Abstract
Background:
Cholesterol 7α-hydroxylase (CYP7A1) catalyzes the rate-limiting step in bile acid biosynthesis from cholesterol, a main pathway for cholesterol removal from the body. CYP7A1 SNPs are associated with total cholesterol and LDL levels, risk of cardiovascular diseases, and other phenotypes; however, results are inconsistent, and causative variants remain uncertain, except for a frequent promoter SNP (rs3808607).
Methods:
We employed chromatin conformation capture (4C assay), chromatin immunoprecipitation (ChIP-qPCR assay) in hepatocytes, and CRISPR-mediated genome editing in HepG2 cells to identify regulatory regions for CYP7A1. We then screened for SNPs located in regulatory regions, testing effects on reporter gene assays and on hpatic CYP7A1 expression by measuring allelic mRNA expression imbalance.
Results:
4C assays showed several regions interacting with CYP7A1promoter. CRISPR-mediated genome editing in HepG2 cells revealed a novel CYP7A1 enhancer and a repressor region, located >10 kb downstream of the CYP7A1 promoter. SNP screening with an allelic mRNA expression imbalance in human livers and reporter gene assays identified a frequent functional SNP (rs9297994) located in the downstream CYP7A1 enhancer region. SNP rs9297994 is in high linkage disequilibrium with promoter SNP rs3808607, but has opposite effects on CYP7A1 mRNA expression. Their combined effects using a 2-SNP model robustly associate with hepatic CYP7A1 mRNA expression, ranging over two orders of magnitude. Moreover, only the 2-SNP model, but not each single SNP alone, is significantly associated with LDL levels, risk of CAD, statin response, and diabetes in several clinical cohorts, including CATHGEN and Framingham.
Conclusion:
Two interacting regulatory SNPs modulate CYP7A1 expression and are associated with risk of coronary artery disease and diabetes.
Keywords: cholesterol, polymorphism myocardial infarction, diabetes mellitus, gene expression, cardiovascular disease, CYP7A1, association, enhancer, repressor
Keywords: Genetic, Association Studies; Gene Expression and Regulation; Biomarkers; Coronary Artery Disease; Myocardial Infarction
Cholesterol 7α-hydroxylase (CYP7A1) is the first and rate-limiting enzyme in bile acid biosynthesis from cholesterol 1, a principal cholesterol removal pathway in the body. CYP7A1 is tightly regulated by bile acids, lipids, and transcription factors, all critical to cholesterol catabolism and bile acid homeostasis, thereby, regulating lipids, glucose, and energy metabolism 1. In genome wide association studies (GWAS), CYP7A1 single nucleotide polymorphisms (SNPs) associate with total cholesterol and LDL levels 2, 3, gallstone diseases 4, and blood deoxycholic acid 5. Candidate gene studies also reveal associations between CYP7A1 and total cholesterol and triglyceride levels 6, 7, response to statins 8–10, atherosclerosis 11, ischemic stroke 12, hypertension 13, coronary artery disease (CAD) 14, colorectal cancer 15, biliary cirrhosis 16, gallbladder cancer 17, diabetes 5, and anti-tuberculosis drug-induced hepatotoxicity 18. These associations highlight the biological relevance of cholesterol-bile acid homeostasis, with apparently opposite effects of CYP7A1 activity on CAD and diabetes.
In contrast to numerous reported CYP7A1 associations, causative genetic variants and underlying mechanisms remain only partially understood. Multiple studies 7–9, 11–13, 15, 17, 19 implicate a frequent CYP7A1 promoter SNP, rs3808607, with the G allele shown to exhibit higher transcriptional activity than the T allele in reporter gene assays 6, 16. However, increased promoter activity of the rs3808607 G allele is inconsistent with expected phenotypic results in CYP7A1 association studies. For example, the rs3808607G allele is associated with higher lipid levels 7, 8, 19, increased risk of atherosclerosis 11, and protection against colorectal cancer 15, results expected for low CYP7A1 activity. Moreover, rs3808607 fails to show a GWAS-significant association with lipids (p=6 × 10−6) 2; rather, SNPs near the 3ꞌ end of CYP7A1 are significant 2, 3 (rs2081687, p=2 × 10−12; imputed rs983812, p=1.8 × 10−12, GRASP search: http://apps.nhlbi.nih.gov/grasp) (Supplemental Table 1). Furthermore, a metabolomics study identifies SNP rs8192870 as more significantly associated with blood deoxycholic acid levels than promoter rs3808607 5. These results indicate that either rs3808607 is not the causative variant in vivo, or more than one functional variant regulates CYP7A1 expression.
A high linkage disequilibrium (LD) block carrying rs3808607 extends from a distal promoter to intron 2 (~7 kb) in all HapMap populations 20. High LD between multiple CYP7A1 SNPs confounds detection of causative SNPs in association studies. To identify regulatory CYP7A1 regions and their interactions, we employed chromatin conformation capture assays (4C), followed by CRISPR-mediated genome editing. We then screened for SNPs located in regulatory regions, testing effects on hepatic CYP7A1 expression by measuring allelic mRNA expression imbalance. This approach revealed a novel CYP7A1 enhancer and a repressor region and identified a potent enhancer SNP, rs9297994. Interactions between rs9297994 and promoter rs3808607 robustly determine CYP7A1 mRNA expression in human livers. Moreover, a 2-SNP model (rs9297994/rs3808607) is associated with total cholesterol and LDL levels, statin response, and risk of cardiovascular diseases (CAD) and diabetes in several clinical cohorts.
Materials and Methods:
The study materials are available from D. Wang upon request. Datasets supporting the conclusions of this article are available in the dbGaP repository as shown at the end of Supplemental Methods.
The institutional review committee at OSU approved the study of human tissue and use of clinical data.
Detailed experimental procedures are in supplemental methods.
Results
The authors declare that all supporting data are available within the article and its online supplementary files.
