Abstract
Whole-genome sequencing is increasingly used to identify Mendelian variants in clinical pipelines. These pipelines focus on single-nucleotide variants (SNVs) and also structural variants, while ignoring more complex repeat sequence variants. Here, we consider the problem of genotyping Variable Number Tandem Repeats (VNTRs), composed of inexact tandem duplications of short (6–100 bp) repeating units. VNTRs span 3% of the human genome, are frequently present in coding regions, and have been implicated in multiple Mendelian disorders. Although existing tools recognize VNTR carrying sequence, genotyping VNTRs (determining repeat unit count and sequence variation) from whole-genome sequencing reads remains challenging. We describe a method, adVNTR, that uses hidden Markov models to model each VNTR, count repeat units, and detect sequence variation. adVNTR models can be developed for short-read (Illumina) and single-molecule (Pacific Biosciences [PacBio]) whole-genome and whole-exome sequencing, and show good results on multiple simulated and real data sets.
Next-generation sequencing (NGS) is increasingly used to identify disease causing variants in clinical and diagnostic settings, but variant detection pipelines focus primarily on single-nucleotide variants (SNVs) and small indels and, to a lesser extent, on structural variants. The human genome contains repeated sequences such as segmental duplications, short tandem repeats, and minisatellites which pose challenges for alignment and variant calling tools. Hence, these regions are typically ignored during analysis of NGS data. In particular, tandem repeats correspond to locations where a short DNA sequence or Repeat Unit (RU) is repeated in tandem multiple times. RUs of length less than 6 bp are classified as short tandem repeats (STRs), whereas longer RUs spanning potentially hundreds of nucleotides are denoted as Variable Number Tandem Repeats (VNTRs) (Shriver et al. 1993; Wright 1994).
VNTRs span 3% of the human genome and are often found in coding regions where the repeat unit length is a multiple of 3 resulting in tandem repeats in the amino acid sequence. More than 1200 VNTRs with an RU length of 10 or greater exist in the coding regions of the human genome (Tyner et al. 2016). Compared to STRs, which have been extensively studied (Ummat and Bashir 2014; Gymrek et al. 2016; Dolzhenko et al. 2017; Liu et al. 2017; Willems et al. 2017), VNTRs have not received as much attention. Nevertheless, multiple studies have linked variation in VNTRs with Mendelian diseases, for example, Medullary cystic kidney disease (Kirby et al. 2013), Myoclonus epilepsy (Lalioti et al. 1997), FSHD (Lemmers et al. 2002), and complex disorders such as bipolar disorder (Table 1). In some cases, the disease-associated variants correspond to point mutations in the VNTR sequence (Ræder et al. 2006; Kirby et al. 2013), but in other cases, changes in the number of tandem repeats (RU count) show a statistical association (or causal relationship) with disease risk. For example, the insulin gene (INS) VNTR has an RU length of 14 bp with RU count varying from 26 to 200 (Pugliese et al. 1997). Variation in this VNTR has been associated with expression of the INS gene and risk for type 1 diabetes (OR = 2.2) (Durinovic-Belló et al. 2010). Notwithstanding these examples, the advent of genome-wide SNP genotyping arrays led to VNTRs being largely ignored. They have been called “the forgotten polymorphisms” (Brookes 2013).
Table 1.
Disease-linked VNTRs are generally distinguished from STRs by a longer length (≥6) of the repeating unit
VNTRs were originally used as markers for linkage mapping since they are highly polymorphic with respect to the number of tandem repeats at a given VNTR locus (Gelfand et al. 2014). Traditionally, VNTR genotyping required labor-intensive gel-based screens which limited the size of large population-based studies of VNTRs (Orita et al. 1989). Whole-genome sequencing has the potential to detect and genotype all types of genetic variation, including VNTRs. However, computational identification of variation in VNTRs from sequence remains challenging. Existing variant calling methods have been developed primarily to identify short sequence variants in unique DNA sequences that fall into a reference versus alternate allele framework, which is not well suited for detecting variation in VNTR sequences.
Genotyping VNTRs in a donor genome sequenced using short (Illumina) or longer single-molecule reads, requires the following: (1) recruitment of reads containing the VNTR sequence; (2) counting RUs for each of the two haplotypes; (3) identification of indels within VNTRs; and (4) identification of mutations within the VNTR. Mapping tools such as BWA (Li and Durbin 2009) and Bowtie 2 (Langmead and Salzberg 2012) can work for read recruitment for STRs, but are challenged by insertion/deletion of larger repeat units. Mapping issues also confound existing variant callers, including realignment tools such as GATK IndelRealigner (DePristo et al. 2011) if the total VNTR length is larger than the read length. This is because reads contained within the VNTR sequence have multiple equally likely mappings and therefore will be mapped randomly to different locations with low mapping quality (Kirby et al. 2013). Detection of point mutations in long VNTRs requires integrating information across the entire VNTR sequence. For VNTRs whose total sequence length (RU count times the RU length) is much longer than the read length, detection of SNVs and indels is not feasible using existing variant callers. We focus mainly on problems (1) and (2) relating to recruitment and RU counting. For problem (3), we focus on the difficult case of large (≥250 bp) VNTRs within coding regions where the indel shifts the translation frame. We do not tackle problem (4) in this manuscript.
Other tools have addressed the problem of RU count estimation, focusing on the related problem of STR genotyping. Some of these tools do not accept large repeating patterns as input (Liu et al. 2017; Willems et al. 2017). Others require all repeat units to be nearly identical (Dolzhenko et al. 2017; Ummat and Bashir 2014). In particular, ExpansionHunter (Dolzhenko et al. 2017) looks for exact matches of short repeating sequence within flanking unique sequences, and works for STRs, but not as well with the larger VNTRs with variations in RUs (see Results). VNTRseek (Gelfand et al. 2014) detects a VNTR-like pattern in reads and aligns it to tandem repeats but uses a complex alignment process, making it difficult to run the tool. Alignment-based tools need to align reads at both unique ends, which may not be possible for short (Illumina) reads. Single-molecule reads, for example, Pacific Biosciences (PacBio) (Eid et al. 2009) and Nanopore (Clarke et al. 2009), can span entire VNTR regions, but it is difficult to estimate the RU count directly because the distance between the flanking regions varies dramatically from read to read due to an excess of indel errors. For example, 14 reads spanning the SLC6A4 VNTR in the PacBio sequencing data of the NA12878 individual from Genome in a Bottle (Zook et al. 2016) included fifteen distinct lengths between 292 and 385 bp, leading to length-based RU count estimates 13, 14, 15, 16, and 18 for the diploid genome.
