Abstract
Impaired immunity in late stage cancer patients is not limited to anti-tumor responses, as demonstrated by poor vaccination protection and high susceptibility to infection1–3. This has been largely attributed to chemotherapy-induced impairment of innate immunity such as neutropenia2, whereas systemic effects of tumors on hematopoiesis and adoptive immunity remain incompletely understood. Here we observed anemia associated with severe deficiency of CD8+ T cell responses against pathogens in treatment-naïve mice bearing large tumors. Specifically, we identify CD45+ erythroid progenitor cells (CD71+TER119+, EPCs) as robust immunosuppressors. CD45+EPCs, induced by tumor growth-associated extramedullary hematopoiesis, accumulate in the spleen to become a major population, outnumbering regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs). The CD45+ EPC transcriptome closely resembles that of MDSCs, and, like MDSCs, reactive oxygen species production is a major mechanism underlying CD45+ EPC-mediated immunosuppression. Similarly, an immunosuppressive CD45+ EPC population was detected in cancer patients with anemia. These findings identify a major population of immunosuppressive cells that likely contributes to the impaired T cell responses commonly observed in advanced cancer patients.
In cancer patients, opportunistic infection is a complication leading to morbidity and mortality especially at their terminal stages1,4–6. Their poor responses to vaccination3 also implicates a deficient adaptive immunity. We investigated whether established tumors dispose patients to infection by producing global T cell immunosuppression. After inoculation of Lewis lung cancer (LLC) cells, we initiated viral infection in tumor-bearing mice using lymphocytic choriomeningitis virus with Armstrong (Supplementary Fig. 1a) or clone 13 (LCMV-Cl13) strain. While tumor-free mice survived infection, by day 9, 80% of tumor-bearing mice succumbed to infection. This susceptibility is not limited to LLC nor LCMV: 90% of B16F10 melanoma-bearing mice succumbed to LCMV infection (Fig. 1a), and death was also induced in LLC or B16F10 tumor-bearing mice after Listeria monocytogenes (Lm) infection (Supplementary Fig. 1b).
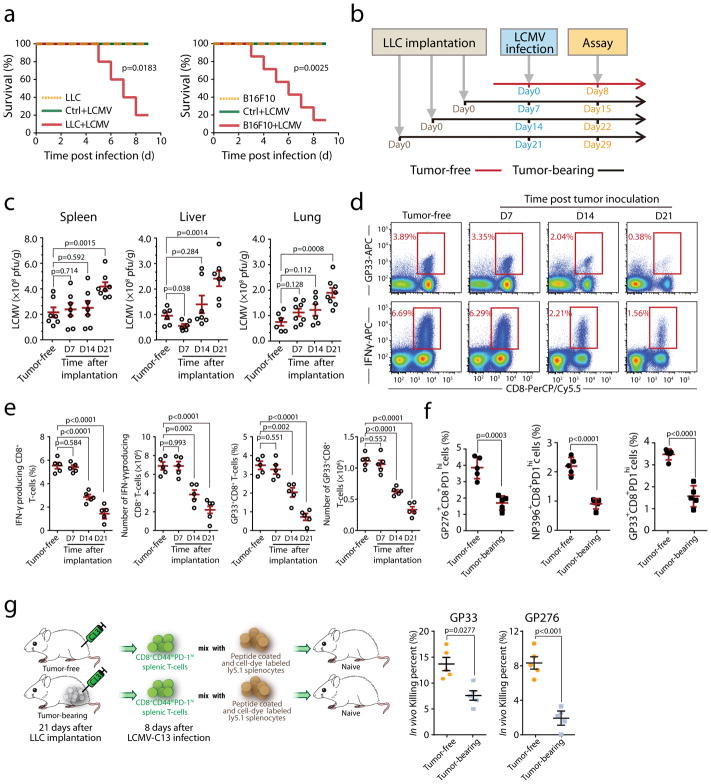
Fig. 1. Increased susceptibility to LCMV-Cl13 infection and decreased immune responses by CD8+T cells in tumor-bearing mice.
a, Survival of tumor-bearing mice (inoculated with LLC or B16F10 cells, n=10), tumor-free mice (n=10) after LCMV-Cl13 infection and uninfected tumor-bearing mice (n=10) was monitored. b–e, Mice were infected with LCMV-Cl13 at different times following LLC inoculation (0, 7, 14 and 21 days) and sacrificed on day 8 post-infection (b). Viral load in the indicated tissues including the spleen, liver and lung at 21 days after tumor implantation, (Tumor free, n=7(spleen), n=6(liver and lung); D7, n=7(spleen), n=6(liver), n=8(lung); D14, n=7(spleen and liver), n=6(lung); D21, n=8(spleen and lung), n=7(liver). (c). Antigen specific CD8+ T cells (top) and production of IFN-γ by splenic CD8+ T cells after stimulation with viral antigen (bottom) were determined by staining for intracellular IFN-γ and LCMV specific tetramers, the frequency and total number of IFN-γ producing and antigen-specific CD8+ T cells in the spleens of tumor-bearing mice (d–e, n=5). f, Mice were infected with LCMV-Cl13 at day21 following LLC inoculation and sacrificed on day 8 post-infection. Antigen specific CD8+CD44+PD-1hi cells recognizing each epitope were determined using LCMV epitope-specific tetramers (n=5). g, The ability of CD8+ T cells isolated from LCMV-Cl13-infected tumor-bearing or control mice to kill viral-peptide pulsed splenocytes in vivo was analyzed(n=5). Each point in (c) and (e) represents data from an individual mouse, and the data are representative of three independent experiments. Two-tailed Student’s t-tests were used for all comparisons, with the exception of survival curves, for which Gehan-Breslow-Wilcoxon tests were used. Two-tailed p-values were reported, Bar graphs denote mean values with SEM.
To determine the impact of tumor establishment on CD8+ T cells during LCMV infection, we infected mice with LCMV-Cl13 at different time points post LLC inoculation and performed analyses on day8 post-infection (Fig. 1b). LCMV viral loads in multiple tissues are well controlled at early stages of tumor bearing, whereas compromised at later tumor stages(Fig. 1c). LCMV infection is controlled by both antibody and T cell-mediated responses. The relative numbers of germinal center B cells and splenic T follicular helper cells were not altered by tumor implantation (Supplementary Fig. 2a–e). However, the number of antigen-specific IFN-γ-producing CD4+ T cells was substantially decreased (Supplementary Fig. 2f–g). With prolonged tumor bearing, both the frequency and absolute number of CD8+ T cells specific for all dominant LCMV class I epitopes and their capacity for IFN-γ production were also significantly reduced (Fig. 1e–f). Similarly, decreases in OVA-specific CD8+ T cell responses were also observed in tumor-bearing mice after infection with OVA-expressing Lm (Supplementary Fig. 3a–c).
To directly assess cancer-associated T cell functional impairment in vivo, effector T cells (CD8+CD44+PD-1high) were purified from LCMV-infected tumor-free or tumor-bearing mice and co-transferred with peptide-loaded splenocytes. We observed compromised killing capacity of effector CD8+ T cells from tumor-bearing mice against two dominant LCMV antigens (Fig. 1g). These results demonstrate that, in tumor-bearing mice, deficient protection against infection is accompanied by impaired CD8+ T cells.
