible R code used to generate the results are provided as Supplemental Data S1. Computational time required for each function is provided in Supplemental Table S8.
R/Bioconductor package
Maftools is implemented as an open source R package and available as a part of the Bioconductor project (Gentleman et al. 2004). MAF file and the associated clinical data along with summary statistics are stored in an S4 class container. For faster data processing and summarization of larger data sets, Maftools uses data.table library, which offers performance of several magnitude higher than base R functions. Package workflow is simple, with every function taking MAF object as an input, and comes with a detailed vignette including a case study describing the usage of available modules.
Visualization
Visualization module in Maftools facilitates generation publication-quality images with easy to use and customizable functions. Plots such as oncoplots and oncostrips are generated using ComplexHeatmap Bioconductor package, whereas plots generated to display results from analysis modules, such as forest plots, lollipop plots, somatic interactions, rainfall plots, among others, are generated using either ggplot2 or base R plot functions (Gu et al. 2016).
Signature and enrichment analyses
Mutational signature analysis in Maftools begins with the process of extracting 5′ and 3′ adjacent bases surrounding the mutated base, thereby constructing a count matrix M of dimension 96 × n, where n is the number of samples available in input MAF file. This process is implemented in the function trinucleotideMatrix which uses BioStrings and GenomicRanges Bioconductor packages for efficiently reading reference genome and extracting adjacent bases while simultaneously classifying mutations into transition and transversion events (Lawrence et al. 2013a).
Once the matrix is generated, the extractSignatures function uses NMF to factorize count matrix M into two smaller matrices—W (96 × r) and H (r × n)—such that product of W and H sufficiently recomposes the original matrix M (Gaujoux and Seoighe 2010).
A key step in factorization is identifying optimal rank, r, used to approximate the target matrix M. Several methods have been described in recent studies such as Bayesian, and Expectation Maximization–based approaches to estimate the ideal value of r (Fischer et al. 2013; Kim et al. 2016). Here, we use the method described by Brunet et al. (2004), wherein NMF is run on a range of incremental values of r, and for each value, the cophenetic correlation coefficient (measure of goodness of fit) is calculated. A final optimal r is chosen such that further increase in r results in decreasing values of the coefficient (Supplemental Fig. S3A,B). Signatures identified following matrix factorization are scaled and compared to known mutagenic processes (COSMIC signatures), and a cosine similarity value is estimated for the best possible match (Alexandrov et al. 2013b). For signature enrichment analysis, we use matrix H, containing signature exposures for every sample in every signature. Using k-means clustering, we group the samples into r clusters, thereby assigning samples to an identified signature.
For APOBEC-based enrichment analysis, we integrated the method described by Roberts et al. (2013) to estimate an enrichment score, which defines the strength of APOBEC-related mutagenic processes for every tumor sample in MAF. Briefly, enrichment of C > T mutations occurring within tCw trinucleotide context over all of the C > T mutations in a given sample is compared to background cytosines and tCw occurring around ±20 bp of mutated bases. We further extended this method to identify genes associated with the APOBEC enrichment by classifying samples as APOBEC-enriched (enrichment score >2) and non-APOBEC-enriched (enrichment score <2), followed by one-way Fisher's exact tests to identify genes overrepresented among APOBEC-enriched samples.
Somatic interactions and pathway analysis
Somatic interactions function in Maftools allows users to identify gene sets mutated in a mutually exclusive or co-occurring manner. For a pair of genes, the pattern of exclusiveness or co-occurrence is estimated by performing a Fisher's exact test on a 2 × 2 contingency table containing frequencies of mutated and nonmutated samples. However, Fisher's exact test is limited by degrees of freedom for larger contingency tables (n > 2; with 2n-n-1 degrees of freedom). To identify pathways involving gene sets (n > 2) mutated in an exclusive manner, we included Combinations of Mutually Exclusive Alterations (CoMEt) algorithm implemented in the cometExactTest R package (Leiserson et al. 2015). CoMEt exact test takes a contingency table of 2n and computes the probability for mutual exclusiveness. As an input to the CoMEt exact test, we first identify all pair of genes that are mutated in a mutually exclusive manner using Fisher's exact test, and CoMEt exact test is run on all unique combination of gene sets of size n. For example, given a gene list of six genes and gene set of size three, the CoMEt exact test is performed on 20 combinations (6C3) to identify gene sets mutated in an exclusive manner. Options are included for users to manually specify the desired gene set size.
Identification of cancer genes
To identify disease-associated cancer genes, we reimplemented OncodriveCLUST algorithm as previously described by Tamborero et al. (2013). Briefly, candidate amino acid positions with mutations above a background threshold (based on binomial distribution accounting for protein length and mutation rate) are compiled for every gene. Candidate positions occurring within a distance of five amino acids are grouped together to form a cluster. Clusters are further refined and extended by merging neighboring amino acid positions occurring within five amino acids. Once cluster formation is completed, a cluster score is calculated, which is a ratio of mutations occurring within the identified clusters to the total number of mutations. Finally, a P-value is estimated based on t-statistic and Z-score.
