Abstract
The massive expansions of odorant receptor (OR) genes in ant genomes are notable examples of rapid genome evolution and adaptive gene duplication. However, the molecular mechanisms leading to gene family expansion remain poorly understood, partly because available ant genomes are fragmentary. Here, we present a highly contiguous, chromosome-level assembly of the clonal raider ant genome, revealing the largest known OR repertoire in an insect. While most ant ORs originate via local tandem duplication, we also observe several cases of dispersed duplication followed by tandem duplication in the most rapidly evolving OR clades. We found that areas of unusually high transposable element density (TE islands) were depauperate in ORs in the clonal raider ant, and found no evidence for retrotransposition of ORs. However, OR loci were enriched for transposons relative to the genome as a whole, potentially facilitating tandem duplication by unequal crossing over. We also found that ant OR genes are highly AT-rich compared to other genes. In contrast, in flies, OR genes are dispersed and largely isolated within the genome, and we find that fly ORs are not AT-rich. The genomic architecture and composition of ant ORs thus show convergence with the unrelated vertebrate ORs rather than the related fly ORs. This might be related to the greater gene numbers and/or potential similarities in gene regulation between ants and vertebrates as compared to flies.
New genes provide abundant raw material for evolution to act upon and are likely instrumental in the phenotypic adaptation of organisms (Long et al. 2003, 2013; Demuth and Hahn 2009; Chen et al. 2013). A variety of processes can generate new genes, including gene duplication, exon shuffling, gene fission-fusion, and de novo origination (Chen et al. 2013). Of these processes, gene duplication has been shown to play a particularly important role in genome evolution and phenotypic adaptation (Demuth et al. 2006; Zhou et al. 2008; Demuth and Hahn 2009). Genes may duplicate through whole genome duplication, segmental duplication, or retrotransposition (Mendivil Ramos and Ferrier 2012). Segmental duplication may be further divided into local (tandem) duplication and dispersed duplication and may arise from a variety of processes leading to structural rearrangements of the genome (Mendivil Ramos and Ferrier 2012). Although the processes leading to gene duplication are fairly well understood, it has been difficult to tease apart the role each process has played in the evolution of different gene families, especially in rapidly evolving gene families (Demuth and Hahn 2009). This is, in part, because repetitive regions of genomes are difficult to assemble, resulting in fragmentary and often inaccurate assemblies (Demuth and Hahn 2009). New long-read sequencing technologies and high-throughput structural mapping techniques have great promise to address this problem (Burton et al. 2013; Huddleston et al. 2014; Chakraborty et al. 2016; Bickhart et al. 2017).
A notable example of adaptive gene duplication is the ant odorant receptor (OR) gene family. Several studies show that the vast majority of the 300–400 OR genes in different ant species are quite young, having arisen by gene duplications since the evolutionary split between ants and their closest relatives, the bees, approximately 150 mya (Zhou et al. 2012, 2015; Engsontia et al. 2015; McKenzie et al. 2016; Branstetter et al. 2017). Rapid expansion of ORs in ants has been associated with pheromone perception (Smith et al. 2011a,b; Zhou et al. 2012; Engsontia et al. 2015; McKenzie et al. 2016; Pask et al. 2017), and two recent functional genetic studies that used CRISPR to knock out the essential OR coreceptor gene Orco have demonstrated that functional ORs are essential for many aspects of social behavior, organismal fitness, and even neural circuit development and/or maintenance (Trible et al. 2017; Yan et al. 2017). This contrasts with studies of Orco knockouts in flies and mosquitos, which show that loss of function of the fewer ORs in these solitary insects has much less severe impacts on behavior, fitness, and neuronal wiring (Asahina et al. 2008; DeGennaro et al. 2013). Despite the demonstrated importance of novel ORs in ant biology, relatively little is known about the molecular mechanisms driving OR duplication.
Poor genomic assembly has plagued studies of ant ORs, and in all published ant genomes, OR loci are predominantly located near contig or scaffold edges, leading to many fragmentary gene models and incomplete pictures of OR genomic structure (Smith et al. 2011a,b; Zhou et al. 2012; Oxley et al. 2014). Researchers have shown that ant ORs are primarily located on tandem arrays (Smith et al. 2011a,b; Zhou et al. 2012; Engsontia et al. 2015); however, the number and size of these arrays and their position within the genome have been impossible to determine. In the corbiculate bees, Brand and Ramirez (2017) found that almost all ORs are located on a few conserved tandem arrays and that local tandem array expansion was nearly the sole driver of receptor repertoire evolution. This finding contrasts starkly with observations from the genus Drosophila, where ORs are scattered throughout the genome and genome transposition plays an important role in OR evolution (Guo and Kim 2007; Conceição and Aguadé 2008). Instead, hymenopteran OR genomic organization resembles that of vertebrate ORs, which belong to a different protein family but are functionally analogous to insect ORs. Vertebrate ORs tend to be located on a few large tandem arrays and duplicate primarily by local tandem array expansion (Niimura and Nei 2005). The genomic organization of vertebrate ORs is important for their nondeterministic gene regulation (Kratz et al. 2002; Clowney et al. 2012), which, in turn, allows for a large OR gene repertoire with a low regulatory burden; i.e., few genes are required to regulate a large gene family (Kratz et al. 2002; Clowney et al. 2011).
The clonal raider ant Ooceraea biroi (formerly Cerapachys biroi [Borowiec 2016]) is an emerging model system for genomic and molecular biological studies of social insects (Oxley et al. 2014). Notably, the first draft genome assembly of the clonal raider ant indicates that it possesses one of the largest chemosensory receptor gene repertoires of all insects (Oxley et al. 2014; McKenzie et al. 2016). To facilitate the study of the genomic evolution of ant chemosensory receptors and genome structure evolution in general, we used third generation sequencing (Pacific Biosciences [PacBio] and Oxford Nanopore) and Hi-C proximity-based scaffolding to assemble a high-quality, chromosome-level genome for the clonal raider ant. We used this genome assembly to study the genomic organization and genomic context of chemosensory genes in the clonal raider ant and compared these data with data from other model insect species. We find that Hymenoptera ORs show convergence in genomic structure and context with the unrelated vertebrate ORs, suggesting similar evolutionary histories and potentially indicating convergent regulatory mechanisms.
