Abstract
Species-specific, new, or “orphan” genes account for 10%–30% of eukaryotic genomes. Although initially considered to have limited function, an increasing number of orphan genes have been shown to provide important phenotypic innovation. How new genes acquire regulatory sequences for proper temporal and spatial expression is unknown. Orphan gene regulation may rely in part on origination in open chromatin adjacent to preexisting promoters, although this has not yet been assessed by genome-wide analysis of chromatin states. Here, we combine taxon-rich nematode phylogenies with Iso-Seq, RNA-seq, ChIP-seq, and ATAC-seq to identify the gene structure and epigenetic signature of orphan genes in the satellite model nematode Pristionchus pacificus. Consistent with previous findings, we find young genes are shorter, contain fewer exons, and are on average less strongly expressed than older genes. However, the subset of orphan genes that are expressed exhibit distinct chromatin states from similarly expressed conserved genes. Orphan gene transcription is determined by a lack of repressive histone modifications, confirming long-held hypotheses that open chromatin is important for new gene formation. Yet orphan gene start sites more closely resemble enhancers defined by H3K4me1, H3K27ac, and ATAC-seq peaks, in contrast to conserved genes that exhibit traditional promoters defined by H3K4me3 and H3K27ac. Although the majority of orphan genes are located on chromosome arms that contain high recombination rates and repressive histone marks, strongly expressed orphan genes are more randomly distributed. Our results support a model of new gene origination by rare integration into open chromatin near enhancers.
Gene regulation is a highly orchestrated process that includes transcription factor binding sites (TFBSs), noncoding RNAs, histone modifications, and chromatin structure (Voss and Hager 2014). The identification and mechanism of these molecular factors have been revealed for several conserved gene networks leading toward a better understanding of development and disease. But how new genes, also referred to as orphan or taxon-restricted, acquire this complex architecture is unknown. For the increasing number of identified new genes that provide important biological function (Burki and Kaessmann 2004; Cai et al. 2008; Rosso et al. 2008; Chen et al. 2010, 2013b; Reinhardt et al. 2013; Mayer et al. 2015; Santos et al. 2017), the evolutionary path from origin to integration into gene networks depends on their precise transcriptional regulation (Carelli et al. 2016). Yet in the majority of cases, it is unclear how even the most fundamental cis-regulatory elements like promoter and termination sequences are obtained (Tautz and Domazet-Lošo 2011; Long et al. 2013). Orphan genes can originate de novo or by duplication, recombination, or horizontal gene transfer into preexisting regulatory architecture (Betrán and Long 2003; Kaessmann et al. 2009; Li et al. 2009; Kaessmann 2010; Chen et al. 2012b; McLysaght and Hurst 2016), but the extent to which this occurs is limited by the potential to disrupt the genes already there (Vinckenbosch et al. 2006). In the few cases in which integration has been observed, the presence of nearby regulatory sequences was largely detected by proximity, or sequence homology with known promoters, CpG islands, or TFBSs (Carvunis et al. 2012; Abrusán 2013; Ruiz-Orera et al. 2015; Li et al. 2016). Given these constraints, the contribution of preexisting regulatory architecture to new gene transcription is still unknown and, with functional genomic information (e.g., chromatin states and enhancers), could potentially be large. Indeed, a recent analysis of mammalian ChIP-seq data sets found 51% of expressed mouse retrogenes (mRNAs that are reverse transcribed and inserted into the genome) exhibit robust H3K4 trimethylation (Carelli et al. 2016), and transcription of the new gene QQS in Arabidopsis thaliana is inversely correlated with DNA methylation at 5′ transposable elements (Silveira et al. 2013), suggesting an important role for chromatin regulation in new gene transcription. We sought to use the rich taxonomic resources of nematodes to first identify young and old genes, and then observe their regulatory architecture by several genome-wide approaches.
The diplogastrid nematode Pristionchus pacificus can be found in a necromenic relationship with beetles, but has been developed in the laboratory as a satellite model for comparative studies to C. elegans (Fig. 1A–D; Sommer and Streit 2011; Sommer and McGaughran 2013). More recent genetic analysis of dimorphic mouth-forms (Fig. 1E–G) has led to P. pacificus emerging as an important model system for phenotypic plasticity in its own right (Bento et al. 2010; Ragsdale et al. 2013; Kieninger et al. 2016; Serobyan et al. 2016). In addition to the vast taxonomic diversity and corresponding genomes of other nematode species, the recent high-quality chromosome-scale genome (Rödelsperger et al. 2017) and reverse genetic tools (Witte et al. 2015) in P. pacificus provide a robust framework for studying new genes (Baskaran et al. 2015; Prabh and Rödelsperger 2016). Here, we probe the gene structure, expression, and regulatory architecture of P. pacificus evolutionary gene classes with long-read Pacific Biosciences (PacBio) transcript sequencing (Iso-Seq), traditional high-depth RNA sequencing (RNA-seq), and chromatin immunoprecipitation (ChIP-seq) of six histone post-translational modifications and assay for transposon-accessible chromatin (ATAC-seq). In addition to our findings, the data sets collected provide the first epigenomic map in P. pacificus, which is only the second comprehensive chromatin state annotation in nematodes, creating a resource for future functional and comparative studies.
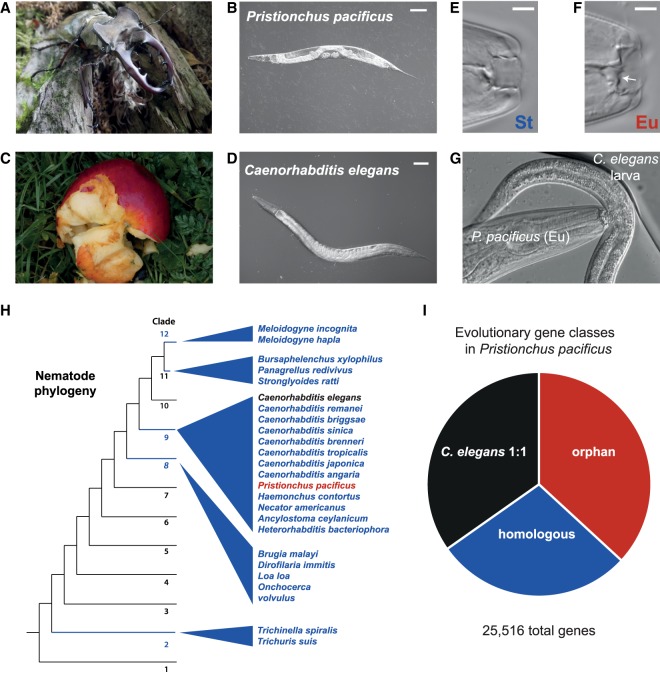
Figure 1.
Comparison of Pristionchus pacificus and Caenorhabditis elegans and phylogenetic relationship. (A,B). P. pacificus is often found in a necromenic relationship with insect hosts, preferentially scarab beetles, in the dormant dauer state. When the beetle dies, worms exit the dauer stage to feed on bacteria that bloom on the decomposing carcass. (C,D) C. elegans, the classic nematode model organism, is often found in leaf detritus and rotting fruits. Example rotting apple photo taken by M.S.W. (E–G) P. pacificus has become an important model for developmental (phenotypic) plasticity. Adults can adopt (E) a narrow mouth form with one tooth (stenostomatous [St]) that makes them strict bacterial feeders. However, the “boom-and-bust” life cycle creates significant competition for resources, and under crowded conditions adults can develop an alternative mouth form (F) with a wider buccal cavity and an extra tooth (eurystomatous [Eu]) that allows them to prey on other nematodes. (G) Shown here is a eurystomatous P. pacificus preying on a C. elegans larva. (H) A schematic phylogeny of nematodes that was generated based on the publications of Holterman et al. (2017) and Van Megen et al. (2009). (I) Breakdown of P. pacificus genes by evolutionary category: One-to-one orthology with C. elegans (C. elegans 1:1) is the most conserved, followed by genes sharing homology with at least one gene from the 24 other nematodes (homologous), and finally genes that are only found in Pristionchus (orphan). All categories were defined by BLASTP homology (e-value ≤0.001) (Methods).
