Abstract
The binding of PRDM9 to chromatin is a key step in the induction of DNA double-strand breaks associated with meiotic recombination hotspots; it is normally expressed solely in germ cells. We interrogated 1879 cancer samples in 39 different cancer types and found that PRDM9 is unexpectedly expressed in 20% of these tumors even after stringent gene homology correction. The expression levels of PRDM9 in tumors are significantly higher than those found in healthy neighboring tissues and in healthy nongerm tissue databases. Recurrently mutated regions located within 5 Mb of the PRDM9 loci, as well as differentially expressed genes in meiotic pathways, correlate with PRDM9 expression. In samples with aberrant PRDM9 expression, structural variant breakpoints frequently neighbor the DNA motif recognized by PRDM9, and there is an enrichment of structural variants at sites of known meiotic PRDM9 activity. This study is the first to provide evidence of an association between aberrant expression of the meiosis-specific gene PRDM9 with genomic instability in cancer.
Proper chromatid segregation and genome stability are dependent on recombination between homologous chromosomes during meiosis (Baker et al. 1976; Purandare and Patel 1997; Baudat and de Massy 2007; Alves et al. 2017). Meiotic recombination events do not occur across most of the human genome but cluster in 1- to 2-kb-wide hotspots (Pratto et al. 2014) whose locations are associated with activity of the protein PR/SET domain 9 (PRDM9) (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). The functional domains of PRDM9 include a Krüppel-associated box (KRAB)–related domain, which facilitates protein–protein interactions; a PR/SET domain, which provides the protein with H3K4me3 and H3K36me3 activity (Blazer et al. 2016; Powers et al. 2016); and a DNA-binding zinc finger (ZnF) array domain. The SET domain places epigenetic marks at sites bearing specific motifs recognized by the ZnF domain, leading to the recruitment of the DNA double-strand break (DSB) and meiotic recombination machinery (Smagulova et al. 2011; Brick et al. 2012; Pratto et al. 2014). PRDM9 expression and associated epigenetic marks disappear during the pachytene stage of meiosis, leading to the inference that PRDM9's biological function is exclusive to gametogenic tissue (Sun et al. 2015). By directing sites of DSBs, PRDM9 contributes to the tight regulation of meiotic recombination processes, which ensures the stability of homologous chromosomes and proper chromosomal disjunction (Baker et al. 1976; Purandare and Patel 1997; Baudat and de Massy 2007; Alves et al. 2017). While PRDM9 expression has previously been observed in cancer cell lines, as well as in five ovarian carcinomas and a lung adenocarcinoma (Feichtinger et al. 2012), a broad characterization of its expression and implications on the transcriptomic and genomic landscape across cancer types is still lacking.
In healthy nongerm cells, genome integrity is dependent on recombination: The homologous recombination repair pathway allows for the conservative repair of DSBs in DNA caused by endogenous or exogenous damage (Andersen and Sekelsky 2010; Symington et al. 2014; Lisby and Rothstein 2016). When DSBs are not immediately repaired, they can result in structural aberrations such as translocations, inversions, and deletions through nonspecific repair mechanisms (Jackson 2002). A high rate of somatic structural variants (SVs) is characteristic of genomic instability, one of the hallmarks of cancer (Hanahan and Weinberg 2011). DSB repair (DSBR) deficiency underlies a mutational signature that has been associated with germline and somatic loss-of-function (LOF) mutations (Alexandrov et al. 2013) in genes involved in the homologous recombination repair pathway (Moynahan and Jasin 2010; Krejci et al. 2012; Lord and Ashworth 2016). However, some cancers exhibit the mutational signatures of DSBR deficiencies despite lacking detectable LOF mutations affecting known regulators in the homologous recombination repair pathway (Connor et al. 2017), indicating the importance of understanding underlying causes of genomic instability.
Here, we describe aberrant expression of PRDM9 among human tumors in vivo in 32 different cancer types (from n = 1879 patients) from the Pan-Cancer Analysis of Whole Genomes Project (PCAWG) and The Cancer Genome Atlas (TCGA) and assess the impact of this aberrant expression on the genomic and transcriptomic landscape among tumors.
Results
Aberrant PRDM9 expression is common in human cancers
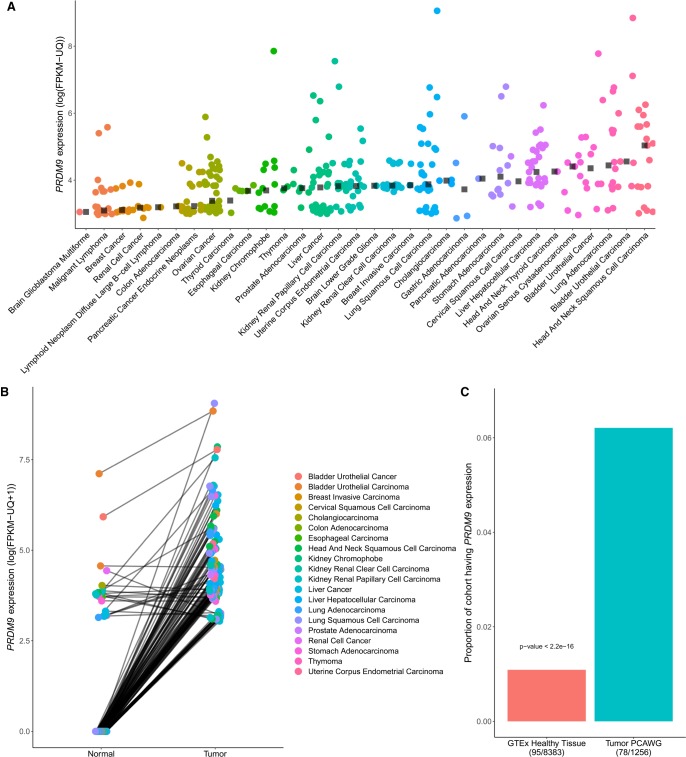
We first evaluated PRDM9 expression in two large cancer cohorts: PCAWG and TCGA (data described in Supplemental Fig. S1) (The Cancer Genome Atlas Research Network et al. 2013). Overall, 365 tumor samples expressed PRDM9 across 32 cancer types (Fig. 1A). Cancers exhibiting the highest expression include head and neck squamous cell carcinoma and bladder urothelial carcinoma, with median expression levels of 153.84 and 95.41 fragments per kilobase of transcript per million mapped reads-upper quartile (FPKM-UQ), respectively. We compared PRDM9 expression levels to other expressed genes within each sample by partitioning gene expression levels into percentiles. PRDM9 expression ranged from the second to the 73rd percentile of gene expression values with a mean of the 10th percentile across all samples. Liver and ovarian cancers in particular had a high proportion of tumors exhibiting PRDM9 expression with, respectively, 46% and 44% of tumors expressing PRDM9 above a threshold of 10 FPKM-UQ (Supplemental Fig. S3). Among most tumor sample pairs expressing PRDM9, higher expression of PRDM9 was found compared to the matching normal sample (one-sided Wilcoxon signed-rank test: P < 2.2 × 10−16) (Fig. 1B), suggesting that PRDM9 expression levels in tumors are higher than those of healthy tissues. To account for any effects possibly originating from tumor infiltration in the surrounding tissue, we next compared expression data in healthy tissues from the Genotype-Tissue Expression (GTEx) project (Melé et al. 2015) to transcriptome data from tumor samples (Fig. 1C). Again, the proportion of samples expressing PRDM9 was significantly higher among tumor transcriptomes compared with the cohort of nongerm healthy tissues, where nearly no PRDM9 expression was observed (one-sided two-sample test for equality of proportions: P < 2.2 × 10−16) (Fig. 1C). Taken together, these analyses confirm that PRDM9 becomes transcriptionally active in many cancers, a feature that is characteristic to cancer in nongerm cells. In the following sections, we focus on the PCAWG cohorts for further analyses to evaluate associations between PRDM9 expression and the genomic and transcriptomic landscape, due to the availability of whole-genome sequencing data for these donors.
