Abstract
The widespread identification of genes without detectable homology in related taxa is a hallmark of genome sequencing projects in animals, together with the abundance of gene duplications. Such genes have been called novel, young, taxon-restricted, or orphans, but little is known about the mechanisms accounting for their origin, age, and mode of evolution. Phylogenomic studies relying on deep and systematic taxon sampling and using the comparative method can provide insight into the evolutionary dynamics acting on novel genes. We used a phylogenomic approach for the nematode model organism Pristionchus pacificus and sequenced six additional Pristionchus and two outgroup species. This resulted in 10 genomes with a ladder-like phylogeny, sequenced in one laboratory using the same platform and analyzed by the same bioinformatic procedures. Our analysis revealed that 68%–81% of genes are assignable to orthologous gene families, the majority of which defined nine age classes with presence/absence patterns that can be explained by single evolutionary events. Contrasting different age classes, we find that older age classes are concentrated at chromosome centers, whereas novel gene families preferentially arise at the periphery, are weakly expressed, evolve rapidly, and have a high propensity of being lost. Over time, they increase in expression and become more constrained. Thus, the detailed phylogenetic resolution allowed a comprehensive characterization of the evolutionary dynamics of Pristionchus genomes indicating that distribution of age classes and their associated differences shape chromosomal divergence. This study establishes the Pristionchus system for future research on the mechanisms that drive the formation of novel genes.
The sequencing of genomes throughout the animal kingdom has shown gene duplication to represent the major driving force for the generation of novel genes, confirming the predictions of Susumu Ohno from his seminal book, Evolution by Gene Duplication (Ohno 1970; Lynch 2007). However, the same sequencing efforts have also shown that up to one-third of genes in a given genome lack homology in any other species and have therefore been called novel, young, taxonomically restricted, pioneer, or orphan genes (Khalturin et al. 2009; Tautz and Domazet-Lošo 2011). Although horizontal gene transfer, rapid divergence, evolution from previously noncoding sequences, as well as genomic artifacts have been proposed to explain their presence (Long et al. 2003; Tautz and Domazet-Lošo 2011; Rödelsperger et al. 2013; Denton et al. 2014), it is still unclear to what extent these mechanisms contribute to their existence. In addition, there are very few studies that comprehensively date their age and characterize the evolutionary forces acting on them (Palmieri et al. 2014; Stein et al. 2018). In the case of the nematode Pristionchus pacificus, which has been established as a model organism for comparative studies with Caenorhabditis elegans (Sommer and Sternberg 1996; Sommer 2015), roughly one-third of genes initially were classified as orphan genes (Dieterich et al. 2008; Borchert et al. 2010). Although extensive comparative genomic studies did show some evidence of horizontal gene transfer (Dieterich et al. 2008; Rödelsperger and Sommer 2011; Meyer et al. 2016), this process could only explain the origin of a handful of orphan gene families. In addition to the high abundance of orphan genes within the genome of P. pacificus, we recently showed that most orphan genes are real (Prabh and Rödelsperger 2016) and they can regulate ecologically relevant traits (Mayer et al. 2015).
The fraction of orphan genes is expected to be higher in the genomes of isolated species that lack genome data from closely related taxa (Tautz and Domazet-Lošo 2011). For example, the divergence time between P. pacificus and C. elegans can be estimated as between 60 and 90 million years ago (Cutter 2008; Dieterich et al. 2008). Thus, deeper taxon sampling is key to understanding gene origin and evolution. P. pacificus belongs to the family Diplogastridae, whereas C. elegans belongs to the family Rhabditidae (Sudhaus 2013). Importantly, P. pacificus was one of two Pristionchus species with a sequenced genome, and no other diplogastrid nematode outside Pristionchus genus has ever been sequenced. Therefore, we decided to create a comprehensive phylogenomic data set to study the genome evolution of Pristionchus nematodes. Phylogenomics is a composite of genome analysis and evolutionary studies (Eisen and Fraser 2003), and it uses the comparative method (Harvey and Pagel 1998) that involves study of phenotypic variation in a given evolutionary framework. Initially, the phylogenomic approach was used to predict the function of a novel protein through common ancestry (DeSalle and Rosenfeld 2012). Further, as more whole-genome data of related species became available, phylogenomic studies started to focus on taxonomically restricted traits (Johnson and Tsutsui 2011; Hunt et al. 2016; Santos et al. 2017; Verster et al. 2017). There are two main requirements for a comprehensive phylogenomic analysis. First, an accurate species tree helps with the selection of species that are best placed to study a particular question. Second, the comparable genome data for the selected species must be available to study the evolution of genomic features within the phylogenetic framework. For the genus Pristionchus, which includes more than 30 culturable species, previous work has established a robust molecular phylogeny for the selection of representative nematode species (Susoy et al. 2016).
In this study, we extend the existing data set of two Pristionchus genomes (Dieterich et al. 2008; Rödelsperger et al. 2014) by sequencing eight additional diplogastrid genomes within and outside of the Pristionchus genus. Together, these are conducive to exploring the dynamics that shape the Pristionchus genome at extremely high phylogenetic resolution. In this study, we focus on (1) establishing our phylogenomic data set, (2) assigning gene families into age classes, and (3) exploring their evolutionary trajectories, whereas we intentionally leave the study of mechanisms of gene formation for future research.
