Abstract
Background and purpose
Polymorphisms in coagulation genes have been associated with early-onset ischemic stroke. Here we pursue an a priori hypothesis that genetic variation in the endothelial-based receptors of the thrombomodulin−protein C system (THBD and PROCR) may similarly be associated with early-onset ischemic stroke. We explored this hypothesis utilizing a multi-stage design of discovery and replication.
Methods
Discovery was performed in the Genetics-of-Early-Onset Stroke (GEOS) Study, a biracial population-based case-control study of ischemic stroke among men and women aged 15–49 including 829 cases of first ischemic stroke (42.2% African-American) and 850 age-comparable stroke-free controls (38.1% African-American). Twenty-four single-nucleotide-polymorphisms (SNPs) in THBD and 22 SNPs in PROCR were evaluated. Following LD pruning (r2≥0.8), we advanced uncorrelated SNPs forward for association analyses. Associated SNPs were evaluated for replication in an early-onset ischemic stroke population (onset-age<60 years) consisting of 3676 cases and 21118 non-stroke controls from 6 case–control studies. Lastly, we determined if the replicated SNPs also associated with older-onset ischemic stroke in the METASTROKE data-base.
Results
Among GEOS Caucasians, PROCR rs9574, which was in strong LD with 8 other SNPs, and one additional independent SNP rs2069951, were significantly associated with ischemic stroke (rs9574, OR = 1.33, p = 0.003; rs2069951, OR = 1.80, p = 0.006) using an additive-model adjusting for age, gender and population-structure. Adjusting for risk factors did not change the associations; however, associations were strengthened among those without risk factors. PROCR rs9574 also associated with early-onset ischemic stroke in the replication sample (OR = 1.08, p = 0.015), but not older-onset stroke. There were no PROCR associations in African-Americans, nor were there any THBD associations in either ethnicity.
Conclusion
PROCR polymorphisms are associated with early-onset ischemic stroke in Caucasians.
Introduction
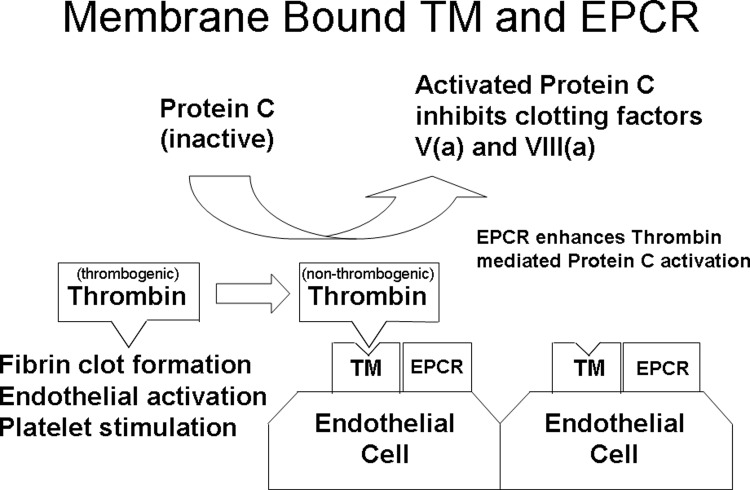
Hemostasis is a dynamic balance between factors that promote clot formation and factors that promote antithrombotic activity and/or fibrinolysis. Central to this balance is the thrombomodulin-protein C antithrombotic system that is located on the endothelial surface, which plays a key role in regulating both coagulation and inflammation. Thrombomodulin forms a 1:1 complex with thrombin on the vascular endothelium, thereby inhibiting the procoagulant actions of thrombin and converting protein C to activated protein C [1]. Activated protein C promotes fibrinolysis, inhibits thrombosis by inactivating coagulation factors Va and VIIIa, and reduces inflammation by decreasing white blood cell and nuclear factor kappa-B activation [2–5]. The activation of protein C by the thrombin-thrombomodulin complex is enhanced when the substrate protein C is presented by the endothelial cell protein C receptor. These relationships are demonstrated in Fig 1. Given the central role that the thrombomodulin-protein C pathway plays in thrombosis and inflammation, the genes encoding these receptor proteins are promising stroke susceptibility candidate genes. Prior genetic studies across the cardiovascular disease (CVD) spectrum have demonstrated increased risk in younger (vs. older) patients [6], including thrombosis [7]. Variants in other prothrombotic genes have also previously been associated with ischemic stroke, again, more consistently with early-onset versus later-onset disease [8,9,10]. As such, an a priori hypothesis to evaluate these 2 genes in the setting of ischemic stroke was developed and successfully funded. To this end we tested the hypothesis that THBD (OMIM 188040) and PROCR (OMIM 600646) variants are associated with early-onset ischemic stroke using a 2-stage discovery and replication design, and then addressed whether the identified variants also associated with older-onset disease.
Fig 1. The thrombomodulin−protein C receptor (TM-EPCR) system located on the endothelial surface.
