Supplemental Digital Content is available in the text
Keywords: Acinetobacter baumannii, antimicrobial peptide, BMAP-28, drug resistant, outer membrane protein
Abstract
Antimicrobial peptides (AMPs) exhibit multiple activities against bacteria and fungi. A bovine myeloid antimicrobial peptide (BMAP-28) belongs to the cathelicidin-derived AMPs and has antimicrobial activity. Due to the rapidly increasing number of infections and outbreaks caused by pan-drug-resistant Acinetobacter baumannii (PDRAB), we sought to determine whether BMAP-28 and its 4 analog peptides (A837, A838, A839, and A840) have antimicrobial activity against PDRAB. Furthermore, we clarified the possible mechanism of inhibition by which of BMAP-28 acts against PDRAB. In the current study, we examined the inhibitory effect of BMAP-28 and its 4 analog peptides on the growth of PDRAB through minimal inhibitory concentration (MIC) analysis and short time killing assays. We also evaluated the effects of BMAP-28 and its analogs on the bacterial cell surface through the use of field emission scanning electron microscopy (FE-SEM). In order to determine the inhibitory mechanism of BMAP-28, we examined the interaction between BMAP-28 and outer membrane proteins (OMPs), especially the interaction between BMAP-28 and A. baumannii OmpA (AbOmpA), which is the main component of OMPs, by using a quartz crystal microbalance (QCM). BMAP-28 and its 4 analogs were effective in inhibiting the growth of PDRAB and had rapid killing ability. BMAP-28 showed exceptionally strong and rapid inhibitory effects on PDRAB when compared to the other peptides and was also shown to cause damage to the cell surface of PDRAB. Moreover, QCM analysis provided evidence of potential interaction between BMAP-28 and AbOmpA. These data indicate that BMAP-28 is a promising candidate for the treatment of PDRAB infections and that its inhibitory effects were related with its binding to AbOmpA.
1. Introduction
Acinetobacter baumannii, a gram-negative coccobacillus, is a nosocomial pathogen that has emerged as a major cause of infection in critical patients with suppressed immunity, major trauma, or burns.[1–4] Because of the inappropriate use of broad-spectrum antibiotics in empirical treatment of nosocomial infections, there have been outbreaks of pan-drug-resistant A baumannii (PDRAB) infections all over the world over the past 20 years or more.[5,6] The rapid emergence of pan-drug-resistant in Acinetobacter genomic species makes PDRAB infections are difficult to treat.[5] Thus, there is an urgent need to develop effective antimicrobial agents against emerging PDRAB.
Antimicrobial peptides (AMPs) are a part of the host defense generated by a wide variety of organisms and act as an important component of innate immunity.[8,9] Cathelicidins are one of several families of AMPs that are found in humans and other species, such as cattle, horses, and pigs.[10] A bovine myeloid antimicrobial peptide (BMAP-28) belongs to the cathelicidin family and exerts potent antimicrobial activity in order to protect animals from bacterial and fungal infections.[11] Brogden et al[12] report that BMAP-28 has antimicrobial activity against Pasteurella multocida. Takagi et al[13] describe the antimicrobial activity of BMAP-28 and its analogs against methicillin-susceptible and methicillin-resistant Staphylococcus aureus (MRSA) and Lynn et al[14] show the antimicrobial effects of BMAP-28 on Leishmania growth. Moreover, it has been indicated that the antimicrobial effect of BMAP-28 can be enhanced by altering its cationic and hydrophobic amino acid residues. To assess this possibility, we have designed 4 analogs of BMAP-28, A837, A838, A839, and A840, which possess a different positive charge when compared to BMAP-28. In the current study, BMAP-28 and its 4 analogs were tested to determine their antimicrobial effects against PDRAB. Specifically, the effect of these peptides on bacterial cell integrity and bacterial activity was assessed.
Moreover, previous researchers have found the AMPs interact with negatively charge outer membrane proteins (OMPs) of bacteria,[15,16] leading to a destabilization and permeabilization of the cell membrane and leakage of the cell contents. It has been reported that the outer membrane protein A of A baumannii (AbOmpA), which is a multifunctional protein, is involved in eukaryotic cell adhesion, invasion, biofilm formation, and apoptosis.[7,17] Here, the interaction between BMAP-28 and AbOmpA was determined by quartz crystal microbalance (QCM) analysis in order to understand the possible inhibitory mechanism of BMAP-28 on PDRAB.