Figure 1.
Characterization of aberrant PRDM9 expression across cancer types. (A) A total of 365 samples expressing PRDM9 above 10 FPKM-UQ across 39 cancer types from the PCAWG and TCGA data sets (n = 1879), after a correction to account for the high homology between PRDM9 and PRDM7 (Methods; see Supplemental Fig. S2). Black squares represent the median of PRDM9 expression above a threshold of 10 FPKM-UQ within each cancer type. (B) PRDM9 expression in 128 tumor samples expressing PRDM9 above a threshold of 10 FPKM-UQ and their matching normal samples, across cancer types within the TCGA and the PCAWG cohorts (n = 813 pairs). Data points are colored according to cancer type, and lines connect tumor and normal samples originating from the same patient. Cancer samples have higher PRDM9 expression than their matching healthy tissues (one-sided Wilcoxon signed-rank test: P < 2.2 × 10−16), a result consistent within each cancer type where at least three sample pairs were expressing PRDM9 (Fisher's method with one-sided Wilcoxon signed-rank test: P = 2.28 × 10−20) (Supplemental Table S1). (C) Proportion of samples expressing PRDM9 in the cancer cohorts (PCAWG and TCGA) compared with the proportion of samples expressing PRDM9 in the GTEx cohort, excluding testes. The proportion of samples expressing PRDM9 in cancer samples is significantly higher than that of healthy tissues.
Aberrant PRDM9 expression is associated with differentially expressed genes
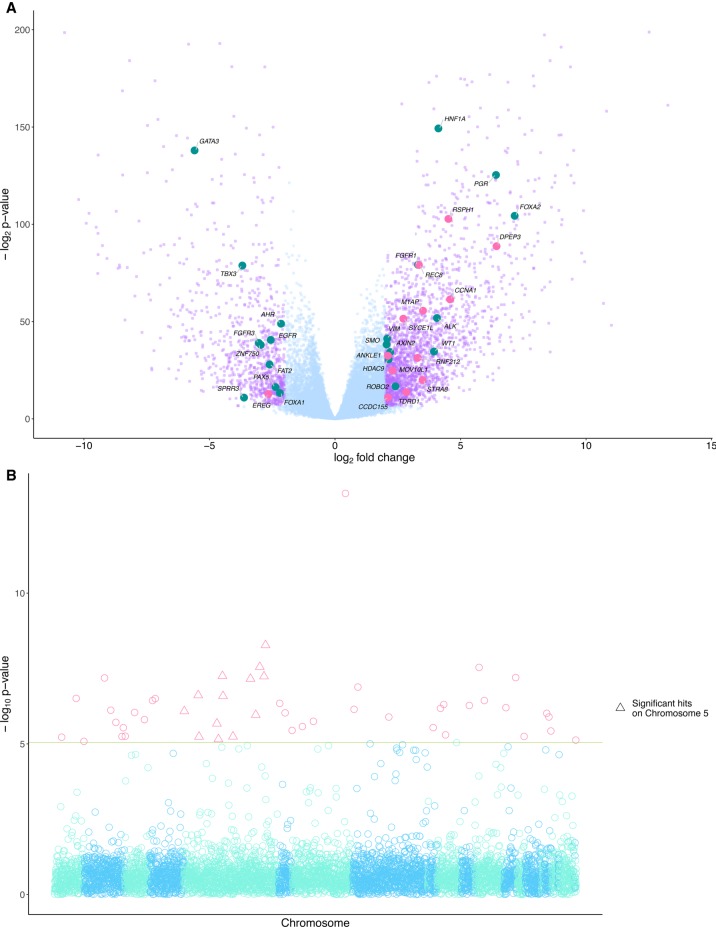
To identify transcripts whose expression was associated with aberrant PRDM9 expression, we performed a differential gene expression (DGE) analysis (Love et al. 2014), comparing tumors expressing PRDM9 to those without any detectable PRDM9 transcripts (Fig. 2A). Overall, 3114 genes were differentially expressed (FDR < 1%) in tumors expressing PRDM9, including 22 cancer driver genes identified in the IntOGen resource (Gonzalez-Perez et al. 2013). Out of 43 genes differentially expressed in cell lines with PRDM9 transfection (Altemose et al. 2017), nine were also differentially expressed in our analysis (Supplemental Data 1.1). Among the differentially expressed genes, we identify 13 genes with functions associated to meiotic processes, including the up-regulation of REC8, a gene coding for a meiosis-specific member of the cohesin complex (Watanabe and Nurse 1999), and of M1AP, whose protein likely contributes to the coordination of meiotic processes (Arango et al. 2013). As well, Gene Ontology (GO) biological processes involving cell differentiation, G-protein signaling, and nucleosome assembly were enriched in pan-cancer highly interacting modules of differentially expressed genes associated with PRDM9 (Supplemental Fig. S4), similar to biological processes enriched in modules of genes with testes-specific expression, which are suggestive of meiotic processes. The overlap in biological processes between modules built from these two different sets of genes suggests common processes between testes and cancers expressing PRDM9. Additionally, 13 out of the 241 genes associated with the meiosis biological process were differentially expressed in our analysis (Supplemental Data 1.1).
Figure 2.