In contrast to methods like VNTRseek that seek to discover/identify VNTRs, we describe a method, adVNTR, for genotyping VNTRs at targeted loci in a donor genome. For any target VNTR in a donor, adVNTR reports an estimate of RU counts and point mutations within the RUs. It trains hidden Markov models (HMMs) for each target VNTR locus, which provide the following advantages: (1) It is sufficient to match any portions of the unique flanking regions for read alignment; (2) it is easier to separate homopolymer runs from other indels helping with frameshift detection, and to estimate RU counts even in the presence of indels; and (3) each VNTR can be modeled individually, and complex models can be constructed for VNTRs with complex structure, along with VNTR specific confidence scores. For longer VNTRs not spanned by short reads, adVNTR can still be used to detect indels while providing lower bounds on RU counts. Also, exact estimates for RU counts could be made for shorter VNTRs. Using simulated data as well as whole-genome sequence data for several human individuals, we demonstrate the power of adVNTR to genotype VNTR loci in the human genome.
Results
Our method, adVNTR, requires training of separate HMM models for each combination of target VNTR and sequencing technologies. The detailed training procedure is described in Methods. Given trained models, adVNTR genotypes the VNTRs in three stages: (1) selection of reads that contain VNTR locus (read recruitment); (2) RU count estimation; and (3) variant detection. We report results on performance of adVNTR in each of these stages using simulated and read data sets based on short-read (Illumina) and single-molecule (PacBio) technologies.
HMM training
Initial HMMs were trained using multiple alignments of RU sequences from the reference assembly hg19 (International Human Genome Sequencing Consortium 2001), as described in Methods. Similarly, HMMs were trained for the left flanking and right flanking regions for each VNTR. The HMM models were augmented using data from Genome in a Bottle (GIAB) project (NA12878 WGS). VNTR models were trained for VNTRs in coding and promoter regions of the genome, for both Illumina (1755 models) and PacBio (2944 models) (Supplemental Material, “Selecting Target VNTRs”). Subsequently, we tested performance for (1) read recruitment, (2) counting of repeat units, and (3) detection of indels.
Test data
To evaluate performance for PacBio, we simulated haplotypes for each of the 2944 VNTRs, revising the RU count to be ±3 of the RU count in hg19, and setting 1 as the minimum RU count. We simulated haplotype reads (15× coverage) using SimLoRD (Stöcker et al. 2016) and aligned those reads to hg19 using BLASR (Chaisson and Tesler 2012). For Illumina sequencing, we used ART (Huang et al. 2011) to simulate haplotype WGS (shotgun 150 bp) reads at 15× coverage for each VNTR and simulated VNTR haplotype with changes in RU counts similar to PacBio. Pairs of haplotypes were merged to get (30× coverage) diploid samples. The resulting data sets were called PacBioSim and IlluminaSim, respectively (Supplemental Material, “Test Datasets”; Supplemental Table S1). To evaluate performance of frameshift identification, we collected a set of 115 VNTRs (Supplemental Material, “Selecting Target VNTRs”). For each VNTR, we simulated haplotypes that contain a deletion or an insertion in the VNTR (Supplemental Material, “Test Datasets”). We simulated reads from each of these haplotypes and merged pairs of haplotypes to obtain diploid samples. We denote this data set as IlluminaFrameshift.
Read recruitment
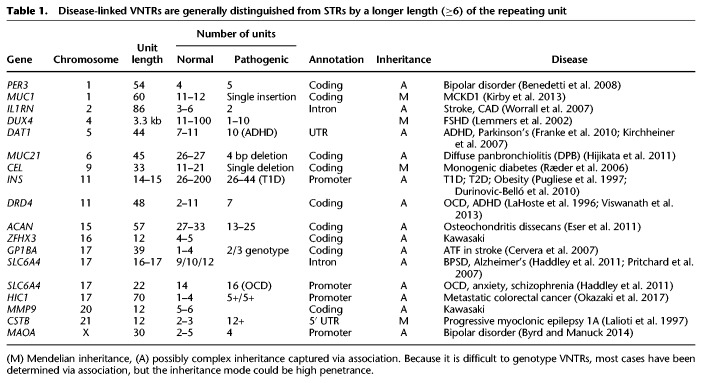
adVNTR takes a collection of VNTR models as input, and as a first step, recruits reads that map to any of the VNTRs in the list. In testing recruitment for PacBio, we found that alignment tools such as BLASR perform well in recruiting VNTR reads even in the presence of deletions and insertions, and we used BLASR for all read recruitment. For Illumina reads, we tested adVNTR read recruitment for all 1775 VNTRs using IlluminaSim and compared against mapping tools BWA-MEM, Bowtie 2, and BLAST. adVNTR achieves much greater recall while maintaining or exceeding the precision of other tools (Fig. 1; Supplemental Fig. S3). Specifically, adVNTR recall was 100% for 99.9% of the VNTRs, whereas the next best tool (BWA-MEM) achieved this only for 68.2% of the VNTRs. The other mapping tools lose mapping sensitivity when RU counts are increased or decreased (large indels) and perform best when the RU counts are the same as reference (Supplemental Fig. S2A–C), partially explaining their lower recall.
Figure 1.
Read recruitment quality on Illumina reads. (A) Comparison of the recall (number of true recruited reads/number of true reads) of adVNTR read recruitment against BWA-MEM and Bowtie 2, as a function of VNTR length for 1775 VNTRs with different counts (31,788 tests). Each dot corresponds to a separate test. (B) Precision of read recruitment (number of true recruited reads/number of recruited reads).