Results
Partitioning of P. pacificus genes into evolutionary classes
The first P. pacificus draft genome published in 2008 (Dieterich et al. 2008) had a large number of genes with undetectable homology. Although the confidence in these gene predictions was initially low, every subsequent refinement of both the genome and gene annotation continually detected 20%–40% of genes that appear as new, orphan, or taxon-restricted (Sinha et al. 2012; Baskaran et al. 2015; Baskaran and Rödelsperger 2015; Prabh and Rödelsperger 2016). Using our most recent chromosome-scale PacBio genome (Rödelsperger et al. 2017) and 24 other nematode species, we reevaluated the relative abundance of evolutionary gene classes (Fig. 1H). We defined the most highly conserved genes as having 1:1 orthology with C. elegans (BLASTP e-value ≤ 0.001), which is estimated to have diverged from P. pacificus between 60 to 90 million years ago (Cutter 2008; Rota-Stabelli et al. 2013; Hedges et al. 2015). We also defined an intermediate conserved class as “homologs” if they display homology with at least one gene in the other 24 nematode species (Methods)—which could represent either relatively young genes or old genes that have been lost. Finally, we define “orphan” genes as having no homology with genes in the other 24 queried species. The resulting partition of genes approximates the “30% rule” of new gene composition (Fig. 1I; Khalturin et al. 2009). We then applied several genomic approaches to molecularly characterize each evolutionary gene class.
Characterization of gene structure by long-read RNA sequencing (Iso-Seq)
We sought to improve the overall gene annotation in P. pacificus and then characterize the genetic structure of each evolutionary gene class using PacBio Iso-Seq on mixed-developmental stage RNA (Supplemental Methods; Supplemental Fig. S1A–C). After alignment, we obtained 640,664 reads with a median insert size of 1363 nucleotides (Supplemental Fig. S1D). Despite low read depth compared to conventional RNA-seq, our Iso-Seq data covered 17,307 genes (68% of genes in the reference annotation “El Paco”) (Rödelsperger et al. 2017).
Relative to the current reference annotation, Iso-Seq identified a tighter distribution of gene lengths (median Iso-Seq = 1452 compared to median reference = 1599, P < 2.2 × 10−16, Wilcoxon rank-sum test) (Fig. 2A). This difference appears to be due to a more narrow distribution of exons, with 96.5% of Iso-Seq gene annotations containing between 1 and 20 exons, compared to 85.7% for the reference annotation (P = <2.2 × 10−16, Wilcoxon rank-sum test) (Fig. 2B). The tighter distribution is also more consistent with the highly curated gene annotation of C. elegans in which 98.0% of genes contain between 1 and 20 exons (Supplemental Fig. S1E,F; Deutsch and Long 1999). This potential improvement in accuracy appears to result from fragmentation of excessively long gene predictions into distinct transcripts (for example, see Supplemental Fig. S1G).
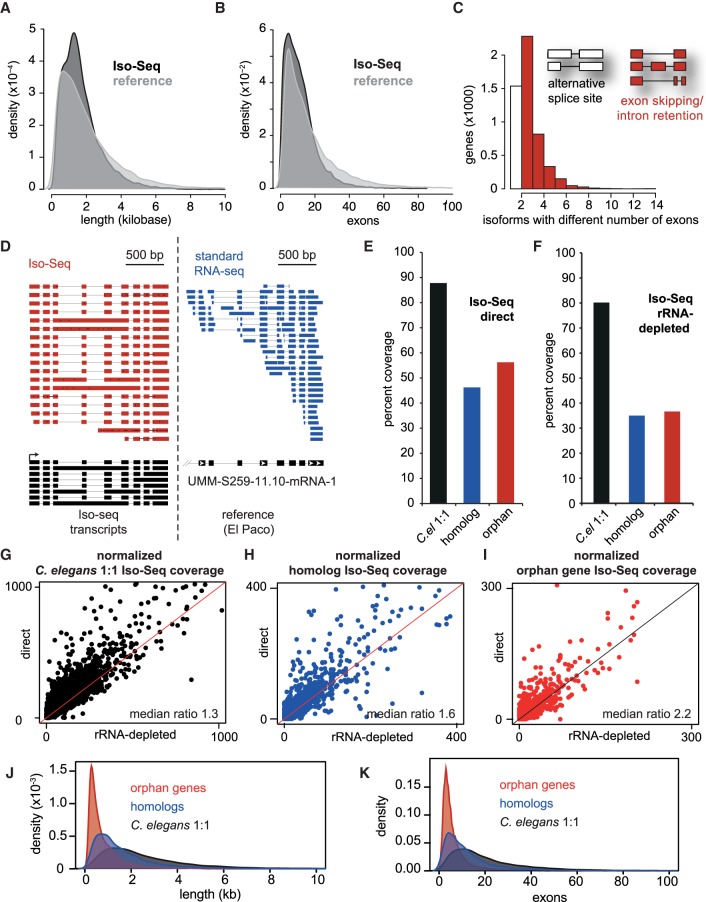
Figure 2.
Long-read RNA sequencing (Iso-Seq) improves gene annotation, identifies alternative splicing, and can distinguish different evolutionary gene classes by gene structure. (A) Density distribution of cDNA gene lengths between the El Paco reference (gray) and Iso-Seq annotation (black). The Iso-Seq annotation was derived from guided assembly using StringTie (Pertea et al. 2016; Methods), and plots were created using the density function in R. (B) Density distribution of exons per gene between El Paco reference and Iso-Seq annotations. Method and color scheme are similar to A. (C) Alternatively spliced isoforms, defined as having multiple detected isoforms with the same start and stop coordinates. The white column represents genes containing isoforms that have the same exon–intron structure but different splice sites, and red columns represent genes containing isoforms with different numbers of exons due to intron retention or exon inclusion/exclusion. (D) Example locus of Iso-Seq reads compared to standard short-read RNA-seq. Also shown are Iso-Seq-assembled isoforms compared to the single reference gene umm-S259-11.10-mRNA-1, visualized using Integrated Genome Viewer (IGV). (E,F) Percent coverage of evolutionary gene classes by Iso-Seq with either the “direct” method (E) or rRNA-depleted “total RNA” (F). (G–I) Iso-Seq coverage per gene of each evolutionary class in direct (y-axis) compared to total (x-axis). Coverage was determined by BEDTools, and median ratios of direct/total RNA are presented. Lines (slope = 1, y intercept = 0) represent equal coverage between methods. (J,K) Similar density distributions of cDNA length and exon number as in A and B, but for the three evolutionary gene classes.
Long-read Iso-Seq also provides more robust identification of isoforms, which are historically difficult to assemble from standard short-read RNA sequencing (Conesa et al. 2016). Approximately half (50.6%) of expressed genes in P. pacificus exhibit greater than one isoform, and roughly a third (30.9%) exhibit greater than three isoforms (Supplemental Fig. S1H,I). However, some of these transcripts could be artifacts biased by incomplete coverage of 5′ ends.
Hence, we conservatively defined alternatively spliced isoforms as transcripts with the same start and stop coordinates, but differential exon inclusion or exclusion, intron retention, or differential splice site. Under this classification we observed 3861 (24%) of expressed genes exhibit alternative splicing in P. pacificus (Fig. 2C), similar to the ∼25% of genes estimated in C. elegans (Ramani et al. 2011). As an example, we highlight gene umm-s259-11.10-mRNA, where the majority of Iso-Seq reads (17/19) cover the entire transcript yielding eight isoforms, in stark contrast to standard short-read sequencing, which rarely covers more than three exons per read (Fig. 2D). Collectively, a tighter distribution of transcript lengths and exon numbers, and diversity of isoforms, suggests that Iso-Seq improves the quality and quantity of gene annotation in P. pacificus.
