Abstract
The use of genome-scale sequencing allows for identification of genetic findings beyond the original indication for testing (secondary findings). The ClinGen Actionability Working Group’s (AWG) protocol for evidence synthesis and semi-quantitative metric scoring evaluates four domains of clinical actionability for potential secondary findings: severity and likelihood of the outcome, and effectiveness and nature of the intervention. As of February 2018, the AWG has scored 127 genes associated with 78 disorders (up-to-date topics/scores are available at www.clinicalgenome.org). Scores across these disorders were assessed to compare genes/disorders recommended for return as secondary findings by the American College of Medical Genetics and Genomics (ACMG) with those not currently recommended. Disorders recommended by the ACMG scored higher on outcome-related domains (severity and likelihood), but not on intervention-related domains (effectiveness and nature of the intervention). Current practices indicate that return of secondary findings will expand beyond those currently recommended by the ACMG. The ClinGen AWG evidence reports and summary scores are not intended as classifications of actionability, rather they provide a resource to aid decision makers as they determine best practices regarding secondary findings. The ClinGen AWG is working with the ACMG Secondary Findings Committee to update future iterations of their secondary findings list.
Keywords: secondary findings, exome sequencing, genome sequencing, clinical utility, incidental findings
Introduction
The use of genome-scale sequencing (i.e., exome and genome sequencing) in clinical practice and research settings is increasing due to diagnostic utility, decreasing costs, and improved access to testing. As this use increases, providers, patients, and policy-makers must decide how to act upon results beyond the original indication for testing (secondary findings). The return of relevant secondary findings allows patients and providers the opportunity to implement medical interventions that could improve future health outcomes. This concept is referred to as “clinical actionability” (Hunter et al., 2016).
While most stakeholders, including patients and clinicians, endorse that secondary findings should be offered to patients undergoing clinical sequencing, the criteria for determining which results to return remains largely unclear (Mackley, Fletcher, Parker, Watkins, & Ormondroyd, 2017). To address this issue, the American College of Medical Genetics and Genomics (ACMG) developed a list of secondary findings recommended to be offered for return to patients, with plans for updating and refinement of this list over time (Green et al., 2013). This list was most recently updated in 2017, and is referred to as the ACMG SF v2.0 (Kalia et al., 2017).
To create a centralized resource for summarizing and assessing evidence related to clinical actionability, the Clinical Genome Resource (ClinGen) created the Actionability Working Group (AWG). The ClinGen AWG defines clinical actionability of secondary findings as the implementation of an available clinical intervention in asymptomatic or previously undiagnosed adults with the goal of ameliorating or preventing a future negative health outcome. The ClinGen AWG previously published its methods development and actionability scores for the genes/disorders originally recommended for return by the ACMG (Green et al., 2013; Hunter et al., 2016). Since this time, scoring by the ClinGen AWG has continued and expanded beyond those genes and conditions included in the ACMG SF v2.0 list. The findings of the AWG are a publicly-available resource that can be accessed and used by a variety of stakeholders to guide the implementation and evaluation of policy-making and recommendations surrounding the return of secondary findings to patients.
In this paper we present updated findings of the ClinGen AWG and compare the scoring metric results for genetic disorders on the ACMG SF v2.0 list versus those not on the list (either because they have not yet been evaluated by the ACMG or because they were excluded from the list).