To elucidate mechanisms underlying this impaired T cell responses, we analyzed mouse spleens at different durations of tumor bearing. While we observed splenomegaly at the later stages, this spleen size increase was not associated with lymphocytes: instead, B cell, CD4+ and CD8+ T cell frequencies were reduced (Fig. 2a). Consistent with previous reports7,8, MDSCs and tumor associated macrophages increased with tumor progression (Fig. 2a and Supplementary Fig. 4). Surprisingly, spleen size increase was mainly attributed to CD71+TER119+ EPCs: CD71+TER119+ cells comprised 5–10% of splenocytes in tumor-free mice, whereas they accounted for 40–60% of total splenocytes with advanced tumor (Fig. 2a). During tumorigenesis, EPCs also steadily accumulated in the bone marrow, liver and blood, but not in lymph nodes or tumor tissues (Supplementary Fig. 5a–b). Notably, in spleens, the quantity of EPCs was ~3- and 30- fold higher than MDSCs and Tregs, respectively, two established immunosuppressive populations (Fig. 2b).
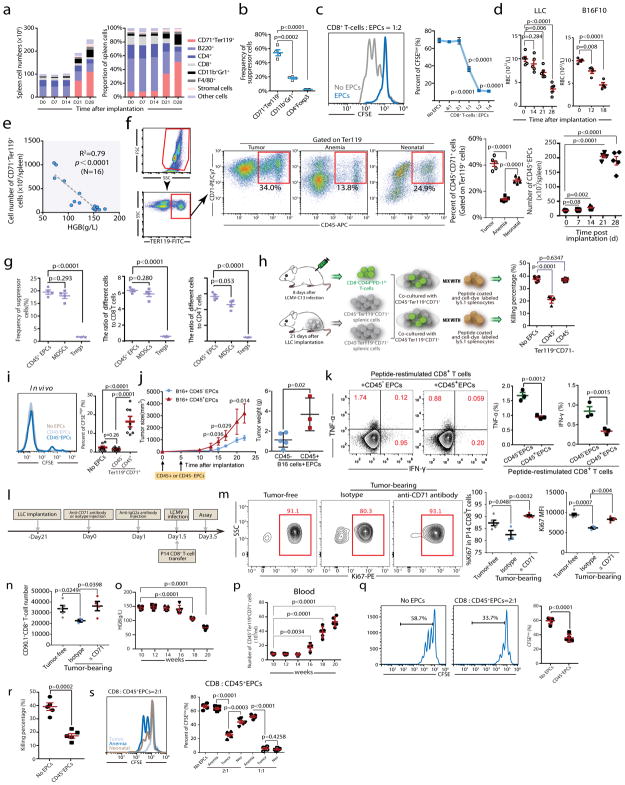
Fig. 2. CD45+CD71+TER119+erythroid progenitor cells accumulate in tumor-bearing mice and exert immunosuppressive effects on CD8+T cells.
a, The number (left) and frequency (right) of MDSCs (CD11b+Gr1+), erythroid progenitor cells (CD71+TER119+), stromal cells (CD45−TER119−), macrophages (F4/80+), CD4+T, CD8+T and B (B220+) cells in the spleens of C57BL/6 mice were quantified at different time points (0, 7, 14, 21 and 28 days) after LLC cell inoculation (n=4). b, The frequencies of MDSCs, CD71+TER119+ cells and Tregs in the spleen 28 days after LLC inoculation were determined (n=4). c, Representative flow cytometry (left) and cumulative composite data (right) showing the proliferation of CFSE-labeled CD8+ T cells after co-culture with CD71+TER119+erythroid progenitor cells isolated from the spleens of tumor-bearing mice at different CD8+ T cell: erythroid progenitor cell ratios (n=4). d, Mature red blood cell (RBC) counts were measured in tumor-bearing mice at different time points (n=4 or 5) after LLC or B16F10 inoculation. Each point represents data from an individual mouse, and the data are representative of at least three independent experiments. e, The correlation between the total number of CD71+TER119+ erythroid progenitor cells in the spleens of tumor-bearing mice and hemoglobin (HGB) concentration was analyzed by Pearson’s correlation coefficient. (n=16). f, Gating strategy: after excluding doublets or larger aggregates, DAPI-positive cells, which are likely membrane-permeable apoptotic cells, were excluded from further analysis. Next, very small events, likely nuclei or debris, were excluded. Finally, we selected TER119+ cells for further analysis. Representative flow cytometry and cumulative composite data show the frequency of CD45+CD71+ cells within the TER119+ population in the spleens of tumor-bearing (21d), anemic(4d) and neonatal mice. Right panel: cumulative composite data show the total number of CD45+CD71+TER119+ cells in the spleens of tumor-bearing mice at the indicated days after tumor inoculation (n=5). g, Cumulative composite data show the frequencies of CD45+CD71+TER119+ EPCs, MDSCs and Tregs in the spleen of mice with advanced tumors (28 days after LLC inoculation)(left). The ratio of immunosuppressive cells, CD45+CD71+TER119+ EPCs, MDSCs or Treg cells, against CD8+(middle) or CD4+(right) T cells in the spleen of mice with advanced tumors (28 days after LLC inoculation) (n=4).
h, CD45+CD71+TER119+ or CD45−CD71+TER119+erythroid progenitor cells isolated from the spleens of tumor-bearing mice were co-cultured with sorted CD8+T cells and the T cell ex vivo killing efficiency was determined after 6 h (n=5). i,2×105 CFSE-labeled P14 CD8+ T cells were mixed with 2×106 sorted CD45+CD71+TER119+ or CD45−CD71+TER119+ cells. The mixed cells were then adoptively transferred into naïve B6 mice immediately (n=8), which were subsequently infected with LCMV-Armstrong. These mice were sacrificed on day 3 post infection and CFSEhigh CD8+ T cells were analyzed by flow cytometry. j–k, A total of 2×105 B16F10-Ova melanoma cells were subcutaneously injected into C57BL/6 mice on day 0. Next, 2×106 CD45+CD71+TER119+ or CD45−CD71+TER119+ cells were intravenously injected on days 0 and 5. j, Tumor growth was monitored every 2 or 3 days (left). Mice were sacrificed on day 22and tumors were collected and weighed (right). Each point represents data from an individual mouse (CD45+CD71+TER119+ group n=3, CD45−CD71+TER119+ group n=5), and data were analyzed by two-tailed unpaired t-test. k, Tumor infiltrating leukocytes were enriched and loaded with the OVA257–264 (SIINFEKL) peptide in vitro for 24-hour restimulation. Frequencies of IFN-γ and TNF-α producing T cells were analyzed by intracellular cytokine staining. Each point represents data from an individual mouse (n=3), and data were analyzed by two-tailed unpaired t-test. l–n, A total of 1×106 Lewis lung cancer cells were subcutaneously injected into C57BL/6 mice (PBS was used as control). Anti-CD71 antibody (1 mg/mouse) was intravenously injected at day 21 after tumor cell inoculation (IgG was used as control, 1 mg/mouse). To attenuate the anti-CD71 antibody, anti-IgG2a antibody (3 mg/mouse) was intravenously injected 24 h later. Finally, we adoptively transferred P14 CD8+ T cells (CD90.1, 2×106 cells/mouse) into mice and infected with LCMV cl13 simultaneously 36 h after administration of anti-CD71 antibody. All mice were sacrificed at day 2 after LCMV infection (l). Representative flow cytometry (m, left) and cumulative composite data (m, middle) show the frequency of Ki67+ cells among P14 CD8+ T cells. Cumulative composite data show the Ki67 MFI in P14 CD8+ T cell (m, right). Cumulative composite data show the total number of CD90.1+CD8+ P14 cells in the spleen (n). o–q, The hemoglobin (HGB)concentration (o) and number of CD45+CD71+TER119+ cells (p) in the peripheral blood of MMTV-PyMT female mice which developed palpable mammary tumors at 12 weeks old were determined at the indicated weeks. The proliferative capacity of CFSE-labeled CD8+ T cells in response to anti-CD3 and anti-CD28 was analyzed after co-culture with CD45+CD71+TER119+ EPCs isolated from the spleens of 20 week old MMTV-PyMT female mice at a CD8+ T cell/EPC ratio of 1:2 (q); CD45+CD71+TER119+ EPCs isolated from spleens of 20 week old MMTV-PyMT females mice were co-cultured with sorted CD8+ T cells and the ex vivo T cell killing efficiency was determined after 6 h (r). s, The proliferative capacity of CFSE-labeled CD8+ T cells in response to anti-CD3 and anti-CD28 was analyzed after co-culture with CD45+CD71+TER119+ erythroid progenitor cells isolated from the spleens of tumor-bearing, anemic or neonatal mice at the indicated CD8+ T cell:EPC ratios (n=5). Each point in (b–e) and (h–m) represents data from an individual mouse. Data are representative of three independent experiments and were analyzed by two-tailed unpaired t-test. Two-tailed p-values were reported. Bar graphs denote mean values with SEM.