VNTR genotyping (RU count estimation) with PacBio reads
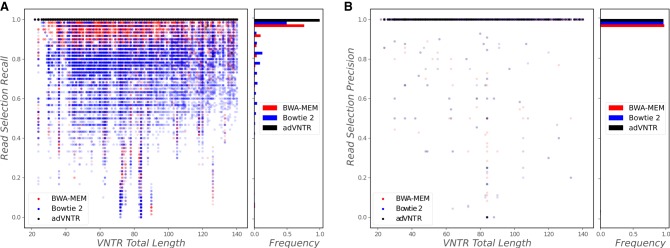
Recall that sequencing (particularly homopolymer) errors can cause lengths to change, particularly for short RU lengths and larger RU counts. To test adVNTR performance on PacBioSim, we compared against a naïve method that estimates RU counts based on read length between the flanking regions from the consensus of reads that cover VNTR. Detailed performance on three exemplars (genes INS, CSTB, and HIC1) showed high genotype accuracy for adVNTR over a wide range of RU counts and coverage (Fig. 2A). Similar results were obtained for all 2944 VNTRs (Fig. 2B). Overall, 98.45% of adVNTR estimates were correct, whereas 26.45% of estimates made by the naïve method were correct. Because it is difficult for the naïve method to call heterozygotes, we also compared the subset of test data with homozygous RU counts: 97.95% of adVNTR estimates were correct, whereas the consensus method was correct in 66.16% of samples (Supplemental Fig. S4). adVNTR estimates were uniformly good except at low sequence coverage. To test for accuracy with changing RU counts, we simulated different RU counts for individuals at three VNTRs (Supplemental Table S4). adVNTR RU counts showed 100% accuracy in each of the 52 different samples tested.
Figure 2.
VNTR genotyping using PacBio data. (A) RU count estimation on simulated PacBio reads as a function of RU count and coverage for three medically relevant VNTRs: INS (RU length 14 bp), CSTB (12 bp), and HIC1 (70 bp). adVNTR performance is compared to a naïve method. (B) The effect of RU length on count accuracy over 2944 VNTRs (30418 tests). (C) Mendelian consistency of genotypes at four VNTR loci in the Chinese Han and Ashkenazi trios. Note that MAOA results are consistent with its location on Chr X. (D) LR-PCR–based validation of genotypes at five disease-linked VNTRs in NA12878. Red arrows correspond to VNTR lengths estimated by multiplying predicted RU counts with RU lengths. (E) Fraction of consistent calls and number of calls across 2944 VNTRs in Ashkenazi Jew (AJ) and Chinese trios from GIAB and NCBI SRA. (F) Fraction of consistent calls allowing for off-by-one errors.
To test performance on real data for which the true VNTR genotype was not known, we checked for Mendelian inheritance consistency in the Ashkenazi Jew (AJ) trio from Genome in a Bottle (GIAB) (Zook et al. 2016) and a Chinese Han trio from NCBI SRA (accession PRJEB12236). On four disease-related VNTRs, adVNTR predictions were consistent in each case (Fig. 2C). On the 2944 genic VNTRs, the trio consistency of adVNTR calls was correlated with coverage. At a posterior probability threshold of 0.99, 86.98% of the calls in the AJ trio and 97.08% of the calls in the Chinese trio, were consistent with Mendelian inheritance (Fig. 2E). Many of the discrepancies could be attributed to low coverage and missing data. Increasing sequence coverage threshold from 5× to 10× increased the average posterior probability from 0.91 to 0.98 and resulted in improved RU count accuracy (Supplemental Fig. S5). Also, many of these discrepancies in RU counts were off-by-one errors (Supplemental Fig. S6). These off-by-one discrepancies could be acceptable for Mendelian disease testing because the pathogenic cases often have large changes in RU counts. Treating the off-by-one counts as correct, we found that 98.66% and 99.91% of the high confidence calls in AJ and Chinese trios, respectively, were consistent (Fig. 2F). Finally, some of the off-by-one counts could be natural genetic variation.
We also performed a long range (LR) PCR experiment on the individual NA12878 to assess the accuracy of the adVNTR genotypes using PacBio data (Supplemental Tables S2, S3). The observed PCR product lengths (black bands in Fig. 2D) were consistent with the adVNTR predictions (red arrows), while being different from the hg19 reference RU count. adVNTR correctly predicted all VNTRs to be heterozygous with the exception of SLC6A4, which was predicted to be homozygous.
Although we could not get the VNTR discovery tool VNTRseek (Gelfand et al. 2014) to run on our machine, we observed that the authors had predicted 125 VNTRs in the Watson sequenced genome (Wheeler et al. 2008) and 75 VNTRs in two trios as being polymorphic. In contrast, analysis of the PacBio sequencing data identified >500 examples of polymorphic VNTRs that overlap with coding regions. The results suggest that variation in RU counts of VNTRs and their role in influencing phenotypes might be greater than previously estimated.