Materials and Methods
The ClinGen AWG’s protocol for standardized evidence synthesis and generation of consensus scores has been published elsewhere (Hunter et al., 2016). Briefly, the AWG uses a standardized protocol to generate a synthesis of available evidence regarding clinical actionability for each topic (i.e., the genetic disorder and associated gene(s)). Nominations for topics considered by the AWG come from a variety of sources (e.g., AWG members, other ClinGen collaborators, and external groups, including other NIH-funded projects). The AWG then applies a scoring metric to generate actionability scores based on the information in these reports. For each topic, the AWG may score multiple outcome–intervention pairs. For example, hereditary breast and ovarian cancer was scored using three outcome and intervention pairs: (1) breast cancer and surveillance, (2) breast cancer and risk-reducing surgery, and (3) ovarian cancer and risk-reducing surgery. For each outcome–intervention pair, AWG consensus scores are generated for four domains related to clinical actionability on a scale of 0-3: (1) severity of the outcome; (2) likelihood that the outcome will occur (penetrance); (3) effectiveness of the intervention to modify the outcome; and (4) nature of the intervention (a measure of the burden and risk to the patient) (Table 1). In addition, the domains of likelihood and effectiveness are assigned a letter score to represent the evidence base. Initial scoring is performed by multiple members of the AWG independently. These scorers include a core team of clinical geneticists and genetic counselors, as well as invited clinicians with topic-specific expertise, such as cardiology. Following group discussion, AWG members then provide a final score with the consensus scores representing the most frequent final score (i.e., mode of the score distribution). Final evidence reports and consensus scores for topics assessed to date are available on the ClinGen website (www.clinicalgenome.org).
Table 1:
ClinGen Actionability Working Group semi-quantitative score metric (Originally published in Hunter et, 2016, scoring metric adapted from Berg et al, 2016)
| Domain | Scores |
|---|---|
| Severity: What is the nature of the threat to health to an individual carrying a clearly deleterious allele in this gene? | 3 = Reasonable possibility of sudden death 2 = Reasonable possibility of death or major morbidity 1 = Modest morbidity 0 = Minimal or no morbidity |
| Likelihood of disease: What is the chance that a serious outcome will materialize given a deleterious variant (akin to penetrance)? | 3 = >40% chance 2 = 5–39% chance 1 = 1–4% chance 0 = <1% chance |
| Effectiveness of specific interventions: How effective is the selected, specific intervention for preventing or significantly diminishing the risk of harm? | 3 = Highly effective 2 = Moderately effective 1 = Minimally effective 0 = Controversial or unknown effectiveness IN = Ineffective/No intervention† |
| Nature of intervention: How risky, medically burdensome, or intensive is a given intervention? | 3 = Low risk, or medically acceptable and low-intensity interventions 2 = Moderate risk, moderately acceptable or intensive interventions 1 = Greater risk, less acceptable and substantial interventions 0 = High risk, poorly acceptable or intensive interventions |
| State of the knowledge base: What is the level of evidence?‡ | A = Substantial evidence, or evidence from a high tier (Tier 1) B = Moderate evidence, or evidence from a moderate tier (Tier 2) C = Minimal evidence, or evidence from a lower tier (Tier 3 or 4) D = Poor evidence, or evidence not provided in the report E = Evidence based on expert contributions (Tier 5) |
† Do not score the remaining categories.
‡ Tier 1: Evidence from a systematic review/meta-analysis or a clinical practice guideline clearly based on a systematic review; Tier 2: Evidence from clinical practice guidelines using expert consensus; Tiers 3 and 4: Evidence from a non-systematic review of evidence with additional primary literature cited (Tier 3) or not cited (Tier 4); Tier 5: Expert contribution or non-systematically identified evidence.
Selection of outcome–intervention pairs for analysis
For the present study, we compiled all the AWG outcome–intervention consensus scores generated through February 2018. For the purposes of this analysis, we selected only a single outcome–intervention pair for each topic to give each topic equal weight in the analysis. To prioritize pairs for the analysis, we computed a summary score for each outcome and intervention by adding scores across the four domains of actionability. For each topic, we used the outcome–intervention pair with the highest summary score. When multiple outcome–intervention pairs tied for the highest summary score, we considered the individual domains to determine which pair to use in our analysis, as follows. If scoring for the four domains was consistent across outcome–intervention pairs, we included that scoring pattern in the analyses once, though it could be attributed to more than one outcome–intervention pair. For example, while two outcome–intervention pairs (recurrent serositis-colchicine and joint problems-colchicine) tied for the highest summary score in Familial Mediterranean Fever (autosomal dominant), both pairs had the same scoring pattern (severity=2, likelihood=3, effectiveness=3, and nature of the intervention=2). Thus, we only included these scores once in the analysis. If the individual domains of the outcome–intervention pairs differed, we prioritized the pair with the highest score in the following domains as tie breakers: effectiveness, followed by severity, followed by likelihood.