Among evolutionary gene classes, most C. elegans 1:1 orthologs (88%), and approximately half of homologous and orphan genes (46% and 56%, respectively) exhibit Iso-Seq coverage, demonstrating that our Iso-Seq data are sensitive enough to detect thousands of transcripts from each evolutionary gene class (Fig. 2E). We also performed Iso-Seq on rRNA-depleted “total RNA” (Supplemental Methods) to assess whether young genes are un-, or under-polyadenylated, which is typical of noncoding RNAs (Derrien et al. 2012). We found a similar percent coverage from the direct and total RNA methods (Fig. 2E,F) and a consistent polyadenylation read bias for all gene classes (Fig. 2G–I). Hence most young genes, or at least transcribed young genes, are polyadenylated. Because polyadenylation is an important component of transcriptional and translational regulation (Proudfoot et al. 2002; Proudfoot 2011), this argues that most young genes have retained, or already acquired, 3′ termination and processing sequence architecture.
We then used our Iso-Seq annotation to characterize gene length and exon number between evolutionary gene classes. Consistent with other systems (Ruiz-Orera et al. 2015; Stein et al. 2018), we found a strong bias of C. elegans 1:1 orthologs to be longer and contain more exons than homologs, which in turn were longer and contained more exons than orphan genes (P = <2.2 × 10−16, Wilcoxon rank-sum test) (Fig. 2J,K). The intermediate gene structure of intermediate conserved genes (homologs) is also consistent with a transitional evolutionary path between young and old genes proposed by Carvunis et al. (2012) (Abrusán 2013; Neme and Tautz 2013). In the following sections we seek to characterize and compare the chromatin regulation of young versus old genes.
The P. pacificus epigenome
To identify regulatory regions and expression levels of orphan, homolog, and C. elegans 1:1 orthologs, we performed two to three replicates of ChIP-seq on nine histone modifications and two replicates of RNA-seq in P. pacificus adults, and two replicates of ATAC-seq on mixed-stage cultures (Supplemental Fig. S2; Supplemental Table S3; Supplemental Methods). All data sets showed good correlations between biological replicates (Pearson’s correlation between 0.70–0.93 for ChIP-seq, 0.88 for ATAC-seq, and 0.98 for gene FPKMs in RNA-seq) (Supplemental Fig. S3). We identified enriched regions (i.e., peaks) for each replicate of ChIP-seq and ATAC-seq using MACS2 (Methods; Supplemental Table S1; Zhang et al. 2008). H2Bub, H3K9ac, and H3K79me2 exhibited <50% peak reproducibility and were excluded from further analysis (Supplemental Fig. S4A). The majority of remaining samples exhibited >70% overlap between replicates, except for H3K9me3 (54% reproducibility). However, H3K9me3 is a broadly distributed histone modification that is challenging for peak-finding software (Wang et al. 2013), and although most H3K9me3 antibodies are of low specificity (Nishikori et al. 2012; Hattori et al. 2013), they can nevertheless distinguish constitutive versus facultative heterochromatin (Trojer and Reinberg 2007).
We also performed ATAC-seq for identifying regions of open chromatin (Buenrostro et al. 2013). Although the standard protocol led to reproducible peaks, initially we could not identify nucleosomal read density, perhaps suggesting a difficulty of obtaining higher resolution fragments from highly differentiated and heterogeneous cell populations. Yet the new Omni-ATAC method (Corces et al. 2017) yielded nucleosomal and subnucleosomal read densities (Supplemental Fig. S2E), which we used for subsequent analysis.
We clustered the six high-confidence histone marks and Omni-ATAC-seq data using a hidden Markov model (ChromHMM) (Ernst and Kellis 2012) into eight chromatin states (Fig. 3A). Each chromatin state is enriched in histone modifications that define specific functional domains, such as actively transcribed regions, heterochromatin, and regulatory loci. We assigned putative chromatin state annotations based on established classifications (Supplemental Table S2; Fig. 3B; Ernst et al. 2011; Rada-Iglesias et al. 2011). We find that, at least at the whole-animal level, approximately half (57%) of the genome is repressed, approximately a fifth (16%) represents actively transcribed genes, and more than a quarter (27%) is regulatory (including 6785 promoters, 13,648 active enhancers, and 3853 “poised” enhancers) (Fig. 3C).
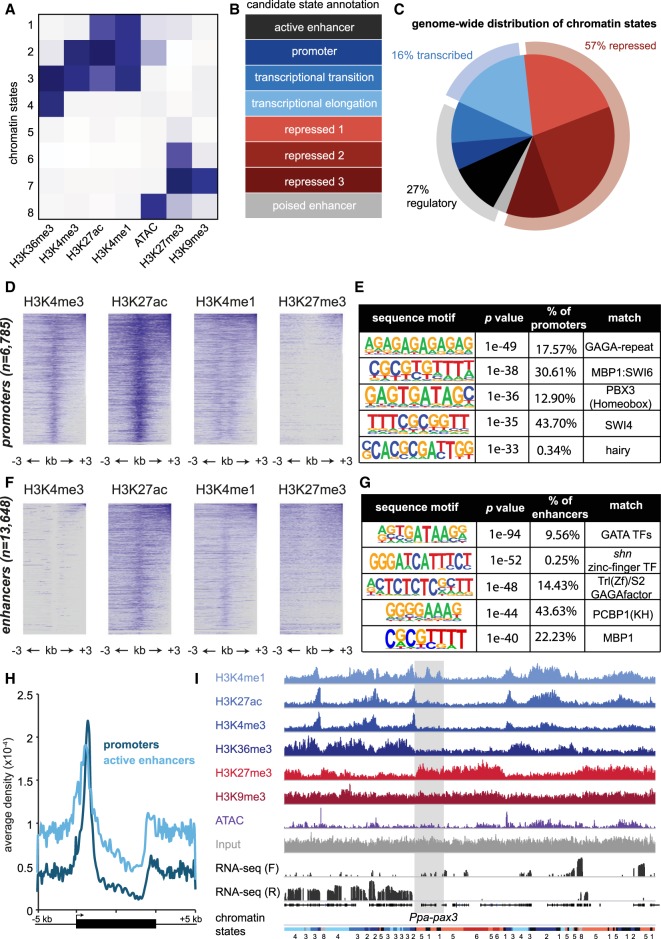
Figure 3.
The epigenome of Pristionchus pacificus. (A) Chromatin states determined through a hidden Markov model (ChromHMM) clustered by histone modifications and ATAC-seq, normalized by coverage. Darker blue represents greater enrichment. (B) Candidate annotation of each chromatin state according to ENCODE/modENCODE data sets (Ernst et al. 2011; Roadmap Epigenomics Consortium et al. 2015). Repressive chromatin states are divided into three categories according to standard definitions of constitutive (repressed 3) and facultative (repressed 1 and 2) heterochromatin. Poised enhancers are defined according to previous annotations of loci containing H3K27me3 and DNase sensitivity. (C) Genome-wide distribution of chromatin states, and further clustering into three categories: repressive, transcribed, or regulatory. (D) Heatmap of indicated histone modifications for promoter chromatin states, in which each line represents a single 6-kb locus centered on the promoter. Heatmap matrices were generated in HOMER, clustered from highest to lowest enrichment, and plotted in R. (E) Position weight matrices of de novo sequence motifs in promoters, queried using HOMER. The table also includes the percentage of promoters containing motif, P-value, and matches to known transcription factors. (F,G) Similar to D,E, but for enhancer chromatin states. (H) Average density plots of promoter (dark blue) and enhancer (light blue) locations relative to gene bodies, extended 5 kb in each direction from their 5′ and 3′ ends. Density values measured using HOMER and plotted in Excel. (I) Epigenomic data of histone modification ChIP-seq, ATAC-seq, and RNA-seq surrounding the Ppa-pax3 gene. Input is included as a reference, and chromatin state annotations are included at the bottom matching the colors in C. ChIP-seq and ATAC-seq coverage are autoscaled per sample, and RNA-seq forward (F) and reverse (R) read coverage is in log-scale.