Results
Sequencing and assembly
Over 21 Gbp of PacBio long reads and 500 Mbp of Oxford Nanopore long reads were assembled using Canu (Koren et al. 2017) and quickmerge (Chakraborty et al. 2016), yielding an assembly containing 227 Mbp on 694 contigs (contig N50 = 3.3 Mbp, N75 = 1.5 Mbp). These contigs were scaffolded with 19 Gbp of Hi-C reads (Fig. 1A; Lieberman-Aiden et al. 2009; Burton et al. 2013). Bacterial contaminants were identified with BLAST and removed, leaving ∼222 Mbp—over 98.9% of the assembled nonbacterial sequence—on 14 large scaffolds ranging in size from 8.8 to 24.6 Mbp. These scaffolds appear to correspond to the 14 clonal raider ant chromosomes (Imai et al. 1984). Scaffolds were gap-filled with the original PacBio and Nanopore reads using the PBJelly program (English et al. 2012), corrected with the PacBio reads using Quiver (github.com/PacificBiosciences/GenomicConsensus) as well as with 33 Gbp of Illumina NextSeq reads using Pilon (Walker et al. 2014) and then further polished with the Illumina data using a custom pipeline to correct spurious indels at heterozygous sites (see Methods). The final assembly consisted of 224 Mbp on 530 contigs (contig N50 = 3.7 Mbp, N75 = 2.1 Mbp) (Fig. 1A). Whole-genome alignment of the new assembly with the previous clonal raider ant genome assembly (Oxley et al. 2014) showed that the two are congruous over most of the genome (Fig. 1B).
Figure 1.
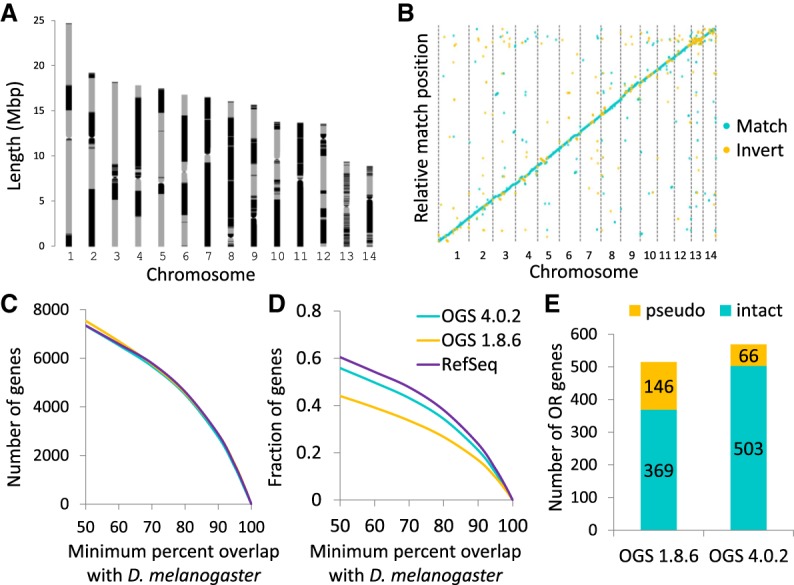
Genome assembly and annotation of the clonal raider ant. (A) Map of contigs within clonal raider ant chromosomes. Alternating gray and black indicate different contigs. Large stretches of uninterrupted gray or black indicate very large contigs, often containing most of a chromosome arm. Putative centromeres were identified based on Hi-C interaction (see Methods). (B) Whole-genome alignment of the second generation draft assembly (Oxley et al. 2014) with the new assembly. Largely continuous, noninverted alignment across the genome indicates little large-scale structural variation between the two assemblies. (C,D) BLAST hit overlap between clonal raider ant gene sets and the Drosophila melanogaster official gene set (v6). The good performance of OGS 4.0.2 in terms of number of genes, and intermediate performance in terms of fraction of genes, indicates high sensitivity and intermediate specificity. (E) Number of odorant receptors found in the annotation based on the previous assembly (OGS 1.8.6) vs. the current annotation (OGS 4.0.2). The large increase in intact ORs and decrease in predicted pseudogenes shows that the previous OR annotations were compromised by poor assembly and misassembly.
Annotation
Manual annotation of chemosensory genes yielded 661 gene models, and automated gene prediction with hint-guided AUGUSTUS (Stanke et al. 2006; see Methods) predicted an additional 12,471 gene models, yielding an official gene set (OGS) of 13,132 gene models (OGS v4.0.2). To evaluate the quality of this annotation, we examined the similarity of this and previous clonal raider ant gene sets to Drosophila melanogaster genes using BLAST (Fig. 1C,D). Similar numbers of OGS v4.0.2 genes had good matches to D. melanogaster genes compared with the previous OGS (OGS v1.8.6) and the NCBI RefSeq v101 gene set (Fig. 1C). In terms of percentages of genes with D. melanogaster matches, OGS v4.0.2 showed intermediate performance between the stringent NCBI RefSeq gene set (which has less than 12,000 gene models) and the previous OGS (Fig. 1D). Across 29 published clonal raider ant RNA-seq libraries (McKenzie et al. 2014, 2016; Oxley et al. 2014; Libbrecht et al. 2016), 10,800 genes had RNA-seq support as defined by >2 FPKM (fragments per kilobase per million reads) in at least one experiment.
Manual annotation of the gustatory receptors (GRs), ionotropic receptors (IRs), odorant binding proteins (OBPs), and chemosensory proteins (CSPs) revealed similar numbers of genes as the previous assembly, although one OBP was missing (Obp12), two CSPs were duplicated (Csp3/3.2 and CSP10/10.2), and one additional OBP pseudogene and GR gene were found (Obp15PSE, Gr26). Unlike in the previous annotation, however, all gene models for these families were complete. In contrast, manual annotation of the odorant receptors revealed 569 gene models, 54 more than in the previous assembly (Fig. 1E). Even more drastic, 503 of these are predicted to encode intact genes (putatively functional), while in the previous annotation only 369 genes were predicted to be intact. Many of the predicted pseudogenes in the previous annotation did not have exact matches in the new annotation, indicating that they were chimeric models predicted over misassembled genomic sequence. Of the 503 putatively functional OR gene models, 495 had RNA-seq support as defined by >2 FPKM in at least one published RNA-seq experiment.