Differentially expressed genes and recurrently mutated regions are associated with PRDM9 expression in cancer. (A) A total of 3114 genes were differentially expressed (in purple) above a threshold of 2 log2 fold change and a P-value adjusted for multiple testing using the Benjamini-Hochberg procedure (FDR < 0.05) between groups of cancers partitioned on PRDM9 expression. Of these genes, 2224 were overexpressed in cancers expressing PRDM9, and 890 were underexpressed. Twenty-two known cancer driver genes (Gonzalez-Perez et al. 2013) were differentially expressed in tumors expressing PRDM9 (in teal), and 13 had functions related to meiosis (in pink). (B) P-values from the associations between PRDM9 expression levels and recurrently mutated regions. The red line shows P < 0.05, with a Bonferroni correction for multiple testing for the number of tested regions (n = 1,507,106). Forty-nine recurrently mutated regions were significantly associated with aberrant PRDM9 expression across cancers. Highlighted with pink triangles are loci significantly associated with aberrant expression located on Chromosome 5, where the PRDM9 locus is located.
When a distinct DGE analysis was conducted separately for each cancer type, the median number of differentially expressed genes associated with PRDM9 expression across cancer types was 122.5, with the lowest being three (renal cell cancer) and the highest being 808 (liver hepatocellular carcinoma). A total of 245 genes were differentially coexpressed with PRDM9 in two or more unique cancer types (Supplemental Fig. S5; Supplemental Data 1.2).
Additionally, we performed DGE analyses to determine whether there was a sex-biased expression of genes correlated with PRDM9 expression. We found no significant association between the sex of patients and PRDM9 expression (χ2 test: P = 0.90). However, when we performed similar DGE analyses using sex as a cofactor (again including cancer types; see Methods), 1178 genes were differentially expressed (FDR < 1%) (Supplemental Data 1.3), including 864 that were not differentially expressed in the previous analysis that did not include sex as a cofactor. Some of these genes have expression specific to testis, such as TEX41, TSKS, BRDT, and TSGA10IP, and others specific to meiotic processes, such as DMC1 and EME1. In Supplemental Data 1.4, we show 507 genes that were differentially expressed in at least two different cancer types when each cancer type was analyzed separately and sex was still included as a cofactor. These results indicate that, in cancer, PRDM9 expression was associated with the sex-biased expression of other loci.
Recurrently mutated regions are associated with PRDM9 expression in tumors
We next investigated whether somatic mutations were associated with PRDM9 expression. Because identical recurrent somatic variants are extremely rare, we identified regions where SNVs cluster across all tumor samples: Such regions had at least two SNVs located no more than 100 bp apart. We identified 1,507,106 such regions and concentrated on the 5548 that exhibited a mutation in at least 5% of the samples (Lamaze et al. 2017). We tested for associations between PRDM9 expression levels and each of the 5548 recurrently mutated regions in order to identify somatic alterations that may contribute to the aberrant expression of PRDM9 (Methods). Forty-nine recurrently mutated regions were significantly associated with aberrant PRDM9 expression (Fig. 2B; Supplemental Data 2). Of these 49 regions, 13 were located on Chromosome 5, where PRDM9 is also located, and of them, eight were located within 5 Mb of the PRDM9 locus. Several genes proximal to these eight recurrently mutated regions have testes-specific expression in data from the GTEx Consortium (Chr 5: 24,349,767–24,353,800, CTD-2074D8.1; Chr 5: 24,852,561–24,856,027, RP11-730N24.1; Chr 5: 28,123,525–28,127,367, RP11-560A7.1; Chr 5: 29,117,954–29,121,731, RP11-42L13.3), similar to PRDM9 (Supplemental Data 2).
PRDM9 binding motif locations are significantly associated with somatic structural breakpoints
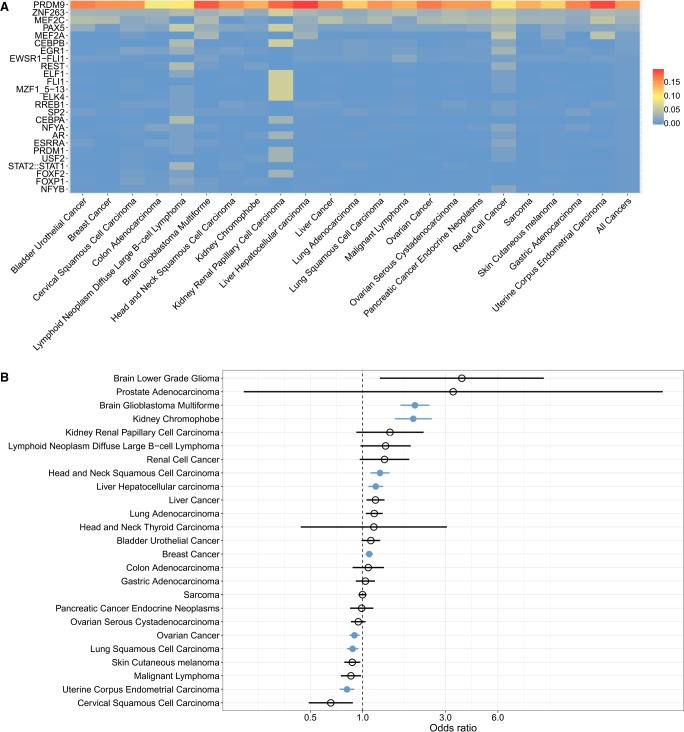
We investigated the downstream effects of PRDM9 expression among tumors by testing for associations of PRDM9 expression with the landscape of somatic SVs. Because the binding of PRDM9 to chromatin during meiosis leads to the recruitment of the DSB machinery, we hypothesized that PRDM9-expressing cancers would exhibit an enrichment of SV breakpoints, which may be indicative of DSBs near PRDM9 binding sites. To explore this hypothesis, we investigated whether the DNA motif known to be recognized by PRDM9 (Pratto et al. 2014) was overrepresented in the flanking sequences of somatic SVs (SV breakpoint sequences [SVBSs]) (Supplemental Fig. S6; Li et al. 2017). For samples expressing PRDM9, SVBSs were queried using a curated set of sequence motifs recognized by transcription factors (Mathelier et al. 2016), including the motif recognized by the A-variant of PRDM9 (Pratto et al. 2014). The allelic frequency of the A allele is 86% in Europeans and other non-African populations, and is 50% within African populations (Parvanov et al. 2010; Ponting 2011). Non-African individuals comprised the majority of the PCAWG cohort at 86.3% of the samples with expression data. We found that the motif recognized by the PRDM9 A-variant significantly matched the most SVBSs consistently across all cancer types (Fig. 3A). The proportion of SVBSs matching the PRDM9 A-variant binding motif was significantly higher than the proportion of SVBSs matching the ZNF263 binding motif, the second motif most frequently matching SVBSs (Wilcoxon signed-rank test P = 2.38 × 10−7). The proportion of SVBSs significantly matching the PRDM9 motif was also significantly higher in samples expressing PRDM9 than in samples not expressing PRDM9 (χ2: P = 2.2 × 10−16). Additionally, we performed a discriminative motif capture to identify motifs whose presence discriminated between PRDM9-expressing and non-PRDM9-expressing tumors (Redhead and Bailey 2007) to assess whether the presence of PRDM9 binding sites in SVBSs was dependent on its gene expression. This analysis identified a motif enriched in SVBSs from PRDM9-expressing samples, which partially matched the motif recognized by PRDM9 (Supplemental Fig. S8). We searched for the complete PRDM9 binding motif in the SVBSs containing the enriched motif from samples expressing PRDM9 and found that 27% of these SVBSs also matched the entire PRDM9 motif with an FDR < 1%.