1. 4C chromatin conformation analysis identified regions interacting with the CYP7A1 promoter.
With the CYP7A1 promoter as an anchor, 4C assays in human hepatocytes as described 21, identified three clusters of 4C signals 13–28 kb downstream of the CYP7A1 promoter (3–18 kb downstream of 3ꞌ UTR), namely R1, R2, and R3 (Figure 1a & b and Supplemental Table 2). R1 and R3 span ~1 kb, while R2 spans 4 kb. Judging by histone marks, we divided R2 into two sub-regions, R2a and R2b, separated by 2 kb (Figure 1b). All regions overlapped with H3K4me1 and H3K27ac histone marks indicative of active enhancers 22 (Supplemental Figure 1). R2 also overlapped with a p300 ChIP-seq signal, indicative of enhancer elements. The results of ChIP-qPCR performed with a p300 antibody in hepatocytes suggest p300 interactions with all three regions, with R2 and R3 displaying stronger signals than R1 (21-fold vs 10-fold enrichment, p < 0.05) (Figure 1c & Supplemental Figure 2), supporting active regulatory functions.
Figure 1.
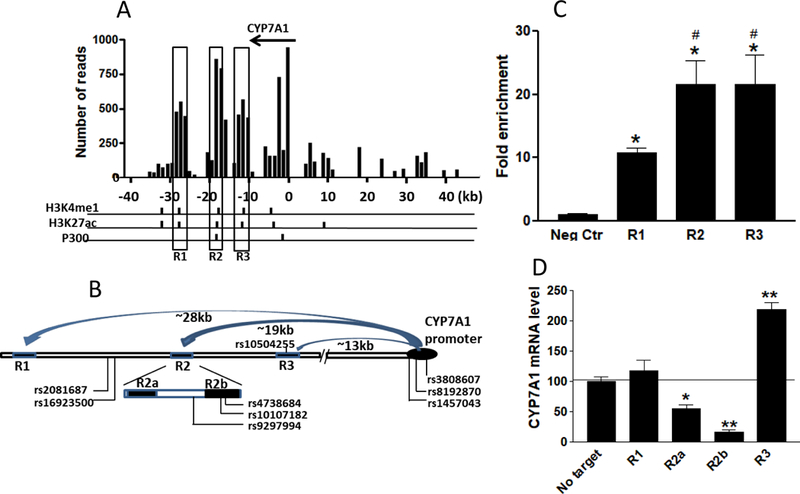
a. 4C assay signals anchored to the CYP7A1 promoter. The x-axis represents relative distance from the CYP7A1 promoter. b. Diagram of interactions between R1, R2, and R3 and the CYP7A1 promoter. c. ChIP-qPCR with p300 antibody in human hepatocytes. Highest signals are shown for each region tested with 2–4 primer pairs (Supplemental Table 2). d. CYP7A1 mRNA expression in HepG2 cells after CRISPR-deletion of regulatory regions, using two combinations of gRNA-lentiCRISPR and 5-day puromycin selection. CYP7A1 mRNA was measured with real-time PCR (CYBR Green) and CYP7A1-specific primers (Supplemental Table 2). Data are represented as mean ± SD. Compared to no target control (NC), * P < 0.05; ** P < 0.01, one-way ANOVA with Dunnett’s post-hoc test.
2. CRISPR-mediated genome editing reveals novel enhancer and repressor regions of CYP7A1.
To delete genomic regions in live HepG2 cells, we used the CRISPR-cas9 system by delivering two gRNAs simultaneously with a lentiviral-based vector (Lenti-CRISPRV2) 21. For each region/sub-region, we designed one or two sets of gRNAs at the 5ꞌ and 3ꞌ sides bracketing the regulatory regions (Supplemental Table 3 for gRNA sequences), deleting 700–1000 bp. Each gRNA combination yielded 50–70% deletion as determined with an agarose gel analysis after PCR amplification (Supplemental Figure 3). Contrary to the no-target control gRNA 23 (Supplemental Table 3), the deletion of R1 did not change the expression of CYP7A1 mRNA, whereas the deletion of R2a, and more strongly R2b, decreased CYP7A1 expression (Figure 1d & Supplemental Figure 4). In contrast, the deletion of R3 increased the expression of CYP7A1. These results indicate that R2 serves as an enhancer and R3 a repressor for CYP7A1 in HepG2 cells.
3. Characterizing regulatory regions/SNPs using reporter gene assays.
DNA fragments (~1000 bp) surrounding R2a, R2b, and R3 were cloned into a reporter gene vector containing a minimal promoter (pGL4.23) (see Supplemental Table 3 for primer sequences and cloning sites). Compared to the empty vector control, R2a increased luciferase activity whereas R3 reduced activity (Figure 2a), consistent with CRISPR-mediated deletion effects (Figure 1d). In contrast, the R2b fragment alone did not alter reporter activity (Figure 2a), despite strongly reducing CYP7A1 expression after CRIPSR-deletion (Figure 1d). Reconciling this discrepancy, a DNA fragment containing both R2a and R2b (R2aR2b, ~3.2 kb) further enhanced luciferase activity 4-fold over R2a alone, indicating that the entire R2 region is required for robust enhancer activity.
Figure 2.
