Abstract
3′ UTRs play important roles in the gene regulation network via their influence on mRNA stability, translational efficiency, and subcellular localization. For a given gene, 3′ UTRs of different lengths generated by alternative polyadenylation (APA) may result in functional differences in regulation. The mechanistic details of how length changes of 3′ UTRs alter gene function remain unclear. By combining APA sequencing and polysome profiling, we observed that mRNA isoforms with shorter 3′ UTRs were bound with more polysomes in six cell lines but not in NIH3T3 cells, suggesting that changing 3′ UTRs to shorter isoforms may lead to a higher gene translational efficiency. By interfering with the expression of TNRC6A and analyzing AGO2-PAR-CLIP data, we revealed that the APA effect on translational efficiency was mainly regulated by miRNAs, and this regulation was cell cycle dependent. The discrepancy between NIH3T3 and other cell lines was due to contact inhibition of NIH3T3. Thus, the crosstalk between APA and miRNAs may be needed for the regulation of protein translational efficiency.
The flow of genetic information from the genome to the proteome is mediated by the mRNA transcriptome, which is highly regulated at specific biological stages during the life cycle. Precise regulation of gene expression networks includes not only the regulation of mRNA quantity but also of mRNA isoforms by alternative initiation, alternative splicing, and alternative polyadenylation (APA). APA is a wide-spread phenomenon in the transcription of eukaryotic genes and produces different mRNA isoforms as well as increases the complexity of gene regulation networks. APA can be classified into two types (Di Giammartino et al. 2011): (1) CR-APA, which can change coding region sequences and then generate different protein isoforms; and (2) UTR-APA, which is usually localized in the last exon and can produce mRNA isoforms with different 3′ UTR lengths.
3′ UTR length changes due to UTR-APA are associated with cell proliferation and transformation as well as specific biological functions. Global shortening of 3′ UTRs has been observed in tumor cells (Mayr and Bartel 2009; Fu et al. 2011; Lin et al. 2012; Xia et al. 2014) and activated T cells (Sandberg et al. 2008; Gruber et al. 2014). Zebrafish embryo development shows a “V”-shape trend of global 3′ UTR length changes (Li et al. 2012), and mouse embryo development displays a trend of a progressive lengthening of 3′ UTRs (Ji et al. 2009). Significantly longer and shorter 3′ UTRs were observed in the fruit fly brain and testis, respectively (Smibert et al. 2012), and lengthening of 3′ UTRs was also observed in the human brain (Derti et al. 2012). Furthermore, shortening of 3′ UTRs due to APA switching was also found in response to stimuli in neurons (Flavell et al. 2008) and to viral infections in macrophages (Jia et al. 2017). Although APA was found to be associated with many biological processes, the mechanistic roles that APA plays in these biological processes are still not well understood.
Bound by RNA-binding proteins and miRNAs, 3′ UTRs exert important roles in mRNA stability, translational efficiency, and subcellular localization. It is known that the initiation of translation requires the communication between the 5′ cap and 3′ poly(A) tail of mRNA (Wells et al. 1998). miRNAs can interact with the mRNA loop and suppress protein translation by binding to the 3′ UTR. In this process, TNRC6A (also known as GW182) protein plays a central role as a hub that bridges the interaction between AGO proteins and several downstream factors, such as PABPC1 and CNOT1 (Iwakawa and Tomari 2015). RNA-binding proteins, for example, ELAVL1 (Poria et al. 2016) and CPEB1 (Stebbins-Boaz et al. 1999), can also contribute to translational regulation by competing with miRNAs for binding to the 3′ UTR. UTR-APA is assumed to affect the translational efficiency via gain or loss of cis-factors on 3′ UTRs.
Several studies using luciferase reporter assays of several genes have shown that mRNA isoforms with proximal APA sites yielded higher protein production than those with distal APA sites (Sandberg et al. 2008; Mayr and Bartel 2009). Using APA sequencing and quantitative mass spectrometry, Gruber et al. (2014) found that 3′ UTR shortening was not accompanied by a corresponding change in protein levels during the activation of T cells. However, this result does not indicate a limited effect of APA on translational efficiency since both the mRNA level and translational efficiency can contribute to the protein level, and their effects could not be separated in that study. Polysome profiling, which fractionates mRNAs based on the number of bound ribosomes, is normally used to monitor the translational activity of mRNA in cells. A combination of polysome profiling and APA sequencing techniques provides an adequate way to evaluate the effect of APA on the mRNA translational efficiency by comparing the distribution of mRNA isoforms among polysome fractions. Using polysome profiling and APA sequencing, Spies et al. (2013) found that isoforms with longer 3′ UTRs had a slightly increased translational efficiency in the mouse NIH3T3 cell line. This contradictory result suggests an urgent need to further study the relationship between APA and translational efficiency as well as its mechanism.
