Supplemental Digital Content is available in the text
Keywords: breast cancer, liquid biopsy, lymph node metastasis, meta-analysis, survival
Abstract
Background:
Liquid biopsies using circulating tumor DNA (ctDNA) and cell-free DNA (cfDNA) have been developed for early cancer detection and patient monitoring. To investigate the clinical usefulness of ctDNA aberrations and cfDNA levels in patients with breast cancer (BC), we conducted a meta-analysis of 69 published studies on 5736 patients with BC.
Methods:
The relevant publications were identified by searching PubMed and Embase databases. The effect sizes of outcome parameters were pooled using a random-effects model.
Results:
The ctDNA mutation rates of TP53, PIK3CA, and ESR1 were approximately 38%, 27%, and 32%, respectively. High levels of cfDNA were associated with BCs rather than with healthy controls. However, these detection rates were not satisfactory for BC screening. Although the precise mechanisms have been unknown, high cfDNA levels were significantly associated with axillary lymph node metastasis (odds ratio [OR] = 2.148, P = .030). The ctDNA mutations were significantly associated with cancer recurrence (OR = 3.793, P < .001), short disease-free survival (univariate hazard ratio [HR] = 5.180, P = .026; multivariate HR = 3.605, P = .001), and progression-free survival (HR = 1.311, P = .013) rates, and poor overall survival outcomes (HR = 2.425, P = .007).
Conclusion:
This meta-analysis demonstrates that ctDNA mutation status predicts disease recurrence and unfavorable survival outcomes, while cfDNA levels can be predictive of axillary lymph node metastasis in patients with BC.
1. Introduction
Liquid biopsies using circulating tumor DNA (ctDNA) and cell-free DNA (cfDNA) have shown great potential as biomarkers for early detection, drug resistance, tumor relapse, and for predicting clinical outcomes in patients with cancer.[1,2] The ctDNA originates mainly from apoptotic and necrotic cancer cells, which release fragmented DNA into the circulation.[1,2] The ctDNA is known to be a specific type of cell-free DNA (cfDNA), which may also be released by dying nonmalignant host cells. The alterations in ctDNA include aberrant mutations, hypermethylation, and copy number variations.[1,2] With advances in molecular diagnostics, clinicians can screen ctDNA and cfDNA for monitoring patients with cancer. Liquid biopsies monitoring ctDNA or cfDNA are expected to be superior to currently widely used plasma biomarkers, such as cancer-implanted antigen, cancer antigen 15-3 (CA15-3), and cancer antigen 19-9 (CA19-9), in terms of test's sensitivity and clinical correlations.
Breast cancer (BC) is the most common cancer in women worldwide. There have been many efforts to find better biomarkers for early detection and treatment monitoring.[2] Liquid biopsy studies using ctDNA and cfDNA have been conducted in patients with BC.[3–71] However, the clinical sensitivity and specificity of ctDNA or cfDNA is unsatisfactory probably due to the complex genetic heterogeneity of BC. The issue of whether a panel of genes should be tested for liquid biopsy has become very important. The prediction of axillary lymph node metastasis has an important factor on whether postoperative adjuvant chemotherapy should be performed. However, no consensus has been reached regarding the prediction of axillary lymph node metastasis or cancer recurrence using liquid biopsy in patients with BC. Therefore, the clinical utility of liquid biopsy using ctDNA and cfDNA in patients with BC has not yet been established.
To address these problems, we investigated the clinical utility of liquid biopsy to analyze ctDNA mutations or hypermethylation and cfDNA levels in patients with BC by conducting a meta-analysis of existing primary studies.
2. Methods
2.1. Selection of published studies
Systematic literature searches of PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and Embase (www.embase.com) databases were conducted. The search strings were generated by combining keywords “circulating cell-free DNA” or “plasma cell-free DNA” or “serum cell-free DNA” or “liquid biopsy,” and “breast cancer” or “breast neoplasm.” The selection process of the articles is shown in Figure 1. All the eligible studies were reviewed and extracted independently by 2 authors (JHL and YSK), and the disagreement was resolved by discussion. There were no geographic or language restrictions. We also manually searched through the reference lists of the identified articles. The ethical approval and patient consent are not required, because this study is a meta-analysis of previously published studies.