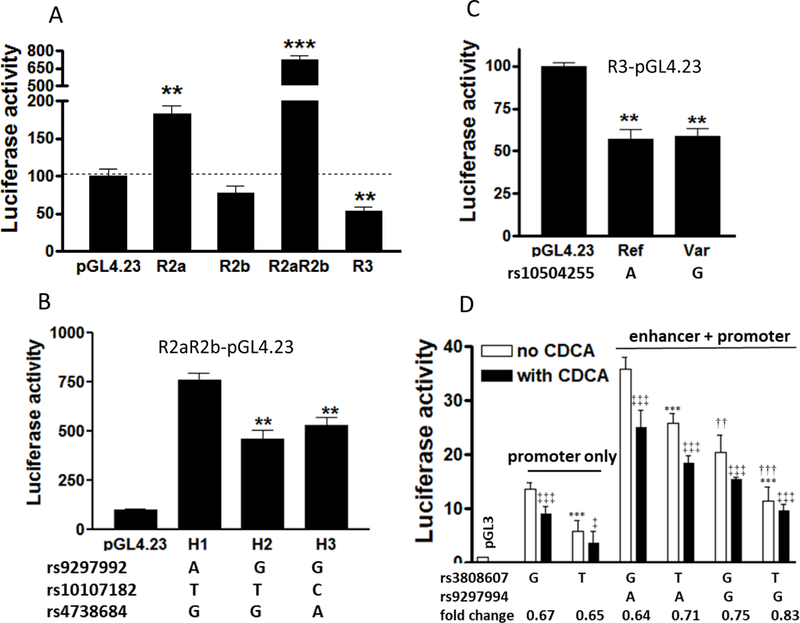
Reporter gene assays (pGL4.23 or pGL3 vector). a. Luciferase activities of regulatory regions 48 h post-transfection. b. Luciferase activities of R2aR2b-pGL4.23 harboring combinations of rs9297994, rs10107187, and rs4738684 as shown. c. Luciferase activities of R3-pGL4.23 reporter with rs10504255 reference A or variant G allele. d. Luciferase activities of reporter constructs with promoter only, enhancer and promoter fusion, and enhancer-promoter allele combinations as indicated in pGL3 vector. Data are from three independent experiments with duplicates, mean ± SD. Panel a-c: Compared to pGL4.23 empty vector, ** P < 0.01. *** P < 0.001; Panel d: rs3808607 G vs T, *** P < 0.001; rs9297994 G vs A, ††P < 0.01, †††P < 0.001; compared to no chenodeoxycholic acid (CDCA), ‡‡‡ P<0.001, ‡P<0.05. One-way ANOVA with Dunnett’s or Tukey post-test.
We then searched for regulatory SNPs located within R2 and R3. Among candidate SNPs, we focused on rs9297994 A>G 24, a top GWAS hit for CYP7A1 (Supplemental Table 1), and SNPs in high LD, including rs10107182 T>C and rs4738684 G>A in R2, and rs10504255 A>G in R3. R2aR2b reporter constructs were generated containing rs9297994, rs10107182, and rs4738684. We tested three haplotypes harboring different combinations of rs9297994, rs10107187, and rs4738684 to identify the causal variant. Shown in Figure 2b, haplotypes H2 and H3 containing variant G alleles of rs9297994 have lower enhancer activity than the reference A allele (haplotype H1), whereas haplotypes containing different alleles for rs10107182 or rs4738684 have similar levels of enhancer activity (H2 vs H3). Furthermore, R3 harboring the rs10504255 reference A or variant G alleles failed to differ in terms of repressor activity (Figure 2c).
The rs3808607 minor G allele has higher transcriptional regulatory activity than the T allele 6, 16. The high activity promoter rs3808607 G allele is in high LD with the low activity enhancer rs9297994 G allele (R2 = 0.715, Dꞌ = 0.954), but with a different minor allele frequency (0.43 vs 0.37) in Europeans. To test the combined effects of rs3808607 and rs9297994, we joined the R2aR2b enhancer fragment to the 5ꞌ end of the CYP7A1 promoter fragment, and tested pGL3 reporter activity in the presence and absence of chenodeoxycholic acid (CDCA), a regulator of CYP7A1 expression. As expected, the reporter gene activity was strongest with a fragment combining enhancer rs9294994 A with promoter rs3808607 G (both high activity alleles), and lowest with G plus T alleles (both low activity alleles). Chenodeoxycholic acid treatment further reduced reporter gene expression, consistent with a negative feedback (Figure 2d). The effects of CDCA did not differ between genotypes (P > 0.05), suggesting that genotype does not interact with bile acid, a main regulator of hepatic CYP7A1.
4. Combined effect of promoter rs3808607 and enhancer rs9297994 SNPs on CYP7A1 mRNA expression in human livers:
To test in vivo effects of rs9297994 and rs3808607 on CYP7A1 expression, we measured the hepatic mRNA allelic expression imbalance (AEI) 25 in 50 samples heterozygous for a frequent marker SNP, rs8192879, located in the CYP7A1 3ꞌUTR. Fifty percent of tested samples displayed AEI (allelic RNA ratio >1.3 fold, compared to DNA ratio, p<0.05), evidence for the presence of frequent regulatory variants. Consistent with reporter gene assay results, of 33 SNPs genotyped (Supplemental Table 4, also see Supplemental Figure 5 for haplotypes), rs9297994 and two SNPs in strong LD (rs10107182 and rs10504255, R2 = 1 and 0.99 in EUR, and 0.78 and 1 in AFR, respectively) showed the strongest association with AEI (k = 0.77), while promoter rs3808607 showed a moderate association (k = 0.45). Except for two samples (marked by arrow, both are from African Americans), all AEI positive samples are heterozygous for rs9297994 (Figure 3a). All samples heterozygous for rs9297994 are also heterozygous for rs3808607, with the exception of two samples (Figure 3a, marked by #). This result indicates enhancer SNP rs9297994 had the strongest impact on CYP7A1 expression, while no single SNP alone completely accounts for the AEI pattern, supporting the presence of more than one regulatory variant. Moreover, the result also suggests the presence of functional variants unique to African Americans.
Figure 3.