Genomic organization and evolution of ant odorant receptors
Odorant receptors were found distributed across the genome, on every chromosome except Chromosome 13. Most putatively functional odorant receptors (469 of 503, 93%) were found in 41 tandem arrays, which contained between two and 89 genes (Fig. 2A). Only 34 putatively functional genes were found outside of tandem arrays (singletons), making a total of 75 genomic loci encoding putatively functional ORs (Fig. 2A). An additional 14 loci encoded only pseudogene singletons.
Figure 2.
Phylogeny and genomic organization of odorant receptors in four Hymenoptera species. Branches from the five largest ant OR subfamilies are colored by subfamily. OR loci indices are assigned by ordering loci as they appear in the phylogeny from top to bottom. Sets of vertically aligned points represent a single tandem array, while breaks in a line represent transposition events. Loci indices assigned this way are not homologous between species. Loci ages are indicated by color and calculated via phylostratigraphy and synteny analysis (see Methods). The few large, contiguous, and old tandem arrays in the top half of A and C and throughout B indicate tandem duplications as the predominant drivers of gene expansion in these gene clades. The many short species-specific tandem arrays in the bottom half of A and C and throughout D indicate that genomic transpositions followed by tandem duplications drove expansions in these clades.
To examine the genomic evolution of ant OR loci, putatively functional OR annotations from the red fire ant Solenopsis invicta (the only ant with most scaffolds mapped to chromosomes), the honeybee Apis mellifera, and the jewel wasp Nasonia vitripennis (Robertson et al. 2010; McKenzie et al. 2016) were mapped onto chromosome assemblies (Desjardins et al. 2013; Wang et al. 2013; Elsik et al. 2014), and then genomic loci were mapped onto the phylogeny of these ORs. This information was used to determine the distribution of tandem arrays in each species. We also calculated the minimum ages of tandem arrays via phylostratigraphy and synteny analyses (Fig. 2A–D). Like the clonal raider ant, the ORs of the fire ant and the honeybee were mostly organized in tandem arrays with few singletons. The honeybee had 138 genes in 15 tandem arrays and 14 genes as singletons, while the fire ant had 354 genes in 26 tandem arrays and 24 genes as singletons. The jewel wasp had smaller tandem arrays and a larger proportion of singletons, with 151 genes on 35 tandem arrays and 54 singletons. In the honeybee and the fire ant, over 64% and 66% of genes, respectively, are found on tandem arrays that were present in the most recent common ancestor (MRCA) of bees and ants, while significantly fewer (54%) clonal raider ant ORs were found on similarly old tandem arrays (χ2 test, P < 0.0001; pair-wise Fisher's exact post-hoc tests, P < 0.05). Correspondingly, the clonal raider ant has significantly more genes present on lineage-specific tandem arrays than the fire ant (35% and 6%, respectively; Fisher's exact test, P < 0.0001; data from the honeybee are not comparable because it represents an older lineage in our analysis). The vast majority (161 of 174, or 93%) of the genes on lineage-specific tandem arrays and singletons in the clonal raider ant belong to the nine-exon OR subfamily (Fig. 2A).
To compare the role of genomic transposition vs. local tandem array expansion in generating OR diversity, we performed gene-tree species-tree reconciliation analysis using NOTUNG (Chen et al. 2000) to calculate total gene duplications in each lineage. The number of transposition events was estimated by counting species-specific loci (singletons and tandem arrays), which result from genomic transpositions in the lineage leading to that species. Array ages are likely somewhat conservative, which leads to a somewhat liberal estimation of transposition events. NOTUNG found 337 gene duplications specific to the clonal raider ant lineage, while we found 42 clonal raider ant species-specific loci (singletons and tandem arrays) indicating transposition events (Supplemental Tables S1, S2). This suggests that, at most, 12.4% of gene duplication events in the clonal raider ant may be transposition events. The fire ant and the honeybee showed even lower putative transposition to duplication ratios (6.6% and 11.2%, respectively), while the jewel wasp showed the highest at 37.8%.
Genomic context of clonal raider ant ORs
In order to gain mechanistic insights into the ant OR repertoire expansion, we investigated the genomic context of clonal raider ant ORs and compared this to the remaining clonal raider ant genes. It has previously been reported that, in the ant Cardiocondyla obscurior, ORs are enriched in areas of high transposable element density (“TE islands”) (Schrader et al. 2014) which could facilitate either local tandem array expansion (via unequal crossing over) or genomic transposition (Kazazian 2004; Schrader et al. 2014). To assess whether this may be a general trend in ants, we searched for TE islands in the clonal raider ant following the methods of Schrader et al. (2014). We found that these procedures actually annotated many duplicated OR loci as novel repeat families, and thus we limited our analysis to classified transposon families. We found that TE islands were significantly depauperate in ORs (Yates corrected χ2 test, P < 0.001) (Fig. 3A). However, OR loci do have approximately a 1.5× higher transposon density than both the entire genome and other gene-dense regions of the genome (Fig. 3B).
Figure 3.
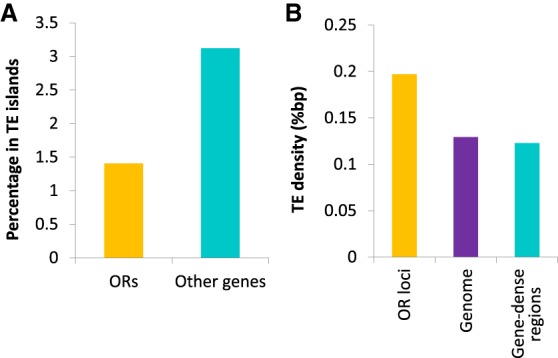
Genomic colocalization of transposable elements (TEs), odorant receptor genes, and other genes. (A) Percentage of OR genes and other genes within TE islands. (B) Density of TEs in OR loci and other gene-dense regions (genomic stretches with at least one OR gene [OR loci] or one non-OR gene [gene-dense regions] per 10 kbp) and within the genome as a whole. These data show that TE islands are depauperate in ORs, but OR loci are enriched for TEs relative to other gene-rich loci and the genome as a whole.