Figure 1.
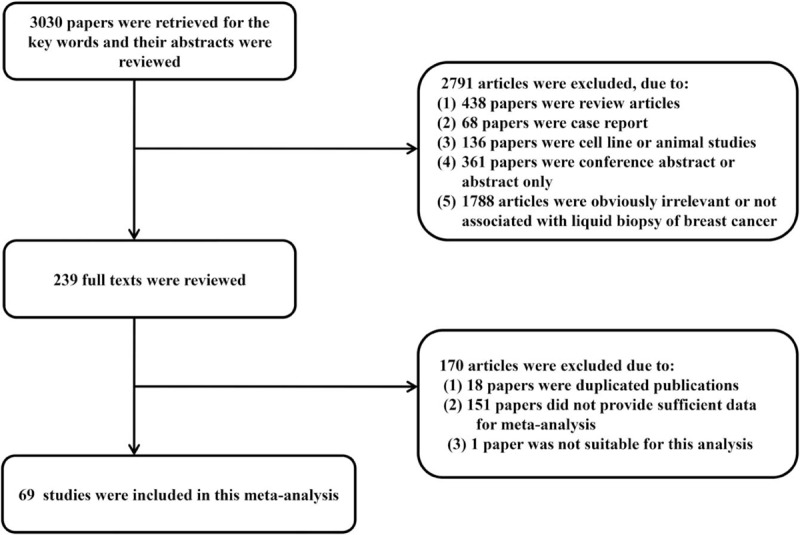
Flow diagram for article selection.
2.2. Selection criteria
Original articles published before February 2017 were included. We retrieved 3030 articles using the keywords. Articles review articles without original data, conference abstracts, case reports, and articles that dealt with cell line or animal studies were excluded. We also excluded the articles that are not explicitly related to the subject. After the excluding process, the full text of 239 articles was thoroughly reviewed. Duplicate data or overlapping articles were excluded by examining author names and affiliations. When multiple articles were published by the same authors or institutions, the most recent or single informative article was selected. We also excluded the articles that provided insufficient data for meta-analysis. As a result, 69 studies were included in this meta-analysis.
2.3. Data extraction and quality assessment
The following data were extracted from each study: sample size, ethnicity, detection methods, liquid biopsy results, and clinicopathologic parameters including axillary lymph node metastasis, clinical stage, tumor recurrence, and survival data. The quality of the selected studies was assessed with the Newcastle–Ottawa scales (NOSs).
2.4. Statistical analysis
Meta-analysis was performed as previously described.[72] The effect sizes and 95% confidence intervals (CIs) of each study were calculated using the inverse variance method and combined using the random-effects model (DerSimonian–Laird method). The choice of model was based on a conceptual understanding of whether the studies included in the meta-analysis, rather than homogeneity tests, all share the same population effect size.[73] The summary effects were presented as the prevalence rate, odds ratio (OR; stage, axillary lymph node metastasis, and recurrence), weighted mean difference (WMD; cfDNA levels between controls and patients with BC), or HR (survival data). Heterogeneity among studies was evaluated using the Cochrane Q test and I2 values. The I2 refers to the percentage of variation across studies that is due to heterogeneity rather than chance, and does not inherently depend on the number of studies considered [I2 = 100% × (Q − df)/Q]. We evaluated the cutoff values for I2s for assignment of low (<25%), moderate (25–50%), and high (>50%) heterogeneities. If the I2 value was >50%, subgroup analysis was done.[74] Sensitivity analyses were performed to examine the influence of each study on the pooled prevalence rate, OR, or HR, by serially omitting an individual study and pooling the remaining studies. Publication bias was examined by funnel plots, and Egger tests were employed for evaluating the degree of asymmetry. A P-value of <.1 was designated as an indicator of publication bias. Pooled analysis was performed using Comprehensive meta-analysis software (version 2.0; Biostat, Englewood, NJ).
3. Results
Sixty-nine studies with a total of 5736 cases were included in this meta-analysis (Fig. 1).[3–71] The main features of the chosen studies are described in Table 1. The number of patients in each study was between 4 and 541.