Results
Shortened 3′ UTRs in the polysome-bound fraction
With our newly developed method of polysome profiling and APA sequencing (IVT-SAPAS) (Fu et al. 2015), we explored the effect of APA on translational efficiency in five human cell lines and two mouse cell lines. We performed polysome profiling according to the previously reported protocol (Gandin et al. 2014): (1) Immobilize ribosomes on mRNA with CHX + media; (2) fractionate RNAs with sucrose gradient ultracentrifugation; and (3) extract total RNAs from each fraction. Then, we mixed the polysome-bound and -unbound fractions and performed APA sequencing with the IVT-SAPAS method (Supplemental Fig. S1). Three biological replicates for each cell line, except MCF10A, were conducted in the experiments. On average, we obtained approximately 8.3 million reads that were uniquely mapped to the genome after filtering internal priming for each sample. A summary of the sequencing data is shown in Supplemental Table S1.
To investigate whether 3′ UTR length changes attributable to APA can affect translational efficiency, we first standardized the 3′ UTR length by designating the longest 3′ UTR as 1.0 and then calculated the weighted average of the 3′ UTR length for each gene in each sample. A notched box plot across genes (Fig. 1) shows that the 3′ UTRs in the polysome-bound fraction are shorter than those in the polysome-unbound fraction for each replicate of each cell line, except the mouse NIH3T3 cell line. To test whether this result is statistically significant, we fit the data to a mixed linear model with genes and replication as random effects. The ANOVA F-test reveals that the shortening of 3′ UTRs in the polysome-bound fraction in human MCF-7, MCF10A, NCM460, SW480, and RKO cells and mouse L929 cells is statistically significant, with P < 2.2 × 10−16, suggesting that the mRNA isoforms with shorter 3′ UTRs may have a higher translational efficiency. However, for the mouse NIH3T3 cell line, the 3′ UTRs in the polysome-bound fraction are longer, with weak significance (P = 0.01014). This result is consistent with a previous report that showed that mRNA isoforms with longer 3′ UTRs exhibited a slightly greater translational efficiency in the mouse NIH3T3 cell line (Spies et al. 2013).
Figure 1.
Notched box plot of the weighted mean of the 3′ UTR length shows the translational efficiency difference of mRNAs with different 3′ UTR lengths. For each gene with UTR-APA, the length of each 3′ UTR isoform was normalized to the longest 3′ UTR, and the weighted mean of 3′ UTR length was calculated. P-values were obtained with the linear mixed model and F-test.
To find genes that have different 3′ UTR lengths in the polysome-bound and -unbound fractions in each cell line, we combined replicate data of each cell line and performed a test of linear trend alternative to independence (Agresti 2002). In the mouse NIH3T3 cell line, there are 86 and 92 genes with significantly shorter and longer 3′ UTRs, respectively, in the polysome-bound fraction than the polysome-unbound fraction (Fig. 2). However, the other six cell lines have considerably more genes with shorter 3′ UTRs in the polysome-bound fraction.
Figure 2.

Numbers of genes with significant 3′ UTR length differences between fractions with and without bound polysomes. The data for different biological replicates for each cell line were combined. A test of linear trend alternative to independence was performed, and FDR < 0.01 and |r| > 0.1 corresponds to a significant threshold.
Taken together, the preceding results suggest that mRNA isoforms with shorter 3′ UTRs may have a higher translational efficiency than those with longer 3′ UTRs in the tested cell lines, except for the mouse NIH3T3 cell line.
To validate these results, we first performed polysome profiling and qRT-PCR. Twelve genes in MCF-7 cells and five in MCF10A cells with shorter 3′ UTRs in the polysome-bound fraction were chosen (Supplemental Fig. S2). We calculated the proportion of mRNA for each fraction using the primers located in the common and extended regions. For all genes, the mRNAs with shorter 3′ UTRs shifted to the heavier fraction (Fig. 3A). Next, we performed a dual-luciferase experiment to directly measure the differences in the protein/mRNA ratios between longer and shorter 3′ UTRs (Fig. 3B). Five genes in MCF-7 cells with shorter 3′ UTRs and six genes in NIH3T3 cells with longer 3′ UTRs in the polysome-bound fraction were chosen. We fused the 3′ UTRs of the tested genes downstream from the Renilla luciferase open reading frame. After normalization to the firefly luciferase on the same plasmid, we measured the translational efficiency as the protein/mRNA ratio. Three genes in MCF-7 cells and four genes in NIH3T3 cells showed significantly higher and lower translational efficiencies, respectively, with shorter 3′ UTRs (Fig. 3C). The other two genes in NIH3T3 also showed a lower translational efficiency with shorter 3′ UTRs, although they were not statistically significant.