Results
Assemblies of 10 diplogastrid genomes as a platform for comparative phylogenomics
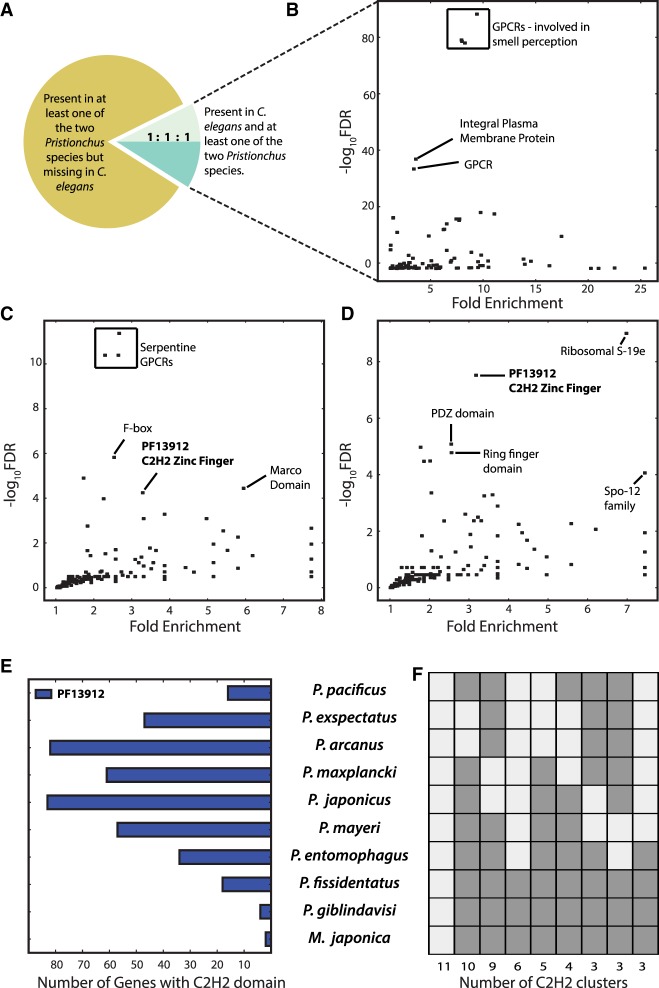
To gain insight into the dynamics of gene gain and loss within the Pristionchus lineage, we complemented the two existing draft genomes of the sister species P. pacificus (Rödelsperger et al. 2017) and P. exspectatus (Rödelsperger et al. 2014) by sequencing eight more diplogastrid genomes. In particular, we sequenced genomes of three gonochoristic (P. arcanus, P. maxplancki, and P. japonicus) and three hermaphroditic (P. mayeri, P. entomophagus, and P. fissidentatus) nematodes of the genus Pristionchus along with the gonochoristic Parapristionchus giblindavisi and Micoletzkya japonica (Susoy et al. 2013, 2016). In addition, we reassembled the genome of P. pacificus based on Illumina data alone to increase the comparability (see Methods for details). Each species was carefully chosen to create a deep taxon sampling of the Pristionchus genus based on our current understanding of the molecular phylogeny (Susoy et al. 2016), and the two non-Pristionchus species were selected as outgroup (Fig. 1A; Supplemental Fig. S1). The genome sizes of Pristionchus nematodes in the scaffolded assemblies varied between 151 and 297 Mb (Table 1). Mode of reproduction in Caenorhabditis nematodes was reported to cause a reduction in genome size of hermaphroditic species (Wang et al. 2010; Fierst et al. 2015; Slos et al. 2017; Yin et al. 2018). We note that gonochorists do not generally have larger genomes than hermaphroditic Pristionchus species. However, when comparing the hermaphroditic P. pacificus with its two closest relatives, P. exspectatus and P. arcanus, the trend for smaller genomes in hermaphrodites holds true (Fig. 1A; Table 1).
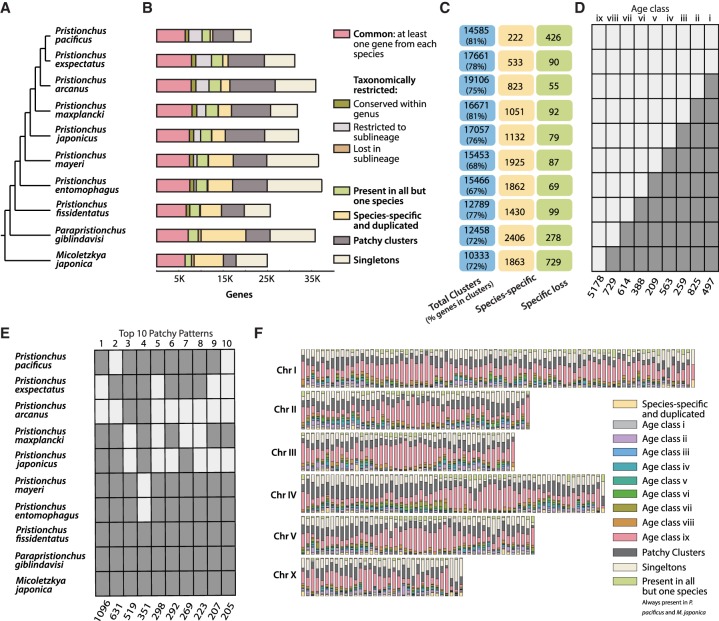
Figure 1.
Gene classes of Pristionchus nematodes and their distribution on P. pacificus chromosomes. (A) Overview of phylogenetic relationship among the 10 diplogastrid species. (B) Distribution of genes within orthology classes across 10 diplogastrid genomes. (C) Numbers of total clusters per species and the percentage of all genes within these clusters, followed by the number of species-specific clusters and clusters that have been exclusively lost in the given species. (D) Graphical representation of the age classes; a light rectangle indicates presence of a gene family in the given species, and a dark rectangle indicates absence of this gene family. The roman numerals at the top of the box indicate the relative age of the age class. (E) Top 10 species distribution patterns in patchy clusters. (F) Distribution of all orthology classes in nonoverlapping 500-Mb windows across chromosomes suggests that older genes are overrepresented at the chromosome centers. Chromosome II, III, IV, and V have their centers at the middle, Chromosome I has two chromosome centers, and Chromosome X has no obvious center.
Table 1.
Summary of basic assembly features
To assess the quality of the draft assemblies, we calculated measures of contiguity (N50, numbers of scaffolds), completeness (BUSCO, percentage of raw reads represented in the assembly), and correctness (paired ends in proper orientation, ambiguous fraction). The largest differences were caused by the switch to a less aggressive assembly strategy during the course of this study. Specifically, the older ALLPATHS-LG strategy, which was based on an initial assembly of overlapping read pairs, yielded substantially fewer contigs at the cost of much higher levels of ambiguous base calls (MacCallum et al. 2009). The more recent approach, as implemented in the software DISCOVAR de novo (https://software.broadinstitute.org/software/discovar/blog/), generates an initial assembly based on a PCR-free library. These assemblies tend to have overall higher number of contigs, but also substantially reduced levels of ambiguity (Table 1). However, it is important to note that these differences between assembly strategies do not seem to have an effect on either the N50, BUSCO, or any of the other measures of assembly quality. Therefore, we conclude that all our assemblies are of comparable quality.
The majority of gene families can be explained by a single evolutionary event
The ladder-like phylogenetic tree (Fig. 1A) allowed the tracing of the phylogenetic origin of genes on nine ancestral nodes and the assignment of genes into age classes. We generated gene annotations based on protein homology information and RNA-seq data for all 10 species (Supplemental Table S1) and computed orthologous gene clusters with orthAgogue (Fig. 1B; Ekseth et al. 2014), which represents a faster reimplementation of the widely used OrthoMCL pipeline (Li et al. 2003). In total, 38,639 clusters having two or more genes were generated, which contained 68%–81% of genes in a given genome (Fig. 1C). More than 5000 clusters were found to have at least one gene from all 10 species; hence, their origin could be traced back to the common ancestor of all studied diplogastrid nematodes (Fig. 1D). Such clusters were designated as “Age class ix” in our analysis (Fig. 1B). Clusters that were missing from M. japonica, but had at least one gene in each Pristionchus species and P. giblindavisi, were classified as “Age class viii” (Fig. 1D). It is important to note that these clusters represent either an M. japonica–specific loss or a taxon-restricted gain. Further, multiple clusters were found to be restricted within a monophyletic sublineage and were designated as “Age class vii–i” (Fig. 1D). Thus, the lower the cluster age class, the more recent is the origin of the genes in it.