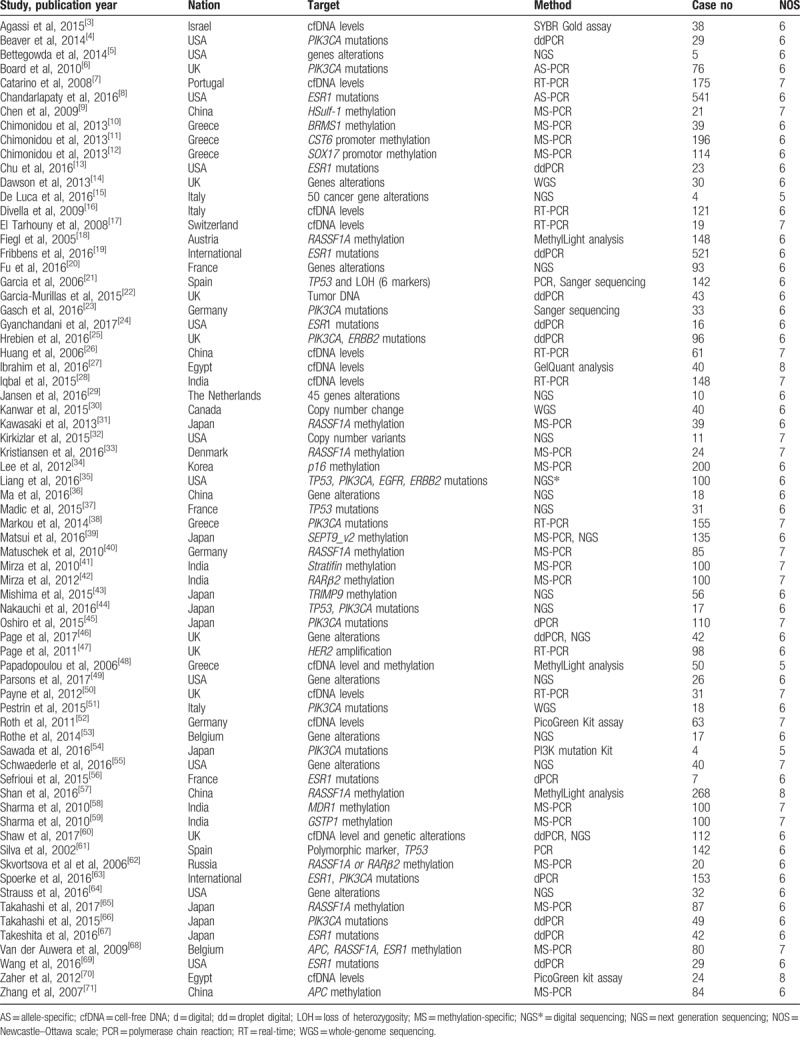
Table 1.
Characteristics of liquid biopsy studies with circulating tumor DNA or cell-free DNA in breast cancer.
3.1. Cancer screening and early detection
The pooled analysis of 37 studies on 2748 patients with BC revealed that the overall prevalence rate of ctDNA mutations in patients with BC was 44.3% (95% CI: 38.2–50.6%) (Supplementary Table 1). The ctDNA mutation frequencies of the TP53, PIK3CA, and ESR1 genes were 37.8% (95% CI: 25.4–52.1%), 26.6% (22.4–31.2%) and 32.4% (26.8–38.5%) in the 11,19, and 10 studies consisting of 338, 1015 and 1379 patients with BC, respectively.[4,6,8,13,19,23–25,29,35–38,44–46,49,53–56,60,63,64,66,67,69] Among the studies that presented the frequency of ctDNA mutations in the TP53 and PIK3CA genes, eleven and nineteen studies were able to distinguish between patients with early surgery and patients with metastasis. However, The TP53 and PIK3CA mutation rates were not different between the early operable patients and the advanced patients with BC (Q = 0.553, P = .57; and Q = 0.160, P = .689, respectively). Surprisingly, the detection rate of ctDNA mutations using next generation sequencing (NGS) techniques was 100% in patients with BC regardless of target genes.[5,15,36]
The pooled analysis of 21 studies involving 2046 patients with BC showed that the overall prevalence rate of ctDNA hypermethylation in patients with BC was 32.8% (95% CI: 26.8–39.4%) (Supplementary Table 2). However, the prevalence of ctDNA hypermethylation was significantly lower than that of ctDNA mutations in patients with BC (32.8% vs. 44.3%, P = .013).