In neonatal mice, EPCs compromise innate immune function, resulting in susceptibility to infection9. To test whether EPCs are responsible for impaired CD8+ T cell responses, we performed an in vitro suppression assay against CD8+ T cells from tumor-free mice. Splenic CD71+TER119+ EPCs from day21 tumor-bearing mice are robustly immunosuppressive: T cell proliferation was suppressed by ~50% at a suppressor: effector ratio of 1:1; and a 2:1 ratio reliably ablated CD8+ T cell proliferation (Fig. 2c). EPCs also inhibited CD4+ T cell proliferation and Th1 differentiation (Supplementary Fig. 6).
Normally generated in the bone marrow, stresses such as anemia10, pregnancy11 and infection12 require a rapid increase of erythrocytes. Through a process called “extramedullary erythropoiesis”, erythroid differentiation can be initiated in the liver and spleen13–15. We collected blood post tumor cell inoculation and found that hematocrit and hemoglobin (HGB) levels and mature RBC counts decrease with prolonged tumor bearing (Fig. 2d and Supplementary Fig. 7). Moreover, blood HGB inversely correlates with splenic EPCs (Fig. 2e). Thus, splenic EPC enrichment may result from tumor-initiated anemia and compensatory extramedullary erythropoiesis.
Extramedullary erythropoiesis is also induced during early embryonic development or by acute anemia10,16 (Supplementary Fig. 5a). While phenyl hydrazine induced acute anemia within 3 days and caused CD71+TER119+ cell accumulation in multiple organs including spleen, antigen-specific CD8+ T cell responses against infection remained unimpaired (Supplementary Fig. 8). The number of splenic EPCs is smaller in acutely anemic mice relative to mice carrying established tumors (Supplementary Fig. 5a), and, these EPCs are also less suppressive at the per cell level (Supplementary Fig. 9a). Whereas, the suppressive capacity of EPCs in neonatal spleens falls between EPCs from acutely anemic and tumor-bearing mice (Supplementary Fig. 9b–c).
We hypothesized that the varying immunosuppressive capacity of EPCs from these three conditions results from distinct stages of EPC maturity. During erythroid development, CD45 stalls CD71+TER119+ precursors at undifferentiated stages and its expression is a hallmark of early precursors17,18. Accordingly, within the splenic EPC population, CD45+ progenitors were more abundant in tumor-bearing and neonatal mice than in mice with acute anemia (Fig. 2f). Comparing to other immunosuppressive cells in spleen, the ratio of CD45+CD71+Ter119+ cells to CD4+ and CD8+ T cells is comparable to MDSCs and much higher than Tregs (Fig. 2g). Using erythropoietic tracking19, we confirmed that the CD45−CD71+TER119+ population contains more terminally differentiated stage III–V erythroid cells whereas CD45+CD71+TER119+ cells are enriched with stage I–III precursors (Supplementary Fig. 10). To test whether CD45+ EPCs are more potent suppressors of CD8+ T cells, we collected LCMV-reactive CD8+ T cells (CD8+CD44+PD-1high) and performed in vitro cytotoxicity assays with CD45+ or CD45− EPCs isolated from tumor bearing mice. Indeed, cytotoxic T lymphocyte (CTL) suppression resides in the CD45+ EPC population (Fig. 2h). The superiority of CD45+ EPCs in suppressing CD8+ T cells was validated using anti-CD3/CD28 antibody-stimulated and cognate antigen-induced in vitro proliferation assays (Supplementary Fig. 11a–b). Additionally, after equal numbers of CD45− or CD45+ EPCs were co-transferred with P14 transgenic CD8+ T cells into recipient mice, LCMV-specific T cell proliferation in vivo was only inhibited by CD45+ EPCs (Fig. 2i). Quantitatively, the suppressive capacity of CD45+ EPCs falls between Tregs and MDSCs (Supplementary Fig. 11c).
Compared to anemic and neonatal conditions, tumor establishment leads to much higher accumulation of CD45+ EPCs, which are selectively enriched in the spleen and bone marrow and possess strong suppressive capacity (Supplementary Fig. 11d–g). When transferred into recipient mice at an early stage of tumorigenesis, CD45+ EPCs accelerated B16 melanoma tumorigenesis (Fig. 2j), which was associated with impaired activation of infiltrating tumor antigen-specific CD8+ T cells (Fig. 2k). Similarly, using Ova as a surrogate tumor antigen, we observed that both proliferation and cytotoxicity of OT-I T cells were suppressed in the spleen, suggesting that CD45+ EPCs also suppress tumor antigen-elicited CTL priming (Supplementary Fig. 12). We further validated the relevance of this population in vivo through anti-CD71 antibody-aided EPC depletion. While CD71 is also expressed in activated CD8+ T cells (Supplementary Fig. 13a–g), to preserve CTL activity we blocked the Fc region of the anti-CD71 antibody (IgG2a) upon EPC depletion (Supplementary Fig. 13h–i). Following IgG2a blockade, P14 CD8+ T cells were transferred, and recipient mice were infected with LCMV-Armstrong simultaneously (Fig. 2l). Efficient EPC depletion (Supplementary Fig. 13j) rescued the proliferation of LCMV-specific CD8+ T cells (Fig. 2m–n), providing direct evidence that EPCs suppress anti-viral CTL responses in tumor-bearing mice. We further monitored the development of CD45+ EPCs in MMTV-PyVT transgenic mice20, in which breast tumors developed spontaneously in 12 weeks. At week20, CD45+ EPCs were significantly enriched in the peripheral blood, liver and spleen of mice accompanied with anemia (Fig. 2o–p). These CD45+ EPCs were also robust suppressors of CTL proliferation and killing (Fig. 2q–r). These findings pinpoint the origin of systemic immunosuppression in tumor bearing mice to CD45+ EPCs.
Additionally, we observed that established tumors not only induce anemia, but also arrest the development of CD45+ EPCs (Supplementary Fig. 14). Although carrying the same surface marker, CD45+ EPCs from tumor-bearing mice are more robust suppressors than those from neonatal mice, and much more powerful than those from acute anemic mice (Fig. 2s).