Figure 3.
Validation of the translation efficiency difference between longer and shorter 3′ UTRs. (A) Heatmap of the proportion of mRNAs in polysome fractions measured by qRT-PCR. For each gene, two pairs of primers located in the common (C) and extended (E) regions of the 3′ UTR were designed to measure the expression levels of total and longer mRNA, respectively. (B) Scheme of the dual-luciferase reporter assay using the psiCHECK-2 Vector. Longer and shorter 3′ UTRs were cloned downstream from the Renilla ORF. Blue lines represent the inserted 3′ UTR. (C) Quantification of the Renilla protein/mRNA ratio (mean ± SE). The Renilla fluorescence and mRNA level were normalized to firefly fluorescence and the mRNA level, respectively. Student's t-test was performed to test the difference between longer and shorter 3′ UTRs.
miRNAs contribute to the translational efficiency difference
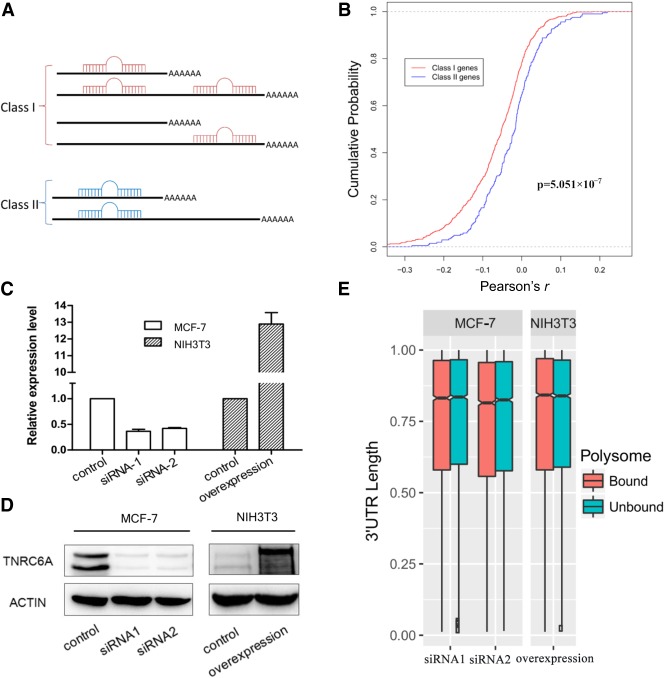
miRNAs play important roles in the post-transcriptional gene regulation network. miRNAs mediate mRNA decay and translational repression in animals by recognizing the 3′ UTRs of target mRNA (Fabian et al. 2010; Iwakawa and Tomari 2015). Therefore, mRNAs with shorter 3′ UTRs may escape the translational repression effect of miRNAs. To test whether miRNAs contribute to the aforementioned finding that mRNAs with shorter 3′ UTRs have a higher translational efficiency, we must simultaneously consider both the position of the miRNA binding site on the 3′ UTR and miRNA expression in the cells. The AGO2-PAR-CLIP method provides an appropriate way to identify the target sites of endogenous miRNAs in a genome. We downloaded the AGO2-PAR-CLIP data of the MCF-7 cell line (Farazi et al. 2014), which provided 3597 canonical miRNA target interactions with 2863 targets. We mapped them to our data and found that 1070 binding sites mapped to the 3′ UTRs of 771 genes with UTR-APA. To check the contribution of miRNAs to the translational differences of mRNA isoforms, we classified these genes into two classes (Fig. 4A) based on the position of the miRNA target and APA sites: (1) a 3′ UTR extended region with miRNA binding sites (558 genes); and (2) a common but not extended region with miRNA binding sites (213 genes). If miRNAs can generally repress translation, we would expect that mRNAs with shorter 3′ UTRs should have a higher translational efficiency in Class I genes compared to Class II genes. A negative Pearson correlation coefficient (r) of the 3′ UTR length and polysome fraction (calculated in the test of linear trend alternative to independence) means shorter 3′ UTRs in the polysome-unbound fraction. We plotted the empirical cumulative distribution of the r-value for the genes in each class (Fig. 4B). The Mann-Whitney U test shows that APA in Class I genes has significantly greater effects on polysome binding than those in Class II genes (Mann-Whitney U test, P = 5.051 × 10−7). Therefore, the higher translational efficiency of mRNA isoforms with shorter 3′ UTRs may benefit from escaping the repression by miRNAs.