Additionally, we identified clusters in which the species distribution could most parsimoniously be explained by gene loss restricted to a monophyletic group (“Lost in sublineage”) (Fig. 1B). There were multiple clusters that had at least one gene from all but one species, and we categorized such clusters as “species-specific loss” (Fig. 1C). Finally, there were gene clusters with two or more genes from only one species, and such clusters were labeled as “species-specific” clusters. They were composed of species-specific genes that were duplicated and thus formed clusters made of paralogs (Fig. 1C). Consistent with the phylogeny that underlies our study design, longer branches between extant taxa and more ancestral inner nodes (Susoy et al. 2016) show higher numbers of species-specific duplications and gene losses. Because it can be difficult to differentiate true losses from missing evidence (Gilabert et al. 2016; Rödelsperger 2018), the numbers of species-specific gene losses within most of the sampled Pristionchus species seem to be rather stable and only increase in the two outgroups (Fig. 1C).
In addition to the already described cluster categories, we were left with genes from every species that did not cluster with any other genes; thus, such genes were called “singletons.” Although we suspect that some of the singletons can be gene annotation artifacts, our previous report suggests that the majority of singletons are real protein coding genes (Prabh and Rödelsperger 2016). However, lack of homologous sequence data prohibits any type of selection analysis. Therefore, we focused on gene families with members from at least two species and left the characterization of singletons for future research. Taken together, the analysis of orthologous cluster types showed that up to 67% of P. pacificus gene families can most parsimoniously be explained by a singular evolutionary event such as a gain or a loss.
Young gene families have a higher propensity of being lost
The orthologous cluster types that we have defined above can be explained by a single evolutionary event. However, 38% of all clusters can only be explained by more than one gain or loss and were labeled as “patchy clusters.” When these patchy clusters were analyzed for most common species distribution patterns, we found that most of the top 10 species patterns can be parsimoniously explained by just two evolutionary events, i.e., a gain at one of the internal nodes within the Pristionchus genus, followed by a loss either in an extant species or at one of the derived internal nodes (Fig. 1E). More precisely, nine of the 10 most abundant patchy cluster types were not older than the common ancestor of P. pacificus and P. japonicus. This finding indicated that younger gene families are more prone to gene loss. Further, we found that none of the most abundant patchy clusters distinguish the two different modes of reproduction. Thus, we conclude that the majority of observed changes are better explained by phylogeny.
A chromosome-scale assembly of the P. pacificus genome (Rödelsperger et al. 2017) allowed us to map the genes from different cluster categories onto the six chromosomes. We created nonoverlapping windows of 500 kb for each chromosome and calculated the fraction of genes falling into different cluster categories or age class within a given window (Fig. 1F). The majority of chromosomes showed enrichment for old cluster categories, i.e., clusters common in all species (Age class ix) or present in all but one species, located at the chromosome centers. Note that chromosome centers are not related to centromeres because Pristionchus nematodes have holocentric chromosomes (Melters et al. 2012). Instead, chromosome centers were defined previously based on characteristic genomic signatures such as high gene density, low repeat content, and low levels of nucleotide diversity (Rödelsperger et al. 2017). Consequently, P. pacificus Chromosome I appears to have two center-like regions. The finding that patchy clusters are preferentially located at chromosome arms is consistent with the fact that they represent young gene families, which have been secondarily lost in one of the species (The C. elegans Sequencing Consortium 1998; Parkinson et al. 2004; Thomas 2006).
Genes increase in expression levels with age
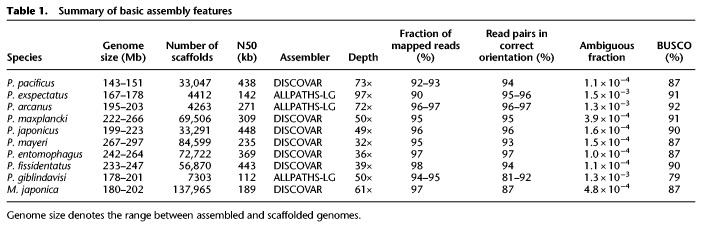
To study how gene expression evolves over time, we compared different age classes with gene expression profiles from multiple developmental stages of P. pacificus (Baskaran et al. 2015). We observed that in all samples, the older age classes (mostly Age class ix and viii) are expressed at a higher level than the younger age classes, and expression levels increase with gene age (Fig. 2A; Supplemental Fig. S2). Also, the genes from Age class ix are expressed at relatively high levels in all the samples (Fig. 2B). Although the correlation between age classes and expression levels is relatively weak (Spearman's rho = 0.33, P < 2−64) (Fig. 2A), this can be improved by calculating the mean expression value of all genes in all 10 samples (Spearman's rho = 0.46, P < 2−64). When mapping gene expression levels in 500-kb nonoverlapping windows on each chromosome, we observed that genes under the highest expression category (mean FPKM ≥10) are also enriched at the chromosome centers (Fig. 2C). Incidentally, some windows at the chromosome centers also had the highest fractions of genes without any expression evidence (mean FPKM = 0), which is most likely due to the presence of old genes with high spatio-temporally restricted expression. In summary, the analysis of expression data shows that young genes usually have either low or spatio-temporally restricted expression, and that their expression tends to increase or become broader over time.
Figure 2.
Expression increases over time. (A) Expression values for P. pacificus genes from different age classes in an RNA-seq data set of late larvae and adults (Late 1) indicate that older age classes are expressed at higher levels. (B) Age class ix genes are expressed at a constitutively high level in all 10 developmental transcriptomes. (C) Distribution of expression classes across the P. pacificus chromosomes.