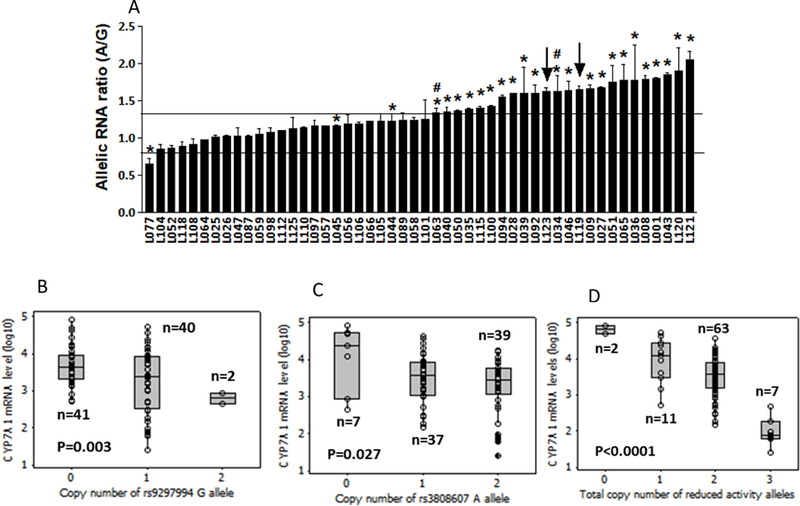
Panel a. Allelic CYP7A1 mRNA expression ratios in human livers measured with a primer extension assay (SNaPshot) using rs8192879 as marker. Allelic RNA ratios are normalized to genomic DNA ratios = 1. Data represent the mean of three measurements (± SD). The star marks 25 livers heterozygous for rs9297994 and rs3808607, # plus * marks two AEI-positive samples heterozygous only for rs9297994, and arrows indicate two AEI-positive samples homozygous for both rs9297994 and rs3808607. Horizontal lines indicate 1.3-fold AEI-cut-off ratios. Panel b-d. CYP7A1 mRNA levels in the liver (log10-scale) grouped by number of reduced activity alleles of (b) rs9297994, (c) rs3808607, or (d) sum of rs9297994 and rs3808607. Statistical analyses were performed using multiple linear regression model. Box and horizontal lines: 25th-75th percentiles and mean, whiskers: minimum and maximum values, circles: individual values.
To assess individual and combined effects of rs9297994 and rs3808607 on CYP7A1 mRNA expression, we tested the association between CYP7A1 mRNA levels and the total number of reduced activity alleles rs3808607T and rs9297994G alone or in combination, in 83 human livers. Both rs3808607 and rs9297994 alone were significantly associated with CYP7A1 mRNA levels (p = 0.027 and 0.003, respectively), with each reduced activity allele accounting for a 2.9 and 5.5-fold reduction in mRNA level, respectively (Figure 3b & c). As expected, the combination of rs3808607 and rs9297994 (2-SNP model, total number of reduced activity alleles ranging from 0 to 4) profoundly affected CYP7A1 mRNA levels, with each reduced activity allele resulting in a 6.9-fold reduction in CYP7A1 mRNA (Figure 3d; no subject had 4 reduced activity alleles), higher than the attributable reduction of either alleles when analyzing the two SNPs separately. The mRNA levels differed by 2.5 orders of magnitude between livers with 0 versus 3 reduced activity alleles, but the number of livers with 0 and 3 reduced activity alleles was low (2 and 7, respectively), limiting the ability to estimate mRNA expression range accurately. Nevertheless, these results demonstrate that rs3808607/rs9297994 profoundly affects hepatic CYP7A1 expression.
5. Association of rs3808607 and rs9297994 with clinical phenotypes in the OSU CAD cohort.
To test the association between CYP7A1 SNPs and lipid levels, we genotyped rs3808607 and rs9297994 in a cohort of 485 subjects with diagnosed cardiovascular disease, having ≥75% angiographic luminal stenosis (newly diagnosed or established), requiring percutaneous coronary intervention 26 (cohort demographics are provided in Supplemental Table 5a). Over 80% of individuals have two copies of reduced activity alleles, the remainder carrying 0 or > 2 reduced activity alleles. We divided patients into two groups: high activity, subjects carrying 0 or 1 reduced activity allele; and low activity, subjects carrying 2 or more reduced activity alleles. Shown in Table 1a, a high activity CYP7A1 status is associated with lower levels of total cholesterol (P=0.05) and LDL (P=0.04) when compared to a low activity status, after adjusting for age, diabetes, tobacco use, sex, race, statin use, and interaction between statin use and CYP7A1 activity status. A portion of patients (n=196) were on statins (atorvastatin, lovastatin, simvastatin, fluvastatin, pravastatin, and rosuvastatin), with doses titrated to reach an optimal cholesterol target goal as described 27. Of the 196 patients, 33% (63/196) did not reach the target cholesterol goal. High activity CYP7A1 status is associated with reduced likelihood to reach cholesterol target goal with statin treatment compared to low activity status (95% confidence interval 0.18–0.99, P = 0.05) (Table 1a), after adjusting for statin dose, tobacco use, diabetes, hypertension, and sex. In contrast, rs3808607 or rs9297994 alone were not significantly associated with lipid levels and reaching statin target goal (P>0.05) (Table 1a).
Table 1.
Association between CYP7A1 SNPs and clinical phenotypes in the OSU CAD cohort (a) and CATHGEN cohort (b).