Next, we verified that histone marks are enriched at the center of promoters and active enhancer annotations, and we performed a de novo motif search (Fig. 3D–G; Heinz et al. 2010). Both promoters (30.6%) and enhancers (22.2%) were enriched in a recognition sequence for MBP1, a yeast transcriptional activator that controls cell-cycle progression (Koch et al. 1993). There is weak homology (BlastP, e = 2 × 10−4) with MBP1 in P. pacificus (UMM-S233-5.4-mRNA-1), and in the future it will be interesting to see if this gene is also involved in cell-cycle control. There were also notable differences between enhancers and promoters, including binding site matches to human homeobox, Drosophila GAGA, and eukaryotic GATA transcription factors, demonstrating the precision of promoter and enhancer annotations, and hinting at the existence of deeply conserved regulatory elements.
As expected, promoter annotations were strongly enriched at the 5′ end of genes (Fig. 3H). There was also another peak near the 3′ end of genes. Enhancers were also enriched at both 5′ and 3′ ends, although they are more evenly distributed throughout gene bodies. The existence of promoter/enhancer elements at the 3′ ends of genes has been observed in other species, and although their functions are still unclear, there are several reports supporting promoter-3′-end chromatin looping to facilitate successive rounds of transcription and enforce directionality (O'Sullivan et al. 2004; Lainé et al. 2009; Grzechnik et al. 2014; Werner et al. 2017).
To verify that our chromatin states correlate with a dynamically regulated gene, we looked at Ppa-pax-3, which our laboratory has shown to be expressed in early juvenile stages but is repressed during development (Yi and Sommer 2007). Indeed, we found Ppa-pax-3 is in a large H3K27me3-repressed domain in adults (Fig. 3I). However, we also noticed two putative enhancers after the first exon and 3′ end, perhaps suggesting preparation for activation in developing embryos. Collectively, these data represent the first genome-wide annotation of chromatin regulation in P. pacificus and, to our knowledge, represents only the second comprehensive data set in nematodes.
Chromatin regulation corresponds to gene expression
We extended the previous single gene example to genome-wide high-depth RNA-seq and binned the adult transcriptome into four expression categories (Fig. 4A), then assessed the chromatin states of each. As predicted, gene bodies (exons and introns) of the highest expressed categories (groups 1 and 2) exhibited enrichment in chromatin states designated as “transcriptional transition” and “elongation.” Conversely, repressive chromatin states were virtually absent from genes in the top two categories. In contrast, the two lowest expression categories (groups 3 and 4) exhibited proportionally greater enrichment in repressive chromatin states and decreased enrichment in transcriptional transition and elongation states (Fig. 4B). Although there was a minor enrichment in promoter chromatin states at 5′ ends and 5′ UTRs among high versus low expression categories, there was a larger difference in repressive chromatin states, especially at the 5′ ends. There was also an increase in enhancer enrichment at 5′ ends and 5′ UTRs in the low expression categories, perhaps reflecting a “poised” chromatin state that is reactive to environmental influence. Although promoters and enhancers exhibit a relatively small portion of the genome (15.6%), they comprise the majority of intergenic regions (Fig. 4B), hinting at a large and mostly unexplored regulatory circuitry in the compact nematode genome.
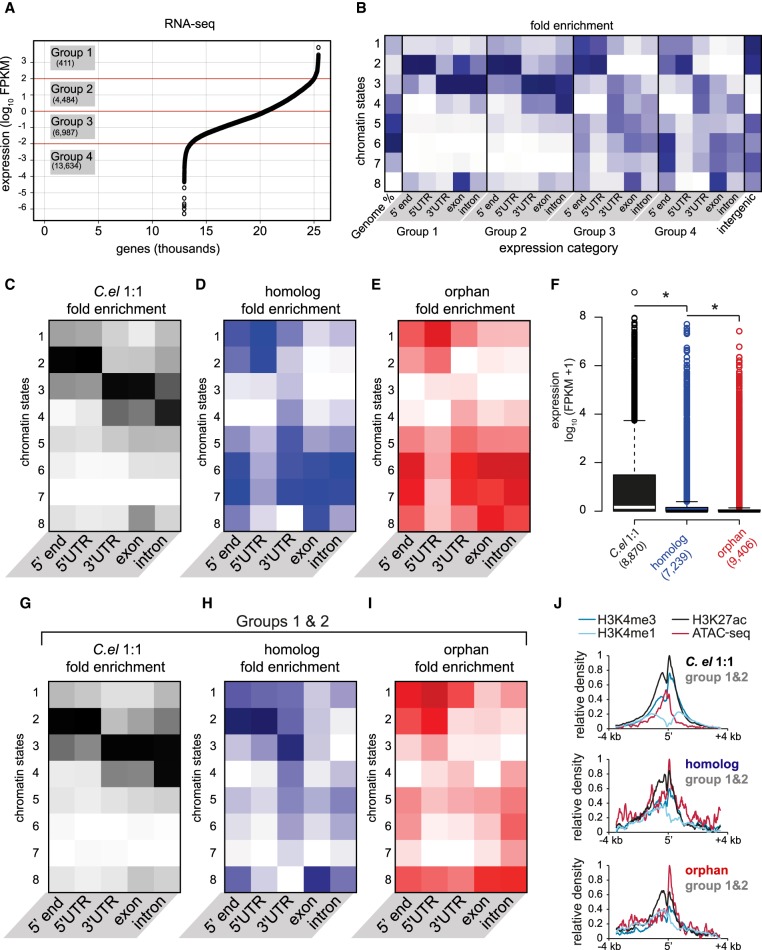
Figure 4.
Chromatin states correlate with expression, but expressed young genes exhibit distinct profiles. (A) Average expression (FPKM) from two biological replicates of RNA-seq, plotted for each gene from highest to lowest along the x-axis. Expression categories were binned according to approximate inflection points. (B) Chromatin state enrichment of each expression category broken down by genetic element (i.e., TSSs, UTRs, exons, and introns). (C–E) Similar to B, but for each evolutionary gene class. (F) Expression of each evolutionary gene class determined from average RNA-seq FPKMs: (*) P-value <0.05, Welch's t-test (two-tailed). (G–I) Similar to B–E, but only for highly expressed (groups 1 and 2) genes belonging to each category. (J) Normalized average densities of H3K4me3, H3K4me1, H3K27ac, and ATAC-seq over a 7-kb window centered at 5′ ends. Densities were measured in HOMER and normalized to the highest and lowest values in each gene class.
Chromatin regulation of evolutionary gene classes
Next, we assessed the chromatin states of evolutionary gene classes. C. elegans 1:1 orthologs resembled the highest expression categories (groups 1 and 2), whereas conserved and orphan genes more closely resembled the lower expression categories (groups 3 and 4) (Fig. 4C–E). These histone patterns reflect the higher expression of C. elegans 1:1 orthologs compared to less conserved gene classes (Fig. 4F). Nevertheless, we noticed a significant number of orphan and homologous expressed gene outliers (Fig. 4F) and wondered whether their chromatin signatures resembled that of expressed C. elegans 1:1 orthologs. Here, we found differences. Specifically, strongly expressed (groups 1 and 2) orphan and homologous genes, which represent only 9.3% and 12.8% of their respective categories, broadly resembled the general chromatin state pattern of their classes except for having reduced repressive histone marks (Fig. 4G–I). Second, chromatin states 3 and 4, representing transcriptional transition and elongation, are more highly represented in C. elegans 1:1 orthologs compared to expressed orphan and conserved genes. Third, C. elegans 1:1 orthologs exhibit little to no signature of active enhancers (chromatin state 1) at their 5′ ends or 5′ UTRs, which are instead dominated by the promoter chromatin state consisting of H3K4me3 and H3K27ac. However, expressed homologous and orphan genes exhibited both promoter and enhancer enrichment at their 5′ ends and 5′ UTRs, and orphan genes, in particular, exhibited greater enrichment in enhancer versus promoter chromatin states.