For situations where the AWG scored topics separately by categories defined by gene (e.g., BRCA1 and BRCA2 were scored separately for hereditary breast and ovarian cancer) or zygosity (e.g., familial hypercholesterolemia), we used one pair for each category.
Analysis approach
Each topic may include more than one gene and/or more than one condition. Thus, for the purposes of this analysis, it was necessary for us to decide how to classify each topic in terms of its inclusion on the ACMG SF v2.0 list (Kalia et al., 2017). There were some topics assessed by the AWG where only a subset of the genes was included by the ACMG (e.g., the AWG assessed MLH1, MSH2, MSH6, PMS2, and EPCAM for Lynch syndrome while EPCAM was not included on the ACMG SF v2.0 list). For this analysis, these topics were still considered to be listed on the ACMG SF v2.0 list, even if all the genes scored by the AWG were not listed. For the summary score and each of the four domains of scoring, we compared the mean scores (t-test) by their presence or absence on the ACMG SF v2.0 list. We compared the distribution of summary scores using a χ2 test. Domains that demonstrated any potential violations to the assumption of normal distribution underwent sensitivity analyses using non-parametric testing. All statistical analyses were conducted using the statistical program Stata (Version 15.1, StataCorp, College Station, Texas).
Results
The AWG has scored a total of 78 topics associated with 127 genes, where 14 of these topics have been updated since the original publication of the actionability scores (Supplemental Table 1) (Hunter et al., 2016). Across these topics, the AWG has scored a total of 213 outcome–intervention pairs. Applying the selection criteria outlined above, we determined 88 outcome–intervention pairs were eligible for analysis, which accounts for the 78 topics, 4 of which had gene-specific scoring (Table 2; Supp. Table S1). Thirty-six of these pairs represent genes/disorders currently included on the ACMG SF v2.0 list.
Table 2:
Summary of 88 outcome-intervention pairs
| All (n=88) | ACMG (n= 36) | Non-ACMG (n= 52) | ||||
|---|---|---|---|---|---|---|
| Range | Mean (SD) | Range | Mean (SD) | Range | Mean (SD) | |
| Summary score | 6-12 | 9.6 (1.2) | 8-12 | 10.0 (0.9)† | 6-11 | 9.2 (1.2)† |
| Severity | 1-3 | 2.1 (0.5) | 1-3 | 2.3 (0.5)† | 1-3 | 2.0 (0.4)† |
| Likelihood | 0-3 | 2.5 (0.7) | 2-3 | 2.7 (0.5)‡ | 0-3 | 2.3 (0.9)‡ |
| Effectiveness | 1-3 | 2.4 (0.6) | 1-3 | 2.4 (0.6) | 1-3 | 2.3 (0.7) |
| Nature of the Intervention | 0-3 | 2.6 (0.6) | 1-3 | 2.6 (0.5) | 0-3 | 2.6 (0.6) |
† Mean ACMG and non-ACMG scores different at p<0.01 significance threshold.
‡ Mean ACMG and non-ACMG scores different at p<0.05 significance threshold.
Comparison of summary scores
The mean summary score (added across the 4 domains) of the pairs included on the ACMG SF v2.0 list was significantly higher than those not currently included on the list (10.0 vs. 9.2, p<0.01) (Table 2). In addition, the distribution of the summary scores is skewed higher for topics on the ACMG SF v2.0 list (p<0.05) (Table 2, Figure 1) than for topics not on the list.