To discover molecular mechanisms underlying immunosuppression by CD45+ EPCs, we performed RNA sequencing (RNAseq) using CD45+ and CD45− EPCs generated by different anemia induction. We included MDSCs, another bone marrow-derived immunosuppressor21, from spleens of tumor-bearing mice as a reference. Although both populations contain heterogeneous erythroid cells across developmental stages, the global gene expression patterns of CD45+ EPCs differ significantly from their CD45− counterparts (Fig. 3a), especially for signature genes defining the erythrocyte lineage and immunosuppression (Supplementary Fig. 15). Within the CD45+ EPC pool, cells from tumor-bearing, neonatal and anemic mice are further separated, whereas CD45+ EPCs and MDSCs from tumor-bearing mice closely resemble one another (Fig. 3b). Indeed, for the MDSC immunosuppression signature genes, CD45+ cells isolated from tumor-bearing or neonatal mice are indistinguishable from MDSCs (Supplementary Fig. 15b).
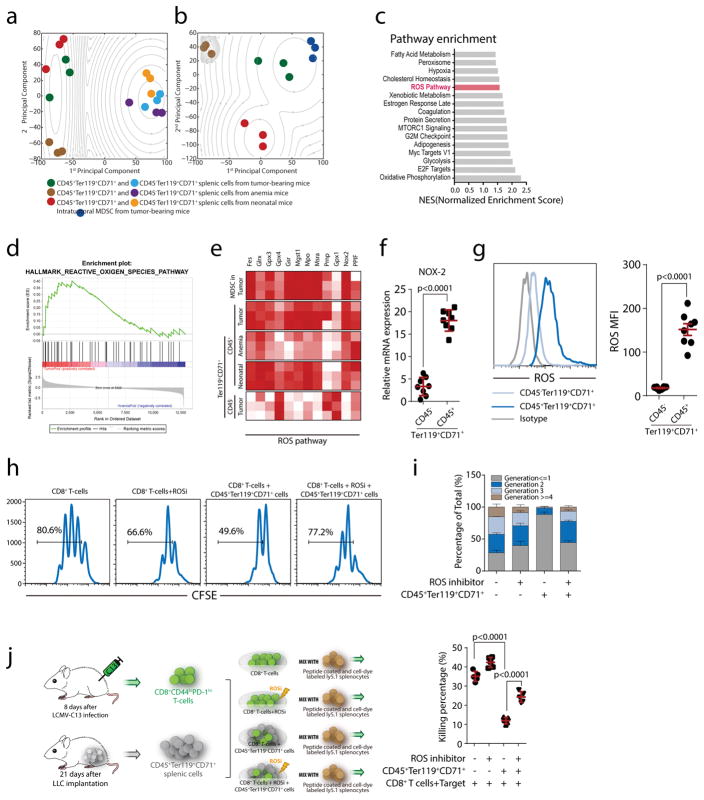
Fig. 3. ROS play a dominant role in the immunosuppressive ability of CD45+CD71+TER119+ cells from tumor-bearing mice.
a–b, Principal component analysis of RNA sequencing data reveals similarity among the CD45+CD71+TER119+ and CD45−CD71+TER119+ cells (a) and similarity among MDSCs and CD45+CD71+TER119+cells (b) from tumor-bearing(n=3), anemic (n=3) and neonatal (n=3) mice. An expectation maximization algorithm was used to perform clustering under Gaussian mixture models. The contour shows the estimated probability density for each group. Log-likelihood is shown. c, Pathway enrichment analysis was performed using Gene Set Enrichment Analysis (GSEA). Significantly enriched (nominal p value <0.05) items in CD45+CD71+TER119+ cells derived from tumor-bearing mice compared with those from anemic mice are shown with enrichment scores (n=3). d, Enrichment plot of the HALLMARK ROS pathway for the comparison between CD45+CD71+TER119+ cells from tumor-bearing mice and anemic mice. e, Heat map illustrating the relative expression of ROS pathway genes in MDSCs and CD45−CD71+TER119+ cells from spleens of tumor-bearing mice and CD45+CD71+TER119+ cells from the spleens of tumor-bearing, anemic and neonatal mice. f, Cybb (Nox2) mRNA expression in CD45+CD71+TER119+ and CD45−CD71+TER119+ cells from tumor-bearing mice (n=8) was quantified by RT-qPCR. g, ROS production in CD45+CD71+TER119+ and CD45−CD71+TER119+ cells from tumor-bearing mice was quantified by flow cytometry(n=8). h–i, CD45+CD71+TER119+erythroid progenitor cells isolated from the spleens of tumor-bearing mice(n=8) were co-cultured with CFSE-labeled CD8+ T cells at a ratio of 1:1 (CD8+ T cells:EPCs). CD8+T cell proliferation was evaluated in the presence or absence of apocynin (ROSi), an NADPH oxidase inhibitor that blocks ROS production. j, CD45+CD71+TER119+erythroid progenitor cells isolated from the spleens of tumor-bearing mice(n=5) were co-cultured with sorted CD8+ T cells in the presence or absence of apocynin, and, after 6 h, the ex vivo killing efficiency of sorted CD8+ T cells was determined. Each point in (f) represents data from an individual mouse. Data are representative of three independent experiments and were analyzed by two-tailed unpaired t-test. Two-tailed p-values were reported. Bar graphs denote the mean value with SEM.
Subsequent analysis of these datasets identified a significant enrichment in the reactive oxygen species (ROS) pathway (Fig. 3c–d). In a comprehensive comparison, the expression of signature genes for ROS production and removal were very similar between CD45+ EPCs and MDSCs (Fig. 3e). Excessive ROS production is a well-established mechanism for MDSC-mediated T cell immunosuppression22,23, where the NOX family of ROS-generating NADPH oxidases, including Nox2, play an essential role23,24. Indeed, compared to their CD45− counterparts, Nox2 expression is markedly higher in CD45+ EPCs (Fig. 3f). Accordingly, ROS levels are significantly elevated in CD45+ EPCs (Fig. 3g). While intracellular ROS are necessary for optimal TCR activation25, excessive intracellular26 and environmental21 ROS is detrimental to effector T cell responses. Consequently, apocynin, a NADPH oxidase inhibitor25,27, released the CD45+ EPC blockade to restore CTL proliferation (Fig. 3h–i) and killing (Fig. 3j). These results suggest that, like in MDSCs, ROS represent a major mechanism for CD45+ EPCs to execute immunosuppression.