Methods
Discovery population
The Genetics of Early Onset Stroke (GEOS) Study is a population-based case-control study designed to identify genes associated with early-onset ischemic stroke and to characterize interactions of identified stroke genes and/or SNPs with environmental risk factors. Participants (921 stroke cases and 941 controls) were recruited from the greater Baltimore-Washington area over 4-time periods between 1992–2008 [11]. The population is primarily composed of two self-reported ethnic groups, European-Americans (Caucasians) (EA; 54.5%) and African-Americans (AA; 40.4%), with the remaining 5.1% of individuals comprising other ethnicities including Chinese, Japanese, other Asians, and other unspecified. Stroke cases were hospitalized with a first cerebral infarction identified by discharge surveillance from one of the 59 hospitals in the greater Baltimore-Washington area and direct referral from regional neurologists. Cases were enrolled in either the sub-acute or chronic post-stroke phases as based on previously described case identification and enrollment procedures [8,11]. Ischemic strokes with the following characteristics were excluded from participation: stroke occurring as an immediate consequence of trauma; stroke within 48 hours after a hospital procedure, stroke within 60 days after the onset of a non-traumatic subarachnoid hemorrhage, and cerebral venous thrombosis. The abstracted hospital records of cases were reviewed and adjudicated for ischemic stroke subtype by a pair of neurologists per previously published procedures [12,13], with disagreements resolved by a third neurologist. The ischemic stroke subtype classification system retains information on all probable and possible causes, and is reducible to the more widely used TOAST system [14] that assigns each case to a single category. All cases had age of first stroke between 15–49 years and were recruited within three years of stroke.
For these genetic analyses, we included only Caucasians and African-Americans, and excluded cases with known single-gene or mitochondrial disorders recognized by a distinctive phenotype (e.g. cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL), mitochondrial encephalopathy with lactic acidosis and stroke-like episodes (MELAS), homocystinuria, Fabry disease, or sickle cell anemia). Additional exclusions included: mechanical aortic or mitral valve at the time of index stroke; untreated or actively treated bacterial endocarditis at the time of the index stroke; neurosyphilis or other CNS infections; neurosarcoidosis; severe sepsis with hypotension at the time of the index stroke; cerebral vasculitis by angiogram and clinical criteria; post-radiation arteriopathy; left atrial myxoma; major congenital heart disease; and cocaine use in the 48 hours prior to their stroke.
Control participants without a history of stroke were identified by random-digit dialing. Controls were balanced to cases by age and region of residence in each study period and were additionally balanced for race/ethnicity in the latter two participant collection periods.
Traditional stroke risk factors and other study variables, including age, race/ethnicity, history of hypertension, diabetes, myocardial infarction (MI) and current smoking status (defined as use within one month prior to event for cases and at a comparable reference time for controls), were also collected during a standardized interview. Age, race/ethnicity, and cigarette smoking status were determined by subject reports (or proxy report, if a participant was unable to answer). Hypertension, diabetes mellitus, and MI were determined by asking study participants (or a proxy) whether a physician had ever told them that they had the condition. This study was conducted with the consent of all study subjects and was approved by the University of Maryland at Baltimore Institutional Review Board.
Genotyping
Genomic DNA was isolated from a variety of sample types, including cell line (55.2%), whole blood (43.1%), mouthwash (0.4%) and buccal swab (0.05%). Whole genome amplification (Qiagen REPLI-g kit, Valencia, CA, USA) was used to obtain sufficient DNA for genotyping in 1.3% of samples. The genotype data implemented in this study was obtained from two fixed-content SNP panels developed by Illumina (Illumina, San Diego, CA, USA), a genome-wide association (GWA) genotyping array, the HumanOmni1-Quad_v1-0_B BeadChip, and a cardiovascular disease (CVD) SNP panel, the ITMAT-Broad-CARe array, that included THBD and PROCR. Genotyping quality from both arrays was excellent with individual SNP call rates > 98% and a between-panel concordance rate of 99.996% based on study duplicates (for further details please see S1 File) [11].
SNP Selection and inclusion criteria
From each array, we extracted all SNPs in the THBD (chr 20: 22,974,270–22,978,301 bp) and PROCR (chr 20: 33,223,435–33,228,826 bp) genes (NCBI Build 37) and then added all additional SNPs within 10kb upstream and downstream to capture regulatory regions. In total, we identified 24 SNPs in THBD (17 GWA; 18 CVD; 11 overlap) and 22 SNPs in PROCR (22 GWA; 4 CVD; 4 overlap). Across both genes there were nine SNPs unique to European-Americans (EA; Caucasians) and six SNPs unique to African-Americans. There was an overlap of 15 SNPs between the two genotype sources with an average SNP call concordance rate between platforms of >99%. Other predefined selection and quality control criteria included required; 1) HWE p-values >0.01, 2) Call rate > 98%, and MAF >0.01 in race-stratified samples.
Analyses
Genetic association analyses were performed using the PLINK statistical software program [15]. Prior to the association analysis we pruned the genotyped SNPs on the basis of linkage-disequilibrium (LD), such that for any SNPs in high LD (r2≥0.8) we retained only a single representative SNP. This LD pruning was performed within each ethnic group separately using PLINK. Within each ethnic group separately, we then used an additive logistic regression model to test for association of genotype with stroke, adjusting for age and gender, and population structure (principal components from GWAS array or CVD panel). Secondary analyses were performed to determine if any observed associations were more prominent in those with cardiovascular risk based on the presence of the traditional risk factors as described above and previously [11]. For all association analyses, we defined a significant Boferroni-corrected p-value as p<0.05 divided by the number of gene- and ethnicity-specific independent (LD-pruned) SNPs (i.e., p = 0.05 / # independent LD-Pruned SNPs).