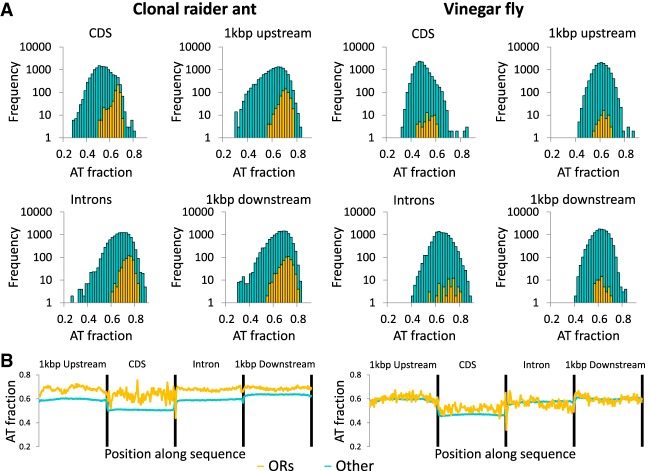
In vertebrates, rapidly expanding gene families have been associated with genomic regions of high AT content, although it is unclear if this is a cause or consequence of gene family expansion or potentially even associated with the unique challenges of regulating large multigene families (Clowney et al. 2011). Vertebrate odorant receptors are unrelated to insect odorant receptors and in fact belong to a different gene superfamily. However, like ant ORs they are organized in large tandem arrays (Niimura and Nei 2005). In the mouse, these arrays are associated with high AT content (Glusman et al. 2001; Clowney et al. 2011). We found that ant odorant receptors have higher AT content in exons, introns, and flanking intergenic regions than most other genes (Fig. 4). In two other insects with large OR repertoires, the fire ant Solenopsis invicta and the flour beetle Triboleum castaneum, OR loci also showed enrichment for AT content relative to other genes (Supplemental Fig. S2). This is not the case for the vinegar fly Drosophila melanogaster or the silk moth Bombyx mori, which have many fewer ORs, mostly organized as singletons or pairs throughout the genome (Fig. 4; Supplemental Fig. S2; Robertson et al. 2003).
Figure 4.
Nucleotide composition of odorant receptors (ORs) vs. other genes in the clonal raider ant and the vinegar fly (Drosophila melanogaster). (A) Frequency distribution of AT content for coding sequence (CDS), introns, downstream, and upstream sequence of OR genes (orange) and other genes (cyan). Y-axes are log-scaled to allow visualization of fly ORs, which make up a small proportion of fly genes. (B) Mean nucleotide composition along the CDS, intronic, and flanking sequences of OR genes or other genes in the clonal raider ant (left) and the vinegar fly (right). These data show that clonal raider ant ORs are AT-rich relative to most other genes, while fly ORs possess typical AT content relative to other genes.
Discussion
Highly contiguous, chromosome-level assembly of genomes provides unparalleled insights into the genomic structure and evolution of organisms. By providing such an assembly for the clonal raider ant, we have revealed significant, previously hidden diversity in the odorant receptor gene family. We have also been able to map out the genomic evolution of this gene family in the Hymenoptera in unprecedented detail. We found that the extraordinary rates of OR gene duplication previously reported (Zhou et al. 2012, 2015; Engsontia et al. 2015; McKenzie et al. 2016) largely arise by local tandem array expansion. We also observed higher rates of transposition in the most rapidly expanding clonal raider ant OR clade and enrichment for transposons in OR loci. However, we found that regions of the genome with especially high transposon density (“transposon islands”) are not enriched for OR genes in the clonal raider ant. Additionally, high AT content may play an important role in ant OR gene expansion and/or regulation. Both the genomic architecture and context of Hymenoptera ORs are highly reminiscent of vertebrate ORs, rather than resembling fly ORs.
The number of ORs we found in our new assembly of the clonal raider ant genome is the highest of any insect examined to date (Sánchez-Gracia et al. 2011; Engsontia et al. 2015; Zhou et al. 2015; McKenzie et al. 2016). Although the previous assembly of the clonal raider ant also contained a large number of ORs (Oxley et al. 2014), a large fraction of the gene models were predicted to encode pseudogenes, and the number of putatively functional ORs was smaller than in the ants Atta cephalotes, Acromyrmex echinatior, and Solenopsis invicta (Engsontia et al. 2015; Zhou et al. 2015; McKenzie et al. 2016). This was enigmatic, as the clonal raider ant has more antennal lobe glomeruli than A. cephalotes (Kelber et al. 2009; McKenzie et al. 2016) (the glomerulus counts for A. echinatior and S. invicta are unknown), and in the vinegar fly Drosophila melanogaster there is almost one-to-one correspondence between AL glomeruli and expressed ORs (Laissue and Vosshall 2008). In general, there was a fairly poor correlation between glomerulus number and identified putatively functional ORs in the first generation of ant genomes (Zhou et al. 2012; McKenzie et al. 2016), suggesting that perhaps the one-to-one OR-glomerulus correspondence observed in flies did not hold true in ants. However, our new OR gene set contains almost exactly as many putatively functional ORs as glomeruli in the female adult AL (493–509) (McKenzie et al. 2016; Trible et al. 2017). The poor correlation between intact OR number and glomerulus number is therefore likely an artifact caused by incomplete genome assembly in most ants. Based on glomerulus number, we do expect that the number of ORs in the clonal raider ant is exceptional even among ants; however, it is likely that better assembly of other ant genomes will likewise show larger OR repertoires than currently reported. For instance, 354 putatively functional ORs are reported for Camponotus floridanus, compared with 434 glomeruli in worker ALs (Zube et al. 2008; Zhou et al. 2012).
A variety of molecular mechanisms lead to gene duplications that can be in tandem or interspersed across the genome (for review, see Mendivil Ramos and Ferrier 2012). Furthermore, genomic rearrangement and transposition can disrupt and scatter genes which duplicated in tandem (Mendivil Ramos and Ferrier 2012). Although evidence from many mammal species suggests that tandem duplication is the predominant form of gene duplication (e.g., She et al. 2008; Liu et al. 2009), most gene duplications in humans and Drosophila appear to be interspersed intra-chromosomally or inter-chromosomally, respectively (Zhang et al. 2005; Fiston-Lavier et al. 2007; Zhou et al. 2008; Meisel 2009). Although some of these translocations may result from tandem duplication followed by genomic rearrangement, there is evidence for abundant nontandem duplication as well (Bailey et al. 2003; Zhang et al. 2005; Fiston-Lavier et al. 2007; Yang et al. 2008; Meisel 2009; Ezawa et al. 2011). In Drosophila, it has been hypothesized that translocations may play an important role in OR evolution (Guo and Kim 2007; Nozawa and Nei 2007; Conceição and Aguadé 2008). In contrast, within mammals and corbiculate bees, little to no translocation of ORs was observed (Niimura and Nei 2005; Brand and Ramirez 2017). However, most mammalian tandem arrays resulted from ancient transpositions (between the MRCA of all bony fish and the MRCA of mammals) followed by subsequent tandem duplication (Niimura and Nei 2005).