2. Materials and methods
2.1. Bacterial strains and ethics approval
Ten strains of PDRAB were isolated from the skin swabs of ten separate patients with wound infection at the First Affiliated Hospital of Xi’an Jiao Tong University according to the guidelines of the Centers for Disease Control and Prevention (CDC). The isolates were identified using the VITEK 2 system (bioMérieux, Marcy l’Etoile, France) and further confirmed using specific PCR with primers sp2F, sp4F, and sp4R. Antibiotics susceptibility testing was performed using VITEK 2 system. PDRAB isolates are resistant to all antibiotics, including the carbapenems, while sensitive to colistin. The ATCC standard strain of A baumannii (19606) was used as a positive control. The bacteria were cultured in brain heart infusion broth (BHI; OXOID, Hampshire, UK) at 37 °C for 24 hours. All experiments were carried out in accordance with the ethical standards of the Helsinki declaration and were approved by the ethics committee of the medical school of Xi’an Jiao Tong University.
2.2. Synthesis of BMAP-28 and its analogs
BMAP-28 and its 4 analogs (A837, A838, A839, and A840) were synthesized as previously described by Isogai et al.[18] Each of the 4 analogs differs from the original BMAP-28 sequence by one to 3 amino acid residues and has a net charge ranging from +7 to +9. Their sequences are shown in Table 1. These peptides were purified by reverse-phase high-performance liquid chromatography with purity levels over 98%. The final products were identified by electrospray ionization mass spectrometry, suspended in Hank's balanced salt solution, and stored at −80 °C.
Table 1.
Amino acid sequences and physical information of BMAP-28 and its 4 analogs.

2.3. Bacterial growth inhibition test
The optical density (OD) at 595 nm of the precultured bacteria was measured using SkanIt software. The bacterial suspension was diluted to an OD595 of 0.02 and then further diluted to a final concentration of 103 to 104 CFU/mL in BHI broth. The peptide solution was prepared by 2-fold serial dilutions in BHI broth. Next, 1 mL of adjusted bacteria and 1 mL of peptide solution were mixed and incubated at 37 °C. The bactericidal activity of each peptide was evaluated by measuring the minimum inhibitory concentration (MIC) over 24 hours.[19]
2.4. Short time killing assay
PDRAB suspension was inoculated to a final concentration of 1 × 104 cells/mL into 100 μL of sodium phosphate buffer (PBS) in the presence or absence of 40 μg/mL BMAP-28, A837, A838, A839, or A840 and then incubated aerobically for 15, 30, or 60 minutes at 37 °C. The bacterial mixtures were then diluted and inoculated onto BHI agar plates and the CFU was calculated for each. A control group was prepared by plating the reaction mixture that had not received peptide treatment.
2.5. Field emission scanning electron microscopy (FE-SEM)
Ten microliters of BMAP-28 (80 μg/mL) and 10 μL of bacterial suspension were placed on a glass slide and incubated at 37 °C for 15, 30, and 60 minutes, and then air-dried for a minimum of 24 hours. After being dried, the slide was saturated first with 70% ethanol for 5 minutes and then 100% ethanol for 5 minutes. The slide was then coated with a palladium alloy and observed by FE-SEM (SU8000; HITACHI, Tokyo, Japan) in order to assess the effect of BMAP-28 on PDRAB.
2.6. Extraction of A baumannii outer membrane proteins (AbOMPs) and preparation of AbOmpA
Extraction of AbOMPs was performed according to the method previously described by Cuenca et al.[20] Briefly, overnight cultures of A baumannii were subcultured into 100 mL of LB medium and cultured at 37 °C overnight. Cell pellets were centrifuged at room temperature and washed with PBS twice. The cells pellets were then resuspended in 20 mL of 10 mM phosphate buffer (pH 7.2) with 1 mM PMSF. The cells were destroyed by sonication for 12 min on ice. To extract the proteins, the cell debris was isolated by centrifugation and then the supernatant was collected. The supernatant was solubilized using 2% sodium lauryl sarcosinate in 10 mM phosphate buffer for 30 min at room temperature. Finally, AbOMPs were centrifuged, re-suspended in 62.6 mM Tris–HCl buffer, and stored at −20 °C.