Replication and extension to older onset stroke
We sought to replicate any associated SNPs identified in the GEOS Study in an independent set of early-onset stroke studies (the Genetics of Early Onset Stroke Consortium) previously reported by Cheng et al. [16] after excluding the GEOS samples from the replication set, as meta-analyzed implementing the GWAMA program. The studies included in the replication were: CADISP, Cervical Artery Dissection and Ischemic Stroke Patients [17]; MILANO, Besta Stroke Study; RACE, Risk Assessment of Cerebrovascular Events Study; SIFAP, Stroke in Young Fabry Patients; and WTCCC2, Wellcome Trust Case–Control Consortium 2 [16]. The details of each of replication cohort are available in the supplementary data of Cheng et al. [16]. In short, only confirmed ischemic strokes, first ever or recurrent, were included in these studies, TIAs and hemorrhagic strokes were excluded. SNPs whose associations replicated in the Genetics of Early Onset Stroke Consortium were then tested for association with later- or older-onset stroke via in silico lookup in the METASTROKE Consortium [17]; the mean age of stroke onset ranged from 57.3–81.6 years among the 14 contributing cohorts of METASTROKE (not including GEOS). Further details regarding the data collection, organization, and relationships between METASTROKE and the other studies involved can be found in the S1 File and S1 Dataset.
The aggregated data that support the findings of this study are available from the corresponding author and participating studies upon reasonable request as listed in the S1 Dataset. Further, each study can be contacted to attain their data individually, and for the NIH funded studies, study data is available via request from the database of Genotypes and Phenotypes (dbGaP) @ https://www.ncbi.nlm.nih.gov/gap/.
Results
Characteristics of the young-onset stroke discovery and replication studies are provided in Table 1. After exclusions, the GEOS Discovery Stage included 448 ischemic stroke cases (mean age stoke-onset = 41.0 yrs) and 498 controls of EA ancestry, and 381 ischemic stroke cases (mean age stroke-onset = 41.9 yrs) and 352 controls of AA ancestry. Further demographic and risk factor characteristics by case–control status for the GEOS Discovery Stage are described in Table A in S1 File.
Table 1. Characteristics of participating studies.
| Study | Cases | Controls | Ancestry | Country | |||||
|---|---|---|---|---|---|---|---|---|---|
| Subjects, n | Age, mean (SD) | Male, n (%) | Subjects, n | Age, mean (SD) | Male, n (%) | External Control | |||
| Stage 1: Discovery Stage | |||||||||
| GEOS EA | 448 | 41.0 (7.0) | 275 (61.4) | 498 | 39.5 (6.7) | 282 (56.6) | No | EA | USA |
| GEOS AA | 381 | 41.9 (6.8) | 207 (54.3) | 352 | 40.0 (6.8) | 196 (55.7) | No | AA | USA |
| Total | 829 | 850 | |||||||
| Stage 2: Replication Stage | |||||||||
| CADISP | 555 | 43.7 (9.9) | 339 (61.1) | 9259 | N/A | N/A | No | EA | Belgium, France, Germany, Italy, Switzerland, and Finland |
| MILANO | 201 | 45.0 (10.4) | 120 (60.9) | 407 | 50.8 (8.1) | 357 (87.8) | No | EA | Italy |
| RACE 1 | 1218 | 50.1 (9.9) | 638 (52.4) | 1158 | 51.9 (7.9) | 613 (53) | PROMIS | South Asian | Pakistan |
| RACE 2 | 339 | 50.2 (9.2) | 272 (80.4) | 3295 | 60.9 (13.2) | 1838 (55.8) | PROMIS | South Asian | Pakistan |
| SIFAP | 981 | 41.7 (7.4) | 599 (61.1) | 1824 | 55.2 (11.6) | 899 (49.3) | KORA | EA | Germany |
| WTCCC2-UK | 382 | 51.9 (7.3) | 228 (59.7) | 5175 | 52 | 2611 (50.5) | British Birth Cohort and UK Blood Service Control | EA | UK |
| Total | 3,676 | 21,118 | |||||||
AA indicates African ancestry; CADISP, Cervical Artery Dissection and Ischemic Stroke Patients; EA, European ancestry; GEOS, Genetics of Early-Onset Stroke; MILANO, Besta Stroke Study; RACE, Risk Assessment of Cerebrovascular Events Study; SIFAP, Stroke in Young Fabry Patients; and WTCCC2, Wellcome Trust Case-Control Consortium 2.
LD pruning resulted in 13 THBD SNPs in EAs and 13 THBD SNPs in AAs, and an additional 4 THBD SNPs in EAs on the CVD chip; and 5 PROCR SNPs in EAs and 11 PROCR SNPs in AAs (see Table 2and Table B in S1 File).