OncodriveCLUST has originally been implemented in Python framework requiring users to prepare input data containing gene symbols and amino acid positions. Its reimplementation as a part of the Maftools package serves two purposes. The package takes care of file parsing and input data preparation allowing novice users to run the program easily. Its implementation in R programming language exposes it to the larger bioinformatic community and also allows visualization of results in an intuitive manner (Fig. 5C).
Cohort comparison and enrichment analysis
mafCompare function in Maftools allows comparison of two independent cohorts to identify differentially mutated genes or to perform association between clinical features. A 2 × 2 contingency table of frequencies of mutation is calculated for every gene from the input cohorts followed by Fisher's exact test to identify genes showing significant differences in their mutation frequencies. Similarly, for clinical enrichment analysis, once again contingency tables are generated for every categorical variable followed by Fisher's exact test to calculate P-values. Results from cohort comparison and enrichment analysis are visualized as forest plots or frequency bar plots (Figs. 4A, 5E,F).
Pfam domain summarization
Inspired by the Pfam annotation module of MuSiC, we implemented pfamDomains function to identify and group mutations by affected protein domains. However, the approach for grouping mutations is much simpler. MuSiC requires a large database containing protein foci translated to genomic loci, which is later queried by external tools such SAMtools tabix to identify affected protein domains. Here, we use protein change information (in HGVSp format) to parse the protein position and transcript annotations. These positions are later mapped onto the Pfam domain database via rapid data.table foverlaps function for summarization. Plotting options are available to display frequently affected domains by means of bubble plots (Fig. 4C,D).
Change-point detection for identification of hypermutated genomic regions
Hypermutated genomic regions are segments along the chromosome where the mutation rate is significantly higher than the average mutation rate across the genome. To identify such regions, we utilize change-point detection method in which genomic boundaries with the sharp decrease in distance between consecutive mutations are identified. Briefly, mutations are ordered for every chromosome based on the position, and distance between consecutive mutations are calculated followed log transformation. Log transformed inter-event distances are later fed into cpt.mean function implemented in the changepoint R package to identify potential change points (Killick and Eckley 2014). Consecutive change points are merged into genomic segments and annotated as Kataegis if the segment contained six or more consecutive mutations with an average inter-mutation distance of <1000 bp.
Software availability
Maftools is implemented as an R package, released under MIT license. Source code is available on GitHub (https://github.com/PoisonAlien/Maftools) and can be installed via Bioconductor project (https://bioconductor.org/packages/release/bioc/html/maftools.html) (R Core Team 2018). R package source code is also available in the Supplemental Data S2. In addition, we also provide an R data package containing ready-to-use, precompiled somatic variants from Broad Firehose, and Multi-Center Mutation Calling in Multiple Cancers project for all 34 TCGA cohorts along with the relevant clinical information (https://github.com/PoisonAlien/TCGAmutations) (Ellrott et al. 2018).
Supplementary Material
Acknowledgments
We thank the users who provided useful suggestions, bug reports, and feature requests during the development of the package. This research is supported by the National Research Foundation Singapore under its Singapore Translational Research (STaR) Investigator Award (NMRC/STaR/0021/2014) and administered by the Singapore Ministry of Health's National Medical Research Council (NMRC), the NMRC Centre Grant awarded to National University Cancer Institute of Singapore, the National Research Foundation Singapore, and the Singapore Ministry of Education under its Research Centers of Excellence initiatives. This research is also supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education's Tier 3 grants, grant number MOE2014-T3-1-006. D.-C.L. is supported by the DeGregorio Family Foundation, the Price Family Foundation, as well as Samuel Oschin Comprehensive Cancer Institute (SOCCI) at Cedars-Sinai Medical Center through the Translational Pipeline Discovery Fund.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.239244.118.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013a. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. 2013b. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 3: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Trobridge P, Romero-Gallo J, Billheimer D, Myeroff LL, Willson JK, Markowitz SD, Grady WM. 2008. Mutational inactivation of TGFBR2 in microsatellite unstable colon cancer arises from the cooperation of genomic instability and the clonal outgrowth of transforming growth factor β resistant cells. Genes Chromosomes Cancer 47: 95–106. [DOI] [PubMed] [Google Scholar]
- Brunet JP, Tamayo P, Golub TR, Mesirov JP. 2004. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci 101: 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012a. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012b. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2017. Integrated genomic characterization of oesophageal carcinoma. Nature 541: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. 2012. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J Physiol 590: 2577–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antonio M, Tamayo P, Mesirov JP, Frazer KA. 2016. Kataegis expression signature in breast cancer is associated with late onset, better prognosis, and higher HER2 levels. Cell Rep 16: 672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, Mooney TB, Callaway MB, Dooling D, Mardis ER, et al. 2012. MuSiC: identifying mutational significance in cancer genomes. Genome Res 22: 1589–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Wendl MC, McMichael JF, Raphael BJ. 2014. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet 15: 556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, et al. 2013. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 45: 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, et al. 2018. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst 6: 271–281 e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichter CD, Gudernatsch V, Przypadlo CM, Follo M, Schmidt G, Werner M, Lassmann S. 2014. ErbB targeting inhibitors repress cell migration of esophageal squamous cell carcinoma and adenocarcinoma cells by distinct signaling pathways. J Mol Med (Berl) 92: 1209–1223. [DOI] [PubMed] [Google Scholar]