AbOmpA was kindly given from Dr Lei (Xi’an Jiaotong University, Xi’an, China). The preparation of AbOmpA was described as following: full-length AbOmpA gene (1317 bp) was amplified by PCR. A chromosomal preparation of A baumannii ATCC 19606T was used as a PCR substrate. The upstream primer 5′-ACAGGATCCATGAAATTGAGTCGTATT-3′ was designed to carry BamHI restriction site and the downstream primer 5′ -ACAAGCTTTTATTGAGCTGCTGCA-3′ carried the HindIII restriction site. PCR products digested with BamHI and HindIII were ligated into the pET28a expression vector (Novagen). Escherichia coli BL21 (DE3)/pET28a harboring a ompA gene was grown in LB medium at 37 °C and recombinant proteins were overexpressed with 1 mM IPTG at 25 °C for 4 hours. After sonication of bacterial cells, the pellet containing the inclusion bodies was discarded and the supernatant containing the soluble form of AbOmpA was collected by centrifugation. The protein samples were loaded on a 5 ml HiTrapTM FF column (Amersham Biosciences) equilibrated with binding buffer (20 mM sodium phosphate, 500 mM NaCl and 5 mM imidazole). His-tagged OmpA was eluted by elution buffer (20 mM sodium phosphate, 500 mM NaCl and 500 mM imidazole). The samples were massively dialyzed against elution buffer without imidazole and then dialyzed with phosphate-buffered saline (PBS, pH 7.4) to reduce salt concentrations. AbOmpA was incubated with endotoxin removal resin for overnight to remove LPS and concentrated by Centricon (2,000 MW cut-off; Millipore). AbOmpA was analyzed for purity by SDS–PAGE after denaturation with sample buffer (60 mM Tris–HCl, pH 6.8, 25% glycerol, 2% SDS, 14.4 mM 2-mercaptoethanol, and 0.1% bromophenol blue) at 100 °C for 5 minutes. After electrophoresis, the gels were stained with 0.1% (w/v) Coomassie blue. Also, endotoxin was assayed under endotoxin-free experimental conditions using a Limulus Amebocytes Lysate (LAL) pyrogen kit (Biowhittaker, Walkersville, MD). The experiments were conducted according to the manufacturer's protocol.
2.7. Western blotting analysis
AbOMPs or AbOmpA (20 μg/lane) were mixed with sample buffer and heated at 100°C for 10 minutes, and then separated by 12% SDS-PAGE and transferred to a PVDF membrane (Bio-Rad Laboratories, Hercules, CA). After incubation in 3% skim milk blocking solution, the membrane was probed overnight at 4°C using anti-OmpA (1:5000; a kind gift from Dr Liu Xi’an Jiaotong University, Xi’an, China). Subsequently, the membrane was incubated at room temperature for 1 h with HRP-labeled goat anti-rabbit IgG (Merck-Millipore, Darmstadt, Germany; 1:10,000), and the immunoreactivities were visualized by reacting with enhanced chemiluminescence (ECL) solution (Beyotime, Shanghai, China). The photos were taken with a Fusion FX5 camera system, and the densities of the protein bands were analyzed using Image J software (National Institute of Health, Bethesda, MA).
2.8. Quartz crystal microbalance (QCM)
BMAP-28 containing an additional C-terminal cysteine was used as a ligand and OmpA from A baumannii as a receptor. QCM measurements were performed with an AFFINIX QN. The quartz crystal sensor surfaces were washed 2 times with concentrated sulfuric acid and hydrogen peroxide solution (3:1) for 5 min. Immobilization of the receptor was achieved using the AFFINIX kit according to the method described by Lahiri et al.[21] After BMAP-28 was bound to the sensor, 5 μL of OmpA solution was applied. OmpA bound to the blocking agent was used as a control at 1 μg/mL. All of these experiments were conducted at 25 °C.
2.9. Statistical analysis
Three independent experiments were performed for each batch of bacterial cells. Results are presented as mean ± standard deviation. The p value was calculated using Dunnett's test. P < .05 was considered to be statistically significant.
3. Results
3.1. Antimicrobial activity of BMAP-28 and its analogs on the growth of PDRAB
The growth inhibition of PDRAB with the treatment of peptides was show in the Supplementary Table 1. The MICs for 10 clinical strains of PDRAB were calculated at 5–10, 20, 5–20, 5–20, and 5–20 μg/mL for BMAP-28, A837, A838, A839, and A840 treatment, respectively (Table 2). For the standard strain of PDRAB, the MICs were calculated at 5, 10, 5–10, 5–10, and 5–10 μg/mL for BMAP-28, A837, A838, A839, and A840, respectively (Table 2).