In ants, we found that the vast majority of gene duplications happened via tandem duplication. We did find evidence of low levels of translocation as well, especially in the largest and most rapidly expanding OR subfamily in the clonal raider ant. Even within these clades, however, most genes are located on tandem arrays. These data suggest that translocation followed by local tandem array expansion is responsible for the most extreme gene duplication events in ants, while local tandem array expansion without translocation is responsible for the vast majority of gene duplications. It is interesting to note that, in contrast to the pattern we observe in ants, the largest honeybee OR subfamily is contained within only one tandem array with a single additional singleton (Fig. 2C; Brand and Ramirez 2017).
Transposable elements (TEs) can promote gene duplications by increasing unequal crossing over, promoting ectopic recombination, or by copy and paste transposition of a gene (e.g., retrotransposition). High transposon densities are correspondingly associated with duplicated genes in a variety of species (e.g., Bailey et al. 2003; Yang et al. 2008; Schrader et al. 2014). Schrader et al. (2014) found that ORs were enriched in TE-dense regions (TE islands) in the genome of the ant Cardiocondyla obscurior and suggested that TEs play an important role in OR duplications in ants. We found that TE islands in the clonal raider ant are, in fact, depauperate in ORs. However, OR loci are enriched for transposons relative to the genome as a whole. This increased transposon density may play a role in OR expansion by providing an additional substrate for ectopic recombination and unequal crossing over (Yang et al. 2008; Mendivil Ramos and Ferrier 2012). We note that there is little evidence for direct involvement of transposon activity in OR expansion. Genomic translocations of ORs are rare and may result from ectopic recombination or genomic rearrangement. We observed no recent intron losses in any ant OR, implying that no OR translocations resulted from reverse-transcription of an OR back into the genome.
The role of sequence content bias in OR evolution is less clear. Genes with AT-rich promoters in the mouse are enriched for multigene families located in tandem arrays with high evolutionary turnover in copy number (Glusman et al. 2001; Clowney et al. 2011). The majority of these proteins are either transmembrane or secreted proteins (Clowney et al. 2011), and many of these genes show nondeterministic gene regulation (e.g., Held et al. 1995; Chess 2005). Clowney et al. (2011) posited that the high AT content of the promotors may be involved in nondeterministic regulation (either because of the nature of transcription factors recruited or because of unique chromosomal architecture [e.g., Segal and Widom 2009]) and that freedom from deterministic gene expression could promote functional diversification of rapidly evolving gene families. However, it is also possible that high AT content is either a cause or consequence of frequent gene duplication and that nondeterministic gene expression of rapidly evolving gene families is incidentally also selected for in vertebrates. OR expression is deterministic in flies (Kaupp 2010), and so far there is no experimental evidence of nondeterministic OR expression in ants or other Hymenoptera. However, we did not observe AT enrichment in fly ORs, and it is possible that different strategies must be employed in the regulation of 60 genes in flies vs. 400–500 in ants.
It is intriguing that, in ants but not in flies, elimination of OR function via knockout of the Orco gene results in a reduction of olfactory glomeruli (Trible et al. 2017; Yan et al. 2017), as seen in OR loss-of-function mutants in mice (Wang et al. 1998). Indeed, Yan et al. (2017) speculated that the high diversity of ORs in the Hymenoptera may require receptor-dependent ORN targeting like that of vertebrates, rather than deterministic, receptor-independent ORN targeting as seen in flies. Vertebrate-like nondeterministic OR gene regulation would strongly support this hypothesis because, under this scenario, OR expression is the only factor differentiating many ORN populations. The many parallels in genomic architecture between ants and vertebrates we observe here are tantalizing but do not conclusively demonstrate vertebrate-like OR regulation in ants. Further functional studies will be required to determine whether OR regulation in the Hymenoptera is, in fact, deterministic or not.
Methods
Sequencing and assembly
Genomic DNA for PacBio sequencing was extracted from 300 ants using the Qiagen Genomic-tip extraction kit, and libraries were prepared and sequenced at the Genomics Core Facility at the Icahn School of Medicine at Mount Sinai (New York, NY). Sequencing was performed on a PacBio RSII with P6-C4 chemistry. Genomic DNA for Nanopore sequencing was extracted from ∼15 mg of ants using the Qiagen MagAttract HMW DNA kit, and libraries were prepared for Nanopore sequencing using the Rapid Sequencing kit (SQK-RAD002) and sequenced on two MinION SpotON flow cells (R9.4). Hi-C sequencing was performed by Phase Genomics (Seattle, WA). Genomic DNA for Illumina sequencing was extracted from five ants using the Qiagen DNeasy extraction kit and sequenced on part of an Illumina NextSeq flow cell at the Rockefeller University Genomics Resource Center. Further details on DNA extraction and sequencing are given in the Supplemental Methods.