|
rs3808607 GT+TT vs GG |
rs9297994 AG+GG vs AA |
rs3808607 + rs92979941 |
||
|
Total cholesterol n=396 |
Beta | −9.19 | 0.85 | −16.59 |
| P value | 0.12 | 0.86 | 0.05 | |
|
LDL level n=383 |
Beta | −3.79 | 3.67 | −13 |
| P value | 0.41 | 0.30 | 0.04 | |
|
HDL level n=396 |
Beta | −2.61 | 0.73 | 3.32 |
| P value | 0.08 | 0.53 | 0.11 | |
|
Triglycerides n=395 |
Beta | −7.09 | −25.51 | −45.89 |
| P value | 0.76 | 0.16 | 0.16 | |
|
Reaching cholesterol reducing target n=196 |
Odds ratio (95% CI) |
1.12 (0.48–2.57) |
0.94 (0.50–1.78) |
0.43 (0.18–0.99) |
| P value | 0.80 | 0.85 | 0.05 | |
|
MI n=477 |
Odds ratio (95% CI) |
0.85 (0.35–2.05) |
0.71 (0.35–1.46) |
0.65 (0.27–1.58) |
| P value | 0.71 | 0.36 | 0.342 | |
|
Hypertension n=477 |
Odds ratio (95% CI) |
0.89 (0.44–1.79) |
1.00 (0.56–1.77) |
0.93 (0.46–1.88) |
| P value | 0.73 | 0.99 | 0.83 | |
|
Diabetes n=477 |
Odds ratio (95% CI) |
1.26 (0.62–2.56) |
0.83 (0.46–1.50) |
0.86 (0.42–1.75) |
| P value | 0.52 | 0.54 | 0.67 |
| rs3808607 GT+TT vs GG |
rs9297994 AG+GG vs AA |
rs3808607 + rs92979941 |
||
| CADINDEX2 n=1140 |
Odds ratio (95% CI) |
1.18 (0.86–1.60) |
1.15 (0.89–1.48) |
0.54 (0.40–0.73) |
| P value | 0.31 | 0.29 | <0.0001 | |
| MI n=1140 |
Odds ratio (95% CI) |
0.90 (0.63–1.29) |
1.29 (0.96–1.73) |
0.56 (0.40–0.79) |
| P value | 0.56 | 0.10 | 0.001 | |
| Diabetes n=1140 |
Odds ratio (95% CI) |
0.87 (0.63–1.20) |
0.78 (0.59–1.02) |
1.48 (1.09–2.01) |
| P value | 0.41 | 0.07 | 0.01 | |
| Hypertension n=1140 |
Odds ratio (95% CI) |
0.71 (0.50–0.99) |
0.99 (0.76–1.29) |
1.88 (1.33–2.64) |
| P value | 0.05 | 0.94 | <0.0001 | |
| Hypercholesterolemia n=1140 |
Odds ratio (95% CI) |
0.99 (0.72–1.36) |
1.14 (0.88–1.48) |
0.84 (0.62–1.14) |
| P value | 0.95 | 0.33 | 0.25 | |
| Death n=1139 |
Odds ratio (95% CI) |
1.03 (0.73–1.47) |
1.02 (0.77–1.36) |
0.79 (0.55–1.12) |
| P value | 0.85 | 0.87 | 0.18 |
Comparing number of reduced activity alleles 0+1 vs 2 or more
CADINDEX comparing subjects with CADINDEX <23 vs CADINDEX >23 (clinically significant).
6. Association of rs3808607 and rs9297994 with clinical phenotypes in CATHGEN and Framingham cohorts.
CATHGEN supports the investigation of genes associated with coronary heart disease and related disorders. We downloaded the rs3808607 genotype data from dbGAP; with rs9297994 data not available, rs10504255 served as a surrogate marker (Dꞌ = 1, R2 = 0.885 in EUR). We again applied the 2-SNP model, grouping individuals into high and low activity groups based on the number of reduced activity CYP7A1 alleles (0–1 vs 2 or more). Significantly more myocardial infarction (MI) and death cases, higher CADINDEX values, and significantly less hypertensive and diabetics cases, occurred in the low activity group than the high activity group (see Supplemental Table 5b). As the low activity group included more males, non-whites, and slightly older individuals (Supplemental Table 5b), age, sex, race, and other comorbidities were included as covariates.
The CADINDEX score incorporated coronary angiographic data representing the extent and anatomical distribution of CAD, with scores over 23 considered clinically significant 28. The high activity status of CYP7A1 was associated with a 44% lower likelihood of having a significant CADINDEX score (95% CI 0.42–0.76, P <0.0001), and a 42% reduction in MI risk (95% CI 0.41–0.81, P = 0.05), after adjusting for age, BMI, hypertension, diabetes, race, sex, and smoking (Table 1b). Moreover, the number of reduced activity alleles was associated with increased odds of the presence of >2 diseased vessels in Caucasians in CATHGEN cohort (p=0.01).
We also tested the 2-SNP model in the Framingham heart study cohort, a longitudinal family study active since 1948. We obtained genotype (rs3808607 and rs10504255) and phenotype data from 1888 participants, which were from 653 families with 1 to 88 members (Supplemental Table 5c for demographics). Due to extensive family structures, we used the GENESIS package in R for analysis (see Supplemental Methods). Consistent with CATHGEN results, each reduced activity allele of CYP7A1 is associated with a 31% increased risk of MI (p = 0.03), after adjusting for age, sex, BMI, blood glucose and systolic blood pressure, and statin treatment.
In contrast to the CAD phenotype, high activity status is associated with 48% increased risk of diabetes (95% CI 1.09–2.01, P = 0.01) and hypertension (95% CI 1.36–2.69, P < 0.0001) in CATHGEN (Table 1b). Consistent with the OSU CAD results, associations between the CYP7A1 genotype and clinical phenotypes are only apparent when using the 2-SNP model, but not when testing each SNP alone, except for hypertension (Table 1b).