Figure 4.
miRNAs contribute to the translational efficiency difference between longer and shorter 3′ UTRs. (A) Genes were classified into two classes based on the position of miRNA targets and APA sites. (B) Cumulative distribution of Pearson's r in the test of linear trend alternative to independence. Statistically significantly smaller r-values were observed in Class I genes. (C,D) qRT-PCR and Western blot analysis of TNRC6A in the MCF-7 (knockdown) and NIH3T3 (overexpression) cell lines, respectively. (E) Notched box plot of the weighted mean of the 3′ UTR length for cells with the TNRC6A knockdown and overexpression.
Then, to what extent do miRNAs contribute to the higher translational efficiency of mRNA isoforms with shorter 3′ UTRs? It is well-known that the TNRC6A protein plays a key role in translation repression by miRNAs, and disruption of the TNRC6A-AGO interaction can abrogate its repression. Here, we first interfered with the expression of TNRC6A in the MCF-7 cell line using two different siRNAs (Fig. 4C,D) and then performed APA sequencing of the polysome profiling fractions (Supplemental Table S1). The results showed that the 3′ UTR length difference between the two fractions almost disappeared in cells with the TNRC6A knockdown (Fig. 4E) compared to that of control cells (Fig. 1). To detect the statistical significance of the TNRC6A knockdown, we fit the 3′ UTR length data to a linear mixed model. The ANOVA F-test showed that the interaction term of the polysome fraction and TNRC6A knockdown was significant with P < 2.2 × 10−16, suggesting that the difference of the 3′ UTR length between polysome-bound and -unbound fractions in knockdown cells was significantly smaller than that in control cells, which indicates that miRNAs play a very important role in APA regulation of translational efficiency.
We validated the result of TNRC6A knockdown with dual-luciferase experiment for five genes in the MCF-7 cell line. We measured the protein/mRNA ratios for longer and shorter 3′ UTRs, respectively, and then calculated the translational efficiency ratios between the shorter and longer 3′ UTRs. Four genes showed a significantly lower ratio with TNRC6A knockdown than the control (P ≤ 0.001), and another gene also showed the same trend although without statistical significance (P = 0.195) with bootstrap test (Supplemental Fig. S3).
We also overexpressed TNRC6A in NIH3T3 cells (Fig. 4C,D) but only observed a very slight shortening of the 3′ UTR in the polysome-bound fraction compared to the polysome-unbound fraction (Fig. 4E). Thus, determining what makes NIH3T3 cell different from other cell lines in the APA regulation of translational efficiency requires further analysis.
Cell cycle is related to the APA regulation of translational efficiency
It is known that the NIH3T3 cell line has a high degree of contact inhibition in continued culture, which leads it to enter the G0 and G1 phases (Küppers et al. 2010). We cultured the NIH3T3, MCF-7, and L929 cells to ∼70% confluency and analyzed the cell cycle phases with a flow cytometer. A higher proportion of G0/G1 cells for NIH3T3 were observed than for the other two cell lines (Supplemental Fig. S4). To investigate whether the cell cycle phase is related to translational regulation by APA, we collected cells (mainly at G0/G1) by culturing them to 100% confluency (for NIH3T3) or serum starvation (for L929) and cells (mainly at G2/M) with a double thymidine block and release (Fig. 5A; Supplemental Fig. S5). Both the NIH3T3 and L929 cell lines showed lower expression of TNRC6A at the G0/G1 phase than at the G2/M phase (Fig. 5B). Consistent with the up-regulation of TNRC6A, not only L929 but also NIH3T3 cells at the G2/M phase had shorter 3′ UTRs for mRNAs in polysome-bound fraction. An example of the Arf6 gene in L929 was shown in Supplemental Figure S6. The ANOVA F-test, by fitting to a linear mixed model, revealed that the differences in the 3′ UTR length between the polysome-bound and unbound fractions in the G2/M phase were significantly greater than those in the G0/G1 phase (Fig. 5C). Furthermore, we also cultured the NIH3T3 cells to approximately 50% confluency and performed polysome profiling and APA sequencing. The result showed a significantly shorter 3′ UTR for mRNAs in the polysome-bound fraction (P = 2.316 × 10−14 with paired t-test) (Supplemental Fig. S7). We also isolated human naïve T cells in the quiescent G0 phase (Supplemental Fig. S8). Similar to NIH3T3 cells, naïve T cells also had longer 3′ UTRs in the polysome-bound fraction (Fig. 5C). This result is also consistent with the lower expression of TNRC6A in naïve T cells than that in activated T cells (Yang et al. 2004).
Figure 5.