Exceptionally high gene loss along the P. pacificus lineage
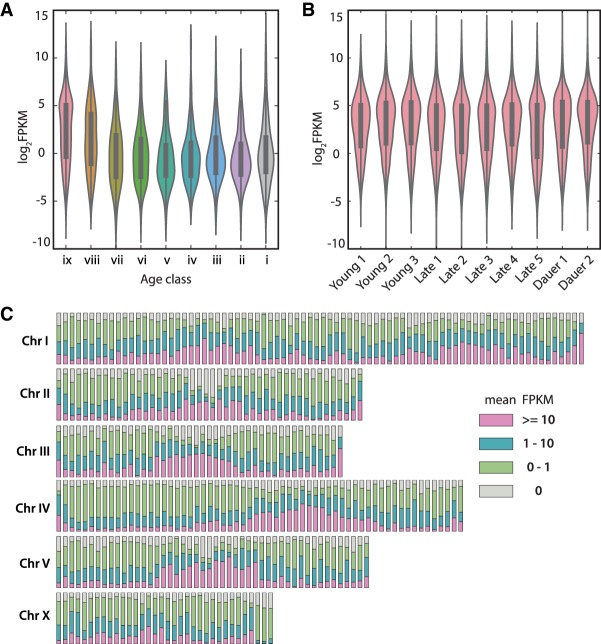
Our previous analysis showed that young genes are preferentially located at chromosome arms, are not highly abundant in transcriptome data, and have a higher probability of getting lost. Along these lines, we observed that P. pacificus shows considerably higher number of species-specific lost clusters as compared with the other Pristionchus species (Fig. 1C). This called for further investigation, and we ascertained the orthology relation among P. pacificus, P. exspectatus, P. arcanus, and C. elegans to functionally characterize the C. elegans orthologs (Fig. 3A). We searched for genes that were lost in P. pacificus but present in other closely related species and C. elegans to perform a Gene Ontology analysis with available C. elegans annotations (Huang et al. 2009). This analysis showed a significant overrepresentation of G-protein coupled receptors (GPCRs) among gene families that have been lost in the P. pacificus lineage (Fig. 3B). However, the majority of the clusters (84%) missing P. pacificus genes are also missing C. elegans genes (Fig. 3A). Therefore, we used P. exspectatus and P. arcanus genes from such clusters to further investigate the P. pacificus losses. Protein domains (PFAM) for all P. exspectatus and P. arcanus genes were annotated based on InterProScan-5.19–58.0 (Finn et al. 2017), and we tested for overrepresentation of protein domains in P. pacificus–specific lost clusters in both species (Fig. 3C,D). C2H2-type zinc finger domain (PF13912) was one of the top candidates that were enriched in both P. exspectatus and P. arcanus. Genes with C2H2 domains in C. elegans, such as lsy-2 and ces-1, are transcription factors that play roles in larval development and programmed cell death (Thellmann et al. 2003; Johnston 2005). Interestingly, in the first draft genome of P. pacificus, this domain was shown as the second most prominent domain expansion in P. pacificus with respect to C. elegans (Dieterich et al. 2008; Finn et al. 2017). However, given our dense phylogenetic sampling, we found that the number of C2H2 domains in P. pacificus (N = 16) has dropped since the separation from P. exspectatus (N = 47) (Fig. 3E), yet it is much higher than in C. elegans (N = 6). Based on the distribution of species patterns among gene clusters that were annotated as having a C2H2 domain (Fig. 3F), we conclude that gene families with this domain have undergone multiple gene losses and gains throughout the Pristionchus genus. This finding highlights the need for dense phylogenetic sampling to accurately describe the evolution of gene families.
Figure 3.
P. pacificus–specific loss. (A) Majority of gene families that have been lost in P. pacificus, but have at least one gene from either P. exspectatus or P. arcanus, do not have any orthologous genes in C. elegans. (B) Gene Ontology analysis of C. elegans orthologs of P. exspectatus and P. arcanus genes whose counterparts were lost in P. pacificus shows an enrichment of G-protein coupled receptors. (C,D) Overrepresentation of protein domains among genes that have been lost in P. pacificus based on orthologs from P. exspectatus (C) and P. arcanus (D). The C2H2-type zinc finger domain (PF13912) shows a consistently significant enrichment in both species. (E) The number of genes with C2H2 domains across all 10 species indicates an expansion of this domain in the Pristionchus lineage. (F) The nine most abundant species distribution patterns in orthologous clusters containing a C2H2 domain show additional expansions and contractions.
All age classes are under evolutionary constraint
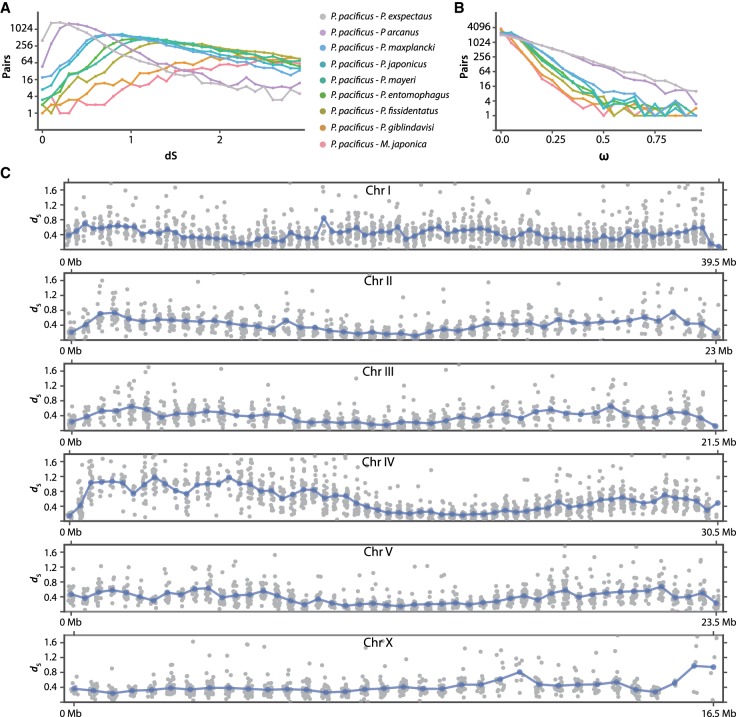
Next, we investigated the evolutionary forces acting on the different age classes. To this end, we calculated rates of nonsynonymous changes (dN), synonymous changes (dS), and ω (dN/dS) for 1:1 orthologs between P. pacificus and each other species. The rate of synonymous changes (dS) obtained from pairwise species comparisons was used as a proxy for divergence time, and it remained consistent with the species phylogeny (Fig. 4A). The two most closely related species to P. pacificus—P. exspectatus and P. arcanus—showed dS peaks between 0.2 and 0.5 substitutions per site. The ω distributions demonstrated that all age classes are indeed under evolutionary constraint (Fig. 4B). Interestingly, the ω distributions also followed the species phylogeny, suggesting that older species pairs were under stronger selection (Fig. 4B). However, it should be noted that the observed patterns of ω distribution might reflect the fact that longer time periods facilitate the removal of more deleterious or slightly deleterious alleles (Thellmann et al. 2003; Johnston 2005; Rödelsperger et al. 2014). Therefore, we decided to narrow our focus on a fixed evolutionary age by only considering P. pacificus and P. exspectatus pairwise data set for further analysis.
Figure 4.
Divergence estimates across different time scales and their chromosomal distribution. (A,B) Pairwise dS (A) and ω (B) distribution between P. pacificus and all other species support the underlying species phylogeny (Susoy et al. 2016). (C) dS value of each 1:1 ortholog between P. pacificus and P. exspectatus were mapped on the P. pacificus chromosomes with a running mean for each window (in blue).