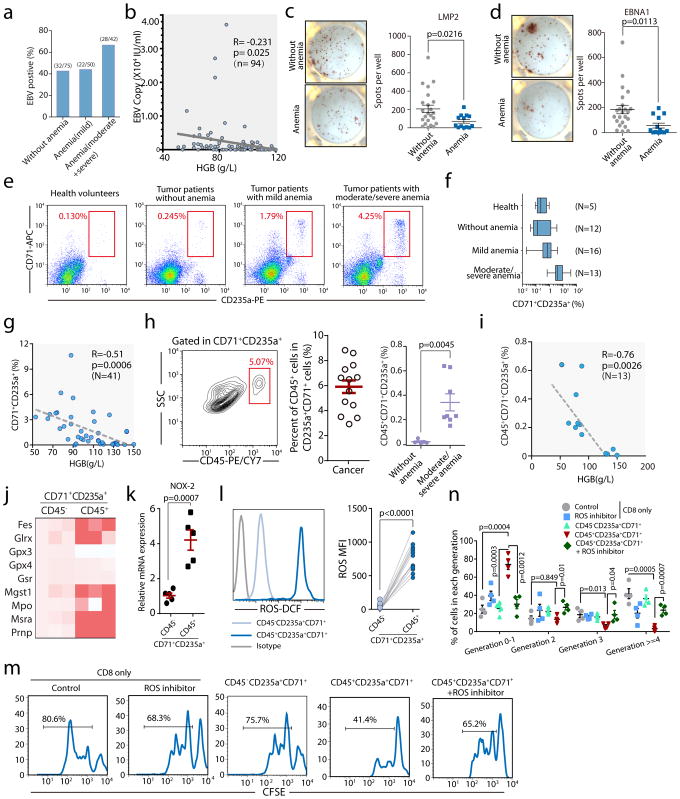
Next, we investigated the clinical relevance of EPCs. Epstein-Barr virus (EBV) is a common latent virus infecting most human populations, and control of viremia relies on EBV-specific CTLs28,29. We collected peripheral blood from 167 cancer patients and examined EBV viremia and EBV-specific CTL responses. For this cohort, EBV reactivation was significantly increased in patients with moderate to severe anemia (Fig. 4a). Moreover, within the subgroup of EBV-reactivated patients, EBV copy number inversely correlated with blood HGB (Fig. 4b). Their CD8+ T cell responses against the major antigenic EBV proteins, LMP2 and EBNA1 were significantly suppressed (Fig. 4c–d). Human EPCs can be identified by CD71+CD235a+ expression9. The frequency of EPCs in peripheral blood is increased in cancer patients with moderate to severe anemia (Fig. 4e–f), which is inversely correlated with their HGB concentrations (Fig. 4g). In patients with moderate to severe anemia, CD45+CD71+CD235a+ cells are detected in every individual with an average frequency of 6% within the EPC pool and 0.4% among all PBMCs (Fig. 4h) and also inversely correlated with blood HGB levels (Fig. 4i). As in mice, the ROS signature (Fig. 4j), and, specifically, NOX2 (Fig. 4k) expression in human CD45+ EPCs is significantly elevated compared to CD45− cells. Mechanistically, CD45+ EPCs are robust ROS producers (Fig. 4l) and suppressors of TCR-stimulated CD8+ proliferation, and this immunosuppression is ROS-dependent (Fig. 4m, n). Although we do not have access to spleens from these cancer patients, we reasoned that human CD45+ EPCs functionally mirror their murine counterparts.
Fig. 4. Erythroid progenitor cells accumulate in cancer patients with anemia, and their inhibitory effects on CD8+T cells can be blocked by a ROS inhibitor.
a–b, EBV DNA loads in the peripheral blood were detected. The percentage of EBV positive patients (EBV DNA>400 copies/mL) without anemia (male: HGB>120 g/L, female: HGB>110 g/L) or with varying degrees of anemia (mild HGB>90 g/L, moderate HGB 60–90 g/L, severe HGB 30–60 g/L) is shown (a). The correlation between hemoglobin (HGB) and EBV DNA loads in the peripheral blood of cancer patients was analyzed by Pearson’s correlation coefficient (b, n=94). c–d, Enzyme-linked immunospot assay (ELISPOT) results show the number of LMP2(c) or EBNA1(d) specific CD8+ T cells in 1×106 PBMCs in cancer patients without (HB>110 g/L, n=23) or with anemia (<110g/L, n=13). e–f, Representative flow cytometry plots (e) and cumulative composite data (f) showing the percentages of CD71+CD235a+erythroid progenitor cells in the peripheral blood of healthy donors and cancer patients without (HB>110 g/L) or with varying degrees of anemia. g, The correlation between hemoglobin (HGB) and the frequency of CD71+CD235a+erythroid progenitor cells in the peripheral blood of cancer patients was analyzed by Pearson’s correlation coefficient (n=41). h, Representative flow cytometry plots and cumulative composite data showing the frequency of CD45+ cells in CD71+CD235a+ cells in the peripheral blood of cancer patients with or without anemia (n=13). Far right panel: frequency of CD45+CD71+TER119+ cells among PBMCs isolated from tumor patients without anemia (n=5) or with moderate and severe anemia (n=8). i, The correlation between hemoglobin (HGB) and the frequency of CD45+CD71+CD235a+erythroid progenitor cells in the peripheral blood of cancer patients was analyzed by Pearson’s correlation coefficient (n=13). j, mRNA expression levels of genes in the ROS pathway in CD45+CD71+CD235a+ and CD45−CD71+CD235a+ cells from the peripheral blood of cancer patients were quantified by RT-qPCR. k, Cybb (Nox2) mRNA expression in CD45+CD71+CD235a+ and CD45−CD71+CD235a+ cells from cancer patients was quantified by RT-qPCR (n=5). l, ROS production in CD45+CD71+CD235a+ and CD45−CD71+CD235a+ cells from cancer patients was analyzed by flow cytometry (left), and the mean fluorescent intensity (MFI)of the fluorescent dye for detecting ROS was quantified (right) (n=11). m–n, CD8+ T cell proliferation after stimulation with anti-CD3 and anti-CD28 was measured using CFSE-labeled CD8+ T cells cultured alone or co-cultured with CD45+CD71+CD235a+or CD45−CD71+CD235a+cells at a 1:1 ratio, with or without the ROS inhibitor apocynin. Representative FACS plat was shown in (m). Two-tailed unpaired t-test of four independent experiments was performed by measuring the distribution of CD8+ T cells in each division(n). Each point in (b–d) and (g–i) represents data from an individual patient. Data are representative of three independent experiments and were analyzed by a two-tailed unpaired t-test. Two-tailed p-values were reported. Error bars denote the SEM.
In summary, stemming from clinical observations, we recapitulated the systemic suppression of CD8+ T cell activation and enhanced susceptibility to infection in animals bearing established tumors. During mouse tumor progression, as recently reported30, an unusual cell type, the CD71+TER119+ EPC, becomes dominant in the spleen; in cancer patients with anemia, CD71+CD235a+ EPCs are enriched. While splenic CD45−CD71+TER119+ EPCs are not immunosuppressive and facilitate metastasis through long-range and direct impact on tumor cells30, the CD45+, less-differentiated progenitor within the EPC population behaves as a potent MDSC-like immunosuppressor impairing both CD8+ T cell priming in the spleen and effector function in peripheral tissues, eventually causing the systemic impairment of CD8+ T cell function. Because this population vastly outnumbers Tregs and is at least equal to MDSCs in size and immunosuppressive activity, we believe that EPCs should not be ignored in therapeutic strategies aimed at enhancing cancer patients’ adaptive immunity.
Online Methods
Cell lines and cell culture
Lewis lung carcinoma (LLC) cells and B16F10 melanoma cells (B16F10) were obtained from the American Type Culture Collection (ATCC), and their identity was verified before experiments were conducted. Cells were cultured in DMEM/H (HyClone) containing 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Beyotime Biotechnology) and incubated at 37°C in a humidified atmosphere with 5% CO2. Cell lines were routinely validated and are free of mycoplasma contamination.
Human sample collection and processing
Peripheral blood was collected from healthy adult volunteers and cancer patients under Xinqiao Hospital Medical Center Institutional review board (IRB) approved protocols (#2013047). Written informed consent was provided from all study participants before sample collection. All peripheral blood collections were performed before any transfusion, even for anemic patients. Mononuclear cells were freshly isolated over lymphocyte separation medium. CD8+ T and CD45+CD71+ CD235a+ cells were sorted as described below. No statistical method was used to predetermine sample size.
Tumor and anemia animal models
OT-1 transgenic C57BL/6J mice were obtained from Jackson Laboratories. P14 (CD90.1) TCR transgenic mice were obtained from Dr. Rafi Ahmed at Emory University. Both male and female mice, ages 6–8 weeks, were included for each experiment. To determine size of animal groups, pilot experiments were performed without statistical estimation. For repeat experiments, we used the JMP statistical package to perform power analysis to arrive at the group size. Based on results from pilot studies, we used parameters of alpha = 0.05; power = 80%; effect size = 50%; and standard deviation = 30% of the mean.