Discussion:
Investigating regulatory CYP7A1 regions in human liver and hepatocytes, we identified three downstream domains (R2a, R2b and R3) that bind to the CYP7A1 promoter and regulate transcriptional activity in the liver over a range exceeding two orders of magnitude. Embedded in domain R2b, a novel enhancer SNP rs9297994 A>G (MAF 0.34) profoundly reduced hepatic CYP7A1 mRNA expression. As the rs9297994 G allele is in high LD (in Europeans) with the minor G allele of promoter rs3308607 T>G (MAF 0.43), the opposing effects of each minor allele contradicted previous interpretations of clinical association studies. Accurate assessments of CYP7A1’s genetic influence requires the interpretation of combined effects attributable to both rs9297994 and rs3808607 (2-SNP model), which occur at different frequencies in different ethnic populations. Testing the effect of the total copy number of reduced activity alleles per subject (possible range from 0 to 4 copies of rs3808607 T and rs9297994 G), we found significant associations with total cholesterol and LDL levels, statin response in OSUMC CAD cohorts, risk of CAD, and diabetes in public clinical Caucasian cohorts (e.g., CATHGEN and Framingham). Similarly, previous single SNP analyses in GWAS have reported associations between lipid levels and CYP7A1 SNPs, but have failed to detect significant associations with cardiovascular disease traits, highlighting the need to consider interactions when more than one causative variant is present in a gene locus. The finding add to a list of genes, including CYP2D6, CETP, CHRNA5, NAT1, DRD2 25, 29–32, that harbor more than one frequent variant regulating expression and function, likely under evolutionary selection pressures to cope with environmental changes or disease risk.
Regulatory domains of CYP7A1
We have identified three domains (R1-R3) that physically interact with the CYP7A1 promoter, located downstream of the transcribed region. While any possible R1 effect was undetectable, R2 displayed robust enhancer activity, and R3 appeared to repress CYP7A expression, in our in vitro model. R2 consists of two candidate enhancer regions (R2a, R2b), each ~1 kb in length, separated by 2 kb. Reporter gene assays indicated that both regions interacted with each other and together, strongly affecting transcription when inserted jointly into the reporter vector, suggesting the entire ~4 kb R2 region is required for full enhancer activity. Together with the nearby repressor region R3, this ~8 kb DNA downstream region appears to serve as a main regulatory switch for CYP7A1 expression, harboring many SNPs in high LD with each other.
Discovery of a robust regulatory variant in enhancer domain R2b
To search for regulatory variants, we measured allelic RNA expression imbalance (AEI) in human livers, a powerful approach for identifying cis-regulatory variants 25, 26, cancelling out trans-acting effects. Regulatory variants in addition to the known promoter rs3808607 SNP can generate complex AEI patterns. With 50% of livers tested displaying AEI (Figure 3a), scanning 33 candidate SNPs reveals three SNPs in high LD (rs9297994, rs10107182, and rs10504255) having the highest AEI association score, two located in R2b and one in R3. Allelic RNA expression imbalance ratios of major over minor alleles greater than 1 indicated a lower expression of the minor allele, consistent with reporter gene assays showing that minor allele rs9297994 G reduced enhancer activity by ~50%. We concluded that rs9297994 decreases hepatic mRNA expression.
Large and frequent LD blocks across the CYP7A1 gene locus are likely a result of evolutionary selection pressures 33, with the minor rs9297994 G allele (MAF 0.37) on an LD block distinct to that of the minor rs3808607 G allele (MAF 0.43), which has been shown to enhance CYP7A1 expression. Therefore, most individuals carrying one variant are also carrying the other variant with opposing effects on mRNA expression, thereby largely canceling each other out. While reduced activity of the enhancer rs9297994 G allele appeared to dominate the effect of the promoter rs3808607 G allele, variability in the net AEI mRNA ratios in the liver suggests further regulation by trans- acting factors, as demonstrated here with CDCA altering expression in the reporter gene assays.
These results emphasized the importance of considering the combined effects of both rs9297994 and rs3808607 (2-SNP model); however, varying allele frequencies and LD between them contradict the assessment of which alleles are paired with each other on the same DNA strand in different ethnic groups. To simplify, we tested the effect of the number of reduced activity alleles carried by any given individual, without considering their phasing and effect sizes. This approach is supported by reporter gene assays showing decreased transcriptional activity with an increasing number of reduced activity alleles (Figure 2d), and further by a robust relationship between total number of reduced activity alleles and CYP7A1 mRNA expression in human livers (Figure 3d). Therefore, we employed the number of reduced activity alleles to assess clinical associations. While we divided subjects into those carrying 0–1 and 2 or more reduced activity alleles for clinical association analysis (2-SNP model), we pointed out the dramatic differences in hepatic CYP7A1 mRNA levels in the small donor groups with 0 and 3 reduced activity alleles (2.5 orders of magnitude; Figure 3d). Owing to their lower frequency, larger clinical cohorts are required for further study.
Clinical associations
Testing the effects of rs3808607 and rs9297994 either exclusively or in combination, we find that the 2-SNP model reveals significant associations with lipid levels, statin response, CAD, and diabetes risks (Table 1), whereas the impact of either single SNP is masked by the other, opposing it in most subjects. These conditions have resulted in failure to detect robust clinical associations or yielding conflict results 7, 8, 19. Several SNPs identified in GWAS (Supplemental Table 1) are either in high LD with the promoter SNP (for example rs8192870 and rs16923500) or with the enhancer SNP (for example rs2081687, rs4738684, rs6471717). The minor G allele of rs3808607 is slightly more frequent than the minor G allele of rs9297994 in a European population (0.43 vs 0.37), with drastically different frequencies in African Americans (rs3808607 MAF=0.61, rs9297994 MAF = 0.035). This suggests that African Americans tend to have higher CYP7A1 activity than Europeans do. However, we cannot rule out additional regulatory variants in African Americans, as the studied liver samples are mostly from European donors. This possibility is supported by the AEI result, where a strong AEI in two samples from African American donors cannot be accounted for by the rs3808607 or rs9297994 genotype (Figure 3a).