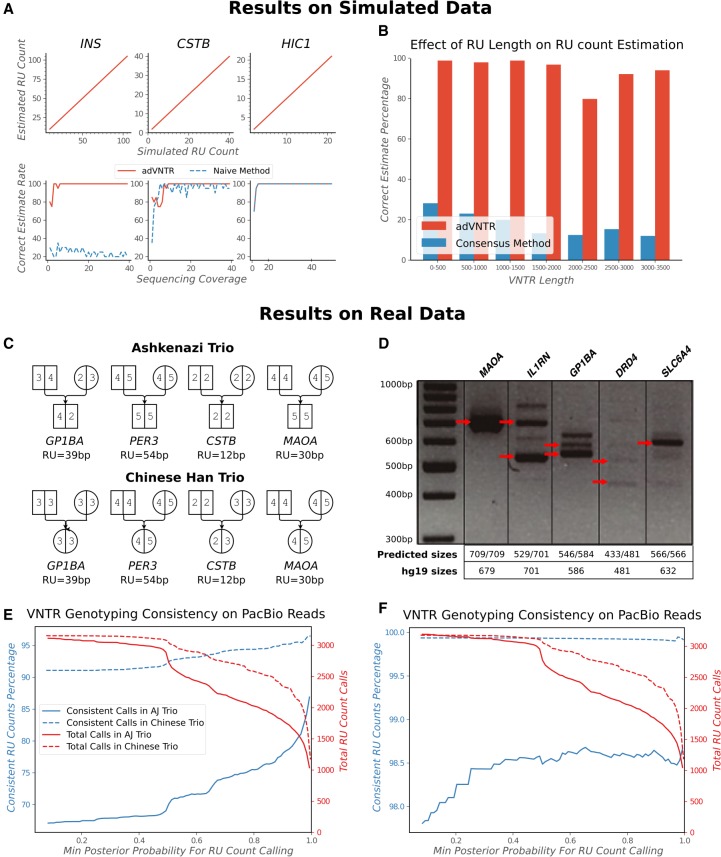
RU counting with Illumina
The adVNTR estimate correctly matched both RU counts in 91.6% of the cases in the IlluminaSim data set (1775 VNTRs with up to 21 diploid RU counts each) and matched at least one RU count in 97% of the cases (Fig. 3A,B). Most of the discrepancies occurred in VNTRs with longer lengths not covered by Illumina reads (Fig. 3C,D). Although there was a drop in accuracy for increasing lengths, 84% of the genic VNTRs are shorter than 150 bp and could be genotyped with 94.6% accuracy. Tools such as VNTRseek require at least 20 bp flanking each side of the VNTR and do not return a result for VNTRs with total length greater than 110 bp, whereas adVNTR could predict the genotype correctly in a majority of those cases (Supplemental Material, “VNTRseek”). ExpansionHunter, a tool designed primarily for STR genotyping (Dolzhenko et al. 2017) provided incorrect estimates in >90% cases from this data set (Supplemental Fig. S7). ExpansionHunter makes the assumption that the different RUs are mostly identical in sequence, which is valid for STRs but not for most VNTRs, and we tested this through 52 samples on three VNTRs. adVNTR predicted the correct genotype in all but six cases, with erroneous calls only in the case of high RU counts where the read length did not span the VNTR perfectly; ExpansionHunter did not return the correct estimate in most cases (Supplemental Table S4).
Figure 3.
VNTR genotyping using Illumina data. (A–D) Correctness of RU count prediction for 1775 coding VNTRs in the IlluminaSim data set, described by RU count discrepancy (A), haplotypes with correct estimates (B), correctness as a function of VNTR length (C), and RU length (D). (E) Consistency of adVNTR calls on the AJ trio WGS data from GIAB. The red line describes the cumulative number of calls made at specific posterior probability cutoffs. (F) Gel electrophoresis–based validation of adVNTR calls on five short VNTRs using WGS of individual NA12878 from GIAB. The red arrows correspond to VNTR lengths estimated by multiplying the RU lengths with the estimated RU counts.
On the AJ trio from GIAB, 98.08% of the high-confidence adVNTR calls were consistent with Mendelian inheritance (Fig. 3E). Note that 95.93% of all calls were high confidence (posterior probability ≥0.99). We validated adVNTR calls on 12 VNTRs using gel electrophoresis (Supplemental Table S3). adVNTR predicted the correct RU counts in all cases, except in two cases for which the PCR primers failed to produce a band (Fig. 3F; Supplemental Fig. S8). We also compared adVNTR against ExpansionHunter on seven disease-related short VNTRs in the AJ trio and obtained similar results (Supplemental Table S5).
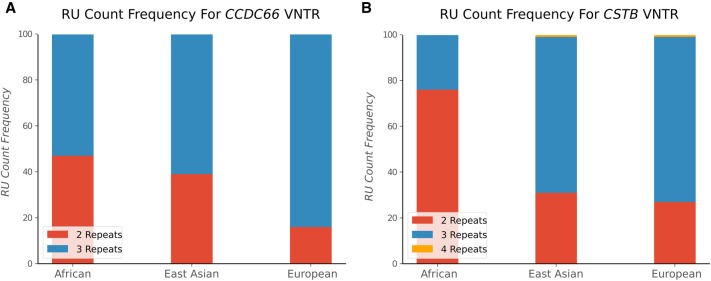
To test adVNTR for population-scale studies of VNTR genotypes using WGS data replacing labor-intensive gel electrophoresis (Cervera et al. 2007; Byrd and Manuck 2014), we scanned the PCR-free WGS data for 150 individuals (50 in each population) obtained from The 1000 Genomes Project Consortium (2015). We observed population-specific RU counts (frequency difference >10%) in 97 of 202 VNTRs tested (Supplemental Table S7). Figure 4 shows the RU count frequencies for a disease-linked VNTR in the coding region of CSTB and a coding VNTR in CCDC66. The results suggest an increase in VNTRs with higher RU counts with an increase in divergence time from Africa. Thus, RU3 is more prevalent in both VNTRs. We also observed RU4 in CSTB VNTR in the Asian and European populations, where RU counts 4 and above have been associated with progressive myoclonal epilepsy (Lalioti et al. 1997).
Figure 4.
Population-scale genotyping of VNTRs. (A) RU count frequencies for the VNTR in CCDC66 gene; (B) CSTB in African, Asian, and European population samples from The 1000 Genomes Project. RU counts of 4 and higher in CSTB are associated with myoclonal epilepsy.
VNTR mutation/indel detection
As a proof of concept for other applications, we tested indel detection, focusing in particular on frameshifts in coding VNTRs. The CEL gene is known to contain a VNTR where a deletion changes the coding frame. We simulated Illumina reads from 20 whole genomes after introducing a single insertion or deletion in the middle of the VNTR region in the CEL gene. As a negative control, we simulated 10 WGS experiments with a range of sequence coverage values. We ran adVNTR, SAMtools mpileup (Li 2011), and GATK Haplotype-Caller (DePristo et al. 2011), which uses GATK IndelRealigner, to identify frameshifts in each of the simulated data sets and the 10 control data sets. On the control data, none of the tools found any variant. On the simulated indels, adVNTR made the correct prediction in each case (Supplemental Table S6), whereas SAMtools and GATK were unable to predict a single insertion or deletion. This result is not surprising because the reads have poor alignment scores, and the indel can be mapped to multiple locations (Supplemental Fig. S9; Robinson et al. 2011). We note that mapping ambiguity in aligning each read made it difficult to pinpoint the location of single indels. However, by integrating the information across all reads, we could predict the occurrence of a frameshift in the VNTR. We next tested adVNTR frameshift prediction on the 115 VNTRs in the IlluminaFrameshift data set, simulating 4090 total cases. Overall, the frameshifts in the VNTR regions were predicted with 51.7% sensitivity and 86.8% specificity, in contrast with the 49.7% sensitivity and 43.5% specificity achieved by GATK. Detailed performance of methods for each VNTR is available in Supplemental Table S7. Note that the performance is model specific and depends on the similarity of different repeat units in a VNTR. For 29 of the 115 VNTRs, adVNTR showed high sensitivity (≥90%) and specificity (100%).