Divergence profiles reflect fast evolving chromosome arms and stable centers
In our previous analysis, we observed that nucleotide diversity is not uniformly distributed throughout the length of the P. pacificus chromosomes and suspected that dS may also vary between different chromosomal regions (Rödelsperger et al. 2017). To investigate dS variation along the chromosome, we plotted dS values for all pairwise comparison between P. pacificus and P. exspectatus for 500-kb nonoverlapping windows and a running mean for each window (Fig. 4C). Median level of dS between P. pacificus and P. exspectatus is 0.33 (interquartile range [IQR] = 0.21–0.51), which would roughly correspond to a divergence time of 1–5 million years ago (Cutter 2008). Similar to the profile of nucleotide diversity across the chromosome (Rödelsperger et al. 2017), we observed that the dS values are lower at the chromosome centers and are higher at chromosome arms. In their analysis of evolutionary rates in Arabidopsis, Yang and Gaut (2011) proposed at least three nonexclusive processes to explain variation in divergence, which are a nonuniformly distributed mutation rate, codon bias, and population genetic processes such as background selection. We ruled out mutation rate and codon bias as the main processes behind this variation, because mutation accumulation line experiments in P. pacificus and other nematodes did not provide evidence for mutation rate biases (Denver et al. 2012; Weller et al. 2014) and the strong positive correlation between dN and dS (Spearman's rho = 0.63 with a P < 2−64) limits the role of codon bias, thus leaving background selection as a plausible explanation. Further, since the spatial distribution of age classes coincided with the distribution of dS and previous analysis of evolutionary constraint suggested old genes to be under stronger selection (Fig. 4B), we hypothesized that differences in proportion of age classes may cause the impression of slower evolving chromosome centers and faster evolving chromosome arms.
Young genes evolve more rapidly
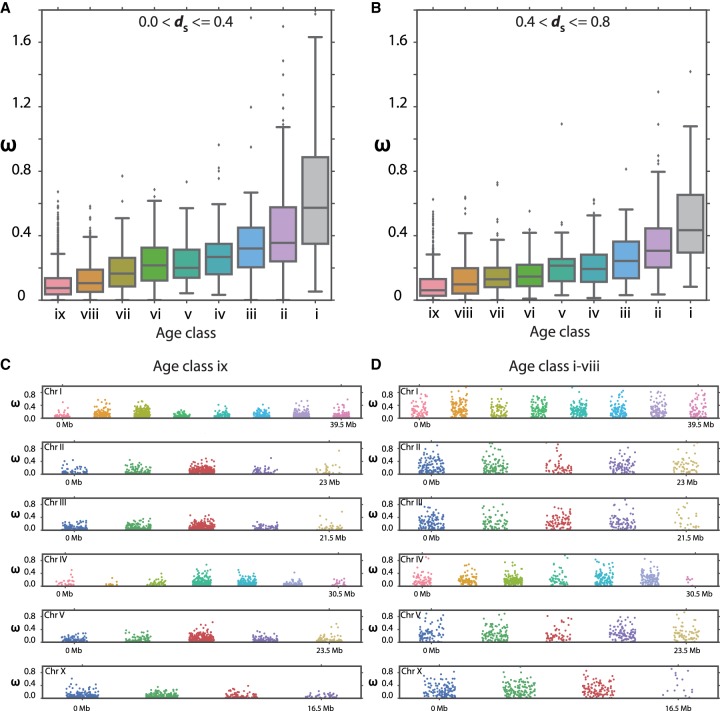
Finally, we wanted to test whether chromosomal location via background selection or the genes themselves determine the level of divergence. Therefore, we tested whether the degree of evolutionary constraint differs between age classes. To this end, we decided to look at the ω distribution for different age classes by separating them into two dS ranges (0–0.4 and 0.4–0.8). Although the lower dS range should largely capture chromosome centers, the upper range represents mostly genes at chromosome arms (Fig. 4C). In both categories, we observe that the old age classes are under strong purifying selection (Spearman's rho = 0.56, P < 2−64) (Fig. 5A,B). Although classification of dS corrected for synonymous divergence, we also directly compared ω distribution for different age classes along the chromosomes. Hence, we divided the age classes into “old” (Age class ix) and “young” (Age classes i–viii) and then plotted their corresponding ω distribution for a 5-Mb nonoverlapping window (Fig. 5C,D). Again, we observed that old genes were under strong purifying selection, whereas young genes could evolve more rapidly, indicating that it is indeed the different composition of age classes within a chromosomal region, which explains the nonuniform divergence across chromosomes (Fig. 4C).
Figure 5.
Young genes evolve more rapidly. (A,B) ω values decrease with age in both the dS ranges, indicating that young genes evolve rapidly and become more constrained over time. The ω values of 1:1 orthologs between P. pacificus and P. exspectatus of Age class ix (C) and Age classes i–viii (D) in 5-Mb windows show that young genes are less constrained irrespective of the chromosomal location. For comparison, in both C and D, corresponding windows on each chromosome have the same color.
Further, we quantified the significance of the comparisons of dS, dN, and ω along the chromosomes (Supplemental Fig. S3). These comparisons were generally highly significant, supporting the idea that selection can act on genes individually. We conclude that at evolutionary time scales, such as the separation between different Pristionchus species, the major determinant of the amount of evolutionary constraint acting on a given gene is the gene itself.
Discussion
This study was designed to bring the comparative method (Harvey and Pagel 1998) to the phylogenomics of Pristionchus nematodes. To our knowledge, this is the first comparative phylogenomic study of nematodes, for which 10 species of a family, including eight of one genus, were chosen to create a unique ladder-like phylogeny so that the focal species always remains under a monophyletic clade. In addition, whole-genome sequencing and gene annotation of each species were done within one laboratory, ensuring that all genomes are of comparable quality and gene annotations were performed using a single protocol. This demonstrates the advantage of nematodes, with their species richness and small genome sizes, in studying various aspects of genome evolution (Dillman et al. 2015; Hunt et al. 2016; Rödelsperger 2018). Our findings result in four major conclusions.
First, the unique ladder-like phylogeny of the 10 diplogastrid genomes enabled us to trace the evolutionary history of the vast majority of P. pacificus genes, including orphan genes that did not show any sequence homology outside the diplogastrid family (Prabh and Rödelsperger 2016). Here, we chose to define age classes based on orthologous clustering rather than using a phylostratigraphy approach (Domazet-Lošo et al. 2007), because orthologous clustering methods are able to split large gene families into broadly shared clusters as well as clusters that arose by recent duplication. Since our aim was to study the evolutionary processes acting on young genes irrespective of their origin, we explicitly include recently duplicated genes that have been shown to undergo distinct evolutionary dynamics (Katju and Lynch 2003; Long et al. 2003, 2013; Chen et al. 2010; Pegueroles et al. 2013; O'Toole et al. 2018). The availability of a chromosome-scale assembly for our focal species allowed us to map the P. pacificus genes on to chromosomes based on their age classes (Rödelsperger et al. 2017), revealing that old genes are concentrated at chromosome centers. This is consistent with the general tendency of novel genes to cluster in certain chromosomal areas, which has been associated with other features such as transposons and late replication timing (Thomas 2006; Juan et al. 2014; Stein et al. 2018).