Figure 3.
Structural variant (SV) breakpoints are enriched at sites of PRDM9 binding and activity. (A) Proportion of SV breakpoint sequences (SVBSs) significantly matching the motifs recognized by proteins in the JASPAR database, in cancer samples expressing PRDM9. We focused on sequences located within 100 bp of SV breakpoints, which is the mean distance separating DSBs and PRDM9 binding sites in meiosis (Baker et al. 2014). Each row shows the proportion of SVBSs matching the given motif per cancer type. The far-right column shows the proportion of SVBSs matching each motif across all cancer types. Supplemental Figure S7 shows the robustness of these results to significance thresholds used (Methods). (B) Enrichment for SV breakpoints at sites of highly recombining regions (HRRs) in samples expressing PRDM9, as shown by odds ratios >1 for each cancer type. Significant cancer types are shown in blue, as determined using Fisher's exact test (P < 0.05 with Bonferroni correction for the number of cancer types tested). Brain glioblastoma multiforme, kidney chromophobe, head and neck squamous cell carcinoma, liver hepatocellular carcinoma, and breast cancer samples all exhibited significant associations between PRDM9 expression and the colocalization of SV breakpoints and meiotic recombination hotspots. Ovarian cancer, lung squamous cell carcinoma, and uterine corpus endometrial carcinoma showed odds ratios <1, indicating significant enrichment for SV breakpoints in non-HRRs in samples expressing PRDM9.
Highly recombining regions (HRRs) are known to be associated with PRDM9 activity (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010) and binding (Myers et al. 2010), even in cell lines unrelated to meiotic functions (Eram et al. 2014; Altemose et al. 2017). We computed odds ratios to assess whether tumors expressing PRDM9 showed enrichment in SVs at HRRs (Hussin et al. 2015) relative to nonexpressing tumors (Fig. 3B). Significant enrichment of SV breakpoints in HRRs of samples expressing PRDM9 were observed in brain glioblastoma, renal cell cancer, kidney chromophobe, head and neck squamous cell carcinoma, liver hepatocellular carcinoma, and breast cancer (Fig. 3B). In these cancer types, we further determined whether there was an enrichment of SV breakpoints in regions associated with PRDM9-specific H3K4me3 marks. To do so, we leveraged regions marked with H3K4me3 that are known to be attributable to PRDM9 binding, following its transfection in HEK293T cell lines (Altemose et al. 2017). In PRDM9-specific H3K4me3 regions, we observed a significant enrichment of SV breakpoints in samples expressing PRDM9 in the cancer types having a significant enrichment of breakpoints in HRRs (OR = 1.12 with 95% confidence interval [1.05–1.19], P = 4.7 × 10−4), suggestive of an association between sites of PRDM9 epigenetic activity and SV breakpoints. Potential confounders are Alu and THE1 elements within HRRs, which often contain PRDM9 binding motifs (Myers et al. 2010) and are known to be prone to somatic structural alterations owing to their repetitive structure (Zhang et al. 2011). After excluding SVs with Alu and THE1 elements, similar results were observed (Supplemental Fig. S9). This trend was observed across all SV classes (Supplemental Fig. S10), although some failed to achieve statistical significance due to reduced N, denoting that no specific class of SVs drove the observed association.
Together, our results indicate that for multiple cancer types there was an association between PRDM9-mediated meiotic recombination sites and the locations of SVs in PRDM9-expressing cancers. On the other hand, ovarian cancer, lung squamous cell carcinoma, and uterine corpus endometrial carcinoma showed the opposite effect, in which SVs were modestly but significantly depleted at meiotic HRRs (OR < 1) (Fig. 3B) in samples expressing PRDM9, implying that other mechanisms may be targeting these regions (Supek and Lehner 2015). Because previous reports suggest that the location of somatic alterations are associated with heterochromatin (Makova and Hardison 2015), we assessed whether there was a difference in the distribution of SV breakpoint sites falling in regions of accessible chromatin in cancer types with HRR versus non-HRR enrichments. By leveraging DNase hypersensitivity data from tissues corresponding to the examined cancer types in the ENCODE Project (ENCODE Project Consortium 2012), we did not detect significant differences in the proportion of SV breakpoints falling in regions of open chromatin (Mann-Whitney U test: P-value = 0.6286). This suggests that chromatin architecture of the corresponding tissue alone cannot explain the distribution of SV breakpoint sites. Of these cancer types, the ovarian cancer and uterine corpus endometrial carcinoma cohorts were enriched for the mutational signature of LOF of the homologous recombination repair pathway, leading to an increase of DSBR through the nonhomologous DSBR machinery. The enrichment of this mutational signature in ovarian cancer and uterine corpus endometrial carcinoma paired with the depletion of SVs in HRRs raises the possibility that DSBs are less accessible to the nonhomologous DSBR machinery in non-HRRs, increasing DSBs unrelated to PRDM9.
Discussion
The function of PRDM9 in meiosis and in recombination is becoming increasingly clear (Walker et al. 2015; Grey et al. 2017; Imai et al. 2017), but its role in cancer still remains ill defined. While PRDM9 expression has previously been observed in some cancer cell lines, in five ovarian carcinoma samples, and in a single lung adenocarcinoma (Feichtinger et al. 2012), we show an aberrant expression of PRDM9 among human tumors in vivo in 32 different cancer types and report SV breakpoint enrichments at PRDM9 sites of binding and activity, providing evidence of potential mechanisms through which it may play a role in cancer biology. Previous studies have identified a correlation between acquired SV breakpoints and sites of meiotic crossovers, across recurrent breaks in cohorts of retinoblastomas (Hagstrom and Dryja 1999) and colorectal cancer (Howarth et al. 2009), as well as within individual meiotic and somatic recombination maps (Paulsson et al. 2011). Hussin et al. (2013) and Woodward et al. (2014) postulated that specific PRDM9-defined sites of meiotic recombination in parents possibly increase genomic instability of the offspring, creating genetic susceptibility to cancer in pediatric cases. Furthermore, following the observation of PRDM9 expression in cancer, Feichtinger et al. (2012) hypothesized this expression may be involved in chromatic lesions.