To establish an in vivo tumor model, 1×106 LLCs or 5×105 B16F10 cells were resuspended in 150 μl PBS and subcutaneously injected into the right flank of C57BL/6 mice. Tumor-bearing mice were infected with pathogens at different time points post-inoculation. For LLC tumors, average tumor volumes were 67.14±20.10 mm3 at Day 7, 195.01±55.39 mm3 at Day 14, 621.53±155.14 mm3 at Day 21, and1888.85±402.95 mm3 at Day 28. For B16F10 tumors, average tumor volumes were 110.11±24.16 mm3 at Day 7, 610.60±141.44 mm3 at Day 14, and 2411.24±346.66 mm3 at Day 21.
To induce anemia, C57BL/6 mice were injected intraperitoneally (i.p.) on day 0 with 50 mg/kg phenylhydrazine hydrochloride solution in 200 μl PBS. Blood (10 μl) was obtained from the tail vein on day 3 for hematocrit, hemoglobin and RBC count measurements. Mice infected with lymphocytic choriomeningitis virus (LCMV) and Listeria monocytogenes (Lm). All mice were used in accordance with the institutional biosafety regulations and guidelines of the Institutional Animal Care and Use Committees of the Third Military Medical University.
Animal infection models
The LCMV-Armstrong and clone 13 (Cl13) strains were gifts from Dr. Rafi Ahmed at Emory University. Recombinant Lm expressing OVA (rLmOVA) and actA-deficient rLmOVA (ΔactArLmOVA) were kindly provided by Dr. John Harty (University of Iowa, Iowa City, IA).
Mice were infected intravenously (i.v.) with LCMV-Armstrong (2×105 PFU) or intraperitoneally (i.p.) with LCMV-Cl13 (2×106 PFU) at different time points (days 0, 7, 14, 21) after LLC cell inoculation. All mice were sacrificed on day 8 after LCMV infection. To assess susceptibility to Lm infection in tumor-bearing mice, mice were intravenously inoculated with a sublethal dose of 4×106CFU ΔactArLmOVA per mouse at different time points (days 0, 7, 14, 21) after LLC cell inoculation. All mice were sacrificed on day 7 after ΔactArLmOVA infection. To assess immune responses to Lm infection in anemic mice, mice were intravenously injected with a dose of 4×106CFUΔactArLmOVA per mouse at different time points. All mice were sacrificed on day 7 after ΔactArLmOVA infection. For both LCMV and Lm infection, tumor-bearing mice were allocated into different groups based on computer-generated randomization.
Virus titration
The viral loads of LCMV-Armstrong were titrated by plaque assay as previously described (28). For quantification of LCMV-Cl13 loads, the weight of harvested tissues from infected mice was measured. After homogenization, total RNA was then extracted from the tissue homogenate using TIANAMP Virus RNA Kit (TIANGEN, China) and was subjected to reverse transcription using the RevertAid Minus First Strand cDNA Synthesis Kit (Thermo, USA) following the manufacturer’s instructions, while LCMV-specific glycoprotein primer (GP-R: GCAACTGCTGTGTTCCCGAAAC) was used for cDNA synthesis. RT-qPCR with LCMV glycoprotein-specific primer pairs (GP-R, GCAACTGCTGTGTTCCCGAAAC, and GP-F CATTCACCTGGACTTTGTCAGACTC) was then used to evaluated viral loads in tissue samples. The Cq (quantification cycle) values from RNA samples of ten-fold serially diluted LCMV-Armstrong that had been previously titrated by plaque assay (28) was used to generate a standard curve. The pfu of LCMV-Cl13 in tissues was calculated according to the formula: lg(pfu)=slope*Cq+y-intercept; pfu/g=pfu calculated from above/tissue weight.
Survival analysis
14 days after LLC cell inoculation, or 10 days after B16F10 melanoma cell inoculation, mice were injected i.v. with 2×105 CFU rLmOVA in 300 ml saline or i.p. with LCMV-Cl13 (2 × 106 PFU). Uninfected tumor-bearing mice and infected C57BL/6 mice without tumor cells were used as controls. All mice were monitored twice daily.
Adoptive transfer
CD8+ OT-1 cells were enriched (>90%) from splenocytes by immunodepletion of CD8− cells using a CD8a+ T Cell Isolation Kit II (Miltenyi Biotec) according to the manufacturer’s instructions. Groups of congenic recipient mice were injected i.v. with equivalent numbers of their respective CD8+ OT-1 cell subpopulations.
Flow cytometry
Antibody staining was performed in PBS containing 2% BSA or FBS (wt/vol). For analysis of cytokine production, splenocytes were first stimulated with GP33 peptide (amino acid sequence: KAVYNFATC, 0.2 μg/ml) in the presence of brefeldin A for 5 hrs at 37 °C. Following surface staining, intracellular cytokine staining was performed with a Cytofix/Cytoperm Fixation/Permeabilization kit (554714, BD Biosciences) according to the manufacturer’s instructions. To detect degranulation, splenocytes were stimulated for 5 h with the indicated peptide (0.2 μg/ml), in the presence of brefeldin A and anti-CD107a/b antibodies (1D4B, BD Biosciences). Major histocompatibility complex (MHC) class I peptide tetramers consisting of H-2Db complexed with LCMV GP33-41 were obtained from Dr. Rafi Ahmed (Emory University). Samples were collected using a FACSCanto system (BD Bioscience) and analyzed with FlowJo software (TreeStar).
Antibodies
Fluorophore- or biotin-conjugated antibodies specific for mouse cell-surface antigens and cytokines were as follows: anti-B220 (RA3-6B2, BioLegend), anti-CD4 (GK1.5, BioLegend), anti-CD8a (53-6.7, BioLegend), anti-CD11b (M1/70, BioLegend), anti-Gr1 (RB6-8C5, BioLegend), anti-CD25 (3C7, BioLegend), anti-Foxp3 (MF14, eBioscience), anti-TER119 (TER-119, BioLegend), anti-CD71 (R17 217.1.3, BioXCell for depletion; C2F2, BD biosciences for FACS analysis), anti-CD45 (I3/2.3, BioLegend), anti-CD90.1 (16-10A1, BioLegend), anti-Gzmb (GL1, BioLegend), anti-IFN-γ (XMG1.2, BioLegend). anti-CXCR5 (L138D7, BioLegend), anti-mouse/human CD44 (IM7, BioLegend), anti-human/mouse Bcl-6 (7D1, BioLegend), anti-T-bet (4B10, BioLegend), anti-mouse Ki-67 (16A8, BioLegend), anti-mouse TNF-α (MP6-XT22, BD). For human studies, the following fluorophore- or biotin-conjugated antibodies specific for cell surface markers or cytokines were used: anti-CD3 (UCHT, BioLegend), anti-CD8 (PRA-T8, BioLegend), anti-CD45 (2D1, BioLegend), anti-CD71 (CY1G4, BioLegend), anti-CD235a (HI264, BioLegend). ROS production was measured by labelling with 2′,7′ –dichlorofluorescin diacetate using the ROS detection kit (S0033, Beyotime).
Cell sorting and in vivo killing assay
Cell sorting was performed on a FACSAriaII sorter (BD Biosciences). The purity of all populations was >95%. In vivo killing assays were performed as previously described (29). Unless otherwise specified, the E:T ratio was set as 4:1 and determined by normalizing all populations to the number of DbGP33 tetramer-positive CD8+ T cells.
Ex vivo killing assay
Ex vivo killing assays were performed as previously described (29). Peptide-pulsed target cells were mixed with sorted CD8+ T cells at a 4:1 (E:T) ratio, which was determined by normalizing all populations to the number of DbGP33 tetramer-positive CD8+ T cells.