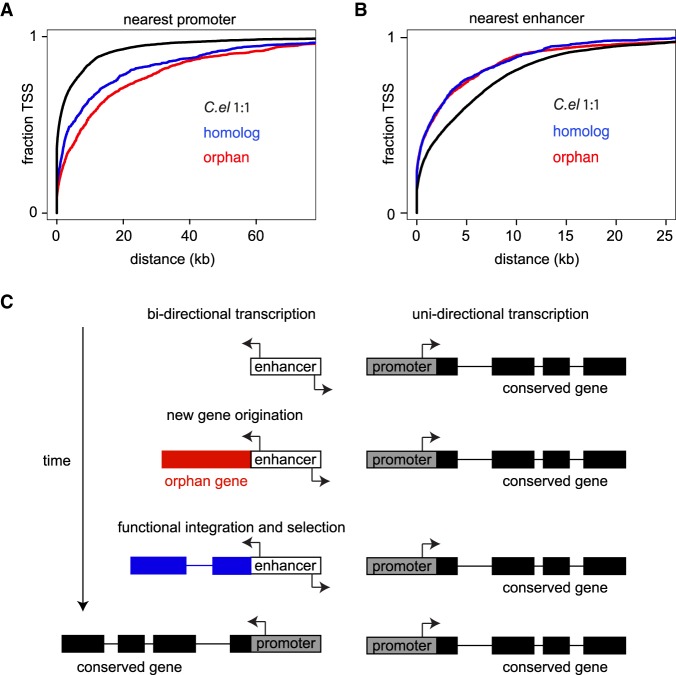
To investigate this difference more closely, we examined the distribution of histone ChIP-seq and ATAC-seq around the 5′ ends of each evolutionary class. Whereas expressed C. elegans 1:1 ortholog TSSs are dominated by H3K4me3 and H3K27ac, expressed orphan and homologous genes exhibit comparatively stronger enrichment of H3K4me1 and ATAC-seq (Fig. 4J). Specifically, C. elegans 1:1 orthologs exhibit an average 5′ H3K4me3/H3K4me1 ratio of 10.1, compared to 2.4 for homologs and 1.4 for orphan genes. Furthermore, although 54% of expressed C. elegans 1:1 ortholog 5′ ends are within 1 kb of an annotated promoter, only 27% of expressed homologous genes and 21% of expressed orphan genes are in similar proximity to promoters (P < 2.2 × 10−16, Wilcoxon rank-sum test) (Fig. 5A). Conversely, 46% of expressed homologous and orphan gene TSSs are within 1 kb of active and poised enhancers, compared to 33% of expressed C. elegans 1:1 ortholog TSSs (P < 2.2 × 10−16, Wilcoxon rank-sum test for both comparisons) (Fig. 5B). Importantly, the expression of groups 1 and 2 orphan and homologous genes are actually higher than C. elegans 1:1 orthologs (P < 2.2 × 10−16, Wilcoxon rank-sum test) (Supplemental Fig. S5), demonstrating that their chromatin architecture is independent of the general correlation with expression. There are two key points from these results. First, the transcription of young genes appears to depend on the absence of repressive heterochromatin, demonstrating a widely held but unconfirmed theory for the requirement of open chromatin in new gene expression. Second, the 5′ end of expressed young genes, especially orphan genes, resemble enhancers rather than canonical promoters.
Figure 5.
Distance of promoters and enhancers to evolutionary gene classes. (A) Distance cumulative frequency distribution of the nearest promoter, or (B) enhancer (active and poised) to transcription start sites (TSSs) from each evolutionary gene category. (C) Model of new gene transcriptional regulation. Enhancers exhibit bidirectional transcription, which can lead to de novo gene expression, or expression of duplications/insertions. If the new gene provides a useful function, selection will occur on not only protein function, but also the gene structure leading to more exons, and on regulatory elements to provide more temporal or spatial control, and more or less transcription. Ultimately, evolution on enhancer sequences will convert it to a traditional promoter.
Genomic position affects chromatin regulation of evolutionary gene classes
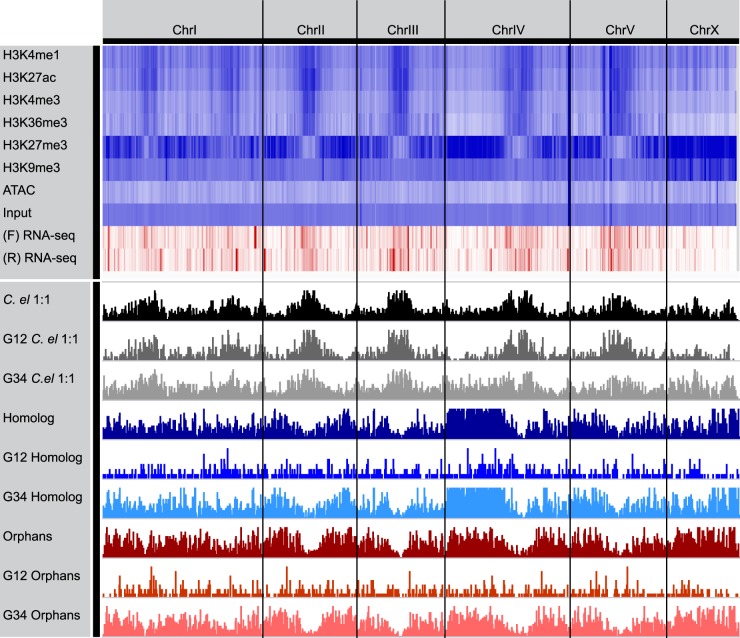
Finally, we analyzed the general pattern of chromatin marks and evolutionary gene classes at the chromosomal level (Fig. 6). We observed strong patterns of activating marks in the center of autosomes II–V and the repressive mark H3K27me3 on the chromosome arms. Conversely the X Chromosome was highly enriched in both repressive marks H3K27me3 and H3K9me3. These patterns have been observed in C. elegans (Liu et al. 2011), and they correspond to general patterns in nematodes of dense clusters of conserved genes and low recombination rates in the center of chromosomes, and species-specific genes and high recombination rates in the chromosome arms (Coghlan 2005). However, Chromosome I in P. pacificus is an exception to other autosomes, in which we observed two bands of activating marks instead of one. Recent analysis suggests that roughly half of P. pacificus Chromosome I is homologous with C. elegans Chromosome X, and the other half is homologous with Chromosome V (Rödelsperger et al. 2017). The P. pacificus chromosome pattern was viewed as ancestral because this organization is also found in the distantly related nematode Bursaphelenchus xylophilus. However, the bipartite presence of histone modifications and conserved genes hints at an ancient chromosomal fusion from an even earlier origin, or frequent and repeated chromosomal fission and fusion events.
Figure 6.
Chromosome-wide distribution of histone modifications reveals distinct patterns for evolutionary gene classes and a double-band pattern on Chr I. Genome-wide patterns of histone modifications from ChIP-seq and ATAC-seq presented as a heatmap with increasing abundance from white to blue, and white to red for RNA-seq (normalized by depth). Also plotted are gene densities of each evolutionary class binned by expressed (groups 1 and 2) or transcriptionally repressed (groups 3 and 4) for each class.
The chromosome-scale distribution of evolutionary gene classes was consistent with histone modification patterns. Specifically, C. elegans 1:1 orthologs, which are strongly expressed, are enriched in the active histone mark chromosome centers. Conversely, the lower expressed homologous and orphan genes are enriched in the chromosome arms, which contain higher recombination rates and a greater density of repressive histone marks. However, these patterns are lost when controlling for expression. Highly transcribed (groups 1 and 2) orphan and homologous genes were more randomly distributed throughout chromosomes, if not slightly biased to be closer toward the centers (Fig. 6). This genome-wide perspective also supports the model that location into open chromatin is a critical factor for origination, or at least transcription, of new genes.