Because frameshifts in the VNTR region of the CEL gene have been linked to a monogenic form of diabetes (Ræder et al. 2006), we tested for frameshifts in CEL using whole-exome sequencing (WES) data from 2081 cases with Type 2 Diabetes (Fuchsberger et al. 2016) and compared the numbers to 2090 control individuals. WES data analysis is challenging because high GC-content makes it difficult to PCR-amplify this VNTR. adVNTR found that although none of the controls had any evidence of a frameshift, eight of the 2081 diabetes cases showed a frameshift in this VNTR region (Supplemental Fig. S10).
Computing requirements for genotyping
adVNTR is multithreaded. In genotyping-mapped PacBio reads at 30× coverage, adVNTR took 6 h using Intel Xeon four-core CPUs (≤24 CPU hours) to genotype all 2944 VNTRs, and 14:15 h (≤57 CPU hours) for 70× coverage. For Illumina reads at 40× coverage, adVNTR took 87:30 CPU hours on a single core to complete read recruitment as well as genotyping of 1775 VNTRs.
Discussion
The problem of genotyping VNTRs (determining diploid RU counts and mutations) is increasingly important for clinical pipelines seeking to find the genetic mechanisms of Mendelian disorders. Because VNTRs have not been extensively studied, existing research is often focused on their discovery. One of the contributions of this paper is the separation of initial VNTR discovery from VNTR genotyping, and a focus on the genotyping problem. adVNTR genotypes VNTRs using a hidden Markov model for each target VNTR, providing a uniform training framework, but still allowing us to tailor the models for complex VNTRs on a case by case basis. The problem of mismapping due to indels introduced by changing RU counts confounds most mapping-based tools but is solved here by collapsing all RU copies and building HMMs that allow for variation in the RUs. adVNTR was tested extensively on data from different sequencing technologies, including Illumina and PacBio. Because some of the data sets used were mapped only to hg19, especially the 150 whole-genome sequencing data set from the Polaris project, we decided to use hg19 as the reference throughout, including simulations. Validation of the data used either orthogonal information (e.g., trios or experiments), or simulations and would not be affected by the use of GRCh38.
Like other STR genotyping tools, adVNTR works best when reads span the VNTR. However, even with this limitation, there are (1) close to 100,000 VNTRs in the genic regions of human genome that can be spanned by Illumina reads; (2) indel detection is possible, even when RU counting is not, for long VNTRs; and (3) lower bounds on RU counts can separate some pathogenic cases from normal cases, particularly when the normal VNTR length is shorter than the read length, while the pathogenic case is much longer (e.g., CSTB). Finally, dropping costs for long-read sequencing (especially PacBio and Nanopore) will allow us to span and genotype over 158,000 genic VNTRs.
The choice between short- and long-read technologies offers some trade-offs. Specifically, long reads allow for the targeted genotyping of a larger set of VNTRs (559,804) and are becoming increasingly cost-effective. However, the large numbers of indels in these technologies reduce the accuracy somewhat, and they are best used when there is a big difference between normal and pathogenic cases in terms of RU counts or when the VNTRs are too long to be spanned by Illumina.
In contrast, short-read Illumina sequencing is increasingly used for Mendelian pipelines and can be easily extended to include VNTR genotyping with higher accuracy than PacBio. Also, the large number of VNTRs (458,158) that can be spanned by Illumina reads makes it the technology of choice for association testing and population-based studies.
In this research, we also provided initial results on genotyping frameshift errors in coding VNTRs, focusing on the easier case when all RUs have the same length. Future work will focus on extending the target VNTRs for RU counting and frameshift detection for VNTRs that are of medical interest, population genetics of VNTRs, and algorithmic strategies for speeding up VNTR discovery and genotyping.
Methods
A VNTR sequence can be represented as SR1R2 … RuP, where S and P are the unique flanking regions, and Ri(1 ≤ i ≤ u) correspond to the tandem repeats. For each i, j, Ri is similar in sequence to Rj, and the number of occurrences, u, is denoted as the RU count. We do not impose a length restriction on S and P, but assume that they are long enough to be unique in the genome. For genotyping a VNTR in a donor genome, we focus primarily on estimating the diploid RU counts (u1,u2). However, many (∼103) VNTRs occur in coding regions, and mutations, particularly frameshift causing indels, are also relevant. Our method, adVNTR, models the problems of RU counting and mutation detection using HMMs trained for each target VNTR. adVNTR requires a one-time training of models for each combination of a VNTR and sequencing technology, although the user has the option to retrain models. Once models are trained, it has three stages for genotyping: (1) read recruitment, (2) RU count estimation, and (3) variant (indel) detection. We describe the training procedure and the three modules below.