Table 2.
Growth inhibition of pan-drug-resistant Acinetobacter baumannii by the treatment of BMAP-28 and its analogs.
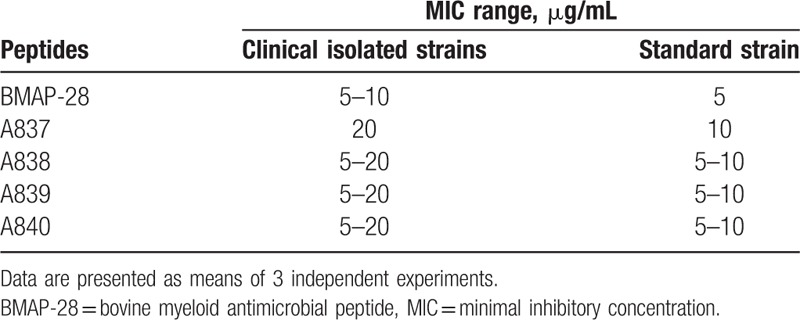
3.2. The short-time antibacterial effect of BMAP-28 and its analogs on the growth of PDRAB
To determine the short time kinetics of the antimicrobial effect of BMAP-28 and its 4 analogs against PDRAB, bacterial suspensions were incubated with 40 μg/mL of peptide for 15, 30, or 60 min. All 5 peptides demonstrated time-dependent antibacterial effects against PDRAB (Fig. 1), with bacterial cells being rapidly killed by BMAP-28 after only 15 mins of incubation. While each of the 4 analogs showed only a minimal effect against bacterial cells when compared to untreated cells, they all showed a gradually reduced trend with the treatment time. The effectiveness of each peptide against PDRAB was ranked as BMAP-28 > A837 > A838 > A839 > A840.
Figure 1.
Short-time bactericidal effect of BMAP-28 and its analogs against pan-drug-resistant Acinetobacter baumannii. Data were mean ± SD of 3 independent experiments. ∗, statistical significance different compared with untreated control (P < .05). ∗∗, statistical significance different compared with untreated control (P < .01).
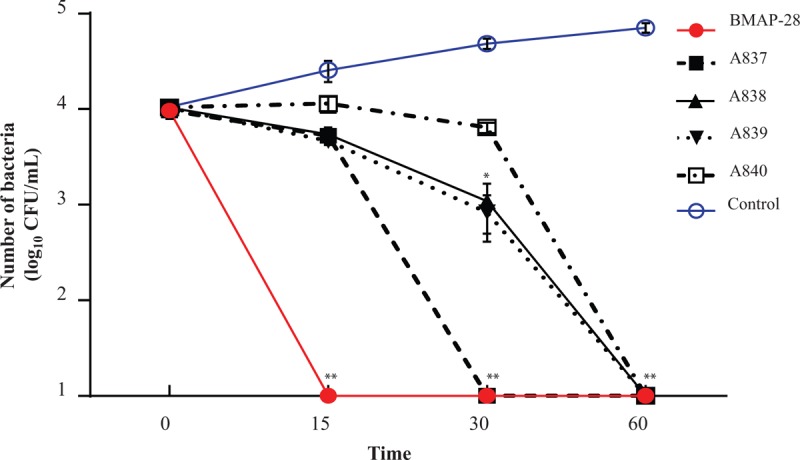
3.3. The effect of BMAP-28 on the cell surface of PDRAB
The effect of BMAP-28 (40 μg/mL) treatment on the cell morphology of PDRAB after 15, 30, or 60 min were observed using FE-SEM. As shown in Figure 2C, the cell morphology changed at 30 minutes with the treatment of BMAP-28, the cell spherical ratio showed an obvious decrease in compared with untreated control. In Figure 2D, some of the bacterial cells treated with BMAP-28 appear to be broken and leaking cytoplasm. Additionally, the surface appears rough compared to untreated PDRAB cells (Fig. 2A), which appear to have mostly smooth outlines and are not aggregated.
Figure 2.
Bacteria cell surface damage after treatment with BMAP-28. The pictures (A–D) are bacteria cells of PDRAB observed by field emission scanning electron microscope after co-culturing with 80 μg/mL BMAP-28 for 0, 15, 30, and 60 minutes, respectively. BMAP-28 = bovine myeloid antimicrobial peptide.