Discussion
Here, we combine the first chromatin state analysis in P. pacificus with taxon-rich nematode phylogenies to analyze the transcriptional regulation of young genes. We identified eight chromatin states that partition the genome into varying levels of repression, transcription, or regulatory elements. Expressed young genes were found in open chromatin states, supporting a widely held model of new gene origination. To our surprise, however, young gene 5′ ends are more similar to enhancers than traditional promoters. We also analyzed young gene transcript structure by long-read Iso-Seq, which revealed a unique signature for each evolutionary gene class. Finally, a bipartite pattern of active histone marks in Chromosome I provides molecular evidence of an ancient chromosomal fusion event ∼180 million years ago. The ability to probe more than 20,000 high-confidence promoters and enhancers will be a valuable resource for future mechanistic studies, especially when combined with the powerful array of genetic, phylogenetic, and ecological tools recently available to P. pacificus.
The origin and subsequent regulation of orphan genes is a widely debated topic that has garnered several theoretical models. Among these are that orphan genes can be transcribed by integrating into open chromatin, or near gene promoters, effectively hijacking their regulatory sequences and thereby mitigating the need to evolve them de novo (Kaessmann et al. 2009; Kaessmann 2010; Tautz and Domazet-Lošo 2011; Chen et al. 2012a,b; Long et al. 2013; McLysaght and Hurst 2016). This model was supported by analyzing the position of transcribed retrogenes in the human genome (Vinckenbosch et al. 2006); however, it was also demonstrated that such integration is often deleterious to the host genes. Indeed, a recent analysis found only ∼14% of mammalian retrogenes utilized preexisting promoters (Carelli et al. 2016). Nevertheless, to date very few tests of these predictions have been performed beyond retrogenes, and the identities of cis-regulatory elements have traditionally been inferred through spatial proximity to genes or known regulatory sequences like TFBSs and CpG islands (Betrán and Long 2003; Carvunis et al. 2012; Chen et al. 2012b; Ni et al. 2012; Ruiz-Orera et al. 2015; Li et al. 2016). The phylogenetic diversity of nematode genomes and our recent chromosome-scale genome (Rödelsperger et al. 2017) allowed us to query all orphan genes in P. pacificus, including but not limited to retrogenes. By applying ChIP-seq and ATAC-seq, we could then interrogate the functional P. pacificus genome, including cis and trans enhancers, and up to eight different chromatin states. The data presented herein support a model of orphan gene integration into open chromatin near enhancers preferentially over promoters.
Enhancers were originally thought of as inactive DNA elements that harbor TFBSs (Wasylyk 1988); however, over the last decade, research from several laboratories has shown that enhancers exhibit bidirectional transcription by RNA polymerase (Chen et al. 2013a; Andersson et al. 2014; Lam et al. 2014). Now there is a growing consensus that enhancers and promoters are similar regulatory elements (Andersson et al. 2015; Kim and Shiekhattar 2015), but promoters have evolved additional sequences to enforce directionality (Grzechnik et al. 2014) or increased expression. Indeed, promoters can even function as enhancers for other genes (Engreitz et al. 2016). Under this paradigm, we propose a model whereby a new gene that originates near an enhancer, and is adaptive, will eventually acquire more sophisticated regulatory architecture, thereby transitioning the enhancer into a promoter (Fig. 5C). This model is complimentary to “proto-promoters” proposed by the Kaessmann laboratory for 8%–9% of rat expressed retrogenes that have H3K4me3 enrichment in rats but H3K4me1 enrichment in syntenic nonexpressed regions in mouse (Carelli et al. 2016). However, here we show that proximity of new genes to enhancers correlates with and presumably drives their transcription, and we extend this argument beyond retrogenes as a general feature of new genes. If these transcripts prove functional, then selection can convert the enhancer to a promoter. This model potentially solves two problems faced by new genes: (1) expression via origination near enhancers, and (2) introduction near enhancers, especially trans enhancers, as opposed to promoters, does not require exchange or competition with the preexisting gene landscape. Nevertheless, we caution that such interpretation is speculative at this point, and examining and then experimentally manipulating H3K4me1/3 at syntenic loci in closely related strains and species is necessary to test these hypotheses.
In principle, this model could operate regardless of the method of new gene origination (de novo, duplication and divergence, or retrotransposition). Transcripts from enhancers or lncRNA promoters generally exhibit less splicing, 3′ processing, and polyadenylation relative to protein coding genes (Derrien et al. 2012) and are often digested by the nuclear exosome (Schlackow et al. 2017). In de novo gene evolution, mutations that recruit sequence-specific splicing factors or 3′ processing factors such as CPSF73 could stabilize enhancer transcripts allowing for their translation and potential functionalization. In some cases, a de novo gene that is acting as a functional ncRNA, referred to as “moonlighting,” could lead to greater expression and a greater window of time to accrue such mutations (Jalali et al. 2016). New genes formed by duplication and insertion, or retrotransposition near an enhancer, could similarly be transcribed, but without the parental regulatory architecture. In this new genomic context, the gene will likely be expressed in different developmental stages or tissues, possibly providing new functions. Although misregulation of genes often coincides with deleterious effects and disease (Lee and Young 2013), in this case, the parental gene is still maintained and operating under normal control, while the copied gene is freed, within limits (Geiler-Samerotte et al. 2011), to explore neofunctionalization.
Compared to the current reference annotation (El Paco), our Iso-Seq annotation identified shorter genes with fewer exons. The distribution was more similar to C. elegans gene structures. Nevertheless, we note that there are still substantially more genes in P. pacificus with more than 10 exons compared to C. elegans (Supplemental Fig. S1F), arguing that further refinement is still required (although an evolutionary divergence in gene length is formally possible). We also explored the genetic structure of young versus old genes. Orphan genes displayed the shortest gene lengths and fewest exons, and C. elegans 1:1 orthologs were the longest and contained the most exons. The result that homologs appear to be intermediate in length and exon number is consistent with a transitional path between old and young genes (Carvunis et al. 2012; Abrusán 2013), but whether this indicates divergence from old genes or de novo evolution from young genes is unknown, likely reflecting examples of both. Iso-Seq also identified that almost a quarter (24%) of expressed genes in P. pacificus have multiple isoforms. Although a subset of observed alternative splicing events can be attributed to splicing errors (Pickrell et al. 2010), there are important examples of alternative isoforms that affect diverse biological processes (Baralle and Giudice 2017). Whether the multiplicity of transcripts observed here are differentially expressed during development or in different environmental conditions, and ultimately if they are functional, will be the focus of future experiments.
Iso-Seq of polyadenylated transcripts and rRNA-depleted total RNA demonstrated that most young genes are polyadenylated. In mammals, noncoding RNAs are un- or underpolyadenylated (Derrien et al. 2012), arguing that most new genes in P. pacificus represent coding transcripts. However, retrogene transcripts that contain their polyadenylation “scar” in the genome may be transcribed directly with a poly(A) tail, and thus appear as polyadenylated regardless of whether they have been pseudogenized or not. Nevertheless, our interpretation that they are mostly coding is consistent with a previous investigation of orphan genes in P. pacificus that found appreciable peptide coverage from mass spectrometry and evidence of negative selection (Prabh and Rödelsperger 2016). Beyond orphan genes, comparing polyadenylated and total RNA Iso-Seq data sets should also be valuable for investigating gene structures of long noncoding RNAs (lncRNAs), including antisense ncRNAs that have been shown to affect phenotypic plasticity in P. pacificus (Serobyan et al. 2016).
Genome-wide, we found most young genes are present in chromosome arms where recombination and repressive chromatin in nematodes is the highest (The C. elegans Sequencing Consortium 1998; Coghlan 2005). However, the ∼10% of young genes that are highly transcribed (expression groups 1 and 2) were more randomly distributed. Thus, although recombination in the arms appears to be a furnace for new gene generation, most of these genes are repressed (expression groups 3 and 4) and have a higher barrier for functionalization. This pattern highlights several unresolved questions. In particular, does the presence in open chromatin reflect rare recombination events or de novo origination? Further, are these transcribed new genes “born” into open chromatin and serve as a template for evolution, or have they already acquired nascent function and their presence in open chromatin is a result of translocation to increase their expression? Additional functional genomic comparisons and synteny analysis may shed light on these questions.