- Fischer A, Illingworth CJ, Campbell PJ, Mustonen V. 2013. EMu: probabilistic inference of mutational processes and their localization in the cancer genome. Genome Biol 14: R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiske JL, Fomin VP, Brown ML, Duncan RL, Sikes RA. 2006. Voltage-sensitive ion channels and cancer. Cancer Metastasis Rev 25: 493–500. [DOI] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. 2010. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Lopez-Bigas N. 2012. Functional impact bias reveals cancer drivers. Nucleic Acids Res 40: e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. [DOI] [PubMed] [Google Scholar]
- Hao JJ, Lin DC, Dinh HQ, Mayakonda A, Jiang YY, Chang C, Jiang Y, Lu CC, Shi ZZ, Xu X, et al. 2016. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat Genet 48: 1500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell 10: 1247–1253. [DOI] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S. 2014. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet 15: 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House CD, Wang BD, Ceniccola K, Williams R, Simaan M, Olender J, Patel V, Baptista-Hon DT, Annunziata CM, Gutkind JS, et al. 2015. Voltage-gated Na+ channel activity increases colon cancer transcriptional activity and invasion via persistent MAPK signaling. Sci Rep 5: 11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. 2003. X-ray structure of a voltage-dependent K+ channel. Nature 423: 33–41. [DOI] [PubMed] [Google Scholar]
- Joshi AD, Parsons DW, Velculescu VE, Riggins GJ. 2011. Sodium ion channel mutations in glioblastoma patients correlate with shorter survival. Mol Cancer 10: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick R, Eckley IA. 2014. changepoint: an R package for changepoint analysis. J Stat Softw 58 10.18637/jss.v058.i03. [DOI] [Google Scholar]
- Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, Rosenberg JE, Van Allen EM, D'Andrea A, Getz G. 2016. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat Genet 48: 600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lada AG, Dhar A, Boissy RJ, Hirano M, Rubel AA, Rogozin IB, Pavlov YI. 2012. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biol Direct 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013a. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013b. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Wu HT, Vandin F, Raphael BJ. 2015. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol 16: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al. 2014. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 46: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Dinh HQ, Xie JJ, Mayakonda A, Silva TC, Jiang YY, Ding LW, He JZ, Xu XE, Hao JJ, et al. 2018a. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut 67: 1769–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Wang MR, Koeffler HP. 2018b. Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology 154: 374–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Wilson RK. 2009. Cancer genome sequencing: a review. Hum Mol Genet 18: R163–R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Lord CJ, Ashworth A. 2010. Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res 16: 5107–5113. [DOI] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. 2011. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149: 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5: 557–572. [DOI] [PubMed] [Google Scholar]
- Parsons BL. 2003. MED1: a central molecule for maintenance of genome integrity and response to DNA damage. Proc Natl Acad Sci 100: 14601–14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KP, Ravandi F, Ma D, Paladugu A, Barkoh BA, Medeiros LJ, Luthra R. 2011. Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features. Am J Clin Pathol 135: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. 2016. deconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönherr R. 2005. Clinical relevance of ion channels for diagnosis and therapy of cancer. J Membr Biol 205: 175–184. [DOI] [PubMed] [Google Scholar]
- Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, MacRae S, Grehan N, O'Donovan M, Miremadi A, et al. 2016. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet 48: 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C, McGranahan N, Starrett GJ, Harris RS. 2015. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov 5: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketani T, Taki T, Takita J, Tsuchida M, Hanada R, Hongo T, Kaneko T, Manabe A, Ida K, Hayashi Y. 2003. AML1/RUNX1 mutations are infrequent, but related to AML-M0, acquired trisomy 21, and leukemic transformation in pediatric hematologic malignancies. Genes Chromosomes Cancer 38: 1–7. [DOI] [PubMed] [Google Scholar]
- Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. 2013. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 29: 2238–2244. [DOI] [PubMed] [Google Scholar]
- Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. 2013. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2: e00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. 2012. De novo discovery of mutated driver pathways in cancer. Genome Res 22: 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendl MC, Wallis JW, Lin L, Kandoth C, Mardis ER, Wilson RK, Ding L. 2011. PathScan: a tool for discerning mutational significance in groups of putative cancer genes. Bioinformatics 27: 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Wang L. 2013. From human genome to cancer genome: the first decade. Genome Res 23: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. 2011. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43: 309–315. [DOI] [PubMed] [Google Scholar]
- Yeang CH, McCormick F, Levine A. 2008. Combinatorial patterns of somatic gene mutations in cancer. FASEB J 22: 2605–2622. [DOI] [PubMed] [Google Scholar]
- Zapata L, Susak H, Drechsel O, Friedlander MR, Estivill X, Ossowski S. 2017. Signatures of positive selection reveal a universal role of chromatin modifiers as cancer driver genes. Sci Rep 7: 13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.