PacBio and Nanopore data were assembled using Canu v1.1 (Koren et al. 2017) with the options “corMhapSensitivity=normal” and “corOutCoverage=80” and all other options set to default. A second assembly was created using only the PacBio data. Both contig assemblies were scaffolded using Hi-C proximity-based scaffolding with the program LACHESIS (Burton et al. 2013). Chimeric contigs were then manually identified by viewing the Hi-C interaction density and either broken at sites with conspicuously low Illumina coverage (PacBio + Nanopore contig assembly) or else removed from the contig set (PacBio-only contig assembly). quickmerge (Chakraborty et al. 2016) was then used to join/extend PacBio + Nanopore contigs using the PacBio-only contigs. The resulting contigs were again scaffolded using Hi-C proximity-based scaffolding. PBJelly (English et al. 2012) was used to gap-fill the resulting assembly with PacBio and Nanopore reads. This assembly was first error-corrected with the PacBio reads using Quiver (github.com/PacificBiosciences/GenomicConsensus) and then with Illumina reads using Pilon (Walker et al. 2014), using BWA-MEM (Li 2013) to align the Illumina reads. We found that Pilon was unable to fix many spurious deletions at heterozygous loci, however, because BWA-MEM often aligned each “insertion” variant to different locations near the spurious deletion, leading Pilon to see two heterozygous insertions instead of a homozygous insertion. To fix this, we used a custom pipeline wherein we identified variants and reference errors using FreeBayes (Garrison and Marth 2012), filtered variants by quality score (quality score >20), phased these variants using WhatsHap (Patterson et al. 2014), and then substituted alleles of a single phase block for the reference alleles using a custom script (Supplemental Scripts: “pvc_pipe.py”). One Hi-C cluster showed suspiciously low coverage and low Hi-C linkage to the rest of the genome (Supplemental Fig. S1), and BLASTX (Altschul et al. 1990) of the contigs therein to the NCBI nonredundant protein database revealed that all of these contigs had high similarity to genes from Sphingomonas bacteria. This cluster was thus removed from our assembly, along with three unassembled contigs which also showed highest BLASTX affinity for bacterial sequences in the NCBI nonredundant protein database. To visualize contig locations relative to chromosome structure (Fig. 1A), we identified putative centromeres based on Hi-C linkage by manually identifying peaks of pan-chromosomal interactions consistent with the Hi-C signal of known centromeres in insects (Supplemental Fig. S1; Dudchenko et al. 2017).
Annotation
Manual annotation of chemosensory genes was conducted as previously described (Oxley et al. 2014). MAKER2 (Holt and Yandell 2011) was used to annotate gene models for nonchemosensory genes using the AUGUSTUS gene prediction software (Stanke et al. 2006). Repeat annotations were generated by using RepeatModeler to generate a library of O. biroi repetitive regions including transposable elements, and then masking the genome using this library with the RepeatMasker software (Smit et al. 2013–2015). Automated annotations which overlapped manual chemosensory gene annotations were removed from the gene set. See Supplemental Methods for further details on manual annotations, gene prediction training, and evidence used for MAKER2 gene prediction.
Genomic organization of odorant receptors
Tandem arrays in all species were defined as loci containing consecutive ORs that are not interrupted by two or more non-OR genes. Manual OR annotations were obtained from Robertson et al. (2010) for the honeybee and jewel wasp and from McKenzie et al. (2016) for the fire ant. NCBI RefSeq annotations were used for other genes. Because manual OR annotations for the honeybee, jewel wasp, and fire ant came from previous, more fragmentary assemblies, the genes were mapped to the new chromosome-scale assemblies (Desjardins et al. 2013; Wang et al. 2013; Elsik et al. 2014) following protocols outlined in the Supplemental Methods.
Phylogenetics and evolutionary analysis
Protein sequences for all putatively functional OR genes from the jewel wasp, honeybee, fire ant, and clonal raider ant were aligned using the alignment software MAFFT with the linsi parameters (Katoh et al. 2005). A maximum likelihood phylogeny was then built using RAxML with the LG + gamma model of protein sequence evolution (Stamatakis 2014). Support was calculated using the RAxML rapid bootstrapping algorithm (Stamatakis et al. 2008) with 100 replicates. To calculate gene births and deaths, we used the resolution plus reconciliation algorithms of the software NOTUNG (Chen et al. 2000) with the 70% bootstrap consensus gene tree and with the species tree topology from Branstetter et al. (2017).
Tandem array age and synteny analyses
To determine the age of tandem arrays, we mapped these arrays onto the terminals of our maximum likelihood OR phylogeny and found the smallest clade containing each tandem array. We assume that OR tandem arrays are formed by local gene duplications and almost never by translocation of ORs next to each other, which is supported by the fact that only two distantly related genes in the clonal raider ant are found on the same tandem array (Orco and Or5-Q2). Based on this assumption, if the smallest clade containing all genes in an array in focal species i contains genes from species ii, the array must date back at least to the divergence between species i and species ii. This provides a minimum array age, though arrays may well be older if (1) genes on this array were in single copy in the common ancestor of the two species, or (2) genes from the array were lost in species ii. In case 1, there should exist an array in species ii with synteny with the array in species I, provided neither array was translocated in the genome. Thus, we supplemented our phylostratographic dating with synteny dating. For our synteny analysis, we ran OrthoMCL (Li et al. 2003) on genes from the clonal raider ant, the fire ant, the honeybee, and the jewel wasp. We then used a custom Python script (Supplemental Scripts: “arraySynteny.py”) to pull the five genes on each side of any given OR locus and then check for orthologs in the five genes on each side of every OR locus in the other species. To maximize sensitivity in light of potential differences in annotation and deletion or translocation of flanking genes, arrays were designated as syntenic if three genes on either side of one array were orthologous to three genes on either side of the other array, or if one gene on each side of one array was orthologous to one gene on each side of the other array. We then assigned dates to syntenic arrays transitively. For example, if an array in S. invicta was syntenic with an array in O. biroi that was also syntenic with an array in N. vitripennis, all these arrays were considered to date to the divergence of N. vitripennis with the Aculeata, even if the S. invicta array did not pass our criteria to be considered syntenic with the N. vitripennis array.
Transposon annotations
Our transposon annotation pipeline followed that of Schrader et al. (2014) with the following exceptions: Only RepeatModeler was used for de novo repeat identification (as opposed to RepeatModeler plus PILER [Edgar and Myers 2005]), only repeats modeled from the clonal raider ant genome were used, and RepeatMasker rather than Censor (Kohany et al. 2006) was used to annotate repeats and transposons based on our de novo repeat libraries. We found that RepeatModeler calls exons within duplicated ORs as novel repeat families, and so for maximum stringency only included repeats belonging to known transposon classes (as annotated by RepeatModeler and the custom scripts of Schrader et al. [2014]). Transposable element islands were defined as in Schrader et al. (2014) as regions of the genome within sliding windows (here, 100 kbp) with TE content in the 95th percentile of all such windows. To compare transposon density in OR loci compared with the genome as a whole or gene-dense genomic regions, we calculated gene density (both for ORs and for other genes) within sliding windows and included regions with at least one non-OR gene per 10 kbp as gene-dense regions and regions with at least one OR gene per 10 kbp as OR loci. TE content within these regions was then calculated using a custom Python script (Supplemental Scripts: “SlidingWindowRepeatCalculations.py”).