Nine studies compared cfDNA levels between healthy controls and patients with BC (Supplementary Table 3). The cfDNA levels were significantly higher in patients with BC than in healthy controls (WMD = 2.598; 95% CI: 1.576–3.621; P < .001, Q = 271.821, I2 = 97.057) (Fig. 2). To explore potential heterogeneity sources, we performed subgroup analyses according to the detection methods and ethnicity. The cfDNA levels were measured by real-time PCR, GelQuant, PicoGreen, and SYBR Gold assay. The WMD of each group according to the test methods was 1.36, 10.81, 5.46, and 1.10, respectively. Since the P-value of Q test for the group differences was <.001, we could reject the null hypothesis that the effect sizes between groups are the same. The subgroup group variable (detection methods) could account for 57.7% of the total actual variance (R2 = 0.577). In addition, the patients were classified into African, Asian, and Caucasian. The WMD of each group according to the ethnicity was 7.78, 0.54, and 1.65, respectively. Similar to the detection method variable, we could reject the null hypothesis that the effect sizes between groups are the same because the P-value of Q test for the group differences was <.001. The ethnicity of moderator variable could account for 55.3% of the total actual variance (R2 = 0.577) (Supplementary Table 4). Thus, different cfDNA detection methods and ethnic differences were considered potential sources of heterogeneity.
Figure 2.

Weighed mean differences with corresponding 95% confidence intervals (CIs) of individual studies and pooled data, in cell-free DNA levels, between healthy controls and patients with breast cancer. The forest plot depicts each study and overall effect sizes and 95% CIs.
3.2. Axillary lymph node metastasis and clinical staging
Four studies described the relationship between ctDNA mutations and axillary lymph node metastasis in 217 patients with BC.[4,23,45,66] No relationship was found between ctDNA mutations and axillary lymph node metastasis (OR = 1.764; 95% CI: 0.877–3.548; P = .112, Q = 3.283, I2 = 8.609). Eight studies investigated the relationship between ctDNA hypermethylation and axillary lymph node metastasis in 819 patients with BC.[10,12,18,41,42,58,59,71] There was no association between ctDNA hypermethylation and axillary lymph node metastasis (OR = 0.829; 95% CI: 0.563–1.220; P = .341, Q = 9.810, I2 = 28.646).
Four studies addressed the association between cfDNA levels and axillary lymph node metastasis in 277 patients with BC (Supplementary Tables 5 and 6). The cfDNA level was significantly associated with axillary lymph node metastasis (OR = 2.148; 95% CI: 1.076–4.290; P = .030, Q = 9.685, I2 = 69.023) (Fig. 3). To explore potential sources of heterogeneity, we conducted a subgroup analysis based on the detection methods. The cfDNA levels were measured by real-time PCR, SYBR Gold, and PicoGreen assays. The OR of each group according to the test methods was 1.58, 12.54, and 1.14, respectively. Since the P-value for Q test for the group differences was 0.01, we could reject the null hypothesis that the effect sizes between groups are the same. The subgroup group variable (detection methods) could account for 100% of the total actual variance (R2 = 1.0) (Supplementary Table 6).
Figure 3.
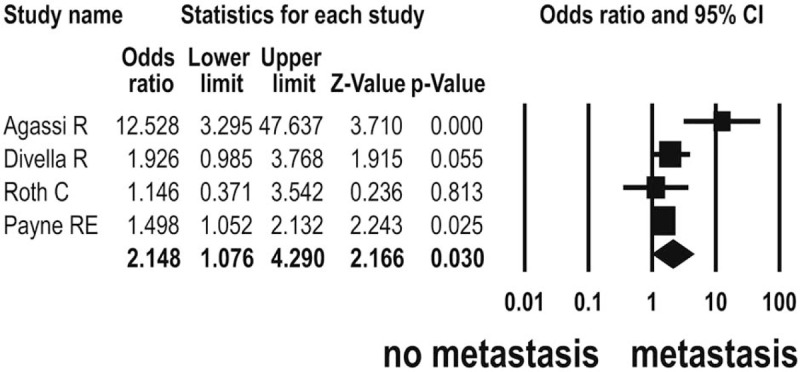
Odds ratios with corresponding 95% confidence intervals (CIs) of individual studies and pooled data for the association between cell-free DNA levels and axillary lymph node metastasis in breast cancer. The forest plot depicts each study and overall effect sizes and 95% CIs.