Table 2. PROCR association results of lead LD SNPs in Discovery GEOS European-Americans for all-stroke detailing allelic variants, effect allele frequencies (EAF) and as stratified by the absence or presence of vascular risk factors, and lead LD SNP young-onset replication results.
| Discovery: GEOS | Replication: Young-Onset Stroke Cohort without GEOS (Meta-analysis of 6 studies) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 448 cases | 498 controls | Primary Model (adjusted for age and gender) |
0 Risk Factors (167 cases / 315 controls) |
≥ 1 Risk Factors (273 cases / 183 controls) |
3671 Cases / 21119 Controls | ||||||||||||
| rsID* | LD_SNPs | BP_ Build37 |
EAF | EAF | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | EAF |
| rs9574 (C/G) (intronic) | rs945960 (intronic) rs1415774 (intronic) rs2069952 (intronic) rs6088753 (intronic) rs2378337 (intronic) rs6087683 (intronic) rs2065979 (intronic) rs6088747 (intronic) |
33764632 | 0.49 | 0.41 | 1.33 | 1.11–1.61 | 0.003 | 1.50 | 1.14–1.99 | 0.0046 | 1.23 | 0.94–1.61 | 0.1367 | 1.08 | 1.02–1.16 | 0.015 | 0.397 |
| rs2069951 (G/A) (intronic) |
NA | 33763764 | 0.96 | 0.93 | 1.80 | 1.18–2.75 | 0.006 | 4.82 | 2.14–10.89 | 0.0002 | 0.80 | 0.43–1.52 | 0.4870 | 1.08 | 0.93–1.27 | 0.331 | 0.956 |
| rs6087682 (A/T) (intronic) |
rs6060278 (intronic) |
33752897 | 0.80 | 0.75 | 1.26 | 1.01–1.58 | 0.050 | 1.33 | 0.96–1.87 | 0.0910 | 1.14 | 0.82–1.6 | 0.4510 | 1.03 | 0.97–1.12 | 0.368 | 0.766 |
| rs867186 (A/G) (missense) |
rs7265317 (intronic) rs11907011 (intronic) rs8119351 (intronic) |
33764554 | 0.91 | 0.90 | 1.18 | 0.86–1.64 | 0.311 | 1.63 | 1–2.69 | 0.0533 | 0.80 | 0.5–1.32 | 0.3789 | 1.10 | 0.98–1.26 | 0.133 | 0.865 |
| rs1415775 (C/T) (intronic) |
NA | 33765771 | 0.76 | 0.74 | 1.09 | 0.89–1.35 | 0.428 | 1.01 | 0.74–1.38 | 0.9694 | 1.23 | 0.92–1.67 | 0.1773 | 0.99 | 0.92–1.07 | 0.745 | 0.759 |
*EAF = Effect allele bolded
Association analyses of the EA revealed a significant association of PROCR rs9574 with ischemic stroke, with the rs9574C allele (MAF case/control = 0.49/0.41) associated with a 1.33-fold increased odds of stroke compared to the G allele (p = 0.003; Table 2). Another independent PROCR SNP rs2069951 was also associated with ischemic stroke significantly in EA (OR = 1.80, P = 0.006). None of the PROCR SNPs were associated with stroke in AA, nor were any associations observed between THBD SNPs and stroke in either ethnic group. An exploratory analyses of stroke subtypes (e.g., TOAST-defined large artery (LA), small artery, cardioembolic, and cryptogenic) did not reveal significant associations with any variants, although the sample sizes were small (range: 33 LA to 230 cryptogenic) in EAs.
To further characterize the EA PROCR associations, we performed a secondary analysis to evaluate the impact of concomitant vascular risk factors. We repeated the association analysis with additional adjustment for vascular risk factors (i.e., hypertension, diabetes mellitus, angina/MI, and current cigarette smoking) and found the association results to be essentially unchanged (data not shown). However, when stratifying for the presence or absence of each vascular risk factor, we observed a stronger, but non-statistically significant, association of rs9574 and rs2069951 with stroke in the absence of each risk factor when considered separately, with the direction of association similar across each risk factor (data not shown). To obtain a more comprehensive picture, we therefore compared subjects with zero vascular risk factors to those with at least one vascular risk factor. In the subset of EA participants without vascular risk factors (n = 167 cases and 315 controls), both SNPs were more strongly associated with stroke (rs9574, OR = 1.50, p = 0.0046; rs2069951, OR = 4.82, p = 0.0002; see Table 2).
Replication of the PROCR rs9574 association with early onset stroke
We sought to replicate the PROCR rs9574 and rs2069951 association in the Early-Onset Stroke Consortium [16], with exclusion of the GEOS study. This replication sample included 3,676 cases and 21,118 controls. Only rs9574 replicated; the effect allele frequency of rs9574C was 0.39, with association analyses demonstrating an OR of 1.08 (p = 0.015) (Table 2). The ischemic stroke replication results of the LD-pruned PROCR SNPs among Caucasians in the young-onset stroke consortium, inclusive and exclusive of GEOS, are shown in Table C in S1 File. There was no significant correlation between stroke subtypes and PROCR rs9574 in the replication samples.
Older-onset stroke
To determine if PROCR rs9574 and/or the other previously identified LD-pruned SNPs were associated with older-onset stroke, these SNPs were evaluated in the METASTROKE cohort [18]. Lookups found no replication of these SNPs with ischemic stroke or in any subtype (data not shown).