HMM training
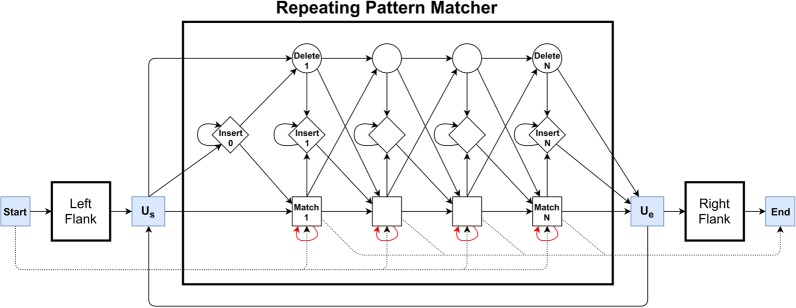
The goal of training is to estimate model parameters for each VNTR and each sequencing technology. Previous works have shown that an HMM with three groups of states could be used to find similarities between biological sequences (Eddy 1996). In this model, a profile HMM can model a group of sequences. Then, a new sequence can be aligned to a profile HMM to discover sequence family (Krogh et al. 1994). We use an HMM architecture with three parts, which have their own three groups of states (Fig. 5). The first part matches the 5′ (left) flanking region of the VNTR. The second part is an HMM that matches an arbitrary number of (approximately identical) repeating units. The last part matches the 3′ (right) flanking region (Supplemental Fig. S1). The RU pattern is matched with a profile HMM (RU HMM), with states for matches, deletions, and insertions, and its model parameters are trained first. To train RU HMM for each VNTR, we collected RU sequences from the reference assembly (International Human Genome Sequencing Consortium 2001) and performed a multiple sequence alignment (Eddy 1995). Let h(i, j) denote the number of observed transitions from state i to state j in hidden path of each sequence in multiple alignment, and hi(α) denote the number of emissions of α in state i. We define permissible transition (arrows in Fig. 5) and match-state emission probabilities as follows:
Nonpermissible transitions have probability 0, and hi(α) = 1/4 for insert state i and 0 for deletions. The pseudocounts b0 and b1 were estimated by initially setting them to the error rate of the sequencing technology, but they (along with other model parameters) were updated after aligning Illumina or PacBio reads to the model. The RU HMM architecture was augmented by adding (1) transitions from Ue to Us to allow matching of variable number of RU; (2) adding the HMMs for the matching of any portions of left and right flanking sequences; and (3) by adding transitions to match reads that match either the left flanking or the right flanking region. In addition, reads anchored to one of the unique regions can jump past the other HMM using dotted arrows.
Figure 5.
The VNTR HMM. The HMM is composed of three profile HMMs, one each for the left and right flanking unique regions, and one in the middle to match multiple and partial numbers of RUs. The special states Us (“Unit-Start”), and Ue (“Unit-End”) are used for RU counting. Dotted lines refer to special transitions for partial reads that do not span the entire region.
Although error correction tools for PacBio have been developed, most do not work for repetitive regions (Au et al. 2012; Hackl et al. 2014; Lee et al. 2014; Salmela and Rivals 2014; Miclotte et al. 2016; Miller et al. 2017), and others assume a single haplotype for error correction (Berlin et al. 2015; Salmela et al. 2016). In contrast, the HMM allows us to model many of the common (homopolymer) errors directly. Insertion deletion errors are common in single-molecule sequencing, particularly in homopolymer runs of length ≥6, and occur mostly as insertions in the homopolymer run (Chaisson and Tesler 2012). Consider a match state i with highest emission probability for nucleotide α. The transition probability T(i, i) from a match state i to itself was set based on the match probabilities of α in previous k = 6 states. The model parameters were further updated using genome sequencing data of NA12878 (Supplemental Material, “Model Structure and Parameter Setting”).
Read recruitment
The first step in adVNTR is to recruit all reads that match a portion of the VNTR sequence. Alignment-based methods do not work well due to changes in RU counts (Results), but the adVNTR HMM allows for variable RU count. To speed up recruitment, we used an Aho-Corasick keyword matching algorithm available as part of the BLAST package (Altschul et al. 1990) to identify all reads that match a keyword from the VNTR patterns or the flanking regions. Note that the dictionary construction is a one-time process, and all reads must be scanned once for filtering. The keyword size and number of keywords were empirically chosen for each VNTR.
Filtered reads were aligned to the HMM using the Viterbi algorithm. Only reads with matching probability higher than a specified threshold were retained. To compute the selection threshold for each VNTR, we aligned nontarget genomic sequences that passed the keyword matching step to the HMM to form an empirical false distribution. Subsequently, we aligned VNTR encoding sequences to the HMM to form the score distribution of true reads. Then, we used a Naïve Bayes classifier to select a threshold.
Estimating VNTR RU counts
All reads covering an RU element are aligned, or ‘matched’ to the HMM using the Viterbi algorithm to create, in effect, a new multiple alignment. Recalling the Viterbi algorithm, let Vk,j denote the highest (log) probability of emitting the first k letters of the sequence s1, s2, … sn and ending in state j of an HMM. Let Prevk,j denote the state j′ immediately prior to j in this optimum parse. then
| (1) |
| (2) |
where k′ = k − 1 for match or insert states; k′ = k otherwise. For each read, the Viterbi algorithm allows for the enumeration of the maximum likelihood (ML) path by going backward from Prev(End, n). Ignoring all but the Us and Ue states in the Viterbi path, we get a pattern of the form with k1, k3 ∈ {0,1}, and k2 ≥ 0. We estimate the RU count of the read as k1 + k2 + k3 and mark it as a lower bound if k1 + k3 > 0 (for an example, see Fig. 6).
Figure 6.
Estimates of RU counts using recruited reads. (A) (k1, k2, k3) = (1,3,1); RU count ≥5. (B) (k1, k2, k3) = (0,3,1); RU count ≥4. (C) (k1, k2, k3) = (0,3,0); RU count = 3.
One of the main reasons for erroneous RU counts is stutter during PCR amplification. The PCR amplification process is similar to replication errors that result on genetic RU count variation during cell division, except that there are multiple rounds of amplification. In each PCR round, the number of copies might change by 1 with some probability. Once a single event has occurred and an erroneous template is generated, the event of having another change is likely to be independent of the previous event (Gymrek 2016). To model errors in read counts, we define parameter r such that r∈Δ is the probability of RU counting error by ±Δ in the estimation of the true count. Thus, the probability of getting the correct count is 1 − r, where
The analysis of reads at a VNTR gives us a multiset of RU counts (or lower bounds) c1, c2, . . . , cn. We assume that the donor genome is diploid but do not require any phasing information in the computation of the multiset. Additionally, we allow the possibility that all reads are sampled from one haplotype with the RU count of the missing haplotype being X. We define C = {c1, c2, … , cn} ∪ {X} and use C to get a list of possible genotypes (ci, cj) with ci ≤ cj. Then, the conditional likelihood of a read with RU count c is given by
Similarly, the likelihood of a read with a lower bound c on the RU count is given by
The likelihood of the data C is given by . The posterior genotype probabilities can be computed using Bayes’ theorem
| (3) |
We generally set equal priors. However, in the event that we only see reads with a single count c′, we choose Pr((c′, c′)) = Pr((c′, X)) = 1/2. The probability of a “missing haplotype” event is modeled as a Bernoulli process because in genome sequencing, sampling from either chromosome is done at random, and so the probability of not observing a haplotype in each read (failure) is 1/2. If we see multiple counts, we set Pr((c′, X)) = 0 for all c′ ∈ C, and give equal priors to all other genotypes.