Cell cycle and translational efficiency. (A) The percentage of cells in a cell cycle phase of the NIH3T3 cell line after cell synchronization with a double thymidine block and release, and contact inhibition. (B) Western blot analysis shows higher expression of TNRC6A in the G2/M phase in the NIH3T3 and L929 cell lines. (C) Notched box plot of the weighted mean of the 3′ UTR length for NIH3T3 and L929 cells obtained by synchronization, and naïve T cells. Cells at the G2/M but not G0/G1 phase show a higher translational efficiency of mRNA with a shorter 3′ UTR.
The preceding results suggest that the different translational efficiencies of mRNA isoforms due to APA are regulated by miRNAs, and this regulation is mediated by TNRC6A expression, which is cell cycle dependent.
Discussion
Recently, genome-wide 3′ UTR length changes due to APA have been found in many biological processes (Elkon et al. 2013), indicating that APA may play an important role in the gene regulation network. Given the canonical functions of the 3′ UTR in mRNA stability, translational efficiency and subcellular localization, it is assumed that APA may affect these functions through a loss or gain of cis-factors in the extended region of the 3′ UTR. Here, through polysome profiling and APA sequencing, we investigated the effects of APA on the translational efficiency in five human and two mouse cell lines. We found a trend of shorter 3′ UTRs in the polysome-bound fraction in six cell lines, except the mouse NIH3T3 line; this result, to some extent, shows that mRNAs with shorter 3′ UTRs have a higher translational efficiency in these cell lines.
In principle, translational efficiency is controlled by both translation initiation and the elongation rate, and these two factors have opposite effects on the number of polysomes on mRNA. However, it has been found that translation is predominantly controlled by translation initiation, with little contribution from the elongation rate (Hoffman et al. 2016), so polysome profiling is still a suitable tool to study the genome-wide translational efficiency. Furthermore, compared to the recently developed ribosome profiling, polysome profiling does not fragment mRNAs, so it is suitable to measure the differences in translational efficiency between mRNA isoforms (Sterne-Weiler et al. 2013). Polysome profiling has been successfully used to study the difference in translational efficiency between alternatively spliced mRNAs (Sterne-Weiler et al. 2013) and APA (Spies et al. 2013). Spies et al. (2013) found a weak but significant bias toward a higher translational efficiency for longer 3′ UTRs in the mouse NIH3T3 cell line using polysome profiling. We also consistently observed this trend for the NIH3T3 cell line in our experiment. This reproducibility also suggests that our finding of a higher translational efficiency with shorter 3′ UTRs in six other cell lines is reliable and informative.
miRNAs can repress translation at the initiation step (Fabian et al. 2010; Huntzinger and Izaurralde 2011; Iwakawa and Tomari 2015), and the extended region of the 3′ UTR contains a larger number of miRNA binding sites than the common region (Sandberg et al. 2008). Thus, we hypothesized that miRNAs may mediate the translational efficiency differences in mRNA isoforms with different 3′ UTR lengths. Combining the AGO2-PAR-CLIP data (Farazi et al. 2014) and our sequencing data of the MCF-7 cell line (Fig. 4), we confirmed that the higher translational efficiency of mRNA isoforms with shorter 3′ UTRs can benefit from escaping the repression effect of miRNAs.
TNRC6A can bind the AGO protein of the miRISC and the other effectors of translational repression and thus mediate the translational repression by miRNAs. To further investigate the contribution of miRNAs to the APA regulation of translational efficiency, we performed knockdown of TNRC6A with siRNAs. This knockdown almost abrogated the difference in the translational efficiency of mRNAs due to APA, suggesting that the difference in translational efficiency due to APA is mainly, if not completely, dominated by miRNAs.
However, upon overexpression of TNRC6A, NIH3T3 still did not display a significantly shorter 3′ UTR in the polysome-bound fraction as did the other cell lines. AGO, TNRC6A, and mRNAs repressed by miRNAs are all enriched in GW or P bodies, and a correlation between miRNA-mediated translational repression and GW bodies was observed (Filipowicz et al. 2008; Fabian et al. 2010). Furthermore, GW bodies are larger and more abundant in the late S and G2 phases but smaller and fewer in the G0 and G1 phases (Yang et al. 2004). NIH3T3 is a mouse embryonic fibroblast cell line with a high degree of contact inhibition in continued culture (Küppers et al. 2010), and a high proportion of NIH3T3 cells are arrested at the G0/G1 phase, which may explain the difference of the NIH3T3 line from the other cell lines regarding the regulation of translational efficiency by APA. Furthermore, we observed shorter 3′ UTRs in the polysome-bound fractions for both the NIH3T3 and L293 cells at the G2/M phase but not the G0/G1 phase, which was consistent with the higher expression of TNRC6A in cells at the G2/M phase than that at the G0/G1 phase. It is well-known that naïve T cells are at quiescent G0 phase. Similar to NIH3T3 cells, naïve T cells also showed slightly longer 3′ UTRs in the polysome-bound faction. Furthermore, the Steitz group showed that although miRNAs repress translation in proliferating cells, they can activate translation in quiescent cells arrested at the G0/G1 phase (Vasudevan and Steitz 2007; Vasudevan et al. 2007, 2008; Truesdell et al. 2012). This result suggests that the difference in translational efficiency between mRNA isoforms with different lengths of 3′ UTRs attributable to APA is regulated by miRNAs and the regulation is cell cycle dependent.