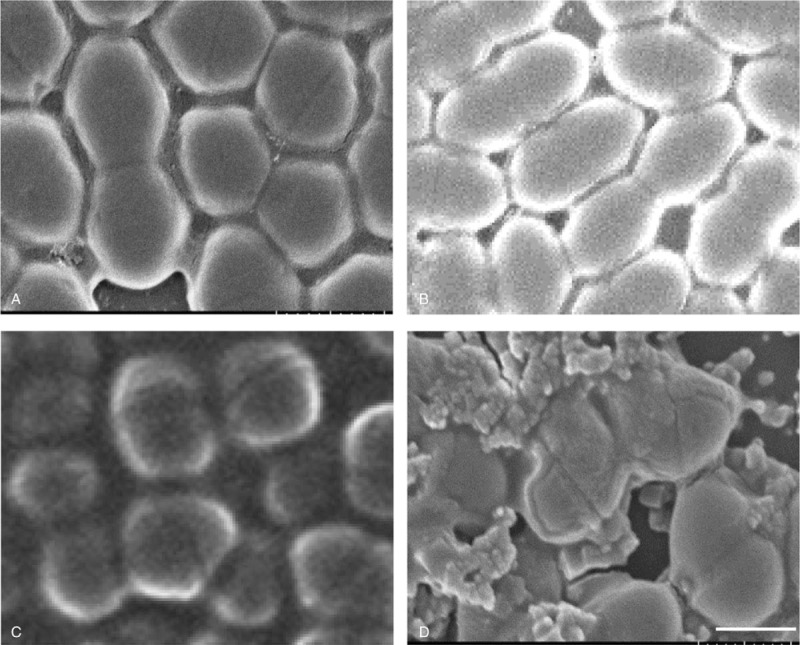
3.4. A baumannii OmpA is the main component of A baumannii OMPs
To verify OmpA is a major porin protein in the OMPs of A baumannii, we observed the composition of AbOMPs by Coomassie blue staining (Fig. 3). The OmpA with a molecular mass of 38 kDa showed the strongest signal in the total OMPs of A baumannii by Coomassie blue staining and verified by western blotting analysis with the anti-OmpA antibody. Here, we use the purified OmpA as a positive control.
Figure 3.
The main component of A baumannii OMPs (AbOMPs). The purificated AbOMPs and AbOmpA were analyzed by electrophoresis, and stained with Coomassie blue (Lane 1 and Lane 2). AbOMPs and AbOmpA in another gel were transferred onto a PVDF membrane and blotted with an anti-OmpA antibody (Lane 3 and Lane 4). The signals were detected using enhanced chemiluminescence (ECL). AbOmpA = Acinetobacter baumannii OmpA, ECL = enhanced chemiluminescence.
3.5. BMAP-28 interacts with the A baumannii OmpA
Because BMAP-28 can inhibit PDRAB growth, we considered that the mode of action may through the interaction of BMAP-28 and PDRAB. Thus, we were interested in identifying potential target for BMAP-28 on the cell surface, especially on the PDRAB outer membrane proteins (OMPs). In this study, we analyzed the interaction of BMAP-28 and AbOMPs by using QCM. Based on the QCM data by plotting the observed frequency shifts in Figure 4, OMPs (1 μg/mL) bound to the blocking agent, as a negative control, showed an insignificant reduction (12 Hz) in the measured frequency. While, OMPs bound to BMAP-28 showed obvious decreases of frequency, the levels of frequency decrease were 198, 427 and 645 Hz after injection of 0.5, 1.0, and 2.0 mg/mL OMPs, respectively. We also analyzed the interaction of BMAP-28 and AbOmpA by using QCM. The level of frequency decrease caused by AbOmpA (1 μg/mL) bound to BMAP-28 was 409 Hz.
Figure 4.