Our study provides evidence explaining the previously observed enrichment of somatic breakpoints at sites of meiotic recombination: We observe enrichment of somatic breakpoints at sites of PRDM9 binding and activity. Multiple cancer types, including glioblastoma multiforme, renal cell, kidney chromophobe, head and neck squamous cell carcinoma, liver hepatocellular carcinoma, and breast, all exhibited consistent associations between PRDM9 expression and the colocalization of SV breakpoints with PRDM9's binding and active sites. In these cancers, assuming PRDM9 transcripts are translated into proteins, PRDM9 binding may lead to the recruitment of the DSB machinery. Although genes coding for SPO11, MRE11, and RAD50, which are known to be recruited by PRDM9 for the creation of meiotic DSBs, were not consistently coexpressed with PRDM9 in the surveyed cancers, other coexpressed genes are relevant to the creation of somatic DSBs: The gene coding for the meiotic structure-specific endonuclease EME1, as well as the gene coding for the meiotic recombination protein DMC1, were overexpressed in cancers expressing PRDM9. Furthermore, our detection of the differential expression of DMC1 and EME1 in cancers expressing PRDM9 only when we incorporated sex as a covariate suggests sex bias in PRDM9 downstream interactors in a cancer context, and possibly in a meiotic context as well. The gene coding for the meiotic cohesin REC8, a component of the synaptonemal complex, was also overexpressed in cancers expressing PRDM9. REC8 and PRDM9 interact during meiosis, leading to the displacement of PRDM9-bound recombination hotspots toward the chromosomal axis (Parvanov et al. 2016). In cancers where PRDM9 and REC8 were overexpressed, the interaction between PRDM9 and REC8 may contribute to the disruption of the mitotic chromosomal architecture, possibly leading to SVs. Replication-based mechanisms (Yang et al. 2013) may also explain a subset of somatic SVs: PRDM9 binding might cause a replicative delay leading to increased SV breakpoints. Under this model, however, we would expect binding sites of other proteins to be enriched in SVBSs. For example, the expression of PAX5 is restricted to the spleen and to the small intestine in healthy tissues, as shown by the GTEx project (Melé et al. 2015), but was expressed at a threshold >10 FPKM-UQ in 1288 cancers in PCAWG originating from neither the spleen nor the small intestine. Even so, we did not find an enrichment of the binding site recognized by PAX5 in SV breakpoint sequences in samples expressing PAX5 relative to samples that do not express it.
Among cancer types where the evidence for PRDM9 having a role in the genomic localization of SVs was weaker, other exogenic or replication-related mechanisms may be leading to SVs (Glodzik et al. 2017), drowning out signals of PRDM9's contribution. The relationship between aberrant PRDM9 expression and increased SV breakpoints at sites associated with PRDM9 activity is nonlinear: In this study, cancer types with consistent associations between PRDM9 sites of activity and the location of SV breakpoints were not those with the highest PRDM9 expression, raising the possibility that PRDM9-related SVs may be balanced by up-regulation of repair pathways in the same tumors. Furthermore, because PRDM9 binding site locations are dependent on the allele expressed, investigating whether there is a shift in the location of SVs in cancers expressing PRDM9 in populations where the A allele is less frequent may lead to a better understanding of the mechanisms involved in the generation of SVs in cancers.
In contrast with its functional role in the recruitment of the DSB machinery at sites of recombination, little is known about upstream mechanisms associated with PRDM9 expression. We observed that aberrant PRDM9 expression in tumors was correlated with the up-regulation of M1AP expression. In mice, M1ap-knockdown males exhibit meiotic arrest as early as during the zygotene/pachytene stage (Arango et al. 2013), and missense variation in PRDM9 in humans has been implicated in male infertility (Miyamoto et al. 2008; Irie et al. 2009). The similarity in downstream effects of LOF of both PRDM9 and M1AP, coupled with the observed association between M1AP up-regulation and PRDM9 expression in tumors, suggests a role for M1AP regulating PRDM9 expression during meiosis. We also identified recurrently mutated genomic regions associated with PRDM9 expression, specifically in a region within 5 Mb of the PRDM9 locus. These mutations in the tumor genome potentially point to cis-acting regulatory domains that have been disrupted. The presence of genes having a testis-specific expression neighboring the recurrently mutated regions may also suggest that these somatic mutations disrupt the repression of entire topologically associated domains associated with testis- and meiotic-specific function, including PRDM9.
DSBs generating somatic SVs are a mechanism that contributes to genome instability, which conduces tumorigenesis. Our work provides strong evidence for a novel mechanism underlying genomic instability during tumorigenesis: that aberrant expression of PRDM9 is associated with somatic SVs, raising the intriguing possibility that there are as-yet uncharacterized genomic features and binding sites that lead to SVs. In the future, identification of features that are overrepresented near somatic breakpoints will provide a more complete picture of the endogenous processes associated with genomic instability.
Methods
Data
We retrieved RNA-sequencing samples from 1256 tumors, of which 162 had matched healthy tissue RNA-sequenced (May 2016 version 1.1) from the PCAWG project (https://dcc.icgc.org/pcawg). Paired healthy and tumor transcriptome samples were retrieved from the TCGA Research Network (n = 651 pairs) (http://cancergenome.nih.gov/). TCGA samples already present in the PCAWG cohort were discarded. An additional 8555 RNA-sequencing samples from healthy tissue were retrieved from the GTEx Consortium (version 6p) (Melé et al. 2015). Sample distributions across cancer types are shown in Supplemental Figure S1. Raw sequencing FASTQ files were aligned with STAR (Dobin et al. 2013) using pipelines available at https://github.com/ucscCancer/icgc_rnaseq_align. Our conclusions would not be significantly altered through realignment of the data on the most recent version of the reference genome: Our results focus on genomic regions that are not targeted by the major improvements between GRCh37 and GRCh38. SVs were identified by the PCAWG-6 group using the intersect of three different pipelines (Campbell et al. 2017; Li et al. 2017; Yung et al. 2017): The Sanger pipeline used BRASS (https://github.com/cancerit/BRASS), the DKFZ/EMBL pipeline used DELLY (Rausch et al. 2012), and the Broad pipeline used SNOWMAN (https://github.com/broadinstitute/SnowmanSV/) and DRanger (Supplemental Fig. S6; http://www.broadinstitute.org/cancer/cga/dranger). Details on the calling of consensus SVs is detailed in the PCAWG-6 marker paper (Li et al. 2017).