In short, we found a higher translational efficiency of mRNAs with shorter 3′ UTRs in proliferating cells, but not quiescent cells. These results are consistent with the fact that proliferating cells usually have shorter 3′ UTRs than quiescent cells. Our mechanistic study here further indicates a crosstalk between APA and miRNA.
Methods
Cell cultures
A breast cancer cell line, MCF-7, was cultured in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% fetal bovine serum (FBS), and 10 µg/mL human recombinant insulin; a normal human mammary epithelial cell line, MCF10A, was cultured in DMEM supplemented with 5% horse serum, 20 ng/mL EGF, 0.5 µg/mL hydrocortisone, 0.1 µg/mL cholera toxin, and 10 µg/mL human recombinant insulin. A normal human colon mucosal epithelial cell line, NCM460, and two colon cancer cell lines, SW480 and RKO, were maintained in RPMI 1640 supplemented with 10% FBS. A murine embryonic fibroblast cell line, NIH3T3, and another fibroblast cell line, L929, were cultured in DMEM formulated with 10% FBS. All media were supplemented with 100 units/mL penicillin and 100 µg/mL streptomycin.
Isolation of naïve T cells and flow cytometer analysis
Human peripheral blood mononuclear cells (PBMCs) were purchased from LDEBIO. CD4+ naïve T cells were isolated from PBMCs using the naïve CD4+ T Cell Isolation Kit II (Miltenyi Biotec, 130-094-131) according to the manufacturer's recommendations. Purified CD4+ naïve T cells were stained by APC Conjugated anti-CD45RA (BD Biosciences, 561884) and FITC Conjugated anti-CD4 antibodies (BD Biosciences, 550628) to analyze the proportion of double-positive cells with a FACSCalibur flow cytometer (BD Biosciences).
Cell synchronization and cell cycle analysis
NIH3T3 and L929 G2/M phase cells were obtained by a double thymidine block and release. In brief, cells at ∼30% confluency were cultured in a medium that contained 2 mM thymidine (Sigma) for 19 h, washed three times in PBS, and replaced with complete DMEM. After 8 h, the complete culture medium was replaced with 2 mM thymidine DMEM for 19 h. Cells were then washed with PBS and released after 9 h for NIH3T3 cells and 8 h for L929 cells to perform polysome profiling. L929 G1/G0 cells were obtained by serum starvation. In short, L929 cells were cultured in DMEM containing 0.1% FBS for 3 d for cell cycle analysis and polysome profiling. Cell cycle analysis was performed on cells fixed in 75% ethanol prior to ribonuclease digestion and staining with 40 mg/mL propidium iodide (Sigma). NIH3T3 cells at the G0 phase were acquired by continuous contact inhibition for 48 h after reaching a 100% confluent monolayer density. To measure the proportion of cells in the G0 phase, cells were stained with a FITC Conjugated Ki-67 Monoclonal Antibody (eBioscience, 11-5698-82) and FITC Conjugated Rat IgG2a kappa Isotype Control (eBioscience, 11-4321-81) and then analyzed with a FACSCalibur flow cytometer (BD Biosciences) according to the manufacturer's recommendations.
Western blot analysis
Cells were washed three times with PBS and lysed in RIPA lysis buffer for 30 min at 4°C. The total protein concentration was measured by the Micro BCA Protein Assay Kit (Thermo Fisher Scientific), and proteins were separated by SDS-PAGE. A wet transfer system (Tanon) was applied to transfer proteins to nitrocellulose membranes for 16 h at 4°C. The membrane was blocked in 5% nonfat milk and incubated with a rabbit anti-TNRC6A antibody (Novus Biologicals, NBP1-28751) for 2 h at room temperature. Then, the membrane was washed with TBS-T and incubated with the corresponding secondary anti-rabbit HRP-antibody (Cell Signaling Technology, 7074S) for 1 h. Protein signals were detected by Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) after TBS-T washing.