At chromosome-scale resolution, we observed a double-banding of active histone marks on Chromosome I, in contrast to all other autosomes in both P. pacificus and C. elegans (Liu et al. 2011). Based on previous phylogeny and synteny analyses (Rödelsperger et al. 2017), we propose this pattern is a remnant from a fusion event that occurred prior to the split between Diplogasterida and Tylenchida, estimated at ∼180 million years ago (Cutter 2008; Hedges et al. 2015). Then more recently, this portion broke off in the Caenorhabditis lineage. This interpretation parsimoniously explains the long-standing conundrum that Chromosome V in C. elegans has unusual chromosome “arm-like” characteristics, including relatively high recombination rates and a low density of conserved genes (Barnes et al. 1995; The C. elegans Sequencing Consortium 1998; Parkinson et al. 2004). In essence, it looks like a chromosome arm because it is, or more precisely, was, prior to breaking off. If true, the remarkable stability of histone mark patterns suggests that chromatin organization per se could serve as a molecular fossil of past genomic rearrangements. Perhaps probing chromatin structure in conjunction with recombination rates could provide a historical record of genome evolution in other nematodes and organisms.
Methods
Evolutionary gene classification
Nematode phylogenies were schematically drawn using data downloaded and analyzed from Holterman et al. (2017) and van Megen et al. (2009). Evolutionary gene classes were defined in a tiered process. First, we defined conserved genes by BLASTP and TBLASTN analyses; we performed all pairwise searches of P. pacificus proteins as query against 24 nematode protein sets as target, and the proteins of each of the 24 nematodes as query against P. pacificus proteins as target. One BLASTP hit (e < 10−3) in any of these 48 comparisons, or one TBLASTN hit (e < 10−5) using P. pacificus proteins as query against any of the 24 nematode species genomes was enough to classify a gene in P. pacificus as conserved. Any gene that did not fit these criteria was defined as a P. pacificus “orphan gene.” Within the conserved gene class, we then defined 1:1 orthologs as having the best reciprocal BLASTP hit (e ≤ 10−3) between C. elegans and P. pacificus (sorted by e-value, and raw scores were used to break ties). Conserved genes that were not in this 1:1 ortholog class but were previously identified by homology in at least one of the 24 nematodes species, were defined as “homologous genes.” We kept the e-value cutoff relatively “high” because of the large phylogenetic distance between C. elegans and P. pacificus, and hence more conservative with respect to our orphan gene lists.
Nematode synchronization and collection
P. pacificus (PS312) cultures for ChIP-, ATAC-, and RNA-seq were synchronized with bleach and grown on agar to young adults following Werner et al. (2017) (Supplemental Methods). Worm pellets were flash-frozen until processing. For Iso-Seq, we used mixed-developmental stage (egg, J2, and J4/young adult) RNA at equimolar ratios.
Native histone ChIP-seq
Native (non-cross-linked) chromatin immunoprecipitation (ChIP) of histone post-translational modifications was performed by combining nematode nuclear isolation (Steiner et al. 2012) with native ChIP (Brand et al. 2008). Coprecipitated DNA was PCR-amplified and converted to Illumina libraries using the TruSeq Nano kit (Illumina) and sequenced on a HiSeq 3000. See Supplemental Methods for the detailed protocol.
ATAC-seq
Omni-ATAC-seq was performed on mixed-stage purified nuclei following the Corces et al. (2017) protocol, with a few modifications (Supplemental Methods) and sequenced on an Illumina HiSeq 3000.
Iso-Seq
RNA was extracted from different developmental time points separately using TRIzol Reagent (Invitrogen), then after the quality control, equal amounts of RNA from different time points were pooled. cDNA synthesis of “direct” Iso-Seq was performed directly using SMARTer PCR cDNA Synthesis Kit (Clontech Laboratories), and “total RNA” was first rRNA-depleted with Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) (Illumina), then in vitro polyadenylated with Poly(A) Polymerase (New England Biolabs). Direct and rRNA-depleted cDNA were converted into SMRTbell libraries following the guidelines provided by Pacific Biosciences. SMRT Link software version 4.0.0 (Pacific Biosciences) was used to convert subreads to circular consensus sequences and identify full-length nonchimeric reads, which were mapped to the El Paco genome using GMAP (Wu and Watanabe 2005). See Supplemental Methods for the detailed protocol.
RNA-seq
Whole-animal young adult (64–68 h post-bleaching) frozen pellets were freeze-thawed 3× in TRIzol Reagent (Invitrogen) before purification, converted to sequencing libraries with the NEBNext Ultra Directional RNA-seq for Illumina kit, and sequenced on a HiSeq 3000. See Supplemental Methods for the detailed protocol.
Bioinformatics
All sequencing data were mapped to the El Paco genome assembly (Rödelsperger et al. 2017) using GMAP (Wu and Watanabe 2005) for Iso-Seq, Bowtie 2 (Langmead and Salzberg 2012) for ChIP- and ATAC-seq, and HISAT2 (Kim et al. 2015) for RNA-seq. Peaks were obtained by MACS2 (Zhang et al. 2008), and only samples containing 50% overlap between replicates were kept. Overlapping peaks were merged using BEDTools (Supplemental Table S1; Quinlan and Hall 2010). Coverage plots were calculated using BEDTools or HOMER (Heinz et al. 2010) with merged replicate files, and plotted in R (R Core Team 2016). Chromatin states were obtained with ChromHMM (Ernst and Kellis 2012) using merged replicate input files. Distances to nearest promoter or enhancers were performed with BEDTools. See Supplemental Methods for the detailed procedure of all bioinformatic steps.
Data access
Raw and processed data sets from this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number PRJEB24584.
Supplementary Material
Acknowledgments
We thank current members of the Sommer laboratory and Dr. Talia Karasov for thoughtful critique of experiments, results, and interpretations. This study was funded by the Max Planck Society.
Author contributions: M.S.W. and R.J.S. conceived and designed all experiments. M.S.W. conducted ChIP- and ATAC-seq with assistance from T.L.; M.S.W, B.S., and C.L. performed Iso-Seq; N.P. conducted phylogenetic analysis and prepared evolutionary gene category data sets; M.S.W. performed all bioinformatic analysis. M.S.W. wrote the manuscript with assistance from R.J.S.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.234872.118.
Freely available online through the Genome Research Open Access option.