Discussion
We observed a significant association between PROCR rs9574 and early-onset ischemic stroke that replicated in a large independent sample of early-onset ischemic stroke. Prior studies have demonstrated that mutations in PROCR have been associated with venous thromboembolism (VTE) [19] and myocardial infarction [20,21], as well as with late fetal loss during pregnancy [22]. Specific to ischemic stroke, PROCR associations have been inconsistently reported [23, 24], which may in part be related to variations in the age and ethnicity of the populations evaluated. Our study is the first to specifically identify and replicate PROCR associations in a young-onset ischemic stroke population of European descent. Our failure to detect an association in GEOS AA may reflect low power and/or that a true causal variant is not well tagged in African-Americans. Our findings add to the growing evidence that prothrombotic mechanisms may be more important for younger compared to older onset stroke as demonstrated with other established prothrombotic variants including Prothrombin G20210A [8], Factor XI [10] and Factor V Leiden [25]. This is also in line with the lack of association we observed between PROCR rs9574 and older onset stroke in the METASTROKE lookup. Our findings are also consistent with the hypothesis that PROCR (and perhaps by analogy other thrombosis-related genes) may be more relevant, or easier to detect, in the setting of a paucity of standard vascular risk factors, as these factors may induce risk via non-thrombotic mechanisms. This may again also partially explain why we did not see replication in the older-onset METASTROKE population, which also has a greater vascular risk factor burden. Differing genetic mechanisms may also partially explain why African-Americans did not demonstrate the associations seen in their Caucasian counterparts.
Strengths of our study include the well-phenotyped and relatively large young-onset discovery sample size, as well as the large replication sample. Notably, GEOS cases are part of the METASROKE as were other young-onset strokes, yet despite the inclusion of the GEOS samples, there was no association seen in this primarily older-onset cohort. A potential study limitation relates to the replication sample, which is predominantly of European rather than North-American origin, although the MAFs were roughly similar on both sides of the Atlantic. Another limitation is that our discovery population-based design, with recruitment at over 50 regional hospitals, precluded consistent assessment of the presence of patent foramen ovale (PFO) and potential paradoxical embolism among cases, given PROCR genetics are known to increase the risk of venous thromboembolism. This is important because an established mechanism by which PROCR variation could cause ischemic stroke is via venous thrombosis and paradoxical embolization. Our study was also limited to non-fatal ischemic strokes, so the possibility that our findings are due to a survival bias cannot be ruled out; though this is unlikely given the low case-fatality rate in this population [26]. Another limitation is that our study provides no information about the role of PROCR in ischemic stroke among young adults with a personal or family history of prior early-onset thrombotic events. Lastly, while some intronic mutations can affect gene expression levels by introducing novel splice sites, activating novel promoters (which may direct sense or antisense transcription causing alterations in mRNA, miRNA or lncRNA expression), or by introducing/eliminating enhancer activity, our study does not provide any such detailed mechanistic analyses. Despite these shortcomings, we have identified several PROCR variants in strong LD that associate and replicate with ischemic stroke among young Caucasians. While these findings are interesting, it is too early to assess their clinical implications regarding anticoagulation and/or genetic testing, as examples. Further replication and research are required to better understand these findings.
Conclusion
PROCR, but not THBD, polymorphisms are associated with early-onset ischemic stroke in young Caucasians.
Supporting information
(Table A) GEOS Characteristics by case–control status. (Table B) Results of linkage-disequilibrium pruning by ethnic group using the PLINK. SNPs in high LD (r2≥0.8) retained only a single representative SNP. None of the listed SNPs here were associated with all-ischemic stroke (results not shown) in the GEOS Discovery population. (Table C) PROCR SNP all-ischemic stroke replication results for Caucasians in the young-onset stroke replication cohort: a) without GEOS, and b) with GEOS.
(DOCX)
(DOCX)
Data Availability
The aggregated data that support the findings of this study are available from the corresponding author and participating studies upon reasonable request. Further, each study can be contacted to attain their data individually. Specific data access information for each study has been included in the Supporting Information files. For the NIH funded studies, study data is available via request from the database of Genotypes and Phenotypes (dbGaP) @ https://www.ncbi.nlm.nih.gov/gap/.