Polysome profiling
Polysome fractionation was performed based on the previously reported protocol (Gandin et al. 2014). Briefly, cells were treated for 5 min with cycloheximide at a final concentration of 100 µg/mL and then lysed with hypotonic buffer (5 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 100 µg/mL CHX, 2 mM DTT, 0.5% Triton X-100, 0.5% sodium deoxycholate, 1× protease inhibitor cocktail [EDTA-free]and 2 units/µL RNAse inhibitor). Cytosolic extracts were ultracentrifuged with a 5%–50% sucrose gradient buffer (200 mM HEPES, pH 7.6, 1 M KCl, 50 mM MgCl2, 100 µg/mL cycloheximide, 1× protease inhibitor cocktail [EDTA-free], 100 units/mL RNase inhibitor) prepared using the Gradient Station (Biocomp). Fractions were collected with the Gradient Station. We then merged them into polysome-bound and -unbound fractions based on the OD254 values. Total RNA for each merged fraction was extracted from the cells using a QIAGEN RNeasy Mini kit and maintained at equal volumes of RNase-free water. The quality of the samples was checked with agarose gel electrophoresis and the OD260/280 ratios.
APA sequencing and analysis
APA sequencing was performed with the IVT-SAPAS method as described previously (Fu et al. 2015). Sequencing data were analyzed with our in-house pipeline (Fu et al. 2011, 2015). Sequencing reads were mapped to the human hg19 and mouse mm9 reference genomes, and internal priming was filtered. Next, poly(A) sites were defined for each sample by clustering the unique mapped reads, and then, the poly(A) sites were merged together across samples. We realigned the sequencing reads of MCF-7 and L929 to the GRCh38 (hg38) and GRCm38 (mm10), respectively, and observed similar mapping rates and 3′ UTR difference changes, suggesting that the version of genome assembly has little influence on the conclusions.
To reduce the variance of the 3′ UTR length across genes, we standardized the length by designating the longest 3′ UTR as 1.0 and calculated the weighted mean of the 3′ UTR length with multiple APA sites for each gene. The box plots of the 3′ UTR length were drawn with the ggplot2 package (ggplot2.org) in R software v3.3.3 (R Core Team 2017). To test whether there was an overall 3′ UTR length difference between fractions with and without polysome bound for each cell line, we fitted the data to a mixed linear model as follows:
where yijk is the 3′ UTR length for fraction i (i = 0: polysome unbound; i = 1: polysome bound), replicate j (j = 1, 2, 3) and gene k (k = 1…#); µ represents an overall mean value; P is the fixed effect of polysome fraction; R is the random effect due to experiment replication; G is the random effect due to the gene; and ε is the random error. The lme4 package (https://CRAN.R-project.org/package=lme4) in R (R Core Team 2017) was used to fit the model; then, the ANOVA F-test was performed to test the fraction effect. To test the effect of TNRC6A on translational efficiency due to APA, we fit the data into a linear mixed model as follows:
where El is the treatment (l = 0: control, l = 1: knockdown or overexpression) and the interaction effect represents the effect of TNRC6A knockdown or overexpression on the APA-regulated translational efficiency. The linear mixed model was also used to test the difference between cell cycles with the cell cycle phase as the treatment.
A test of the linear trend alternative to independence (Agresti 2002) was used to detect the tandem 3′ UTR length difference between samples (Fu et al. 2011, 2015). We denoted the fraction without polysomes as 0 and the fraction with polysomes as 1, and a negative Pearson correlation coefficient (r) indicates that the fraction with polysomes harbors shorter 3′ UTRs than the fraction without polysomes, and vice versa.
Validation experiments
Two pairs of primers located in the common and extended regions were designed for 17 genes (Supplemental Table S2). An equal volume of RNAs for each fraction was used to perform qRT-PCR. For each pair of primers, we calculated the fold-change of each fraction compared to the first fraction and subsequently calculated the proportion of each fraction for each pair of primers.
A dual-luciferase reporter assay was used to validate the translation difference of the 3′ UTRs. Longer and shorter 3′ UTRs were cloned downstream from the Renilla ORF of psiCHECK-2 Vector (Promega). The luciferase activity was measured by the GloMax Discover System (Promega) according to the manufacturer's recommendations, and the mRNA levels were measured by qRT-PCR. Six replicates were performed for each 3′ UTR isoform. The Renilla fluorescence and mRNA levels were normalized to the firefly fluorescence and mRNA levels, respectively, for each 3′ UTR isoform; then the protein/mRNA ratios were calculated, and the one-tailed Student's t-test was performed. The primers are shown in Supplemental Table S3.
AGO-PAR-CLIP data analysis
We downloaded the AGO2-PAR-CLIP data of the MCF-7 cell line (Supplemental Table S3 in Farazi et al. 2014) and mapped the target sequences to the 3′ UTR. Then, based on the position of the target and APA sites, we classified the genes with UTR-APA into two classes (Fig. 4A). The Pearson correlation coefficient (r) between the 3′ UTR length and polysome fraction in the test of linear trend alternative to independence measured the translational efficiency difference between the fractions with and without polysomes bound. The Mann-Whitney U test was performed to compare the difference in r-values between the two classes of genes.