Two studies reported the association between ctDNA mutations and clinical stage (I–II vs III–IV) in 40 patients with BC.[23,24] There was no association between ctDNA mutations and clinical stage (OR = 0.942; 95% CI: 0.187–4.753; P = .943, Q = 1.251, I2 = 20.050). Five studies addressed the relationship between ctDNA hypermethylation and clinical stage in 487 patients with BC.[41,42,58,59,65] No association was found between ctDNA hypermethylation and clinical stage (OR = 1.095; 95% CI: 0.444–2.703; P = .844, Q = 19.840, I2 = 79.839).
3.3. Cancer recurrence and patient survival
Cancer recurrence was defined as locoregional relapse and distant metastasis after initial treatments. This included cases with recurrence locally in the ipsilateral breast or chest wall, regionally in drainage lymph nodes, or at distant sites.[21,44] Three studies evaluated the association between ctDNA mutations and cancer recurrence in patients with BC.[21,24,44] Genetic aberrations in ctDNA were more frequent in 34 (59%) of patients with 58 BC with recurrence than in 39 (33%) of 117 patients with BC without recurrence. The detection rate of ctDNA mutations was significantly associated with tumor recurrence (OR = 3.793; 95% CI: 1.798–8.003; P < .001, Q = 1.885, I2 = 0.000) (Fig. 4).
Figure 4.
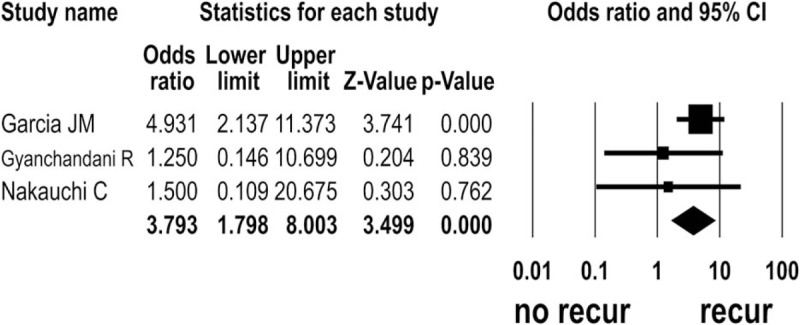
Odds ratios with corresponding 95% confidence intervals (CIs) of individual studies and pooled data for the association between circulating tumor DNA mutations and tumor recurrence in breast cancer. The forest plot depicts each study and overall effect sizes and 95% CIs.
Four studies presented hazard ratios (HRs) and CIs of overall survival outcomes in 782 patients with BC according to genetic mutations in ctDNA.[8,19,21,46] The unadjusted HRs ranged from 1.62 to 25.61. The ctDNA mutations were significantly associated with poor overall survival outcomes in patients with BC (HR = 2.425; 95% CI: 1.270–4.629; P = .007, Q = 10.693, I2 = 71.945) (Fig. 5). We conducted the subgroup analyses based on the mutated gene types (ESR1 mutations vs the other ctDNA mutations; Q = 0.006, P = .941) and ethnicity (Caucasian vs the other races; Q = 0.469, P = .493). However, since the P-values for Q tests for the group differences were >.1, we could not reject the null hypothesis that the effect sizes between groups are the same (Supplementary Table 7).
Figure 5.