Funding Statement
This study was supported in part by NIH grants U01 NS069208, R01 NS100178, and R01 NS105150; an Epidemiology of Aging Training Program Grant, NIH/NIA T32 AG000262; the U.S. Department of Veterans Affairs, and the American Heart Association Cardiovascular Genome-Phenome Study (grant# 15GPSPG23770000), and an American Heart Association Discovery Grant supported by Bayer Group (grant# 17IBDG33700328). Further details regarding the data collection, organization, funding and relationships between METASTROKE and the other studies involved can be found below. Genetics of Early Onset Stroke (GEOS) Study (Baltimore, USA): GWAS data for the GEOS Study was supported by the National Institutes of Health Genes, Environment and Health Initiative (GEI) grant U01 HG004436, as part of the GENEVA consortium under GEI, with additional support provided by the Mid-Atlantic Nutrition and Obesity Research Center (P30 DK072488); and the Office of Research and Development, Medical Research Service, and the Baltimore Geriatrics Research, Education, and Clinical Center of the Department of Veterans Affairs. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University (contract number HHSN268200782096C). Assistance with data cleaning was provided by the GENEVA Coordinating Center (U01 HG 004446; PI Bruce S Weir). Study recruitment and collection of datasets were supported by a cooperative agreement with the Division of Adult and Community Health, Centers for Disease Control and by grants from the National Institute of Neurological Disorders and Stroke (NINDS) and the NIH Office of Research on Women's Health (R01 NS45012, U01 NS069208-01). METASTROKE: METASTROKE is a collaboration of numerous international studies with the aim of validating associations from previous GWAS and identifying novel genetic associations through meta-analysis of GWAS datasets for ischemic stroke and its subtypes. Included studies are as follows: ASGC: Australian population control data were derived from the Hunter Community Study. We also thank the University of Newcastle for funding and the men and women of the Hunter region who participated in this study. This research was funded by grants from the Australian National and Medical Health Research Council (NHMRC Project Grant ID: 569257), the Australian National Heart Foundation (NHF Project Grant ID: G 04S 1623), the University of Newcastle, the Gladys M Brawn Fellowship scheme, and the Vincent Fairfax Family Foundation in Australia. Elizabeth G Holliday was supported by a Fellowship from the National Heart Foundation and National Stroke Foundation of Australia (ID: 100071). BRAINS: Bio-Repository of DNA in Stroke (BRAINS) is partly funded by a Senior Fellowship from the Department of Health (UK) to P Sharma, the Henry Smith Charity and the UK-India Education Research Institutive (UKIERI) from the British Council. HPS: Heart Protection Study (HPS) (ISRCTN48489393) was supported by the UK Medical Research Council (MRC), British Heart Foundation, Merck and Co (manufacturers of simvastatin), and Roche Vitamins Ltd (manufacturers of vitamins). Genotyping was supported by a grant to Oxford University and CNG from Merck and Co. Jemma C Hopewell acknowledges support from the British Heart Foundation (FS/14/55/30806). ISGS: Ischemic Stroke Genetics Study (ISGS)/Siblings With Ischemic Stroke Study (SWISS) was supported in part by the Intramural Research Program of the NIA, NIH project Z01 AG-000954-06. ISGS/SWISS used samples and clinical data from the NIH-NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds), human subjects protocol numbers 2003-081 and 2004-147. ISGS/SWISS used stroke-free participants from the Baltimore Longitudinal Study of Aging (BLSA) as controls. The inclusion of BLSA samples was supported in part by the Intramural Research Program of the NIA, NIH project Z01 AG-000015-50, human subjects protocol number 2003-078. The ISGS study was funded by NIH-NINDS grant R01 NS-42733 (JF Meschia). The SWISS study was funded by NIH-NINDS grant R01 NS-39987 (J F Meschia). This study used the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov). MGH-GASROS: MGH Genes Affecting Stroke Risk and Outcome Study (MGH-GASROS) was supported by NINDS (U01 NS069208), the American Heart Association/Bugher Foundation Centers for Stroke Prevention Research 0775010N, the NIH and NHLBI's STAMPEED genomics research program (R01 HL087676), and a grant from the National Center for Research Resources. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research resources. MILANO: Milano - Besta Stroke Register Collection and genotyping of the Milan cases within CEDIR were supported by the Italian Ministry of Health (grant numbers: RC 2007/LR6, RC 2008/LR6; RC 2009/LR8; RC 2010/LR8; GR-2011-02347041). FP6 LSHM-CT-2007-037273 for the PROCARDIS control samples. WTCCC2: Wellcome Trust Case-Control Consortium 2 (WTCCC2) was principally funded by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z and WT084724MA). The Stroke Association provided additional support for collection of some of the St George's, London cases. The Oxford cases were collected as part of the Oxford Vascular Study which is funded by the MRC, Stroke Association, Dunhill Medical Trust, National Institute of Health Research (NIHR) and the NIHR Biomedical Research Centre, Oxford. The Edinburgh Stroke Study was supported by the Wellcome Trust (clinician scientist award to C Sudlow), and the Binks Trust. Sample processing occurred in the Genetics Core Laboratory of the Wellcome Trust Clinical Research Facility, Western General Hospital, Edinburgh. Much of the neuroimaging occurred in the Scottish Funding Council Brain Imaging Research Centre (www.sbirc.ed.ac.uk), Division of Clinical Neurosciences, University of Edinburgh, a core area of the Wellcome Trust Clinical Research Facility and part of the SINAPSE (Scottish Imaging Network—A Platform for Scientific Excellence) collaboration (www.sinapse.ac.uk), funded by the Scottish Funding Council and the Chief Scientist Office. Collection of the Munich cases and data analysis was supported by the Vascular Dementia Research Foundation. M Farrall and A Helgadottir acknowledge support from the BHF Centre of Research Excellence in Oxford and the Wellcome Trust core award (090532/Z/09/Z). VISP: The GWAS component of the Vitamin Intervention for Stroke Prevention (VISP) study was supported by the United States National Human Genome Research Institute (NHGRI), grant U01 HG005160 (PI Michèle Sale & Bradford Worrall), as part of the Genomics and Randomized Trials Network (GARNET). Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the NIH to the Johns Hopkins University. Assistance with data cleaning was provided by the GARNET Coordinating Center (U01 HG005157; PI Bruce S Weir). Study recruitment and collection of datasets for the VISP clinical trial were supported by an investigator-initiated research grant (R01 NS34447; PI James Toole) from the United States Public Health Service, NINDS, Bethesda, Maryland. Control data obtained through the database of genotypes and phenotypes (dbGAP) maintained and supported by the United States National Center for Biotechnology Information, US National Library of Medicine. WHI: Funding support for WHI-GARNET was provided through the NHGRI GARNET (Grant Number U01 HG005152). Assistance with phenotype harmonisation and genotype cleaning, as well as with general study coordination, was provided by the GARNET Coordinating Center (U01 HG005157). Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH Genes, Environment, and Health Initiative (GEI; U01 HG004424). SiGN: The Stroke Genetics Network (SiGN) study was funded by a cooperative agreement grant from the National Institute of Neurological Disorders and Stroke (NINDS) U01 NS069208. Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which is fully funded through a federal contract from the National Institutes of Health (NIH) to the Johns Hopkins University (contract no. HHSN268200782096C). The Biostatistics Department Genetics Coordinating Center at the University of Washington (Seattle) provided more extensive quality control of the genotype data through a subcontract with CIDR. Additional support to the Administrative Core of SiGN was provided by the Dean’s Office, University of Maryland School of Medicine. This work was supported by grants received from the German Federal Ministry of Education and Research (BMBF) in the context of the e:Med program (e:AtheroSysMed), the FP7 European Union project CVgenes@target (261123), the DFG as part of the CRC 1123 (B3), the Corona Foundation and the Fondation Leducq (Transatlantic Network of Excellence on the Pathogenesis of Small Vessel Disease of the Brain).