Figure 1:
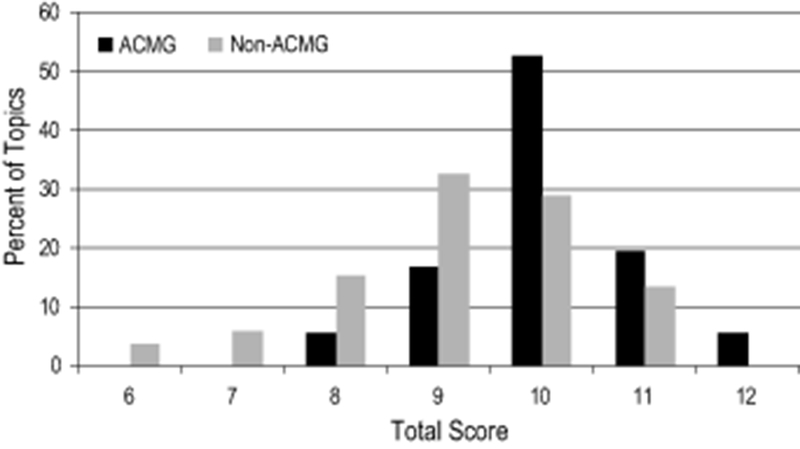
Distribution of summary scores of 88 outcome—intervention scoring pairs
Scores related to the outcome
The outcomes related to topics on the ACMG SF v2.0 list had significantly higher mean domain scores for severity and likelihood of the outcome, compared to those not on the ACMG SF v2.0 list (p<0.05) (Table 2). To account for a non-normal distribution of these scores, a non-parametric test was performed, which did not alter conclusions (data not shown). The range of severity scores (1-3) was the same across both subsets of topics (Figure 2a). Interestingly, none of the topics on the ACMG SF v2.0 list had a likelihood score of 0 or 1 to indicate low or unknown likelihood; however, approximately 10% of non-ACMG SF v2.0 topics had a likelihood score in this lower range (Figure 2b).
Figure 2:
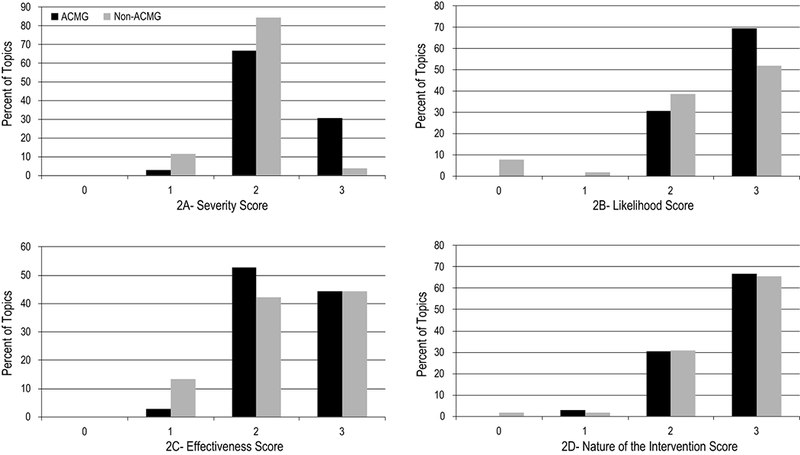
Distribution of summary scores of 88 outcome—intervention scoring pairs by domain
Scores related to the intervention
We found no difference in the mean domain scores for either the effectiveness or nature of the intervention (Table 2). The range of scores for effectiveness was the same across both groups (1-3) (Figures 2c and 2d). Only one topic included in the analysis received a “0” score for nature of the intervention indicating a “high risk, poorly acceptable or intensive intervention”, which was for liver transplantation to prevent morbidity associated with hereditary transthyretin-related amyloidosis, a topic not on the ACMG SF v2.0 list.