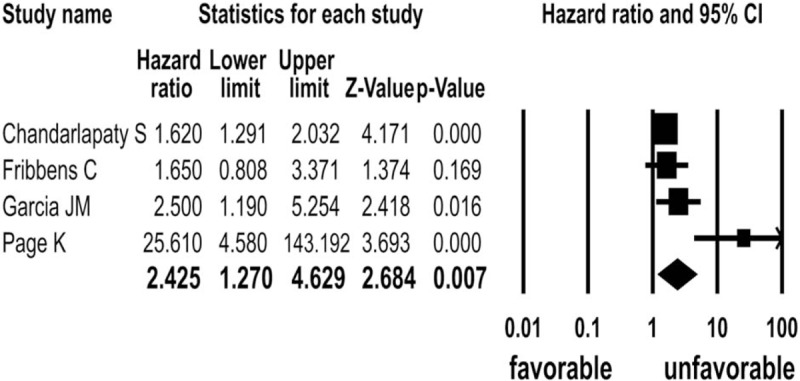
Hazard ratios with corresponding 95% confidence intervals (CIs) of individual studies and pooled data for the association between circulating tumor DNA mutations and overall survival outcomes in breast cancer. The forest plot depicts each study and overall effect sizes and 95% CIs.
Two studies presented HRs and CIs of univariate disease-free survivals in 185 patients with BC according to ctDNA mutations.[21,22] The detection of ctDNA mutations in patients with BC was significantly associated with short univariate disease-free survival outcomes (HR = 5.180; 95% CI: 1.215–22.084; P = .026, Q = 3.934, I2 = 74.578). Three studies described HRs and CIs of multivariate disease-free survivals in 325 patients with BC according to ctDNA mutations.[21,22,26] The adjusted HRs ranged from 2.6 to 9.6. The ctDNA mutation rate in patients with BC was significantly associated with short multivariate disease-free survival rates (HR = 3.605; 95% CI: 1.718–7.562; P = .001, Q = 2.567, I2 = 22.078).
Three studies described HRs and CIs of progression-free survivals in 597 patients with BC according to ctDNA mutations.[8,19,63] The detection of ctDNA mutations in patients with BC was significantly associated with shorter progression-free survival (HR = 1.311; 95% CI: 1.060–1.621; P = .013, Q = 1.089, I2 = 0.000). However, the survival data of investigations using ctDNA hypermethylation and cfDNA levels were insufficient to perform a meta-analysis.
3.4. Sensitivity analyses and publication bias
The sensitivity analyses showed that the study by Roth et al[52] affected the pooled OR between cfDNA levels and axillary lymph node metastasis. If the Roth's study was removed, the summary effect size would be increased from 2.148 to 3.611. None of the other sensitivity analyses affected the summary effect sizes. The funnel plots and Egger regression tests revealed no evidence of publication bias (Fig. 6), except for the meta-analysis of the association between cfDNA levels and the prevalence of patients with BC (Supplementary Table 8).
Figure 6.
Funnel plot of meta-analysis for the association between circulating tumor DNA hypermethylation and axillary lymph node metastasis in breast cancer. Individual studies, indicated by small circles, are uniformly distributed in an inverted V-shape, and indicate no publication bias.
4. Discussion
This meta-analysis was conducted to estimate the clinical usefulness of liquid biopsies to predict axillary lymph node metastasis, cancer recurrence, and patient's survival in BC. The pooled analysis of 69 studies with a total of 5736 cases revealed that ctDNA mutations are useful prognostic markers for recurrence and survival in patients with BC. In addition, cfDNA levels can be predictive of axillary lymph node metastasis.
For the early detection of BC, cfDNA levels and ctDNA mutations of TP53, PIK3CA, and ESR1 genes were not valuable as biomarkers, although the ctDNA mutations were frequently detected in patients with BC. There was no significant difference in mutation frequencies of TP53 and PIK3CA ctDNA between early and advanced patients with BC. The mutations of TP53 and PIK3CA are often selected for liquid biopsy in patients with BC, because they are the 2 most common genetic variants in newly diagnosed BCs.[44] Since the 2 genes are mutually exclusive, the prevalence of mutations in at least one of these genes in all patients with BC increases from 55% to 65%.[44] Moreover, these mutations are generally concentrated in the exons 5 to 9 of TP53 and exon 9/20 of PIK3CA and are conserved between primary tumors and recurrent or metastatic cancers.[44] Advances in NGS have dramatically improved the detection rates of ctDNA mutations in blood.[5,13,15,32,49] The development of a BC-specific gene panel of ctDNA mutations using NGS technology is expected to enable clinicians to detect BC early. In contrast, although cfDNA levels were higher in patients with BC than in healthy controls, there was considerable overlap between the groups. Furthermore, there was a significant variation in cfDNA levels depending on the detection methods. As a marker for early detection of BC, the possibility of cfDNA level testing is reduced.