Second, our data shows that genes of older age classes are either more broadly or more highly expressed compared with younger genes. This trend holds true for every life stage that we looked at, suggesting that expression levels increase or become broader with time. Again, this finding is consistent with previous studies in animals and plants (Baskaran and Rödelsperger 2015; Rogers et al. 2017; Stein et al. 2018).
The third major conclusion of this study is that although chromosome arms and centers show different levels of divergence, this pattern is created by differences in composition of age classes, which themselves show a variable level of evolutionary constraint. More precisely, in agreement with previous studies (Chen et al. 2010; Palmieri et al. 2014; Stein et al. 2018), younger age classes evolve more rapidly than older age classes, indicating that at evolutionary time scales, such as the separation between different Pristionchus species, selection can act on individual genes independent of their chromosomal location.
Finally, we found exceptionally high levels of gene losses in P. pacificus relative to its most closely related Pristionchus species. It has been speculated that the genetic hitchhiking of slightly deleterious alleles along with favorable alleles at linked loci in regions of low recombination can degrade gene function, causing transcriptional silencing (Smith and Haigh 1974; Cutter and Jovelin 2015). Loss of genes due to linked selection can be more pronounced in self-fertilizing nematodes like C. elegans, C. briggsae, and P. pacificus (Thomas et al. 2015) and may at least partially account for the unusually high number of species-specific lost gene clusters in P. pacificus. In the case of C2H2-type zinc finger domain containing proteins (PF13912), we found this gene family to show a statistically significant depletion in P. pacificus when compared with either P. exspectatus or P. arcanus. However, the same domain was previously reported to have undergone the second largest expansion in P. pacificus relative to C. elegans (Dieterich et al. 2008). This result not only supports the overall pattern of gene loss in P. pacificus, but also highlights the necessity of proper taxon sampling for understanding the complete dynamics of gene family size variation at the level of protein domains.
In summary, our study comprehensively characterizes the evolutionary dynamics of novel gene families at an extremely high phylogenetic resolution and integrates it into a global picture of nematode genome evolution. In future, we would like to exploit our phylogenomic framework to further investigate the mechanisms that drive the formation of novel genes and to quantify what fraction of orphan genes can be explained by them.
Methods
DNA extraction, sequencing, assembly, and scaffolding
All nematodes were grown on nematode growth medium (NGM) plates, and gonochoristic species were inbred (10 generations of full-sibling inbreeding) before DNA extraction. We rinsed the plates with M9 buffer and collected worm pellets by slow centrifugation at 1300 rpm for 3 min at 4°C. Then we followed the method described by Rödelsperger et al. (2017) for DNA extraction. Overlapping and mate pair libraries for P. arcanus and P. giblindavisi were sequenced and assembled based on the protocol described by Rödelsperger et al. (2014) for P. exspectatus genome sequencing (MacCallum et al. 2009). For the seven other species, PCR-free libraries were generated with TruSeq DNA PCR-Free Library Prep kit following the manufacturer's protocol, and sequencing was done on Illumina MiSeq. These seven species included P. pacificus itself, which we chose to resequence and assemble to make the data sets more comparable. Initial assemblies were constructed with the DISCOVAR de novo assembler (version r52488; https://software.broadinstitute.org/software/discovar/blog/). We checked for E. coli contamination by BLASTN against in-house and NCBI E. coli genomes and removed contaminated contigs after manual inspection. Finally, scaffolding was done with SSPACE_Basic_ v2.0 (Boetzer et al. 2011) using four mate pair libraries of sizes 1.5, 3, 5, and 8 kb (that were generated with Nextera Mate Pair Sample Preparation Kit).
Assembly evaluation
To assess the completeness of final assemblies, we calculated the fraction of raw reads that is represented in each final assembly. This was done by realigning reads from individual libraries with BWA (version 0.7.12-r1039) and stampy (version v1.0.21 r1713) and extracting the fraction of aligned reads from the output of the SAMtools flagstat program (version 0.1.19-96b5f2294a) (Li and Durbin 2009; Li et al. 2009; Lunter and Goodson 2011). Similarly, the SAMtools flagstat output provided information about the fraction of correctly oriented read paired ends, which can be interpreted as a measure of correctness. In addition, based on the realignments, the ambiguous fraction was defined as the fraction of the genome assembly with apparent heterozygous variant calls (Rödelsperger et al. 2014). Finally, we applied the universal single-copy orthologs benchmarking (BUSCO, version 3.0.1) approach as an additional measure for assembly completeness (Simão et al. 2015). Based on the definition of the BUSCO genes to be conserved as single copy in >90% of genomes, the effective maximum score that should be expected would be slightly above 90% and is reached for the P. arcanus (Table 1) genome as well as the previously published P. pacificus genome (Rödelsperger et al. 2017).
RNA extraction, sequencing, and assembly
Worm pellets for all species were collected by the aforementioned methods and were immediately resuspended in 10 volumes of TRIzol. RNA extraction was done with Direct-zol RNA miniprep kit (Zymo Research), and library preparation was done using Illumina TruSeq RNA Library Prep Kit v2. Libraries were sequenced on Illumina HiSeq 3000. We assembled the transcriptome with Trinity “trinityrnaseq-2.2.0” (Grabherr et al. 2011). For P. pacificus, we additionally generated a strand-specific transcriptome assembly based on previously published RNA-seq data (Rödelsperger et al. 2016; Serobyan et al. 2016).
Gene annotation
Initial prediction of protein coding genes was done using both AUGUSTUS (3.2.2) and SNAP within the MAKER2 (v2.31.8) pipeline (Korf 2004; Stanke et al. 2008; Holt and Yandell 2011). Three iterations of the MAKER2 pipeline were run, in the first run, both gene finders were trained with the transcriptome assembly of the given species. In the second run, we generated joint gene models that were either fully supported by transcriptome data or partially supported by predictions of the gene finders that were trained during the first run (AED_threshold<1). For the final run, we repeated the second run using gene models resulting from the second MAKER run of all other species as additional protein homology data. Additionally, we allowed MAKER2 to retain predicted gene models without transcriptome or homology evidence (AED_ threshold≤1). For MAKER2 runs 2 and 3, we used minimum contig length threshold of 2 kb (min_contig=2000). PFAM domains were annotated by InterProScan-5.19-58.0 (Finn et al. 2017). To visualize the distribution of genomic features across chromosomes, P. pacificus protein annotations were mapped to the El Paco assembly of P. pacificus with the exonerate protein2genome program (version 2.2.0) (Slater and Birney 2005).