T cell proliferation and co-culture
A 96-well plate was coated with a monoclonal antibody to CD3. The antibody was allowed to bind, and then the plate was washed. T cell proliferation was analyzed by labeling with carboxy-fluorescein diacetate succinimidyl ester (CFSE) at a final concentration of 5 μM (Molecular Probes, Invitrogen) according to the manufacturer’s instructions. Briefly, cells were resuspended in warm PBS containing 5 μM CFSE and incubated at 37°C for 10 min. Subsequently, cells were washed twice and resuspended in RPMI 1640 containing 10% FBS. CFSE-labeled human PBMCs were added to the wells along with test compound and anti-CD28 antibody to provide costimulation. For co-culture analysis, different types of sorted cells were added at different ratios. In some co-culture experiments, CD45+ EPCs were treated for 30 min before and during co-culture with 300 mM apocynin (49-hydroxy-39-methoxyacetophenone) to inhibit NADPH oxidase. For all experiments, cells were analyzed by flow cytometry after 72 h in culture. For in vivo proliferation assays, P14 (CD45.1+) CD8+ T cells were labeled with 2 μM CFSE according to the manufacturer’s instructions (Invitrogen), and 2×105 P14 cells were mixed with 2×106 sorted CD45+CD71+TER119+ or CD45−CD71+TER119+ cells. These mixed cell populations were then adoptively transferred into naïve B6 mice immediately, which were subsequently infected with LCMV-Armstrong. Mice were sacrificed on day 3 post-infection and cell populations were analyzed by flow cytometry.
Real-time quantitative PCR
RNA was extracted using the RNeasy Mini kit (Qiagen), and 1 μg total RNA was used for first-strand cDNA synthesis. Reverse transcription polymerase chain reaction (RT-qPCR) was performed using a LightCycler instrument (Roche Diagnostics). Primer used for RT-qPCR analysis is listed in Supplementary Table 4.
RNA sequencing (RNAseq) library construction
Total RNA from sorted CD45+CD71+TER119+ and CD45-CD71+ TER119+ cells from neonatal, anemic and tumor-bearing (LLC) mice and MDSCs from tumor-bearing (LLC) mice was extracted using Trizol reagent (Life Technologies) and then treated with DNase I (Qiagen). The RNAseq library for these RNA samples was constructed according to the strand-specific RNA sequencing library preparation protocol. mRNA transcripts were enriched by two rounds of poly-(A+) selection with Dynabeads oligo-(dT) 25 (Invitrogen) before library construction. The prepared libraries were sequenced on an Illumina HiSeq 2000 sequencer. The full RNAseq dataset has been uploaded into the Gene Expression Omnibus (GEO) repository (Accession code: GSE106384).
Bioinformatic analysis
Raw sequence reads were first aligned to the mouse UniGene transcriptome with bowtie (v1.0.0) to estimate the insert fragment size and the standard deviations needed by TopHat2 to align the reads to the genome. Then, TopHat2 was used to align the reads to the reference mouse genome (GRCm38) with the aligning parameter-bowtie1 and Ensembl annotated transcripts (version 77) as a guide reference. Uniquely mapped reads were used to quantify gene expression, and differential gene expression evaluation was analyzed by Cuffdiff, a subpackage of Cufflinks (v2.1.1) with Ensembl annotated genes (version 77). The abundance of transcripts (including mRNAs, pseudogenes, non-coding RNAs and other predicted RNAs) was calculated and normalized in FPKM as described above from the raw RNAseq data and used for Gene Set Enrichment Analysis (GSEA, Broad Institute).
In vivo depletion of CD71+ cells
1×106 LLC cells were subcutaneously injected into C57BL/6 mice (PBS was injected as a control). Then, anti-CD71 deletion antibody (0.5 or 1.0 mg/mouse, R17 217.1.3 clone, BioXcell, USA) or isotype control rat IgG2a (1.0mg/mouse, BioXcell, USA) was intravenously injected at day 21 after tumor cell inoculation. To attenuate the anti-CD71 deletion antibody, anti-IgG2a antibody (3.0 mg/mouse, BioXcell, USA) was injected intravenously 24 h later. We adoptively transferred P14 CD8+ T cells (CD90.1, 2×106 cells/mouse) into mice and simultaneously infected with LCMV 36 h after administration of anti-CD71 deletion antibody. All mice were sacrificed at day 2 after LCMV infection.
Evaluation of EBV DNA levels
Peripheral venous blood (3 mL) was collected before treatment from each patient into EDTA-containing tubes and centrifuged at 3000 rpm for 5 min. Total plasma DNA was extracted using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Fluorescence polymerase chain reaction (PCR) was carried out using the EBV PCR quantitative diagnostic kit (Da-AnGenetic Diagnostic Center, Guangzhou, China). EBV DNA levels were measured by RT-qPCR with primers specific for the BamHI-W region of the EBV genome. Experimental data were analyzed using Applied Biosystems 7300 SDS software for statistics and an EBV DNA level of <400 copies/mL was defined as negative.
IFN-γ ELISPOT Assay
ELISPOT assays were performed using a human IFN-γ ELISPOT kit according to the manufacturer’s procedure (DAKAWE, China). Briefly, 96 well plates pre-coated with anti-IFN-γ mAb (DAKAWE, China) were washed 3 times with phosphate-buffered saline (PBS) and then blocked with RPMI 1640 for 2 h, followed by removal of blocking medium and washing with PBS. PBMCs (1×106 cells in 100 μl complete RPMI) from cancer patients were seeded into each well and stimulated with 2 μg/ml of PepMix EBV peptide (EBNA1 or LMP2; JPT, USA) for 20 h. A CEF control peptide pool (Mabtech, USA) was included as a positive control. Plates were then washed 4 times with washing buffer, after which biotinylated detection antibody for IFN-γ (DAKAWE, China) was added and incubated for 1 h at 37°C. Following 4 additional washes with washing buffer, avidin-horseradish peroxidase (HRP) (DAKAWE, China) was added and plates were incubated for 1 h at 37°C. Plates were then washed 4 more times with washing buffer, followed by 2 washes with PBS and addition of AEC substrate (BD) to each well. After 15 minutes the reaction was stopped by washing with deionized water, and plates were dried overnight prior to membrane removal. Spots were counted using an ELISPOT reader (ImmunoSpotTM, Cellular Technology, Cleveland, OH).
Th1 differentiation
CD4+ T cells were added to pre-coated anti-CD3 and anti-CD28 plates. Culture media was supplemented with cytokines and blocking antibodies (20 ng/ml rmIL-12, 20 ng/ml hIL-2, and 10ug/ml anti-IL-4). The plate was incubated at 37˚C with 5% CO2 for 4 days prior to analysis of cytokine production by flow cytometry.
Statistical analysis
Statistical analysis was conducted with Prism 6.0 (GraphPad). Two-tailed unpaired Student’s t tests with 95% confidence intervals were used to calculate all P values. For all experiments, no data points or animals were excluded from statistical analysis, and variance was similar between groups being statistically compared. Significance was set at P < 0.05.
Data Availability Statement
All original data used in this study are available upon request. All published non-commercial reagents can be made available upon request.
Supplementary Material
Acknowledgments
Q.-J.L. is a Whitehead Family Foundation Scholar and supported by NIAID (R01-AI091878), NCI(P50-CA190991), and the NIH Beau Biden Cancer Moonshot Initiative (R33CA225328). This work is also supported by the National Nature Science Foundation of China (grant nos. 81472648, 81500089, 81620108023, 81222031, 81773041 and 31600733), by the Research on The Basis and Frontier of Chongqing (grant nos. cstc2016jcyjA0049). MMTV-PyMT mice were kindly provided by Prof. Xiaolong Liu (State Key Laboratory of Cell Biology, CAS Center for Excellence in Molecular Cell Science, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai).