More recently, the ESR1 mutations have been associated with as a resistance mechanism for endocrine therapy, which are clustered in the ligand-binding domain of the receptor, and result in ligand-independent estrogen receptor activity.[24,75] The ESR1 mutations are rare in primary patients with BC without metastasis, but are more commonly found in patients with metastatic BC or metastatic BC after endocrine therapy.[75] In this meta-analysis, the ESR1 mutations were analyzed only in patients with advanced or metastatic BC. Given that ESR1 ctDNA mutations are generally associated with endocrine therapy resistance and poor survival, the mutated ESR1 can be explored as a therapeutic target.
The results of our analysis showed that high cfDNA levels were significantly associated with axillary lymph node metastasis. Conservative approach of axillary staging has been developed in patients with BC such as the examination of a sentinel node, which is the first lymph node draining from a tumor bed. Sentinel node biopsy during surgery is important for staging the status of axillary lymph node involvement. Therefore, the prediction of preoperative axillary lymph node status as well as sentinel node biopsy during surgery is important determinants for postoperative treatments of BC. There has been considerable controversy as to whether ctDNA mutations or high levels of cfDNA could predict axillary lymph node metastasis. In some studies, high levels of cfDNA or ctDNA hypermethylation have been shown to be associated with lymph node metastasis or advanced clinical stage.[3,50,52,65] In contrast, Mirza et al[41] and Sharma et al[58] reported that ctDNA hypermethylation was significantly associated with a low probability of lymph node metastasis and low clinical stages. However, this meta-analysis showed that ctDNA mutations and hypermethylation are not associated with lymph node metastasis or clinical stage. Our meta-analysis has demonstrated a significant prognostic value of ctDNA mutations in tumor recurrence and poor survival in patients with BC. The presence of ctDNA mutations was significantly related to tumor recurrence with a 4-fold OR. Of 58 patients with BC recurrence, 19 (33%) had local recurrence and 39 (67%) had metastatic recurrence.
Unfortunately, because there was not enough data on this aspect, a meta-analysis could not be performed to assess whether ctDNA mutations were associated with local recurrence or remote recurrence. The ctDNA mutation rate was a prognostic indicator of reduced disease-free survival and overall survival with 2-fold and 4-fold HRs, respectively. Thus, this meta-analysis suggests that liquid biopsy may be useful as a decision-making tool for postoperative adjuvant chemotherapy in patients with localized BC.
There are several limitations in this meta-analysis. First, individual primary studies used different target genes, various analytical methods, and heterogeneous clinical samples. Since ctDNA and cfDNA detection methods are very different, we performed pool analysis for each. Second, of the studies included in the meta-analysis, ctDNA and cfDNA tests were recently recognized and developed only as potential biomarkers, so small sample size studies were included. Many of these small studies can distort the results of meta-analysis. In addition, in patients with cancer, ctDNA is a fraction of cfDNA, accounting for approximately 0.01% to as much as 50%, so that there is a limitation that they cannot be completely separable from each other.[76] However, most cfDNAs in healthy individuals are originate from bone marrow with a length of 70 to 200 bp and a concentration of 0 to 100 ng/mL. In contrast, ctDNAs of patients with cancer are 200 bp to >1 kb in length and the half-life is 15 minutes to several hours, which are removed from the liver and kidneys.[76] There is a need to develop laboratory methods to make it easier to distinguish clinically from cfDNA and ctDNA. Finally, we have classified the patient as white, Asia or Africa for subgroup analysis, but in reality there is a possibility of discrepancies between our classification and the original data. These limitations might affect the results of this pooled analysis.
In conclusion, this meta-analysis provides evidence that ctDNA mutations are significantly associated with tumor recurrence and poor survival outcomes in patients with BC. In addition, high cfDNA levels may be indicators of axillary lymph node metastasis, which is an important determinant for postoperative adjuvant chemotherapy.
Author contributions
Conceptualization: Young-Sik Kim.
Data curation: Ju-Han Lee, Young-Sik Kim.
Formal analysis: Ju-Han Lee, Hoiseon Jeong, Jung-Woo Choi, HwaEun Oh.