Our results indicated that a high activity CYP7A1 status is associated with reduced total cholesterol and LDL, reduced risk of a significant CADINDEX score, and reduced risk of MI, consistent with CYP7A1’s key role in cholesterol clearance and a positive relationship between lipid level and CAD risk. Previously reported associations between CYP7A1 SNPs and lipid levels show small effect sizes, detectable only with large cohorts (7,000 to 100,000) 2, 3, 34, and no significant association was detectable between CYP7A1 SNPs and risk of CAD traits (e.g., vascular disease and MI), nor with diabetes, even in GWAS with over 100,000 participants 34. In contrast, use of the 2-SNP model reveals robust effects on total cholesterol and LDL levels with sample size less than 400 subjects. Moreover, we reported associations between high activity CYP7A1 status and reduced risk of coronary disease (CADINDEX) and MI in CATHGEN and Framingham, consistent with the effect on lipid profiles, supporting robustness of the CYP7A1 2-SNP model.
These results also highlight limitations of GWAS in detecting the impact of more than one causative variant per gene locus on complex diseases such as CAD. While the associations with CAD and MI reported here are supported by mechanistic studies on the genetics and underlying biology, further replications are needed to test the validity and clinical utility of the CYP7A1 2-SNP markers, including study of extreme high and low activity CYP7A1 carriers.
In further analyses of a CAD patient cohort maintained at OSUMC 26, we find that a high activity CYP7A1 status was associated with reduced likelihood to reach an optimal cholesterol goal targeted with a titrating statin dosage regimen (Table 1a). This result reinforces previous studies showing the high activity rs3808607 G allele to be associated with lower statin-induced LDL reduction than the A allele 8–10, 35. The mechanism underlying this effect is unknown. Nevertheless, this result suggests that the more robust 2-SNP CYP7A1 activity status can be considered as a biomarker for statin therapy, predicting the efficacy of statin responses and guiding the selection of lipid lowering drugs.
In addition to a potential protective effect against CAD, a high activity CYP7A1 status defined by the 2-SNP model is associated with increased risk of type 2 diabetes mellitus (T2DM) in CATHGEN, consistent with GWAS results showing that SNPs affecting lipid traits inversely affect risk of T2DM 34. With CYP7A1 being a key enzyme in bile acid synthesis, we considered whether the link between CYP7A1 and risk of T2DM arose from bile acid activity. Bile acids activate insulin signaling pathways, thereby regulating lipid, glucose, and energy metabolism 1. However, excess bile acid, especially hydrophobic secondary bile acids synthesized by gut bacteria (for example, deoxycholic acid, DCA), are highly toxic to mammalian cells, causing insulin resistance 36. In support of this hypothesis, CYP7A1 knockout mice are protected against high fat/high-cholesterol diet-induced metabolic disorders and have improved glucose sensitivity 37. Moreover, DCA blood levels are associated with incidence of T2DM 5, while a population study reported higher bile acid levels in insulin-resistant individuals regardless of diabetes status 38.
Hypercholesterolemia is an early and strong predictor for hypertension. However, we found that high activity CYP7A1 status was associated with increased risk of hypertension after adjusting for hypercholesterolemia and other covariates in CATHGEN (Table 1b). This result indicated factors other than dyslipidemia are involved in hypertension, and suggests a possible link between bile acids and hypertension. However, this result was inconsistent with a previous report showing the promoter rs3808607 T allele (reduced activity) is associated with higher blood pressure 13. Therefore, the association between CYP7A1 activity and hypertension requires further study.
There are several limitations in our clinical association study. First, lipid levels in the OSU CAD cohort were obtained at the time of enrollment before treatment was initiated at OSUMC, but lipid lowering therapy before enrollment is uncertain in some cases, potentially confusing the results. Second, the sample size of the statin response cohort is small, requiring replication in larger cohorts. Nevertheless, our results are consistent with other studies 8–10. Similarly, the CAD trait and T2DM associations require replication, even while supported by a strong mechanistic basis and suggestive previous data. Lastly, the enhancer SNP rs9297994 was not directly genotyped in CATHGEN, while the surrogate marker rs10504255 is in high, but not complete LD in all CATHGEN populations, slightly contradictory assessment of rs9297994.
Thus, we have identified enhancer and repressor regions of CYP7A1, and a novel enhancer variant, rs9297994, which robustly reduces CYP7A1 mRNA expression. Together with the previously identified promoter SNP rs3808607, a 2-SNP combination reveals significant associations with clinical phenotypes. Separated by ~20 kb, the low expression CYP7A1 enhancer SNP rs9297994 G allele is in high LD with the high expression promoter rs3808607 G allele, balancing the expression level of CYP7A1. Balancing genetic effects on CYP7A1 expression are likely under evolutionary pressure, since both high and low CYP7A1 activity are associated with increased disease risk.
Supplementary Material
Acknowledgments
Funding Sources: This study was supported by the NIH Pharmacogenetics Research Network grant U01 GM092655 (WS), U01-GM074492 (WS), and NIH grant R01GM120396 (DW). We also acknowledge support from the Ohio Supercomputer Center (grant #PAS0885–2). Human hepatocytes were provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. Liver Tissue Cell Distribution System (LTCDS, Pittsburgh, PA) is funded by NIH Contract # HHSN276201200017C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Center for Research Resources.
Footnotes
Disclosures: OSU has filed a patent application for the use of the CYP7A1 genotype as a biomarker.