Footnotes
Accession Codes
The RNA-seq data are available through GEO repository (Accession code: GSE106384)
Disclosure of potential conflict of interest
The authors declare no competing financial interests.
Author Contribution
L. Z., R. H., H. L., and B. G. perfomed experiments, analyzed data and drafted the manuscript with comments from all authors. Q. J. is responsible for transcriptome analysis. D. Q. and S.-Q. L. performed Lm infection and EPC depletion experiments. Z. W., T. X., J. Z., Y. T., J. H., J. C., F. W., M. X., J. G., X. Y., H. Z., X. W., and C. H. assisted or independently perfromed some experiments. P. B. A., A. L.J. S., J. Y., and Y. W. provided critical reagents, technical support, or clinical samples. They also provided help in writing this manuscript. L. Z. generated the initial hypothesis and Q.-J. L. and B. Z. conceptulated this study. Q.-J. L., L. Y., and B. Z. supervised the project, designed experiments, interpreted data and wrote the manuscript.
References
- 1.Baden LR, et al. Prevention and Treatment of Cancer-Related Infections. Journal of the National Comprehensive Cancer Network. 2012;10:1412–1445. doi: 10.6004/jnccn.2012.0146. [DOI] [PubMed] [Google Scholar]
- 2.Vento S, Cainelli F, Ternesgen Z. Lung infections after cancer chemotherapy. Lancet Oncology. 2008;9:982–992. doi: 10.1016/S1470-2045(08)70255-9. [DOI] [PubMed] [Google Scholar]
- 3.van der Burg SH, Arens R, Ossendorp F, van Hall T, Melief AJM. Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nature Reviews Cancer. 2016;16:219–233. doi: 10.1038/nrc.2016.16. [DOI] [PubMed] [Google Scholar]
- 4.Bodey GP. Infection in cancer patients. A continuing association. The American journal of medicine. 1986;81:11–26. doi: 10.1016/0002-9343(86)90510-3. [DOI] [PubMed] [Google Scholar]
- 5.Kosmidis CI, Chandrasekar PH. Management of gram-positive bacterial infections in patients with cancer. Leukemia & Lymphoma. 2012;53:8–18. doi: 10.3109/10428194.2011.602770. [DOI] [PubMed] [Google Scholar]
- 6.Segal BH, et al. Prevention and treatment of cancer-related infections. Journal of the National Comprehensive Cancer Network : JNCCN. 2008;6:122–174. doi: 10.6004/jnccn.2008.0013. [DOI] [PubMed] [Google Scholar]
- 7.Ugel S, et al. Immune Tolerance to Tumor Antigens Occurs in a Specialized Environment of the Spleen. Cell Reports. 2012;2:628–639. doi: 10.1016/j.celrep.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Cortez-Retamozo V, et al. Origins of tumor-associated macrophages and neutrophils. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elahi S, et al. Immunosuppressive CD71(+) erythroid cells compromise neonatal host defence against infection. Nature. 2013;504:158. doi: 10.1038/nature12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett M, Pinkerton PH, Cudkowicz G, Bannerman RM. Hemopoietic progenitor cells in marrow and spleen of mice with hereditary iron deficiency anemia. Blood. 1968;32:908–921. [PubMed] [Google Scholar]
- 11.Nakada D, et al. Oestrogen increases haematopoietic stem-cell self-renewal in females and during pregnancy. Nature. 2014;505:555. doi: 10.1038/nature12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–U799. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inra CN, et al. A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature. 2015;527:466. doi: 10.1038/nature15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman MH, Saunders EF. Hematopoiesis in the human spleen. American journal of hematology. 1981;11:271–275. doi: 10.1002/ajh.2830110307. [DOI] [PubMed] [Google Scholar]
- 15.Lowell CA, Niwa M, Soriano P, Varmus HE. Deficiency of the Hck and Src tyrosine kinases results in extreme levels of extramedullary hematopoiesis. Blood. 1996;87:1780–1792. [PubMed] [Google Scholar]
- 16.Cheshier SH, Prohaska SS, Weissman IL. The effect of bleeding on hematopoietic stem cell cycling and self-renewal. Stem Cells and Development. 2007;16:707–717. doi: 10.1089/scd.2007.0017. [DOI] [PubMed] [Google Scholar]
- 17.Craig W, Poppema S, Little MT, Dragowska W, Lansdorp PM. CD45 isoform expression on human haemopoietic cells at different stages of development. British journal of haematology. 1994;88:24–30. doi: 10.1111/j.1365-2141.1994.tb04972.x. [DOI] [PubMed] [Google Scholar]
- 18.Harashima A, et al. CD45 tyrosine phosphatase inhibits erythroid differentiation of umbilical cord blood CD34+ cells associated with selective inactivation of Lyn. Blood. 2002;100:4440–4445. doi: 10.1182/blood-2002-03-0864. [DOI] [PubMed] [Google Scholar]
- 19.Chen K, et al. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci U S A. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nature Reviews Immunology. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusmartsev S, Nefedova Y, Yoder D, Gabrilovich DI. Antigen-specific inhibition of CD8(+) T cell response by immature myeloid cells in cancer is mediated by reactive oxygen species (vol 172, pg 989, 2004) Journal of Immunology. 2004;172:4647–4647. doi: 10.4049/jimmunol.172.2.989. [DOI] [PubMed] [Google Scholar]
- 23.Lam GY, Huang J, Brumell JH. The many roles of NOX2 NADPH oxidase-derived ROS in immunity. Seminars in Immunopathology. 2010;32:415–430. doi: 10.1007/s00281-010-0221-0. [DOI] [PubMed] [Google Scholar]
- 24.Sareila O, Kelkka T, Pizzolla A, Hultqvist M, Holmdahl R. NOX2 Complex-Derived ROS as Immune Regulators. Antioxidants & Redox Signaling. 2011;15:2197–2208. doi: 10.1089/ars.2010.3635. [DOI] [PubMed] [Google Scholar]
- 25.Devadas S, Zaritskaya L, Rhee SG, Oberley L, Williams MS. Discrete generation of superoxide and hydrogen peroxide by T cell receptor stimulation: Selective regulation of mitogen-activated protein kinase activation and Fas ligand expression. Journal of Experimental Medicine. 2002;195:59–70. doi: 10.1084/jem.20010659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B, et al. MicroRNA-23a Curbs Necrosis during Early T Cell Activation by Enforcing Intracellular Reactive Oxygen Species Equilibrium. Immunity. 2016;44:568–581. doi: 10.1016/j.immuni.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson SH, Devadas S, Kwon J, Pinto LA, Williams MS. T cells express a phagocyte-type NADPH oxidase that is activated after T cell receptor stimulation. Nature Immunology. 2004;5:818–827. doi: 10.1038/ni1096. [DOI] [PubMed] [Google Scholar]
- 28.Khanna R, Burrows SR. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu Rev Microbiol. 2000;54:19–48. doi: 10.1146/annurev.micro.54.1.19. [DOI] [PubMed] [Google Scholar]
- 29.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 30.Han Y, et al. Tumor-Induced Generation of Splenic Erythroblast-like Ter-Cells Promotes Tumor Progression. Cell. 2018;173:634–648 e612. doi: 10.1016/j.cell.2018.02.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original data used in this study are available upon request. All published non-commercial reagents can be made available upon request.