Sequence content analysis
For analysis of the sequence content of OR loci compared to the genome as a whole, RefSeq genome assemblies and annotations for Tribolium castaneum, Bombyx mori, Drosophila melanogaster, and Solenopsis invicta were downloaded from NCBI. All manually annotated ORs in B. mori and D. melanogaster are included and annotated in the RefSeq gene sets; however, T. castaneum and S. invicta manual OR annotations are not included in the RefSeq gene sets and therefore were obtained from Engsontia et al. (2008) and McKenzie et al. (2016). Because only peptide sequences were provided for T. castaneum, genomic loci for the ORs in this species were found by aligning these sequences to the genome using the protein2genome algorithm in Exonerate with the settings “--bestn 1 --percent 90”. ORs were then excluded from the T. castaneum and S. invicta RefSeq gene sets by blasting genes against the PFAM 7TM_6 model (insect odorant receptors) and removing sequences with an E-value below 0.01. For O. biroi, we used the annotation presented in this manuscript. The gene sets were then split into OR genes and other genes, the longest isoforms for each gene selected, and then the sequence of 1-kbp flanking regions, coding regions, and introns were extracted and AT content calculated across 100 bins over the length of the sequence using a custom Python script (Supplemental Scripts: “ExtractATcontent.py”).
Data access
Raw sequencing data, genome assembly, and genome annotation from this study have been submitted to the NCBI BioProject database (http://www.ncbi.nlm.nih.gov/bioproject/) and are all under accession number PRJNA420369. All custom scripts are available as Supplemental Materials in the Supplemental Scripts gzipped tarball archive.
Supplementary Material
Acknowledgments
We thank Josie Clowney for helpful comments on the analyses and manuscript and Leonora Olivos-Cisneros and Buck Trible for assistance with lab work. We also thank John Wang and Oksana Riba-Grognuz for providing fire ant assembly files, Lukas Schrader and Jay Kim for providing their TE annotation scripts, the Genomics Core Facility at the Icahn School of Medicine at Mount Sinai for help with PacBio sequencing, The Rockefeller University Genomics Resource Center for help with Illumina sequencing, and Phase Genomics for help with Hi-C sequencing and proximity-based assembly. This work was supported by Grant 1DP2GM105454-01 from the National Institutes of Health, a Klingenstein-Simons Fellowship Award in the Neurosciences, a Pew Biomedical Scholar Award, and an HHMI Faculty Scholar Award to D.J.C.K. S.K.M. was supported by NIH National Research Service Award Training Grant GM066699. This is Clonal Raider Ant Project paper #7.
Author contributions: S.K.M. and D.J.C.K. designed the research; S.K.M. performed the research; S.K.M. analyzed the data; S.K.M. and D.J.C.K. wrote the paper; and D.J.C.K. supervised the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.237123.118.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Asahina K, Pavlenkovich V, Vosshall LB. 2008. The survival advantage of olfaction in a competitive environment. Curr Biol 18: 1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 73: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, et al. 2017. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet 49: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec ML. 2016. Generic revision of the ant subfamily Dorylinae (Hymenoptera, Formicidae). Zookeys 608: 1–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, Ramirez SR. 2017. The evolutionary dynamics of the odorant receptor gene family in corbiculate bees. Genome Biol Evol 9: 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branstetter MG, Danforth BN, Pitts JP, Faircloth BC, Ward PS, Buffington ML, Gates MW, Kula RR, Brady SG. 2017. Phylogenomic insights into the evolution of stinging wasps and the origins of ants and bees. Curr Biol 27: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J. 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol 31: 1119–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ. 2016. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res 44: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Durand D, Farach-Colton M. 2000. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol 7: 429–447. [DOI] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet 14: 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A. 2005. Monoallelic expression of protocadherin genes. Nat Genet 37: 120–121. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. 2011. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res 21: 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Legros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou C, Myllys M, Barnea G, Larabell CA, Lomvardas S. 2012. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151: 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição IC, Aguadé M. 2008. High incidence of interchromosomal transpositions in the evolutionary history of a subset of Or genes in Drosophila. J Mol Evol 66: 325–332. [DOI] [PubMed] [Google Scholar]
- DeGennaro M, McBride CS, Seeholzer L, Nakagawa T, Dennis EJ, Goldman C, Jasinskiene N, James AA, Vosshall LB. 2013. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498: 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. 2009. The life and death of gene families. BioEssays 31: 29–39. [DOI] [PubMed] [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. 2006. The evolution of mammalian gene families. PLoS One 1: e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Gadau J, Lopez JA, Niehuis O, Avery AR, Loehlin DW, Richards S, Colbourne JK, Werren JH. 2013. Fine-scale mapping of the Nasonia genome to chromosomes using a high-density genotyping microarray. G3 (Bethesda) 3: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356: 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Myers EW. 2005. PILER: identification and classification of genomic repeats. Bioinformatics 21(Suppl. 1): i152–i158. [DOI] [PubMed] [Google Scholar]
- Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, et al. 2014. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 7: e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P, Sanderson AP, Cobb M, Walden KKO, Robertson HM, Brown S. 2008. The red flour beetle's large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem Mol Biol 38: 387–397. [DOI] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Robertson HM, Satasook C. 2015. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Res Notes 8: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezawa K, Ikeo K, Gojobori T, Saitou N. 2011. Evolutionary patterns of recently emerged animal duplogs. Genome Biol Evol 3: 1119–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiston-Lavier A-S, Anxolabehere D, Quesneville H. 2007. A model of segmental duplication formation in Drosophila melanogaster. Genome Res 17: 1458–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907v2.