Examination of highest scoring outcome-intervention pairs
Two topics scored a “3” across all four domains of actionability (summary score=12), both of which are on the ACMG SF v2.0 list: Loeys-Dietz syndrome and Marfan syndrome. Nearly half (46%) of the 13 topics with a summary score of 11, however, represent genes/disorders that are not currently included on the ACMG SF v2.0 list (Table 3). Comparing all topics with a summary score of 11 across domains reveals that all but one (familial thoracic aortic aneurysm and dissection) have the same scoring pattern across the four domains (severity=2, likelihood=3, effectiveness=3, nature of the intervention=3). This indicates that each of these topics represents gene-disorders with a moderately severe outcome of at least 40% penetrance that could be altered by a highly effective and a lower risk/more medically acceptable intervention.
Table 3:
Topics with high total scores (11-12) on the ClinGen AWG semi-quantitative metric
| Disorder | Disorder OMIM(s) | On ACMG SF v2.0 List | Gene(s) | Outcome | Intervention | Severity | Likelihood | Effectiveness | Nature of the Intervention | Total | Likelihood Evidence Level | Effectiveness Evidence Level |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Loeys-Dietz Syndrome | 609192 610168 613795 614816 615582 |
Yes |
TGFBR1 TGFBR2 SMAD3 TGFB2† TGFB3† |
Clinically Significant Aortic Aneurysm | Surveillance | 3 | 3 | 3 | 3 | 12 | C | B |
| Marfan Syndrome | 154700 | Yes | FBN1 | Clinically Significant Aortic Aneurysms | Surveillance | 3 | 3 | 3 | 3 | 12 | C | B |
| Aortic Dilation Progression | Beta-blockers | 3 | 3 | 3 | 3 | 12 | C | A | ||||
| Adrenoleukodystrophy | 300100 | No | ABCD1 | Neurological/Cognitive decline | Neurological surveillance to plan initiation of hematopoietic cell transplantation | 2 | 3 | 3 | 3 | 11 | C | C |
| Biotinidase Deficiency | 253260 | No | BTD | Neurological complications | Biotin therapy | 2 | 3 | 3 | 3 | 11 | C | C |
| Dopa-responsive dystonia (autosomal dominant) | 128230 | No | GCH1 | Neuromuscular dysfunction | Levodopa therapy | 2 | 3 | 3 | 3 | 11 | C | C |
| Familial Hypercholesterolemia (Heterozygous) | 143890 144010 603776 |
Yes |
LDLR APOB PCSK9 |
High cholesterol | Statins | 2 | 3 | 3 | 3 | 11 | C | A |
| Familial Hypercholesterolemia (Homozygous) | 143890 144010 603776 |
Yes |
LDLR APOB PCSK9 |
High cholesterol | Statins | 2 | 3 | 3 | 3 | 11 | C | A |
| Familial Hyperparathyroidism and Parathyroid Carcinoma | 145000 145001 608266 |
No | CDC73 | Morbidity from primary hyperparathyroidism | Surveillance of parathyroid hormone and calcium | 2 | 3 | 3 | 3 | 11 | C | C |
| Familial Thoracic Aortic Aneurysms and Dissections | 132900 611788 154700 609192 610168 613795 613780 |
Yes |
MYH11 ACTA2 FBN1 TGFBR1 TGFBR2 SMAD3 MYLK† |
Clinically significant aortic aneurysm | Aortic surveillance | 3 | 2 | 3 | 3 | 11 | D | C |
| Aortic dilation progression | Beta blockers | 3 | 2 | 3 | 3 | 11 | D | C | ||||
| Hereditary Hemorrhagic Telangiectasia | 600376 187300 615506 175050 |
No | ENG | Anticipatory treatment to avoid pulmonary arteriovenous malformation-related morbidity | Transthoracic contrast echocardiography | 2 | 3 | 3 | 3 | 11 | E | B |
| SMAD4 | Anticipatory treatment to avoid pulmonary arteriovenous malformation-related morbidity | Transthoracic contrast echocardiography | 2 | 3 | 3 | 3 | 11 | E | B | |||
| Maturity Onset Diabetes of the Young, Type 3 (MODY3) | 600496 | No | HNF1A | Suboptimal glycemic control | Sulfonylureas/ optimal diabetic control | 2 | 3 | 3 | 3 | 11 | C | C |
| Multiple endocrine neoplasia type IIA, Familial medullary thyroid cancer | 171400 155240 |
Yes | RET | Pheochromocytoma | Biochemical screening | 2 | 3 | 3 | 3 | 11 | C | B |
| Multiple Endocrine Neoplasia Type IIB | 162300 | Yes | RET | Pheochromocytoma | Biochemical screening | 2 | 3 | 3 | 3 | 11 | C | B |
| Peutz-Jeghers syndrome | 175200 | Yes | STK11 | Colorectal Cancer | Cancer-Specific Surveillance | 2 | 3 | 3 | 3 | 11 | A | B |
| Von Hippel-Lindau Syndrome | 193300 | Yes | VHL | Renal cell carcinoma | Surveillance | 2 | 3 | 3 | 3 | 11 | C | C |
† Gene not included on the ACMG SF v2.0 list for this disorder.