VNTR mutation detection
It is not difficult to see that alignment-based methods do not work well in VNTRs. Changes in RU counts make it difficult to align reads even for mappers that allow split reads, because the gaps in different reads can be placed in different locations. A similar problem appears with small indels, because there are multiple ways to align reads with an indel in a Repeat Unit. The adVNTR HMM aligns all repeat units to the same HMM, and this has the effect of aligning all mutations/indels in the same column. Consider the case where reads contain a total of v nucleotides matching a VNTR RU of length and RU count u. Moreover at a specific position covered by d repeats, suppose we observe ɩ indel transitions.
For a true indel mutation, we expect fraction of transitions at a location to be an indel, giving a likelihood of the observed data as Binom. Alternatively, for a homopolymer run of i > 0 nucleotides, let εi denote the per-nucleotide indel error rate. We modeled ε1 empirically in non-VNTR, nonpolymorphic regions and confirmed prior results that εi increases with increasing i (Margulies et al. 2005). Thus, the likelihood of seeing ɩ indel transitions due to sequencing error in a homopolymer run of length i is Binom. We scored an indel in the VNTR using the log-likelihood ratio
| (4) |
which follows a χ2 distribution. We select the indel if the nominal P-value is lower than 0.01.
Command line usage of adVNTR for RU count genotyping and frameshift identification is available in Supplemental Material (“Running adVNTR”).
Software availability
adVNTR source code can be found in the Supplemental Material and is also available at https://github.com/mehrdadbakhtiari/adVNTR.
Competing interest statement
Vineet Bafna is a co-founder and has equity interest in Pretzel Therapeutics, Inc. (PT) and Digital Proteomics, LLC (DP), and receives income from DP. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. PT and DP were not involved in the research presented here.
Supplementary Material
Acknowledgments
The T2D-GENES data used in this study were obtained from the NIH dbGaP repository (study accession phs001095.v1.p1, phs001096.v1.p1, and phs001097.v1.p1) and supplied by investigators of the Korea Association Research Project, Singapore Diabetes Cohort Study, and Singapore Prospective Study Program, Albert Einstein College of Medicine and Broad Institute. The T2D-GENES Sequencing study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Human Genome Research Institute (NHGRI). Research was supported in part by a grant from the NIH (HG010149). V. Bafna and M.B. were also supported in part by NIH GM114362 and NSF-DBI-1458557. M.G. and S.S.-B. were supported in part by the Office of the Director, NIH (DP5OD024577).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.235119.118.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Au KF, Underwood JG, Lee L, Wong WH. 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7: e46679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F, Dallaspezia S, Colombo C, Pirovano A, Marino E, Smeraldi E. 2008. A length polymorphism in the circadian clock gene Per3 influences age at onset of bipolar disorder. Neurosci Lett 445: 184–187. [DOI] [PubMed] [Google Scholar]
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 33: 623–630. [DOI] [PubMed] [Google Scholar]
- Brookes K. 2013. The VNTR in complex disorders: the forgotten polymorphisms? A functional way forward? Genomics 101: 273–281. [DOI] [PubMed] [Google Scholar]
- Byrd AL, Manuck SB. 2014. MAOA, childhood maltreatment, and antisocial behavior: meta-analysis of a gene-environment interaction. Biol Psychiatry 75: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera A, Tàssies D, Obach V, Amaro S, Reverter J, Chamorro A. 2007. The BC genotype of the VNTR polymorphism of platelet glycoprotein Ibα is overrepresented in patients with recurrent stroke regardless of aspirin therapy. Cerebrovasc Dis 24: 242–246. [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. 2009. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4: 265–270. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhenko E, van Vugt JJ, Shaw RJ, Bekritsky MA, van Blitterswijk M, Narzisi G, Ajay SS, Rajan V, Lajoie B, Johnson NH, et al. 2017. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res 27: 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinovic-Belló I, Wu R, Gersuk V, Sanda S, Shilling H, Nepom G. 2010. Insulin gene VNTR genotype associates with frequency and phenotype of the autoimmune response to proinsulin. Genes Immun 11: 188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 1995. Multiple alignment using hidden Markov models. Proc Int Conf Intell Syst Mol Biol 3: 114–120. [PubMed] [Google Scholar]
- Eddy SR. 1996. Hidden Markov models. Curr Opin Struct Biol 6: 361–365. [DOI] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323: 133–138. [DOI] [PubMed] [Google Scholar]
- Eser O, Eser B, Cosar M, Erdogan M, Aslan A, Yildiz H, Solak M, Haktanir A. 2011. Short aggrecan gene repetitive alleles associated with lumbar degenerative disc disease in Turkish patients. Genet Mol Res 10: 1923–1930. [DOI] [PubMed] [Google Scholar]
- Franke B, Vasquez AA, Johansson S, Hoogman M, Romanos J, Boreatti-Hümmer A, Heine M, Jacob CP, Lesch KP, Casas M, et al. 2010. Multicenter analysis of the SLC6A3/DAT1 VNTR haplotype in persistent ADHD suggests differential involvement of the gene in childhood and persistent ADHD. Neuropsychopharmacology 35: 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas L, McCarthy DJ, et al. 2016. The genetic architecture of type 2 diabetes. Nature 536: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand Y, Hernandez Y, Loving J, Benson G. 2014. VNTRseek—a computational tool to detect tandem repeat variants in high-throughput sequencing data. Nucleic Acids Res 42: 8884–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M. 2016. PCR-free library preparation greatly reduces stutter noise at short tandem repeats. bioRxiv 10.1101/043448. [DOI]