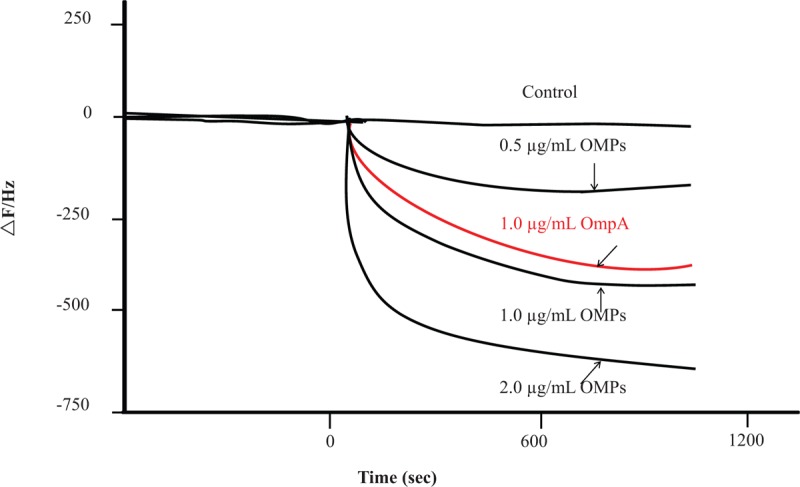
Interaction between BMAP-28 and AbOMPs or AbOmpA. The graphs show the change of delta frequency after injecting AbOMPs or AbOmpA on the sensor of quartz crystal microbalance (QCM). Around 5 μL of AbOMPs (the final concentrations were 0.5, 1.0 and 2.0 μg/mL) or AbOmpA (the final concentrations were 1.0 μg/mL) was injected, respectively. OmpA without the binding of BMAP-28 was injected on the sensor as a control. This graph is one of the most typical data which have done 3 times. AbOmpA = Acinetobacter baumannii OmpA, BMAP-28 = bovine myeloid antimicrobial peptide, QCM = quartz crystal microbalance.
4. Discussion
With the inappropriate usage of broad-spectrum antibiotics and the versatile genetic machinery of PDRAB, outbreaks of pan-drug-resistant A baumannii have become a significant public health threat. Currently, effective treatments for PDRAB infection are limited. Thus, the development of new and effective agents to inhibit PDRAB growth is urgent. In this study, the antimicrobial activity of BMAP-28 and its 4 analogs against PDRAB was examined. We found no significant difference between the MIC of BMAP-28 and its analog peptides. In the short time killing assay, BMAP-28 and its analogs demonstrated rapid killing of PDRAB within 60 min, with the effects of BMAP-28 occurring more quickly than those for its analog peptides. The antimicrobial effect of BMAP-28 against PDRAB did not improve with increased net charge, as seen in A837, A838, and A840; this indicates that there might be a critical threshold of positive charge density or net positive charge on BMAP-28 that governs its antimicrobial activities. [22,23]
Based on the average hydrophobicity, only A839 had a higher value than BMAP-28, with no significant differences observed between the other peptides. Takagi et al. reported that substituting a small number of amino acids on BMAP-28 did not affect its antimicrobial activity against MSSA and MRSA.[24] This indicates that there might be an optimal hydrophobicity window to obtain the antimicrobial activity of AMPs, which may be dramatically compromised by decreases or increases in hydrophobicity.[9]
Furthermore, we found that BMAP-28 has the potential to kill PDRAB through disruption of the integrity of bacterial membranes, as seen under FE-SEM observation. Most studies exhibit that AMPs act on gram-negative bacteria through their surface lipopolysaccharide (LPS) molecules.[25–27] A previous study reported that LL-37 may bind to A baumannii LPS with high affinity, but its bactericidal activity is not LPS-dependent.[28] However, several studies have emphasized the interaction between AMPs and bacterial outer membrane proteins (OMPs). OprI of Pseudomonas aeruginosa has been shown to be the target of cationic AMPs,[16] while OmpF from E coli and lipoteichoic acid (LTA) from Staphylococcus aureus cell walls have been observed to interact with cationic AMPs.[29,30] OmpA is an outer membrane porin protein that has a highly conserved amino acid sequence among gram-negative bacteria.[31] It has been reported that OmpA from A baumannii can interact with LL-37.[32] To understand the possible killing mechanism of BMAP-28 against PDRAB in this study, we isolated OMPs from PDRAB and found their interaction with BMAP-28 through QCM analysis, and furthermore we purified OmpA from PDRAB and found that it can interact with BMAP-28. Based on the above results, our study suggests that OmpA from PDRAB can act as a receptor for BMAP-28 and may involve in the bacteria-killing effect of BMAP-28 on PDRAB.