References
- 1.Esmon CT, Owen WG. Identification of an endothelial cell cofactor for thrombin-catalyzed activation of Protein C. Proc Natl Acad Sci USA. 1981;78:2249–52. https://www.ncbi.nlm.nih.gov/pubmed/?term=7017729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esmon CT. Thrombomodulin as a model of molecular mechanism that modulates protease specificity and function at the vessel surface. FASEB J. 1995;9:946–55. https://www.ncbi.nlm.nih.gov/pubmed/?term=7615164 [DOI] [PubMed] [Google Scholar]
- 3.Esmon CT. The regulation of natural anticoagulant pathways. Science. 1987;235:1348–52. https://www.ncbi.nlm.nih.gov/pubmed/?term=3029867 [DOI] [PubMed] [Google Scholar]
- 4.Barnes PJ, Karin M. Nuclear factor-kappa b: a pivotal transcription factor in chronic inflammatory disease. N Engl J Med. 1997;336:1066–71. 10.1056/NEJM199704103361506 https://www.ncbi.nlm.nih.gov/pubmed/?term=9091804 [DOI] [PubMed] [Google Scholar]
- 5.Esmon CT. Role of coagulation inhibitors in inflammation. Thromb Haemost. 2001;86:51–6. https://www.ncbi.nlm.nih.gov/pubmed/?term=11487041 [PubMed] [Google Scholar]
- 6.Sayed-Tabatabaei FA, Schut AF, Arias Vásquez A, Bertoli-Avella AM, Hofman A, Witteman JC, et al. Angiotensin converting enzyme gene polymorphism and cardiovascular morbidity and mortality: the Rotterdam Study. J Med Genet. 2005;42:26–30. https://www.ncbi.nlm.nih.gov/pubmed/15635071 10.1136/jmg.2004.022756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridker PM, Glynn RJ, Miletich JP, Goldhaber SZ, Stampfer MJ, Hennekens CH. Age-specific incidence rates of venous thromboembolism among heterozygous carriers of factor V Leiden mutation. Ann Intern Med. 1997;126:528–31. https://www.ncbi.nlm.nih.gov/pubmed/9092318 [DOI] [PubMed] [Google Scholar]
- 8.Jiang B, Ryan KA, Hamedani A, Cheng Y, Sparks MJ, Koontz D, et al. Prothrombin G20210A mutation is associated with young-onset stroke: the genetics of early-onset stroke study and meta-analysis. Stroke. 2014;45:961–7. https://www.ncbi.nlm.nih.gov/pubmed/24619398 10.1161/STROKEAHA.113.004063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng YC, Cole JW, Kittner SJ, Mitchell BD. Genetics of ischemic stroke in young adults. Circ Cardiovasc Genet. 2014;7:383–92. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4231871/ 10.1161/CIRCGENETICS.113.000390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson E, Nilsson S, Jood K, Norrving B, Engström G, Blomstrand C, et al. Genetic variants of coagulation factor XI show association with ischemic stroke up to 70 years of age. PLoS One. 2013;8:e75286 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3783404/ 10.1371/journal.pone.0075286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng YC, O'Connell JR, Cole JW, Stine OC, Dueker N, McArdle PF, et al. Genome-wide association analysis of ischemic stroke in young adults. G3 (Bethesda). 2011;1:505–14. 10.1534/g3.111.001164 https://www.ncbi.nlm.nih.gov/pubmed/22384361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson CJ, Kittner SJ, McCarter RJ, Sloan MA, Stern BJ, Buchholz D, et al. Interrater reliability of an etiologic classification of ischemic stroke. Stroke. 1995;26:46–51. https://www.ncbi.nlm.nih.gov/pubmed/?term=7839396 [DOI] [PubMed] [Google Scholar]
- 13.Kittner SJ, Stern BJ, Feeser BR, Hebel R, Nagey DA, Buchholz DW, et al. Pregnancy and the risk of stroke. N Engl J Med. 1996;335:768–74. https://www.ncbi.nlm.nih.gov/pubmed/?term=8703181 10.1056/NEJM199609123351102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams HP Jr, Bendixen BH, Kapelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke: Definitions for use in a multicenter clinical trial: TOAST: Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. https://www.ncbi.nlm.nih.gov/pubmed/?term=7678184 [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81: 559–75. https://www.ncbi.nlm.nih.gov/pubmed/?term=PMC1950838 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng YC, Stanne TM, Giese AK, Ho WK, Traylor M, Amouyel P, et al. Genome-Wide Association Analysis of Young-Onset Stroke Identifies a Locus on Chromosome 10q25 Near HABP2. Stroke. 