Discussion
The use of genomic sequencing is increasing, with stakeholders faced with the challenge of deciding whether to report and how to prioritize the return of secondary findings. Secondary findings offer the opportunity to provide valuable clinical information to implement interventions and minimize or prevent future negative clinical outcomes. However, a centralized resource of available evidence on clinical actionability to guide decision-making by stakeholders has been lacking. The ClinGen AWG was established to address this need.
Among the 78 topics (127 genes) that have been scored by the ClinGen AWG, we found that the topics already on the ACMG SF v2.0 list had higher overall summary scores than the topics not on the list. These differences in scoring tended to be due to aspects of the outcome (higher level of severity and likelihood). These results could indicate prioritization of topics by the ACMG based on the severity and likelihood of outcomes. As part of our analyses, we found several topics that do not appear on the ACMG SF v2.0 list but do appear to fit the pattern of conditions already recommended for return by the ACMG (Table 3). The ClinGen AWG and ACMG Secondary Findings Working Group are coordinating efforts to use this data to determine which genes/disorders should be considered for addition to the ACMG SF v2.0 list.
The need for the standardized assessment of the actionability of secondary findings is increasing as decisions related to returning secondary findings occur in the clinical and research setting. A survey of the practices of 12 United States clinical laboratories in 2014-2015, for example, found that secondary findings were consistently reported back to individuals (8 opt-out, 4 opt-in) (O’Daniel et al., 2017). Of these laboratories, however, only five reported limiting their findings to those on the ACMG SF v2.0 list. Other laboratories reported returning “medically actionable” or “medically important” findings, as defined by the individual laboratories. In contrast, the practice of returning of secondary results appears to be less common internationally. A survey of 24 laboratories in Europe, Canada, and Australasia found that only four laboratories actively searched for secondary findings and multiple laboratories actively took efforts to reduce the identification of these findings though bioinformatic filters and limited analyses (Vears, Senecal, & Borry, 2017). These differences between United States and international practice may be influenced by the fact that international guidelines do not endorse actively seeking and reporting these findings as routine practice (Boycott et al., 2015; van El et al., 2013). Standardized assessments, like those produced by the ClinGen AWG, offer a resource for laboratory directors and other stakeholders during future decision-making regarding the return of secondary findings to patients.
Importantly, perspectives on what results may be considered actionable may differ between patients, providers, and other decision-makers (e.g., laboratory directors, payers). Currently, the ClinGen AWG only considers interventions that can confer medical actionability (i.e., the availability of a clinical intervention) and does not consider personal utility (i.e., non-clinical interventions or benefits beyond clinical utility) when making its recommendations (Kohler et al., 2017). When viewed as a whole, studies of a variety of stakeholders (i.e., patients, members of the public, health care professionals, researchers) report that over half of those surveyed endorsed the return of all secondary findings, regardless of actionability or clinical relevance; however, in some studies participants acknowledged becoming less inclined to receive all findings following further discussion (Mackley et al., 2017). When examined by group, the desire to receive all findings, regardless of medical actionability, was more frequently expressed by patients and based on reasons related to patient empowerment. Providers, on the other hand, were more likely than patients to favor a more conservative approach due to concerns regarding the potential distress and the resources required to follow up secondary findings (Mackley et al., 2017). Given these differences in perspectives from patients and providers or other decision-makers, the work of the ClinGen AWG may serve as an important resource for clinical decision making.