RNA-sequencing data quantification and normalization
RNA-sequencing was quantified for each transcript using HTSeq (version 0.6.1p1) (Anders et al. 2015) over all gene IDs from a modified version of the GENCODE v19 annotation file that only considered the PRDM9 locus to span the PRDM9/PRDM7 low-homology region. Overlaps were resolved using the intersection of all nonempty sets. Read counts for each gene were then normalized into FPKM-UQ, which gives a more uniform distribution across genes than FPKM (Bullard et al. 2010).
PRDM7 homology correction
Owing to the high homology between PRDM9 and its paralog PRDM7 (Supplemental Fig. S2A; Fumasoni et al. 2007; Blazer et al. 2016), we used the PCAWG cohort to evaluate a homology correction based on low-homology regions of the two genes to ensure that the computed PRDM9 read counts actually originated from PRDM9 transcripts. This homology correction added the contribution of a read toward the PRDM9 read count only if it mapped uniquely to a restricted 756-bp region of low homology between PRDM9 and PRDM7, namely, the ZnF array domain of PRDM9 (hg19; Chr 5: 23,526,838–23,528,706). Upon comparing computed PRDM9 expression levels before and after applying the homology correction within each sample (Supplemental Fig. S2B), we found that the two metrics were highly correlated (Pearson's correlation coefficient = 0.98, P < 2.2 × 10−16), although 246 samples expressed PRDM9 above a threshold of 10 FPKM-UQ only in the uncorrected samples. Among reads mapping to the PRDM9 locus, there were no PRDM7-specific substitutions in the SET domain. We interpreted these 246 samples to be false positives due to homologous PRDM7 expression or to other mapping biases and removed these samples in further analyses. It is also highly likely that the homology correction underestimated the expression levels of PRDM9 due to the repetitive nature of the ZnF array, which encompasses most of the low-homology region (Hinch et al. 2011; Hussin et al. 2013).
Comparisons of PRDM9 expression between cohorts
Within analyses including only the PCAWG and TCGA data sets, samples were considered to express PRDM9 if the FPKM-UQ for this gene was above a threshold of 10. In analyses including the GTEx data, we considered a more stringent expression threshold of 50 FPKM-UQ to account for higher levels of technical variation across different cohorts. We tested for differences in PRDM9 expression between matching tumor and surrounding healthy tissue from the TCGA and PCAWG cohorts with a one-sided Wilcoxon signed-rank test. We applied this same test for pairs of samples per cancer type and used Fisher's method to combine P-values from multiple test statistics, employing the metap R package (version 0.8) (https://CRAN.R-project.org/package=metap). We used a one-sided two-sample test for equality of proportions to test for differences in samples expressing PRDM9 between the healthy GTEx cohort, excluding testis samples, and the tumor PCAWG cohort.
DGE analysis
The DGE analyses were performed using DESeq2 (Love et al. 2014). Only genes that had a mean of 10 counts across all samples were considered, resulting in a total of 23,413 genes assessed. We considered samples with FPKM-UQ > 10 to express PRDM9 and samples with FPKM-UQ = 0 to not express PRDM9, both in the corrected and in the uncorrected analyses, resulting in 209 samples expressing PRDM9 and 907 that did not. The PCAWG project codes, representing the cancer type and country of origin of different cancer cohorts, were included as a cofactor to account for tissue- and cohort-specific differences in expression. Reported P-values were adjusted to assess the false-discovery rate (FDR) using the Benjamini and Hochberg procedure (Benjamini and Hochberg 1995). By using these adjusted P-values, we determined significance if they fell below a prescribed threshold for FDR (here, <1%), as well as a log2 fold change >2 in gene expression. For DGE analyses including sex as a cofactor, sex-specific cancer types were not considered. Gene lists associated with the meiosis biological process (GO:0051321) were extracted using QuickGO (Huntley et al. 2015). The gene list enriched in testes was obtained from the Human Protein Atlas (Uhlén et al. 2015). Modules represent highly interacting gene products of differentially expressed genes associated with PRDM9 expression and were defined using the Reactome FI for Cytoscape (Wu et al. 2010). Modules shown included at least 50 nodes in differentially expressed genes and nine nodes in genes enriched in testes. Enriched GO biological processes in each module were defined as having an FDR <5%.
Association between PRDM9 expression and recurrently mutated regions
Recurrently mutated regions are defined as densely mutated regions where at least two SNVs across all tumor samples were located within 100 bp from each other, as described by Lamaze et al. (2017). We tested for linear regression between PRDM9 expression and each recurrently mutated region disrupted in at least 5% of samples, accounting for cancer type. Significance was established with P < 0.05, corrected using a Bonferroni correction for multiple testing for each recurrently mutated region tested (n = 5548). Genes closest to each significant recurrently mutated region were assessed using BEDTools closest (version 2.25) (Quinlan 2014), using the hg19 Ensembl gene list as a reference. Testis-specific expression of genes closest to significant recurrently mutated regions located on Chromosome 5 were manually inspected in the GTEx data portal.
PRDM9 binding motif analyses
There is variation in PRDM9 binding sites within and across individuals carrying different alleles. To uncover a binding site robustly recognized by the A-variant of PRDM9, we used MEME-ChIP from the MEME package (Bailey et al. 2009) to identify a motif shared among sites of DSBs in a previous study using sperm from individuals genotyped as AA (Pratto et al. 2014). SVBSs were identified using BEDTools slop (version 2.25) (Quinlan 2014) by adding 100 bp flanking each SV breakpoint, because this is the approximate distance separating DSBs and PRDM9 binding sites in meiosis (Baker et al. 2014). Motifs enriched in these sequences were identified by a comparison with the JASPAR core vertebrate database (version 2016) (Mathelier et al. 2016), using the R package rtfbs (version 0.3.5). SVBSs were partitioned into four categories based on GC content to reduce the number of false positives. Enrichments were tested by comparing motif occurrence in SVBSs to that in random sequences matched for GC content, and significance was assessed using FDR thresholds of 0.1 and 0.01. Results shown only include human binding sites and are the 25 motifs with the highest proportion of SVBSs matching each respective motif across all cancer types.