There were some limitations to this study. First, the contribution of OmpA to BMAP-28 action in PDRAB was not understood clearly. Second, the contribution of other bacterial cell surface molecules, especially LPS, to BMAP-28 action in PDRAB deserved further investigation. Therefore, we hope to use the ompA deletion strain and LPS-deficient A baumannii to further investigate the contribution of OmpA and LPS to BMAP-28 action in PDRAB.
In conclusion, we have compared the effects of BMAP-28 and its analog peptides against PDRAB and determined that the amino acid substitutions present in the analog peptides do not improve antimicrobial activity. We also observed the damaging effect of BMAP-28 on the cell morphology of PDRAB, and additionally found that BMAP-28 can interact with OmpA of PDRAB which may be related with the bacteria-killing effect of BMAP-28. In this study, it suggests that BMAP-28 and its analogs may provide effective, alternative treatments for PDRAB infections.
Acknowledgments
We are grateful to Dr Jine Lei (Clinical Laboratory, The First Affiliated Hospital of Xi’an Jiao Tong University, Xi’anJiaotong University, China) for collecting bacteria.
Author contributions
Data curation: Jing Han.
Funding acquisition: Yijie Guo.
Methodology: Jing Han.
Project administration: Meng Xun.
Writing – original draft: Yijie Guo.
Writing – review & editing: Yijie Guo.
Supplementary Material
Footnotes
Abbreviations: AbOmpA = Acinetobacter baumannii OmpA, AMPs = antimicrobial peptides, BMAP-28 = bovine myeloid antimicrobial peptide, FE-SEM = field emission scanning electron microscopy, MIC = minimal inhibitory concentration, OmpA = outer membrane protein A, PDRAB = pan-drug-resistant Acinetobacter baumannii, QCM = quartz crystal microbalance.
The project was sponsored by the National Natural Science Foundation of China (No. 31700127), China Postdoctoral Science Foundation Grant (2018T111072), Shaanxi Province Postdoctoral Science Foundation (No. 2016BSHEDZZ84), and Natural Science Foundation of Shaanxi Province (No. 2016JQ8051).
The authors declare that they have no competing interests.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Scott P, Deye G, Srinivasan A, et al. An outbreak of multidrug-resistant Acinetobacter baumannii–calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin Infect Dis 2007;44:1577–84. [DOI] [PubMed] [Google Scholar]
- [2].Visca P, Seifert H, Towner KJ. Acinetobacter infection—an emerging threat to human health. IUBMB Life 2011;63:1048–54. [DOI] [PubMed] [Google Scholar]
- [3].De Pascale G, Montini L, Pennisi M, et al. High dose tigecycline in critically ill patients with severe infections due to multidrug-resistant bacteria. Crit Care 2014;18:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Merino M, Poza M, Roca I, et al. Nosocomial outbreak of a multiresistant Acinetobacter baumannii expressing OXA-23 carbapenemase in Spain. Microbial Drug Resist 2014;20:259–63. [DOI] [PubMed] [Google Scholar]
- [5].Alireza JN, Masoomeh S, Alex van B, et al. Nosocomial outbreak of extensively and pan drug-resistant Acinetobacter baumannii in tertiary hospital in central part of Iran. Jundishapur J Microb 2013;6:e9892. [Google Scholar]
- [6].Dettori M, Piana A, Deriu MG, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol 2014;37:185–91. [PubMed] [Google Scholar]
- [7].Smith SG, Mahon V, Lambert MA, et al. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett 2007;273:1–1. [DOI] [PubMed] [Google Scholar]
- [8].Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 2006;24:1551–7. [DOI] [PubMed] [Google Scholar]
- [9].Huang HW, Charron NE. Understanding membrane-active antimicrobial peptides. Q Rev Biophys 2017;50:e10. [DOI] [PubMed] [Google Scholar]
- [10].Cubeddu T, Cacciotto C, Pisanu S, et al. Cathelicidin production and release by mammary epithelial cells during infectious mastitis. Vet Immunol Immunopathol 2017;189:66–70. [DOI] [PubMed] [Google Scholar]
- [11].Storici P, Tossi A, Lenarcic B, et al. Purification and structural characterization of bovine cathelicidins, precursors of antimicrobial peptides. Eur J Biochem 1996;238:769–76. [DOI] [PubMed] [Google Scholar]
- [12].Brogden KA, Nordholm G, Ackermann M. Antimicrobial activity of cathelicidins BMAP28, SMAP28, SMAP29, and PMAP23 against Pasteurella multocida is more broad-spectrum than host species specific. Vet Microbiol 2007;119:76–81. [DOI] [PubMed] [Google Scholar]