While using the summary score across the four domains may help identify the “most actionable” topics for most users, specific domains (such as severity or effectiveness) may take precedence for some decision makers. For example, a case study reporting the selection of which secondary findings to return to research study participants found that, in addition to information scored from a related semi-quantitative metric, decision makers utilized additional subjective factors (e.g., cost and likelihood of identifying a finding with a low population prevalence) to determine the final list. The conclusion was that the actionability criteria considered in the metric couldn’t be used to “just blindly turn the crank” to determine which findings to return; rather they should be considered a starting point for more nuanced decision making (Lazaro-Munoz, Conley, Davis, Prince, & Cadigan, 2017). In addition, the relative value of domains may differ between decision makers and patients. Within a Danish population-based discrete choice experiment, the population preference for reporting favored the severity and likelihood of the result and gave less weight to the existence of preventive measures and treatment. Therefore, individuals may value more than just findings that could inform clinical decision making and may also focus more on those findings which are most likely to have a negative impact their future health (Ploug & Holm, 2017).
The scope of work of the ClinGen AWG is targeted at evaluating the medical actionability of secondary findings in adult patients (aged 18 and older) undergoing diagnostic testing in a clinical setting. While the criteria to determine actionability may capture some elements that could be relevant for determination of which genetic findings to consider in population-based screening, the ClinGen AWG does not consider factors such as systems-based practice and the availability of population-scale follow-up that would be necessary to evaluate genetic testing on a public-health scale. This analysis represents a snapshot of the work of the ClinGen AWG as of February 2018. The list of topics addressed by the ClinGen AWG to date is not intended to be a comprehensive list of potentially actionable genes; the working group will continue to address new topics as well as update existing reports and scores over time. The current list of AWG topics and scores can be accessed at www.clinicalgenome.org. In addition, while the current work of the ClinGen AWG is focused on secondary-findings in adult populations, a workgroup has been formed to address secondary findings within the pediatric population.
Limitations
There is no true gold standard to evaluate which secondary findings are truly “actionable” at an individual or population level. Therefore, this analysis primarily focuses on a comparison of the currently available information from the ClinGen AWG and the ACMG to guide decision makers. In addition, the use of the summary score for selection of outcome–intervention pairs for the analysis may not represent what some groups would consider the most actionable pair (e.g., selecting the pair based on the highest severity or likelihood). Additionally, the scores that were used for the ClinGen AWG are the final group consensus scores, and do not account for the variation in scores from individual members of the group.
Some components of the semi-quantitative metric require subjective judgement by individual scorers, in particular the severity of the outcome and the nature of the intervention. It is acknowledged by the AWG that individual perspectives, including perspectives among clinicians and patients, may vary regarding these domains. Future work of the AWG will examine how well the consensus scores reflect the views of the general patient population. In addition, while the use of objective criteria to determine the effectiveness of the intervention is desirable, the variability in how these measures may be reported and the heterogeneity of clinical outcomes considered (e.g., mortality, quality of life, severity of symptoms) necessitates the use of relatively subjective criteria (e.g., “moderately effective”).
Lastly, based on the AWG protocol reliance on synthesized evidence, it is difficult for the level of evidence related to likelihood to rise above a level C (or Tier 3 evidence), given that systematic reviews and meta-analyses of penetrance data are rare. This points towards the need for methods development to be able to identify additional sources of data (including unpublished data) to inform the evidence on likelihood, particularly in an unselected population.
Despite these limitations, these findings provide valuable information to illuminate a standardized process for evaluating the complicated issues surrounding secondary findings.