To test whether the high number of significant PRDM9 motif matches overlapped with SVBSs were specific to samples expressing PRDM9, we performed a discriminative regular expression motif search using DREME from the MEME package (Bailey et al. 2009), which searches for motif patterns that can discriminate between two sets of sequences. The primary set of sequences consisted of SVBSs found in samples expressing PRDM9, while the negative control set of sequences consisted of SVBSs identified in samples without PRDM9 expression, both before and after the PRDM7 homology correction. From these results, we searched for the complete PRDM9 binding motif in sequences containing the discriminatory motif enriched in SVBSs from samples expressing PRDM9 using FIMO from the MEME package (MEME/4.9.1_1).
Odds ratios calculations
HRRs of the genome were determined as defined by Hussin et al. (2015). The number of SV breakpoints falling in HRRs and in the rest of the genome was determined using BEDTools intersect (version 2.25) (Quinlan 2014). Odds ratios were computed to quantify the differences in proportions of SVs falling in the HRRs relative to the rest of the genome (Non_HRR). For each cancer type, the odds ratios were computed as OR = (xHRR × yNon_HRR)/(xNon_HRR × yHRR), where xHRR and xNon_HRR represent the number of SVs in samples with PRDM9 expression, and yHRR and yNon_HRR represent the number of SVs in samples that do not express PRDM9. Ninety-five percent confidence intervals were computed as detailed by Szumilas (2010), and significance was established using a Fisher's exact test, with a P < 0.05 threshold after Bonferroni correction for multiple testing for the different cancers tested. PRDM9-specific H3K4me3 marked regions were identified from ChIP-seq data from Altemose et al. (2017). H3K4me3 peaks in PRDM9-transfected cells (GEO sample accession: GSM2643614) that did not overlap any peaks from both replicates of untransfected cells (GEO sample accession: GSM2643608 and GSM2643609) were considered as PRDM9-attributable H3K4me3 peaks.
The locations of Alu and THE1 repeat elements were extracted from the hg19 RepeatMasker (http://www.repeatmasker.org) track on UCSC's genome browser. SV breakpoints and HRRs overlapping Alu and THE1 elements were discarded, and then odds ratios were computed. SV classification was done based on results provided by the PCAWG consortium (Li et al. 2017). Odds ratios, significance, and confidence intervals were determined as previously described for each SV type.
DNase-seq data were retrieved as BED files (broad peaks) from the ENCODE data portal (ENCODE Project Consortium 2012). The tissue correspondences used were as follows: Brain glioblastoma multiforme, head and neck squamous cell carcinoma, liver hepatocellular carcinoma, breast cancer, ovarian cancer, lung squamous cell carcinoma, and uterine corpus endometrial carcinoma were matched, respectively, with cerebellum and frontal cortex, esophagus squamous epithelium, right lobe of liver, breast epithelium, ovary, upper lobe of left lung, and uterus tissue types.
Supplementary Material
Acknowledgments
We acknowledge the use of pre-embargo Pan-Cancer Analysis of Whole Genomes (PCAWG) project data, approved by the PCAWG steering committee (with L.D.S. recused), and we thank the working groups from both the PCAWG and the TCGA for providing the data. We also thank S. Wright and J.D. Hussin for insightful comments on the study. We acknowledge financial support from the Government of Ontario, Ministry of Research, and Innovation Senior Investigator Award (Ontario Institute for Cancer Research, 070055 to P.A.). A.A.H. was supported by an Ontario Graduate Scholarship (Ontario Council on Graduate Studies, Council of Ontario Universities). F.C.L. is a Fond de Recherche en Santé du Québec (FRSQ) Research Fellow.
Author contributions: A.A.H., L.D.S., and P.A. designed the study. A.A.H. performed normalization on sequencing read data and performed bioinformatics and statistical analyses. D.S. contributed to statistical analyses. A.A.H., H.A.E., H.G., M.-J.F., and P.A. wrote the manuscript. H.G. performed bioinformatics validation. F.C.L. provided the list of recurrently mutated regions. M.A. and V.B. retrieved and processed data, and performed quality control on transcriptomic data.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231696.117.
References
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altemose N, Noor N, Bitoun E, Tumian A, Imbeault M, Chapman JR, Radu Aricescu A, Myers SR. 2017. A map of human PRDM9 binding provides evidence for novel behaviors of PRDM9 and other zinc-finger proteins in meiosis. eLife 6: e28383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves I, Ang Houle A, Hussin JG, Awadalla P. 2017. The impact of recombination on human mutation load and disease. Philos Trans R Soc Lond B Biol Sci 372: 20160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Sekelsky J. 2010. Meiotic versus mitotic recombination: two different routes for double-strand break repair. The different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango NA, Li L, Dabir D, Nicolau F, Pieretti-Vanmarcke R, Koehler C, McCarrey JR, Lu N, Donahoe PK. 2013. Meiosis I arrest abnormalities lead to severe oligozoospermia in meiosis 1 arresting protein (M1ap)-deficient mice. Biol Reprod 88: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Carpenter ATC, Esposito MS, Esposito RE, Sandler L. 1976. The genetic control of meiosis. Annu Rev Genet 10: 53–134. [DOI] [PubMed] [Google Scholar]
- Baker CL, Walker M, Kajita S, Petkov PM, Paigen K. 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res 24: 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, de Massy B. 2007. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. [Google Scholar]
- Blazer LL, Lima-Fernandes E, Gibson E, Eram MS, Loppnau P, Arrowsmith CH, Schapira M, Vedadi M. 2016. PR domain-containing protein 7 (PRDM7) is a histone 3 lysine 4 trimethyltransferase. J Biol Chem 291: 13509–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Getz G, Stuart JM, Korbel JO, Stein LD, Net–ICGC/TCGA Pan-Cancer Analysis of Whole Genomes. 2017. Pan-cancer analysis of whole genomes. bioRxiv 10.1101/162784. [DOI]
- The Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor AA, Denroche RE, Jang GH, Timms L, Kalimuthu SN, Selander I, McPherson T, Wilson GW, Chan-Seng-Yue MA, Borozan I, et al. 2017. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol 3: 774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eram MS, Bustos SP, Lima-Fernandes E, Siarheyeva A, Senisterra G, Hajian T, Chau I, Duan S, Wu H, Dombrovski L, et al. 2014. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J Biol Chem 289: 12177–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feichtinger J, Aldeailej I, Anderson R, Almutairi M, Almatrafi A, Alsiwiehri N, Griffiths K, Stuart N, Wakeman JA, Larcombe L, et al. 2012. Meta-analysis of clinical data using human meiotic genes identifies a novel cohort of highly restricted cancer-specific marker genes. Oncotarget 3: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD. 2007. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol 7: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glodzik D, Morganella S, Davies H, Simpson PT, Li Y, Zou X, Diez-Perez J, Staaf J, Alexandrov LB, Smid M, et al. 2017. A somatic-mutational process recurrently duplicates germline susceptibility loci and tissue-specific super-enhancers in breast cancers. Nat Genet 49: 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A, Lopez-Bigas N. 2013. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods 10: 1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, Clément JAJ, Buard J, Leblanc B, Gut I, Gut M, Duret L, de Massy B. 2017. In vivo binding of PRDM9 reveals interactions with noncanonical genomic sites. Genome Res 27: 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom SA, Dryja TP. 1999. Mitotic recombination map of 13cen–13q14 derived from an investigation of loss of heterozygosity in retinoblastomas. Proc Natl Acad Sci 96: 2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, et al. 2011. The landscape of recombination in African Americans. Nature 476: 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth K, Ranta S, Winter E, Teixeira A, Schaschl H, Harvey JJ, Rowan A, Jones A, Spain S, Clark S, et al. 2009. A mitotic recombination map proximal to the APC locus on chromosome 5q and assessment of influences on colorectal cancer risk. BMC Med Genet 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O'Donovan C. 2015. The GOA database: Gene Ontology annotation updates for 2015. Nucleic Acids Res 43: D1057–D1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussin J, Sinnett D, Casals F, Idaghdour Y, Bruat V, Saillour V, Healy J, Grenier J, de Malliard T, Busche S, et al. 2013. Rare allelic forms of PRDM9 associated with childhood leukemogenesis. Genome Res 23: 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussin JG, Hodgkinson A, Idaghdour Y, Grenier J-C, Goulet J-P, Gbeha E, Hip-Ki E, Awadalla P. 2015. Recombination affects accumulation of damaging and disease-associated mutations in human populations. Nat Genet 47: 400–404. [DOI] [PubMed] [Google Scholar]
- Imai Y, Baudat F, Taillepierre M, Stanzione M, Toth A, de Massy B. 2017. The PRDM9 KRAB domain is required for meiosis and involved in protein interactions. Chromosoma 126: 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie S, Tsujimura A, Miyagawa Y, Ueda T, Matsuoka Y, Matsui Y, Okuyama A, Nishimune Y, Tanaka H. 2009. Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl 30: 426–431. [DOI] [PubMed] [Google Scholar]
- Jackson SPP. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23: 687–696. [DOI] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X. 2012. Homologous recombination and its regulation. Nucleic Acids Res 40: gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze FC, Chateigner A, Edgington H, Fave M-J, Ang Houle A, Awadalla P. 2017. Motif disruption domains lead to cancer gene expression rewiring. bioRxiv 10.1101/126359. [DOI]
- Li Y, Roberts N, Weischenfeldt J, Wala JA, Shapira O, Schumacher S, Khurana E, Korbel JO, Imielinski M, Beroukhim R, et al. 2017. Patterns of structural variation in human cancer. bioRxiv 10.1101/181339. [DOI]
- Lisby M, Rothstein R. 2016. Cell biology of mitotic recombination. Cold Spring Harb Perspect Biol 7: a016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. 2016. BRCAness revisited. Nat Rev Cancer 16: 110–120. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova KD, Hardison RC. 2015. The effects of chromatin organization on variation in mutation rates in the genome. Nat Rev Genet 16: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen C, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. 2016. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44: D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. The human transcriptome across tissues and individuals. Science 348: 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Koh E, Sakugawa N, Sato H, Hayashi H, Namiki M, Sengoku K. 2008. Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet 25: 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop REE, Freeman C, MacFie TSS, McVean G, Donnelly P. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov EDD, Petkov PMM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Tian H, Billings T, Saxl RL, Spruce C, Aithal R, Krejci L, Paigen K, Petkov PM. 2016. PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Mol Biol Cell 28: 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson K, Lindgren D, Johansson B. 2011. SNP array analysis of leukemic relapse samples after allogeneic hematopoietic stem cell transplantation with a sibling donor identifies meiotic recombination spots and reveals possible correlation with the breakpoints of acquired genetic aberrations. Leukemia 25: 1358–1361. [DOI] [PubMed] [Google Scholar]
- Ponting CP. 2011. What are the genomic drivers of the rapid evolution of PRDM9? Trends Genet 27: 165–171. [DOI] [PubMed] [Google Scholar]
- Powers NR, Parvanov ED, Baker CL, Walker M, Petkov PM, Paigen K. 2016. The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLoS Genet 12: e1006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. 2014. Recombination initiation maps of individual human genomes. Science 346: 1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purandare SM, Patel PI. 1997. Recombination hot spots and human disease. Genome Res 7: 773–786. [DOI] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO, Stütz AM, Benes V, Korbel JO. 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redhead E, Bailey TL. 2007. Discriminative motif discovery in DNA and protein sequences using the DEME algorithm. BMC Bioinformatics 8: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Fujiwara Y, Reinholdt LG, Hu J, Saxl RL, Baker CL, Petkov PM, Paigen K, Handel MA. 2015. Nuclear localization of PRDM9 and its role in meiotic chromatin modifications and homologous synapsis. Chromosoma 124: 397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Lehner B. 2015. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 521: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Rothstein R, Lisby M. 2014. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198: 795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumilas M. 2010. Explaining odds ratios. J Can Acad Child Adolesc Psychiatry 19: 227–229. [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Proteomics: tissue-based map of the human proteome. Science 347: 1260419. [DOI] [PubMed] [Google Scholar]
- Walker M, Billings T, Baker CL, Powers N, Tian H, Saxl RL, Choi K, Hibbs MA, Carter GW, Handel MA, et al. 2015. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin 8: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461–464. [DOI] [PubMed] [Google Scholar]
- Woodward EL, Olsson ML, Johansson B, Paulsson K. 2014. Allelic variants of PRDM9 associated with high hyperdiploid childhood acute lymphoblastic leukaemia. Br J Haematol 166: 947–949. [DOI] [PubMed] [Google Scholar]
- Wu G, Feng X, Stein L. 2010. A human functional protein interaction network and its application to cancer data analysis. Genome Biol 11: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh C-H, Zhang C, Ren X, Protopopov A, Chin L, et al. 2013. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153: 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung CK, O'Connor BD, Yakneen S, Zhang J, Ellrott K, Kleinheinz K, Miyoshi N, Raine KM, Royo R, Saksena GB, et al. 2017. Large-scale uniform analysis of cancer whole genomes in multiple computing environments. bioRxiv 10.1101/161638. [DOI]
- Zhang W, Edwards A, Fan W, Deininger P, Zhang K. 2011. Alu distribution and mutation types of cancer genes. BMC Genomics 12: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.