- Gymrek M, Willems T, Guilmatre A, Zeng H, Markus B, Georgiev S, Daly MJ, Price AL, Pritchard JK, Sharp AJ, et al. 2016. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet 48: 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl T, Hedrich R, Schultz J, Förster F. 2014. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30: 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddley K, Bubb V, Breen G, Parades-Esquivel U, Quinn J. 2011. Behavioural genetics of the serotonin transporter. Curr Top Behav Neurosci 12: 503–535. [DOI] [PubMed] [Google Scholar]
- Hijikata M, Matsushita I, Tanaka G, Tsuchiya T, Ito H, Tokunaga K, Ohashi J, Homma S, Kobashi Y, Taguchi Y, et al. 2011. Molecular cloning of two novel mucin-like genes in the disease-susceptibility locus for diffuse panbronchiolitis. Hum Genet 129: 117–128. [DOI] [PubMed] [Google Scholar]
- Huang W, Li L, Myers JR, Marth GT. 2011. ART: a next-generation sequencing read simulator. Bioinformatics 28: 593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Kirby A, Gnirke A, Jaffe DB, Barešová V, Pochet N, Blumenstiel B, Ye C, Aird D, Stevens C, Robinson JT, et al. 2013. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet 45: 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchheiner J, Nickchen K, Sasse J, Bauer M, Roots I, Brockmöller J. 2007. A 40-basepair VNTR polymorphism in the dopamine transporter (DAT1) gene and the rapid response to antidepressant treatment. Pharmacogenomics J 7: 48. [DOI] [PubMed] [Google Scholar]
- Krogh A, Brown M, Mian IS, Sjölander K, Haussler D. 1994. Hidden Markov models in computational biology: applications to protein modeling. J Mol Biol 235: 1501–1531. [DOI] [PubMed] [Google Scholar]
- LaHoste G, Swanson J, Wigal S, Glabe C, Wigal T, King N, Kennedy J. 1996. Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder. Mol Psychiatry 1: 121–124. [PubMed] [Google Scholar]
- Lalioti MD, Scott HS, Buresi C, Rossier C, Bottani A, Morris MA, Malafosse A, Antonarakis SE. 1997. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature 386: 847. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Gurtowski J, Yoo S, Marcus S, McCombie WR, Schatz M. 2014. Error correction and assembly complexity of single molecule sequencing reads. bioRxiv 10.1101/006395. [DOI]
- Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJB, Frants RR, van der Maarel SM. 2002. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 32: 235. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang P, Wang D, Gu W, Wang K. 2017. Interrogating the “unsequenceable” genomic trinucleotide repeat disorders by long-read sequencing. Genome Med 9: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclotte G, Heydari M, Demeester P, Rombauts S, Van de Peer Y, Audenaert P, Fostier J. 2016. Jabba: hybrid error correction for long sequencing reads. Algorithms Mol Biol 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Zhou P, Mudge J, Gurtowski J, Lee H, Ramaraj T, Walenz BP, Liu J, Stupar RM, Denny R, et al. 2017. Hybrid assembly with long and short reads improves discovery of gene family expansions. BMC Genomics 18: 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki S, Schirripa M, Loupakis F, Cao S, Zhang W, Yang D, Ning Y, Berger MD, Miyamoto Y, Suenaga M, et al. 2017. Tandem repeat variation near the HIC1 (hypermethylated in cancer 1) promoter predicts outcome of oxaliplatin-based chemotherapy in patients with metastatic colorectal cancer. Cancer 123: 4506–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci 86: 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard AL, Pritchard CW, Bentham P, Lendon CL. 2007. Role of serotonin transporter polymorphisms in the behavioural and psychological symptoms in probable Alzheimer disease patients. Dement Geriatr Cogn Disord 24: 201–206. [DOI] [PubMed] [Google Scholar]
- Pugliese A, Zeller M, Fernandez A, Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisen-barth GS, Bennett ST, Patel DD, et al. 1997. The insulin gene is transcribed in the human thymus and transcription levels correlate with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 15: 293–297. [DOI] [PubMed] [Google Scholar]
- Ræder H, Johansson S, Holm PI, Haldorsen IS, Mas E, Sbarra V, Nermoen I, Eide SA, Grevle L, Bjørkhaug L, et al. 2006. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nat Genet 38: 54. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela L, Rivals E. 2014. LoRDEC: accurate and efficient long read error correction. Bioinformatics 30: 3506–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela L, Walve R, Rivals E, Ukkonen E. 2016. Accurate self-correction of errors in long reads using de Bruijn graphs. Bioinformatics 33: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Jin L, Chakraborty R, Boerwinkle E. 1993. VNTR allele frequency distributions under the stepwise mutation model: a computer simulation approach. Genetics 134: 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöcker BK, Köster J, Rahmann S. 2016. SimLoRD: simulation of long read data. Bioinformatics 32: 2704–2706. [DOI] [PubMed] [Google Scholar]
- Tyner C, Barber GP, Casper J, Clawson H, Diekhans M, Eisenhart C, Fischer CM, Gibson D, Gonzalez JN, Guruvadoo L, et al. 2016. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res 45: D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ummat A, Bashir A. 2014. Resolving complex tandem repeats with long reads. Bioinformatics 30: 3491–3498. [DOI] [PubMed] [Google Scholar]
- Viswanath B, Purushottam M, Kandavel T, Reddy YJ, Jain S, et al. 2013. DRD4 gene and obsessive compulsive disorder: Do symptom dimensions have specific genetic correlates? Prog Neuropsychopharmacol Biol Psychiatry 41: 18–23. [DOI] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876. [DOI] [PubMed] [Google Scholar]
- Willems T, Zielinski D, Yuan J, Gordon A, Gymrek M, Erlich Y. 2017. Genome-wide profiling of heritable and de novo STR variations. Nat Methods 14: 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall BB, Brott TG, Brown RD, Brown WM, Rich SS, Arepalli S, Wavrant-De Vrièze F, Duckworth J, Singleton AB, Hardy J, et al. 2007. IL1RN VNTR polymorphism in ischemic stroke. Stroke 38: 1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JM. 1994. Mutation at VNTRs: Are minisatellites the evolutionary progeny of microsatellites? Genome 37: 345–347. [DOI] [PubMed] [Google Scholar]
- Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, Weng Z, Liu Y, Mason CE, Alexander N, et al. 2016. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data 3: 160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.