Knockdown and overexpression of TNRC6A
Human TNRC6A siRNAs were synthesized (RiboBio) (Supplemental Table S4). The siRNAs were transfected into MCF-7 cells at a final concentration of 50 nM using Lipofectamine RNAiMAX Reagent (Invitrogen). Total RNA extracted from L929 cells using TRIzol Reagent (Life Technologies) was used for cDNA synthesis by SuperScript III Reverse Transcriptase (Invitrogen) and an oligo(dT) primer. The cDNA thus obtained was used as a template to amplify the mouse full-length Tnrc6a gene (NM_144925), which was cloned into the pcDNA3.1-FLAG (Invitrogen) expression vector. The plasmids were transfected using Lipofectamine 3000 (Invitrogen) following the manufacturer's instructions.
Data access
All raw sequence data from this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRP121057.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (No. 2017YFC1308800 to Y. Fu), the National Natural Science Foundation of China (No. 91231206 and No. 81430099 to A.X. and No. 91331113 to Y. Fu), the National Basic Research Program of China (No. 2013CB917801 to A.X.), Guangzhou Municipal Science and Technology Bureau (No. 201607020037 to S.C.), and the Fundamental Research Funds for the Central Universities (to Y. Fu.).
Author contributions: Y. Fu designed the experiment and analysis, analyzed the data, and wrote the manuscript. L.C. performed the experiments and analyzed the data. C.C., Y.G., and J.L. performed the polysome profiling experiments. M.K. analyzed the AGO-PAR-CLIP data. Y. Feng and Z.S. analyzed the sequencing data. M.H., Z.H., and G.H. performed the IVT-SAPAS experiments. S.C. analyzed the data and edited the manuscript. A.X. conceived the study, analyzed the data, and edited and finally approved the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231506.117.
References
- Agresti A. 2002. Categorical data analysis. Wiley-Interscience, New York. [Google Scholar]
- Derti A, Garrett-Engele P, MacIsaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Ten Hoeve JJ, Brown M, Mihailovic A, Horlings HM, van de Vijver MJ, Tuschl T, Wessels LF. 2014. Identification of distinct miRNA target regulation between breast cancer molecular subtypes using AGO2-PAR-CLIP and patient datasets. Genome Biol 15: R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Ge Y, Sun Y, Liang J, Wan L, Wu X, Xu A. 2015. IVT-SAPAS: low-input and rapid method for sequencing alternative polyadenylation sites. PLoS One 10: e0145477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Sikstrom K, Alain T, Morita M, McLaughlan S, Larsson O, Topisirovic I. 2014. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp 10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Müller P, Schmidt A, Gruber AJ, Gumienny R, Mittal N, Jayachandran R, Pieters J, Keller W, et al. 2014. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun 5: 5465. [DOI] [PubMed] [Google Scholar]
- Hoffman Y, Bublik DR, Ugalde AP, Elkon R, Biniashvili T, Agami R, Oren M, Pilpel Y. 2016. 3′UTR shortening potentiates microRNA-based repression of pro-differentiation genes in proliferating human cells. PLoS Genet 12: e1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110. [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. 2015. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 25: 651–665. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Yuan S, Wang Y, Fu Y, Ge Y, Ge Y, Lan X, Feng Y, Qiu F, Li P, et al. 2017. The role of alternative polyadenylation in the antiviral innate immune response. Nat Commun 8: 14605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers M, Ittrich C, Faust D, Dietrich C. 2010. The transcriptional programme of contact-inhibition. J Cell Biochem 110: 1234–1243. [DOI] [PubMed] [Google Scholar]
- Li Y, Sun Y, Fu Y, Li M, Huang G, Zhang C, Liang J, Huang S, Shen G, Yuan S, et al. 2012. Dynamic landscape of tandem 3′ UTRs during zebrafish development. Genome Res 22: 1899–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, et al. 2012. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res 40: 8460–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poria DK, Guha A, Nandi I, Ray PS. 2016. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 35: 1703–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Burge CB, Bartel DP. 2013. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res 23: 2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell 4: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Sterne-Weiler T, Martinez-Nunez RT, Howard JM, Cvitovik I, Katzman S, Tariq MA, Pourmand N, Sanford JR. 2013. Frac-seq reveals isoform-specific recruitment to polyribosomes. Genome Res 23: 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. 2012. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep 2: 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. 2008. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 7: 1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 5: 5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EKL. 2004. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci 117: 5567–5578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.