References:
- 1.Chiang JY. Bile acids: Regulation of synthesis. J Lipid Res. 2009;50:1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adeyemo A, et al. Transferability and fine mapping of genome-wide associated loci for lipids in african americans. BMC Med Genet. 2012;13:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joshi AD, et al. Four susceptibility loci for gallstone disease identified in a meta-analysis of genome-wide association studies. Gastroenterology. 2016;151:351–363 e328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fall T, et al. Non-targeted metabolomics combined with genetic analyses identifies bile acid synthesis and phospholipid metabolism as being associated with incident type 2 diabetes. Diabetologia. 2016;59:2114–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Castro-Oros I, et al. Promoter variant −204a > c of the cholesterol 7alpha-hydroxylase gene: Association with response to plant sterols in humans and increased transcriptional activity in transfected hepg2 cells. Clin Nutr. 2011;30:239–246 [DOI] [PubMed] [Google Scholar]
- 7.Barcelos AL, et al. Association of cyp7a1 −278a>c polymorphism and the response of plasma triglyceride after dietary intervention in dyslipidemic patients. Braz J Med Biol Res. 2009;42:487–493 [DOI] [PubMed] [Google Scholar]
- 8.Poduri A, et al. Common variants of hmgcr, cetp, apoai, abcb1, cyp3a4, and cyp7a1 genes as predictors of lipid-lowering response to atorvastatin therapy. DNA Cell Biol. 2010;29:629–637 [DOI] [PubMed] [Google Scholar]
- 9.Jiang XY, et al. Cyp7a1 polymorphism influences the ldl cholesterol-lowering response to atorvastatin. J Clin Pharm Ther. 2012;37:719–723 [DOI] [PubMed] [Google Scholar]
- 10.Kadam P, et al. Genetic determinants of lipid-lowering response to atorvastatin therapy in an indian population. J Clin Pharm Ther. 2016;41:329–333 [DOI] [PubMed] [Google Scholar]
- 11.Lambrinoudaki IV, et al. Cyp a-204c polymorphism is associated with subclinical atherosclerosis in postmenopausal women. Menopause. 2008;15:1163–1168 [DOI] [PubMed] [Google Scholar]
- 12.Kim SK, et al. Association between cytochrome p450 promoter polymorphisms and ischemic stroke. Exp Ther Med. 2012;3:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu L, et al. Cyp7a1 genotypes and haplotypes associated with hypertension in an obese han chinese population. Hypertens Res. 2011;34:722–727 [DOI] [PubMed] [Google Scholar]
- 14.Iwanicki T, et al. Cyp7a1 gene polymorphism located in the 5’ upstream region modifies the risk of coronary artery disease. Dis Markers. 2015;2015:185969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagiwara T, et al. Genetic polymorphism in cytochrome p450 7a1 and risk of colorectal cancer: The fukuoka colorectal cancer study. Cancer Res. 2005;65:2979–2982 [DOI] [PubMed] [Google Scholar]
- 16.Inamine T, et al. Association of genes involved in bile acid synthesis with the progression of primary biliary cirrhosis in japanese patients. J Gastroenterol. 2013;48:1160–1170 [DOI] [PubMed] [Google Scholar]
- 17.Srivastava A, et al. Role of genetic variant a-204c of cholesterol 7alpha-hydroxylase (cyp7a1) in susceptibility to gallbladder cancer. Mol Genet Metab. 2008;94:83–89 [DOI] [PubMed] [Google Scholar]
- 18.Chen R, et al. Cyp7a1, baat and ugt1a1 polymorphisms and susceptibility to anti-tuberculosis drug-induced hepatotoxicity. Int J Tuberc Lung Dis. 2016;20:812–818 [DOI] [PubMed] [Google Scholar]
- 19.Couture P, et al. Association of the a-204c polymorphism in the cholesterol 7alpha-hydroxylase gene with variations in plasma low density lipoprotein cholesterol levels in the framingham offspring study. J Lipid Res. 1999;40:1883–1889 [PubMed] [Google Scholar]
- 20.Nakamoto K, et al. Linkage disequilibrium blocks, haplotype structure, and htsnps of human cyp7a1 gene. BMC Genet. 2006;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, et al. Functional characterization of cyp2d6 enhancer polymorphisms. Hum Mol Genet. 2015;24:1556–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith RP, et al. Genome-wide discovery of drug-dependent human liver regulatory elements. PLoS Genet. 2014;10:e1004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konermann S, et al. Genome-scale transcriptional activation by an engineered crispr-cas9 complex. Nature. 2015;517:583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surakka I, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, et al. Common cyp2d6 polymorphisms affecting alternative splicing and transcription: Long-range haplotypes with two regulatory variants modulate cyp2d6 activity. Hum Mol Genet. 2014;23:268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, et al. Intronic polymorphism in cyp3a4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11:274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundy SM, et al. Implications of recent clinical trials for the national cholesterol education program adult treatment panel iii guidelines. J Am Coll Cardiol. 2004;44:720–732 [DOI] [PubMed] [Google Scholar]
- 28.Smith LR, et al. Determinants of early versus late cardiac death in patients undergoing coronary artery bypass graft surgery. Circulation. 1991;84:III245–253 [PubMed] [Google Scholar]
- 29.Papp AC, et al. Cholesteryl ester transfer protein (cetp) polymorphisms affect mrna splicing, hdl levels, and sex-dependent cardiovascular risk. PLoS One. 2012;7:e31930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith RM, et al. Nicotinic alpha5 receptor subunit mrna expression is associated with distant 5’ upstream polymorphisms. Eur J Hum Genet. 2011;19:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Polymorphisms in human dopamine d2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci U S A. 2007;104:20552–20557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, et al. Human n-acetyltransferase 1 *10 and *11 alleles increase protein expression through distinct mechanisms and associate with sulfamethoxazole-induced hypersensitivity. Pharmacogenet Genomics. 2011;21:652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadee W The relevance of “missing heritability “ in pharmacogenomics. Clin Pharmacol Ther. 2012;92:428–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, et al. The role of common variants of abcb1 and cyp7a1 genes in serum lipid levels and lipid-lowering efficacy of statin treatment: A meta-analysis. J Clin Lipidol. 2014;8:618–629 [DOI] [PubMed] [Google Scholar]
- 36.Zhou H, et al. Bile acids are nutrient signaling hormones. Steroids. 2014;86:62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrell JM, et al. Cholesterol 7alpha-hydroxylase-deficient mice are protected from high-fat/high-cholesterol diet-induced metabolic disorders. J Lipid Res. 2016;57:1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun W, et al. Insulin resistance is associated with total bile acid level in type 2 diabetic and nondiabetic population: A cross-sectional study. Medicine (Baltimore). 2016;95:e2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