Orthology clustering and inference of gene gain and loss
We ran pairwise BLASTP (e-value <10−5) between all species pairs in our analysis and created orthologous gene clusters with orthAgogue and MCL (both programs were run with default settings) (Enright et al. 2002; Ekseth et al. 2014). Based on the presence and absence of genes from different species, each cluster was segregated into different categories. Based on maximum parsimony, clusters were classified into age classes, each of which corresponds to a single origin at an internal branch of the phylogeny (Fig. 1A).
Expression analysis
We mapped stage-specific transcriptome data (10 samples) generated by Baskaran et al. (2015) to the P. pacificus genome with TopHat2 (Kim et al. 2013). Then, we computed the expression values for our P. pacificus gene annotations in each sample using Cufflinks 2.2.1 (Trapnell et al. 2013). Expression pattern for all age classes in each sample, mean expression for each gene in all samples, and mapping of mean expression pattern on chromosomes in nonoverlapping windows of 500-kb size were generated with custom Python scripts (Supplemental Code).
Estimation of evolutionary constraints
Pairwise 1:1 orthologs between P. pacificus and all other species were extracted by selecting only those clusters that have one gene each from P. pacificus and the other species. We aligned 1:1 orthologs using MUSCLE (Edgar 2004) and converted the protein alignments into codon alignments with pal2nal (Edgar 2004; Suyama et al. 2006). The codon alignments were passed on to PAML to calculate the rate of substitution at synonymous (dS) and nonsynonymous sites (dN), and ω (dN/dS) values (Yang 2007).
Statistical methods
To screen for Gene Ontology term enrichment of C. elegans genes from clusters missing P. pacificus genes but having at least one gene from one of the other two Pristionchus species, we used the functional annotation tool of the DAVID Bioinformatics Resource webserver (Huang et al. 2009). We performed PFAM annotation enrichment analysis on the P. exspectatus or P. arcanus genes from the clusters missing P. pacificus genes by comparing them with all the other genes from the given species (Fisher's exact test). For all enrichment tests, we applied the FDR method for multiple testing correction. We used “spearmanr” function from scipy.stats Python package to calculate correlation between two variables. The “ranksums” function from the same package was used to test whether dN, dS, and ω distributions along the nonoverlapping windows of 500-kb size on the chromosomes between young and old age classes are drawn from the same distribution or show statistically significant differences.
Data access
All raw sequencing reads and assemblies from this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession numbers PRJEB22188 and PRJEB27334, respectively.
Supplementary Material
Acknowledgments
We thank Tess Renahan for proofreading the manuscript. The work was funded by the Max Planck Society.
Author contributions: N.P., R.J.S., and C.R. conceptualized the project. N.P. and C.R. developed the methodology. Formal analysis and visualization were the responsibility of N.P. Experiments were carried out by N.P., W.R., H.W., and G.E. W.R., H.W., G.E., and R.J.S. gathered resources. N.P. and C.R. wrote the original draft, and N.P., R.J.S., and C.R. helped with writing, review, and editing. C.R. supervised the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.234971.118.
Freely available online through the Genome Research Open Access option.
References
- Baskaran P, Rödelsperger C. 2015. Microevolution of duplications and deletions and their impact on gene expression in the nematode Pristionchus pacificus. PLoS One 10: e0131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran P, Rödelsperger C, Prabh N, Serobyan V, Markov GV, Hirsekorn A, Dieterich C. 2015. Ancient gene duplications have shaped developmental stage-specific expression in Pristionchus pacificus. BMC Evol Biol 15: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579. [DOI] [PubMed] [Google Scholar]
- Borchert N, Dieterich C, Krug K, Schütz W, Jung S, Nordheim A, Sommer RJ, Macek B. 2010. Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res 20: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol 25: 778–786. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Jovelin R. 2015. When natural selection gives gene function the cold shoulder. Bioessays 37: 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton JF, Lugo-Martinez J, Tucker AE, Schrider DR, Warren WC, Hahn MW. 2014. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput Biol 10: e1003998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Wilhelm LJ, Howe DK, Gafner K, Dolan PC, Baer CF. 2012. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol Evol 4: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalle R, Rosenfeld JA. 2012. Phylogenomics. Garland Science, New York. [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, et al. 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman AR, Macchietto M, Porter CF, Rogers A, Williams B, Antoshechkin I, Lee MM, Goodwin Z, Lu X, Lewis EE, et al. 2015. Comparative genomics of Steinernema reveals deeply conserved gene regulatory networks. Genome Biol 16: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Brajković J, Tautz D. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet 23: 533–539. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Fraser CM. 2003. Phylogenomics: intersection of evolution and genomics. Science 300: 1706–1707. [DOI] [PubMed] [Google Scholar]
- Ekseth OK, Kuiper M, Mironov V. 2014. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics 30: 734–736. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierst JL, Willis JH, Thomas CG, Wang W, Reynolds RM, Ahearne TE, Cutter AD, Phillips PC. 2015. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLoS Genet 11: e1005323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztányi Z, El-Gebali S, Fraser M, et al. 2017. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res 45: D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilabert A, Curran DM, Harvey SC, Wasmuth JD. 2016. Expanding the view on the evolution of the nematode dauer signalling pathways: refinement through gene gain and pathway co-option. BMC Genomics 17: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. 1998. The comparative method in evolutionary biology. Oxford University Press, New York. [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Hunt VL, Tsai IJ, Coghlan A, Reid AJ, Holroyd N, Foth BJ, Tracey A, Cotton JA, Stanley EJ, Beasley H, et al. 2016. The genomic basis of parasitism in the Strongyloides clade of nematodes. Nat Genet 48: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, Tsutsui ND. 2011. Taxonomically restricted genes are associated with the evolution of sociality in the honey bee. BMC Genomics 12: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ. 2005. A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development 132: 5451–5460. [DOI] [PubMed] [Google Scholar]