2016;47:307–16. http://www.ncbi.nlm.nih.gov/pubmed/26732560 10.1161/STROKEAHA.115.011328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debette S, Kamatani Y, Metso TM, Kloss M, Chauhan G, Engelter ST, et al. Common variation in PHACTR1 is associated with susceptibility to cervical artery dissection. Nat Genet. 2015;47:78–83. 10.1038/ng.3154 https://www.ncbi.nlm.nih.gov/pubmed/?term=25420145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–62. https://www.ncbi.nlm.nih.gov/pubmed/23041239 10.1016/S1474-4422(12)70234-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinds DA, Buil A, Ziemek D, Martinez-Perez A, Malik R, Folkersen L, et al. Genome-wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum Mol Genet. 2016;25:1867–74.https://www.ncbi.nlm.nih.gov/pubmed/26908601 10.1093/hmg/ddw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guella I, Duga S, Ardissino D, Merlini PA, Peyvandi F, Mannucci PM, et al. Common variants in the haemostatic gene pathway contribute to risk of early-onset myocardial infarction in the Italian population. Thromb Haemost. 2011;106:655–64. 10.1160/TH11-04-0247 https://www.ncbi.nlm.nih.gov/pubmed/21901231 [DOI] [PubMed] [Google Scholar]
- 21.Medina P, Navarro S, Corral J, Zorio E, Roldán V, Estellés A, et al. Endothelial protein C receptor polymorphisms and risk of myocardial infarction. Haematologica. 2008;93:1358–63. 10.3324/haematol.13066 https://www.ncbi.nlm.nih.gov/pubmed/18757851 [DOI] [PubMed] [Google Scholar]
- 22.Franchi F, Biguzzi E, Cetin I, Facchetti F, Radaelli T, Bozzo M, et al. Mutations in the thrombomodulin and endothelial protein C receptor genes in women with late fetal loss. Br J Haematol. 2001;114:641–6. https://www.ncbi.nlm.nih.gov/pubmed/11552992 [DOI] [PubMed] [Google Scholar]
- 23.Reiner AP, Carty CL, Jenny NS, Nievergelt C, Cushman M, Stearns-Kurosawa DJ, et al. PROC, PROCR and PROS1 polymorphisms, plasma anticoagulant phenotypes, and risk of cardiovascular disease and mortality in older adults: The Cardiovascular Health Study. J Thromb Haemost. 2008;6:1625–32. 10.1111/j.1538-7836.2008.03118.x https://www.ncbi.nlm.nih.gov/pubmed/18680534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson NC, Raffield LM, Lange LA, Lange EM, Longstreth WT Jr, Chauhan G, et al. Associations of activated coagulation factor VII and factor VIIa-antithrombin levels with genome-wide polymorphisms and cardiovascular disease risk.J Thromb Haemost. 2018;16:19–30. 10.1111/jth.13899 https://www.ncbi.nlm.nih.gov/pubmed/29112333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamedani AG, Cole JW, Cheng YC, Sparks MJ, O'Connell JR, Stine OC, et al. Factor V leiden and ischemic stroke risk: The Genetics of Early Onset Stroke (GEOS) study. J Stroke Cerebrovasc Dis. 2013;22:419–23. https://www.ncbi.nlm.nih.gov/pubmed/22100829 10.1016/j.jstrokecerebrovasdis.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naess H, Nyland HI, Thomassen L, Aarseth J, Nyland G, Myhr KM. Incidence and short-term outcome of cerebral infarction in young adults in western Norway. Stroke. 2002;33:2105–8. https://www.ncbi.nlm.nih.gov/pubmed/?term=12154271 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Table A) GEOS Characteristics by case–control status. (Table B) Results of linkage-disequilibrium pruning by ethnic group using the PLINK. SNPs in high LD (r2≥0.8) retained only a single representative SNP. None of the listed SNPs here were associated with all-ischemic stroke (results not shown) in the GEOS Discovery population. (Table C) PROCR SNP all-ischemic stroke replication results for Caucasians in the young-onset stroke replication cohort: a) without GEOS, and b) with GEOS.
(DOCX)
(DOCX)
Data Availability Statement
The aggregated data that support the findings of this study are available from the corresponding author and participating studies upon reasonable request. Further, each study can be contacted to attain their data individually. Specific data access information for each study has been included in the Supporting Information files. For the NIH funded studies, study data is available via request from the database of Genotypes and Phenotypes (dbGaP) @ https://www.ncbi.nlm.nih.gov/gap/.