Conclusions
The ongoing ClinGen AWG process has thus far created summary reports and scores for a total of 78 topics (including 127 genes). The ClinGen AWG evidence reports and summary scores are intended to aid decision makers as they determine best practices for returning secondary genomic findings to individuals. As a part of this effort, the ClinGen AWG is working closely with the ACMG Secondary Findings Committee to update future iterations of their recommended secondary findings list.
Supplementary Material
Acknowledgements:
Ms. Webber had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
We thank Sameer Paithankar, Andrew R Jackson, Piotr Pawliczek, and Neethu Shah for their work on the development of the Actionability Curation Interface utilized by the AWG during curation and scoring. We also thank Kevin Lutz for editorial support and Michael Leo for statistical consultation on this manuscript. The authors have no conflicts of interest to declare.
Funding: This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. The ClinGen consortium is funded by the National Human Genome Research Institute of the National Institutes of Health through the following grants and contracts: U41HG006834 (Rehm), U41HG009649 (Bustamante), U41HG009650 (Berg), U01HG007436 (Bustamante), U01HG007437 (Berg), HHSN261200800001E. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Boycott K, Hartley T, Adam S, Bernier F, Chong K, Fernandez BA, Canadian College of Medical, G. (2015). The clinical application of genome-wide sequencing for monogenic diseases in Canada: Position Statement of the Canadian College of Medical Geneticists. J Med Genet, 52(7), 431–437. doi: 10.1136/jmedgenet-2015-103144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Foreman AK, O'Daniel JM, Booker JK, Boshe L, Carey T, … Evans JP (2016). A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genetics in Medicine, 18(5). 467–475. 10.1038/gim.2015.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, Biesecker LG (2013). ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med, 15(7), 565–574. doi:gim201373 [pii];10.1038/gim.2013.73 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JE, Irving SA, Biesecker LG, Buchanan A, Jensen B, Lee K, Goddard KA (2016). A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet Med, 18(12), 1258–1268. doi: 10.1038/gim.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, Miller DT (2017). Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet Med, 19(2), 249–255. doi: 10.1038/gim.2016.190 [DOI] [PubMed] [Google Scholar]
- Kohler JN, Turbitt E, Lewis KL, Wilfond BS, Jamal L, Peay HL, Biesecker BB (2017). Defining personal utility in genomics: A Delphi study. Clin Genet, 92(3), 290–297. doi: 10.1111/cge.12998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro-Munoz G, Conley JM, Davis AM, Prince AE, & Cadigan RJ (2017). Which Results to Return: Subjective Judgments in Selecting Medically Actionable Genes. Genet Test Mol Biomarkers, 21(3), 184–194. doi: 10.1089/gtmb.2016.0397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackley MP, Fletcher B, Parker M, Watkins H, & Ormondroyd E (2017). Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: a systematic review of quantitative and qualitative studies. Genet Med, 19(3), 283–293. doi: 10.1038/gim.2016.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Daniel JM, McLaughlin HM, Amendola LM, Bale SJ, Berg JS, Bick D, Rehm HL (2017). A survey of current practices for genomic sequencing test interpretation and reporting processes in US laboratories. Genet Med, 19(5), 575–582. doi: 10.1038/gim.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploug T, & Holm S (2017). Clinical genome sequencing and population preferences for information about ‘incidental’ findings-From medically actionable genes (MAGs) to patient actionable genes (PAGs). PLoS One, 12(7), e0179935. doi: 10.1371/journal.pone.0179935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van El CG, Cornel MC, Borry P, Hastings RJ, Fellmann F, Hodgson SV, Professional Policy C (2013). Whole-genome sequencing in health care. Recommendations of the European Society of Human Genetics. Eur J Hum Genet, 21 Suppl 1, S1–5. [PMC free article] [PubMed] [Google Scholar]
- Vears DF, Senecal K, & Borry P (2017). Reporting practices for unsolicited and secondary findings from next-generation sequencing technologies: Perspectives of laboratory personnel. Hum Mutat, 38(8), 905–911. doi: 10.1002/humu.23259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