- [13].Takagi S, Hayashi S, Takahashi K, et al. Antimicrobial activity of a bovine myeloid antimicrobial peptide (BMAP-28) against methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Anim Sci J 2012;83:482–6. [DOI] [PubMed] [Google Scholar]
- [14].Lynn MA, Kindrachuk J, Marr AK, et al. Effect of BMAP-28 antimicrobial peptides on Leishmania major promastigote and amastigote growth: role of leishmanolysin in parasite survival. PLoS Negl Trop Dis 2011;5:e1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tennessen JA. Molecular evolution of animal antimicrobial peptides: widespread moderate positive selection. J Evol Biol 2005;18:1387–94. [DOI] [PubMed] [Google Scholar]
- [16].Lin YM, Wu SJ, Chang TW, et al. Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein. J Biol Chem 2010;285:8985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 2009;77:3150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Isogai E, Isogai H, Takahashi K, et al. Antimicrobial activity of three tick defensins and four mammalian cathelicidin-derived synthetic peptides against Lyme disease spirochetes and bacteria isolated from the midgut. Exp Appl Acarol 2009;49:221–8. [DOI] [PubMed] [Google Scholar]
- [19].Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18:268–81. [DOI] [PubMed] [Google Scholar]
- [20].Cuenca FF, Pascual A, Marẗanez ML, et al. Evaluation of SDS-polyacrylamide gel systems for the study of outer membrane protein profiles of clinical strains of Acinetobacter baumannii. J Basic Microbiol 2003;43:194–201. [DOI] [PubMed] [Google Scholar]
- [21].Lahiri J, Isaacs L, Tien J, et al. A strategy for the generation of surfaces presenting ligands for studies of binding based on an active ester as a common reactive intermediate: a surface plasmon resonance study. Anal Chem 1999;71:777–90. [DOI] [PubMed] [Google Scholar]
- [22].Jiang Z, Vasil AI, Hale J, et al. Effects of net charge and the number of positively charged residues on the biological activity of amphipathic alpha-helical cationic antimicrobial peptides. Adv Exp Med Biol 2009;611:561–2. [DOI] [PubMed] [Google Scholar]
- [23].Huang Y, Huang J, Chen Y. Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cel 2010;1:143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Takagi S, Nishimura J, Bai L, et al. Susceptibility difference between methicillin-susceptible and methicillin-resistant Staphylococcus aureus to a bovine myeloid antimicrobial peptide (BMAP-28). Anim Sci J 2014;85:174–9. [DOI] [PubMed] [Google Scholar]
- [25].Rosenfeld Y, Shai Y. Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta 2006;1758:1513–22. [DOI] [PubMed] [Google Scholar]
- [26].Rosenfeld Y, Lev N, Shai Y. Effect of the hydrophobicity to net positive charge ratio on antibacterial and anti-endotoxin activities of structurally similar antimicrobial peptides. Biochemistry 2010;49:853–61. [DOI] [PubMed] [Google Scholar]
- [27].Chai H, Allen WE, Hicks RP. Synthetic antimicrobial peptides exhibit two different binding mechanisms to the lipopolysaccharides isolated from Pseudomonas aeruginosa and Klebsiella pneumoniae. Int J Med Chem 2014;2014:809283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Moffatt JH, Harper M, Mansell A, et al. Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect Immun 2013;81:684–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Apetrei A, Asandei A, Park Y, et al. Unimolecular study of the interaction between the outer membrane protein OmpF from E. coli and an analogue of the HP(2-20) antimicrobial peptide. J Bioenerg Biomembr 2010;42:173–80. [DOI] [PubMed] [Google Scholar]
- [30].Takagi S, Bai L, Ozeki T, et al. A bovine myeloid antimicrobial peptide (BMAP-28) kills methicillin-resistant Staphylococcus aureus but promotes adherence of the bacteria. Anim Sci J 2014;85:342–6. [DOI] [PubMed] [Google Scholar]
- [31].Krishnan S, Prasadarao NV. Outer membrane protein A and OprF: versatile roles in Gram-negative bacterial infections. FEBS J 2012;279:919–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lin MF, Tsai PW, Chen JY, et al. OmpA binding mediates the effect of antimicrobial peptide LL-37 on Acinetobacter baumannii. PLoS One 2015;10:e0141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