- Glusman G, Yanai I, Rubin I, Lancet D. 2001. The complete human olfactory subgenome. Genome Res 11: 685–702. [DOI] [PubMed] [Google Scholar]
- Guo S, Kim J. 2007. Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol 24: 1198–1207. [DOI] [PubMed] [Google Scholar]
- Held W, Roland J, Raulet DH. 1995. Allelic exclusion of Ly49-family genes encoding class I MHC-specific receptors on NK cells. Nature 376: 355–358. [DOI] [PubMed] [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, Hon L, Sudmant PH, Graves TA, Alkan C, Dennis MY, et al. 2014. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res 24: 688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai HT, Baroni Urbani C, Kubota M, Sharma GP, Narasimhanna MN, Das BC, Sharma AK, Sharma A, Deodikar GB, Vaidya VG, et al. 1984. Karyological survey of Indian ants. Jpn J Genet 59: 1–32. [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB. 2010. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci 11: 188–200. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. 2004. Mobile elements: drivers of genome evolution. Science 303: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Kelber C, Rössler W, Roces F, Kleineidam CJ. 2009. The antennal lobes of fungus-growing ants (Attini): neuroanatomical traits and evolutionary trends. Brain Behav Evol 73: 273–284. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz E, Dugas JC, Ngai J. 2002. Odorant receptor gene regulation: implications from genomic organization. Trends Genet 18: 29–34. [DOI] [PubMed] [Google Scholar]
- Laissue PP, Vosshall LB. 2008. The olfactory sensory map in Drosophila. In Brain development in Drosophila melanogaster (ed. Technau GM), pp. 102–114. Springer-Verlag, New York. [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997v2.
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht R, Oxley PR, Keller L, Kronauer DJC. 2016. Robust DNA methylation in the clonal raider ant brain. Curr Biol 26: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Ventura M, Cellamare A, Chen L, Cheng Z, Zhu B, Li C, Song J, Eichler EE. 2009. Analysis of recent segmental duplications in the bovine genome. BMC Genomics 10: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. 2003. The origin of new genes: glimpses from the young and old. Nat Rev Genet 4: 865–875. [DOI] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD. 2013. New gene evolution: Little did we know. Annu Rev Genet 47: 307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie SK, Oxley PR, Kronauer DJC. 2014. Comparative genomics and transcriptomics in ants provide new insights into the evolution and function of odorant binding and chemosensory proteins. BMC Genomics 15: 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie SK, Fetter-Pruneda I, Ruta V, Kronauer DJC. 2016. Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proc Natl Acad Sci 113: 14091–14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. 2009. Repeat mediated gene duplication in the Drosophila pseudoobscura genome. Gene 438: 1–7. [DOI] [PubMed] [Google Scholar]
- Mendivil Ramos O, Ferrier DEK. 2012. Mechanisms of gene duplication and translocation and progress towards understanding their relative contributions to animal genome evolution. Int J Evol Biol 2012: 846421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. 2005. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene 346: 13–21. [DOI] [PubMed] [Google Scholar]
- Nozawa M, Nei M. 2007. Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc Natl Acad Sci 104: 7122–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxley PR, Ji L, Fetter-Pruneda I, McKenzie SK, Li C, Hu H, Zhang G, Kronauer DJC. 2014. The genome of the clonal raider ant Cerapachys biroi. Curr Biol 24: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask GM, Slone JD, Millar JG, Das P, Moreira JA, Zhou X, Bello J, Berger SL, Bonasio R, Desplan C, et al. 2017. Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nat Commun 8: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Marschall T, Pisanti N, van Iersel L, Stougie L, Klau GW, Schönhuth A. 2014. WhatsHap: haplotype assembly for future-generation sequencing reads. In Research in computational molecular biology. RECOMB 2014 (ed. Sharan R), Lecture Notes in Computer Science, Vol. 8394, pp. 237–249. Springer, Cham, Switzerland. [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci 100: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Gadau J, Wanner KW. 2010. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol 19(Suppl. 1): 121–136. [DOI] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Almeida FC, Rozas J. 2011. Comparative genomics of the major chemosensory gene families in arthropods. In Encyclopedia of life sciences. Wiley, Chichester, UK. [Google Scholar]
- Schrader L, Kim JW, Ence D, Zimin A, Klein A, Wyschetzki K, Weichselgartner T, Kemena C, Stökl J, Schultner E, et al. 2014. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Commun 5: 5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J. 2009. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zöllner S, Church DM, Eichler EE. 2008. Mouse segmental duplication and copy number variation. Nat Genet 40: 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org. [Google Scholar]
- Smith CD, Zimin A, Holt C, Abouheif E, Benton R, Cash E, Croset V, Currie CR, Elhaik E, Elsik CG, et al. 2011a. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci 108: 5673–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Smith CD, Robertson HM, Helmkampf M, Zimin A, Yandell M, Holt C, Hu H, Abouheif E, Benton R, et al. 2011b. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci 108: 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stanke M, Schöffmann O, Morgenstern B, Waack S. 2006. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trible W, Olivos-Cisneros L, McKenzie SK, Saragosti J, Chang N-C, Matthews BJ, Oxley PR, Kronauer DJC. 2017. orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 170: 727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. 1998. Odorant receptors govern the formation of a precise topographic map. Cell 93: 47–60. [DOI] [PubMed] [Google Scholar]
- Wang J, Wurm Y, Nipitwattanaphon M, Riba-Grognuz O, Huang Y-C, Shoemaker D, Keller L. 2013. A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493: 664–668. [DOI] [PubMed] [Google Scholar]
- Yan H, Opachaloemphan C, Mancini G, Yang H, Gallitto M, Mlejnek J, Leibholz A, Haight K, Ghaninia M, Huo L, et al. 2017. An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell 170: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Arguello JR, Li X, Ding Y, Zhou Q, Chen Y, Zhang Y, Zhao R, Brunet F, Peng L, et al. 2008. Repetitive element-mediated recombination as a mechanism for new gene origination in Drosophila. PLoS Genet 4: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu HHS, Chung W, Yang J, Li W-H. 2005. Patterns of segmental duplication in the human genome. Mol Biol Evol 22: 135–141. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhang G, Zhang Y, Xu S, Zhao R, Zhan Z, Li X, Ding Y, Yang S, Wang W. 2008. On the origin of new genes in Drosophila. Genome Res 18: 1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Slone JD, Rokas A, Berger SL, Liebig J, Ray A, Reinberg D, Zwiebel LJ. 2012. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet 8: e1002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Rokas A, Berger SL, Liebig J, Ray A, Zwiebel LJ. 2015. Chemoreceptor evolution in Hymenoptera and its implications for the evolution of eusociality. Genome Biol Evol 7: 2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zube C, Kleineidam CJ, Kirschner S, Neef J, Rössler W. 2008. Organization of the olfactory pathway and odor processing in the antennal lobe of the ant Camponotus floridanus. J Comp Neurol 506: 425–441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.