- Juan D, Rico D, Marques-Bonet T, Fernández-Capetillo O, Valencia A. 2014. Late-replicating CNVs as a source of new genes. Biol Open 3: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, Lynch M. 2003. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165: 1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. 2009. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet 25: 404–413. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betrán E, Thornton K, Wang W. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet 4: 865–875. [DOI] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD. 2013. New gene evolution: Little did we know. Annu Rev Genet 47: 307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res 21: 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sinauer Associates, Inc, Sunderland, MA. [Google Scholar]
- MacCallum I, Przybylski D, Gnerre S, Burton J, Shlyakhter I, Gnirke A, Malek J, McKernan K, Ranade S, Shea TP, et al. 2009. ALLPATHS 2: small genomes assembled accurately and with high continuity from short paired reads. Genome Biol 10: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MG, Rödelsperger C, Witte H, Riebesell M, Sommer RJ. 2015. The orphan gene dauerless regulates dauer development and intraspecific competition in nematodes by copy number variation. PLoS Genet 11: e1005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SW. 2012. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res 20: 579–593. [DOI] [PubMed] [Google Scholar]
- Meyer JM, Markov GV, Baskaran P, Herrmann M, Sommer RJ, Rödelsperger C. 2016. Draft genome of the scarab beetle Oryctes borbonicus on La Réunion Island. Genome Biol Evol 8: 2093–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Springer, New York. [Google Scholar]
- O'Toole ÁN, Hurst LD, McLysaght A. 2018. Faster evolving primate genes are more likely to duplicate. Mol Biol Evol 35: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlötterer C. 2014. The life cycle of Drosophila orphan genes. eLife 3: e01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, Schmid R, Hall N, Barrell B, Waterston RH, et al. 2004. A transcriptomic analysis of the phylum Nematoda. Nat Genet 36: 1259–1267. [DOI] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Albà MM. 2013. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol 30: 1830–1842. [DOI] [PubMed] [Google Scholar]
- Prabh N, Rödelsperger C. 2016. Are orphan genes protein-coding, prediction artifacts, or non-coding RNAs? BMC Bioinformatics 17: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C. 2018. Comparative genomics of gene loss and gain in Caenorhabditis and other nematodes. Methods Mol Biol 1704: 419–432. [DOI] [PubMed] [Google Scholar]
- Rödelsperger C, Sommer RJ. 2011. Computational archaeology of the Pristionchus pacificus genome reveals evidence of horizontal gene transfers from insects. BMC Evol Biol 11: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Streit A, Sommer RJ. 2013. Structure, function and evolution of the nematode genome. In eLS. Wiley, Chichester: 10.1002/9780470015902.a0024603. [DOI] [Google Scholar]
- Rödelsperger C, Neher RA, Weller AM, Eberhardt G, Witte H, Mayer WE, Dieterich C, Sommer RJ. 2014. Characterization of genetic diversity in the nematode Pristionchus pacificus from population-scale resequencing data. Genetics 196: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Menden K, Serobyan V, Witte H, Baskaran P. 2016. First insights into the nature and evolution of antisense transcription in nematodes. BMC Evol Biol 16: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Meyer JM, Prabh N, Lanz C, Bemm F, Sommer RJ. 2017. Single-molecule sequencing reveals the chromosome-scale genomic architecture of the nematode model organism Pristionchus pacificus. Cell Rep 21: 834–844. [DOI] [PubMed] [Google Scholar]
- Rogers RL, Shao L, Thornton KR. 2017. Tandem duplications lead to novel expression patterns through exon shuffling in Drosophila yakuba. PLoS Genet 13: e1006795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos ME, Le Bouquin A, Crumière AJJ, Khila A. 2017. Taxon-restricted genes at the origin of a novel trait allowing access to a new environment. Science 358: 386–390. [DOI] [PubMed] [Google Scholar]
- Serobyan V, Xiao H, Namdeo S, Rödelsperger C, Sieriebriennikov B, Witte H, Röseler W, Sommer RJ. 2016. Chromatin remodelling and antisense-mediated up-regulation of the developmental switch gene eud-1 control predatory feeding plasticity. Nat Commun 7: 12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slos D, Sudhaus W, Stevens L, Bert W, Blaxter M. 2017. Caenorhabditis monodelphis sp. n.: defining the stem morphology and genomics of the genus Caenorhabditis. BMC Zool 2: 4. [Google Scholar]
- Smith JM, Haigh J. 1974. The hitch-hiking effect of a favourable gene. Genet Res 23: 23. [PubMed] [Google Scholar]
- Sommer RJ. 2015. Pristionchus pacificus: a nematode model for comparative and evolutionary biology. Brill, Leiden, Netherlands. [Google Scholar]
- Sommer RJ, Sternberg PW. 1996. Evolution of nematode vulval fate patterning. Dev Biol 173: 396–407. [DOI] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. [DOI] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, Chougule K, Gao D, Iwata A, Goicoechea JL, et al. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet 50: 285–296. [DOI] [PubMed] [Google Scholar]
- Sudhaus W. 2013. Order Rhabditina: “Rhabditidae”. In Handbook of zoology (ed. Schmidt-Rhaesa A), Vol. 2 Nematoda, pp. 537–556. De Gruyter, Berlin. [Google Scholar]
- Susoy V, Kanzaki N, Herrmann M. 2013. Description of the bark beetle associated nematodes Micoletzkya masseyi n. sp. and M. japonica n. sp. (Nematoda: Diplogastridae). Nematology 15: 213–231. [Google Scholar]
- Susoy V, Herrmann M, Kanzaki N, Kruger M, Nguyen CN, Rödelsperger C, Röseler W, Weiler C, Giblin-Davis RM, Ragsdale EJ, et al. 2016. Large-scale diversification without genetic isolation in nematode symbionts of figs. Sci Adv 2: e1501031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T. 2011. The evolutionary origin of orphan genes. Nat Rev Genet 12: 692–702. [DOI] [PubMed] [Google Scholar]
- Thellmann M, Hatzold J, Conradt B. 2003. The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development 130: 4057–4071. [DOI] [PubMed] [Google Scholar]
- Thomas JH. 2006. Analysis of homologous gene clusters in Caenorhabditis elegans reveals striking regional cluster domains. Genetics 172: 127–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Wang W, Jovelin R, Ghosh R, Lomasko T, Trinh Q, Kruglyak L, Stein LD, Cutter AD. 2015. Full-genome evolutionary histories of selfing, splitting, and selection in Caenorhabditis. Genome Res 25: 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verster AJ, Styles EB, Mateo A, Derry WB, Andrews BJ, Fraser AG. 2017. Taxonomically restricted genes with essential functions frequently play roles in chromosome segregation in Caenorhabditis elegans and Saccharomyces cerevisiae. G3 7: 3337–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen PJ, Wang GJ, Keller L. 2010. Chromosome size differences may affect meiosis and genome size. Science 329: 293. [DOI] [PubMed] [Google Scholar]
- Weller AM, Rödelsperger C, Eberhardt G, Molnar RI, Sommer RJ. 2014. Opposing forces of A/T-biased mutations and G/C-biased gene conversions shape the genome of the nematode Pristionchus pacificus. Genetics 196: 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang L, Gaut BS. 2011. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol 28: 2359–2369. [DOI] [PubMed] [Google Scholar]
- Yin D, Schwarz EM, Thomas CG, Felde RL, Korf IF, Cutter AD, Schartner CM, Ralston EJ, Meyer BJ, Haag ES. 2018. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 359: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.