References
- Abrusán G. 2013. Integration of new genes into cellular networks, and their structural maturation. Genetics 195: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Sandelin A, Danko CG. 2015. A unified architecture of transcriptional regulatory elements. Trends Genet 31: 426–433. [DOI] [PubMed] [Google Scholar]
- Baralle FE, Giudice J. 2017. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 18: 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes TM, Kohara Y, Coulson A, Hekimi S. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran P, Rödelsperger C. 2015. Microevolution of duplications and deletions and their impact on gene expression in the nematode Pristionchus pacificus. PLoS One 10: e0131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran P, Rödelsperger C, Prabh N, Serobyan V, Markov GV, Hirsekorn A, Dieterich C. 2015. Ancient gene duplications have shaped developmental stage-specific expression in Pristionchus pacificus. BMC Evol Biol 15: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento G, Ogawa A, Sommer RJ. 2010. Co-option of the hormone-signalling module dafachronic acid–DAF-12 in nematode evolution. Nature 466: 494–497. [DOI] [PubMed] [Google Scholar]
- Betrán E, Long M. 2003. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics 164: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. 2008. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc 3: 398–409. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Kaessmann H. 2004. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet 36: 1061–1063. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhao R, Jiang H, Wang W. 2008. De novo origination of a new protein-coding gene in Saccharomyces cerevisiae. Genetics 179: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli FN, Hayakawa T, Go Y, Imai H, Warnefors M, Kaessmann H. 2016. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res 26: 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ni X, Krinsky BH, Zhang YE, Vibranovski MD, White KP, Long M. 2012a. Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J 31: 2798–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Spletter M, Ni X, White KP, Luo L, Long M. 2012b. Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell Rep 1: 118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RA, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J. 2013a. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res 23: 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M. 2013b. New genes as drivers of phenotypic evolution. Nat Rev Genet 14: 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A. 2005. Nematode genome evolution. In WormBook (ed. The C. elegans Research Community). 10.1895/wormbook.1.15.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol 17: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14: 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD. 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol 25: 778–786. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussoti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch M, Long M. 1999. Intron–exon structures of eukaryotic model organisms. Nucleic Acids Res 27: 3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, et al. 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet 40: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. 2016. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539: 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci 108: 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik P, Tan-Wong SM, Proudfoot NJ. 2014. Terminate and make a loop: regulation of transcriptional directionality. Trends Biochem Sci 39: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Taft JM, Swist KM, Luo H, Witt H, Slattery M, Koide A, Ruthenburg AJ, Krajewski K, Strahl BD, et al. 2013. Recombinant antibodies to histone post-translational modifications. Nat Methods 10: 992–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman M, Karegar A, Mooijman P, van Megen H, van den Elsen S, Vervoort MTW, Quist CW, Karssen G, Decraemer W, Opperman CH, et al. 2017. Disparate gain and loss of parasitic abilities among nematode lineages. PLoS One 12: e0185445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalali S, Gandhi S, Scaria V. 2016. Navigating the dynamic landscape of long noncoding RNA and protein-coding gene annotations in GENCODE. Hum Genomics 10: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H. 2010. Origins, evolution, and phenotypic impact of new genes. Genome Res 20: 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet 10: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. 2009. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet 25: 404–413. [DOI] [PubMed] [Google Scholar]
- Kieninger MR, Ivers NA, Rödelsperger C, Markov GV, Sommer RJ, Ragsdale EJ. 2016. The nuclear hormone receptor NHR-40 acts downstream of the sulfatase EUD-1 as part of a developmental plasticity switch in Pristionchus. Curr Biol 26: 2174–2179. [DOI] [PubMed] [Google Scholar]
- Kim TK, Shiekhattar R. 2015. Architectural and functional commonalities between enhancers and promoters. Cell 162: 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261: 1551–1557. [DOI] [PubMed] [Google Scholar]
- Lainé JP, Singh BN, Krishnamurthy S, Hampsey M. 2009. A physiological role for gene loops in yeast. Genes Dev 23: 2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Li W, Rosenfeld MG, Glass CK. 2014. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 39: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. 2013. Transcriptional regulation and its misregulation in disease. Cell 152: 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Foster CM, Gan Q, Nettleton D, James MG, Myers AM, Wurtele ES. 2009. Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J 58: 485–498. [DOI] [PubMed] [Google Scholar]
- Li ZW, Chen X, Wu Q, Hagmann J, Han TS, Zou YP, Ge S, Guo YL. 2016. On the origin of de novo genes in Arabidopsis thaliana populations. Genome Biol Evol 8: 2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung M-S, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD. 2013. New gene evolution: Little did we know. Annu Rev Genet 47: 307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MG, Rödelsperger C, Witte H, Riebesell M, Sommer RJ. 2015. The orphan gene dauerless regulates dauer development and intraspecific competition in nematodes by copy number variation. PLoS Genet 11: e1005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet 17: 567–578. [DOI] [PubMed] [Google Scholar]
- Neme R, Tautz D. 2013. Phylogenetic patterns of emergence of new genes support a model of frequent de novo evolution. BMC Genomics 14: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Zhang YE, Nègre N, Chen S, Long M, White KP. 2012. Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome. PLoS Biol 10: e1001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikori S, Hattori T, Fuchs SM, Yasui N, Wojcik J, Koide A, Strahl BD, Koide S. 2012. Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J Mol Biol 424: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018. [DOI] [PubMed] [Google Scholar]
- Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, Schmid R, Hall N, Barrell B, Waterston RH, et al. 2004. A transcriptomic analysis of the phylum Nematoda. Nat Genet 36: 1259–1267. [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pai AA, Gilad Y, Pritchard JK. 2010. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet 6: e1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabh N, Rödelsperger C. 2016. Are orphan genes protein-coding, prediction artifacts, or non-coding RNAs? BMC Bioinformatics 17: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale EJ, Müller MR, Rödelsperger C, Sommer RJ. 2013. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell 155: 922–933. [DOI] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD. 2013. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet 9: e1003860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C, Meyer JM, Prabh N, Lanz C, Bemm F, Sommer RJ. 2017. Single-molecule sequencing reveals the chromosome-scale genomic architecture of the nematode model organism Pristionchus pacificus. Cell Rep 21: 834–844. [DOI] [PubMed] [Google Scholar]
- Rosso L, Marques AC, Weier M, Lambert N, Lambot MA, Vanderhaeghen P, Kaessmann H. 2008. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol 6: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota-Stabelli O, Daley AC, Pisani D. 2013. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol 23: 392–398. [DOI] [PubMed] [Google Scholar]
- Ruiz-Orera J, Hernandez-Rodriguez J, Chiva C, Sabidó E, Kondova I, Bontrop R, Marqués-Bonet T, Albà MM. 2015. Origins of de novo genes in human and chimpanzee. PLoS Genet 11: e1005721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos ME, Le Bouquin A, Crumière AJJ, Khila A. 2017. Taxon-restricted genes at the origin of a novel trait allowing access to a new environment. Science 358: 386–390. [DOI] [PubMed] [Google Scholar]
- Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell 65: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serobyan V, Xiao H, Namdeo S, Rödelsperger C, Sieriebriennikov B, Witte H, Röseler W, Sommer RJ. 2016. Chromatin remodelling and antisense-mediated up-regulation of the developmental switch gene eud-1 control predatory feeding plasticity. Nat Commun 7: 12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira AB, Trontin C, Cortijo S, Barau J, Del Bem LE, Loudet O, Colot V, Vincentz M. 2013. Extensive natural epigenetic variation at a de novo originated gene. PLoS Genet 9: e1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Sommer RJ, Dieterich C. 2012. Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans. BMC Genomics 13: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, McGaughran A. 2013. The nematode Pristionchus pacificus as a model system for integrative studies in evolutionary biology. Mol Ecol 22: 2380–2393. [DOI] [PubMed] [Google Scholar]
- Sommer RJ, Streit A. 2011. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Ann Rev Genet 45: 1–20. [DOI] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, Chougule K, Gao D, Iwata A, Goicoechea JL, et al. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet 50: 285–296. [DOI] [PubMed] [Google Scholar]
- Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S. 2012. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res 22: 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T. 2011. The evolutionary origin of orphan genes. Nat Rev Genet 12: 692–702. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. 2007. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell 28: 1–13. [DOI] [PubMed] [Google Scholar]
- van Megen H, van den Elsen S, Holterman M, Karssen G, Mooyman P, Bongers T, Holovachov O, Bakker J, Helder J. 2009. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 11: 927–950. [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci 103: 3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Hager GL. 2014. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15: 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lunyak VV, Jordan IK. 2013. BroadPeak: a novel algorithm for identifying broad peaks in diffuse ChIP-seq datasets. Bioinformatics 29: 492–493. [DOI] [PubMed] [Google Scholar]
- Wasylyk B. 1988. Enhancers and transcription factors in the control of gene expression. Biochim Biophys Acta 951: 17–35. [DOI] [PubMed] [Google Scholar]
- Werner MS, Sullivan MA, Shah RN, Nadadur RD, Grzybowski AT, Galat V, Moskowitz IP, Ruthenburg AJ. 2017. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat Struct Mol Biol 24: 596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H, Moreno E, Rödelsperger C, Kim J, Kim JS, Streit A, Sommer RJ. 2015. Gene inactivation using the CRISPR/Cas9 system in the nematode Pristionchus pacificus. Dev Genes Evol 225: 55–62. [DOI] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. [DOI] [PubMed] [Google Scholar]
- Yi B, Sommer RJ. 2007. The pax-3 gene is involved in vulva formation in Pristionchus pacificus and is a target of the Hox gene lin-39. Development 134: 3111–3119. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.