Funding acquisition: Young-Sik Kim.
Investigation: Ju-Han Lee.
Methodology: Hoiseon Jeong, Jung-Woo Choi, HwaEun Oh.
Supervision: Young-Sik Kim.
Validation: Hoiseon Jeong, Jung-Woo Choi, HwaEun Oh, Young-Sik Kim.
Writing – original draft: Ju-Han Lee.
Writing – review & editing: Young-Sik Kim.
Supplementary Material
Footnotes
Abbreviations: BC = breast cancer, CA15-3 = cancer antigen 15-3, CA19-9 = cancer antigen 19-9, cfDNA = cell-free DNA, CI = confidence interval, ctDNA = circulating tumor DNA, HR = hazard ratio, NGS = next generation sequencing, OR = odds ratio, PCR = polymerase chain reaction, WMD = weighted mean difference.
This work was supported by Mid-career Researcher Program through National Research Foundation of Korea (NRF) (grant no: 2016 R1A2B4012030) funded by the Ministry of Education, Science, and Technology, and by Korea University Ansan Hospital grants (grant no: K1811011).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Alix-Panabière C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 2016;6:479–91. [DOI] [PubMed] [Google Scholar]
- [2].Openshaw MR, Page K, Fernandez-Garcia D, et al. The role of ctDNA detection and the potential of the liquid biopsy for breast cancer monitoring. Expert Rev Mol Diagn 2016;16:751–5. [DOI] [PubMed] [Google Scholar]
- [3].Agassi R, Czeiger D, Shaked G, et al. Measurement of circulating cell-free DNA levels by a simple fluorescent test in patients with breast cancer. Am J Clin Pathol 2015;143:18–24. [DOI] [PubMed] [Google Scholar]
- [4].Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 2014;20:2643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Board RE, Wardley AM, Dixon JM, et al. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat 2010;120:461–7. [DOI] [PubMed] [Google Scholar]
- [7].Catarino R, Ferreira MM, Rodrigues H, et al. Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol 2008;27:415–21. [DOI] [PubMed] [Google Scholar]
- [8].Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol 2016;2:1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Z, Fan JQ, Li J, et al. Promoter hypermethylation correlates with the Hsulf-1 silencing in human breast and gastric cancer. Int J Cancer 2009;124:739–44. [DOI] [PubMed] [Google Scholar]
- [10].Chimonidou M, Kallergi G, Georgoulias V, et al. Breast cancer metastasis suppressor-1 promoter methylation in primary breast tumors and corresponding circulating tumor cells. Mol Cancer Res 2013;11:1248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chimonidou M, Strati A, Malamos N, et al. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin Chem 2013;59:270–9. [DOI] [PubMed] [Google Scholar]
- [12].Chimonidou M, Tzitzira A, Strati A, et al. CST6 promoter methylation in circulating cell-free DNA of breast cancer patients. Clin Biochem 2013;46:235–40. [DOI] [PubMed] [Google Scholar]
- [13].Chu D, Paoletti C, Gersch C, et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin Cancer Res 2016;22:993–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199–209. [DOI] [PubMed] [Google Scholar]
- [15].De Luca F, Rotunno G, Salvianti F, et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget 2016;7:26107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Divella R, Tommasi S, Lacalamita R, et al. Circulating hTERT DNA in early breast cancer. Anticancer Res 2009;29:2845–9. [PubMed] [Google Scholar]
- [17].El Tarhouny S, Seefeld M, Fan AX, et al. Comparison of serum VEGF and its soluble receptor sVEGFR1 with serum cell-free DNA in patients with breast tumor. Cytokine 2008;44:65–9. [DOI] [PubMed] [Google Scholar]
- [18].Fiegl H, Millinger S, Mueller-Holzner E, et al. Circulating tumor-specific DNA: a marker for monitoring efficacy of adjuvant therapy in cancer patients. Cancer Res 2005;65:1141–5. [DOI] [PubMed] [Google Scholar]
- [19].Fribbens C, O’Leary B, Kilburn L, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol 2016;34:2961–8. [DOI] [PubMed] [Google Scholar]
- [20].Fu Y, Jovelet C, Filleron T, et al. Improving the performance of somatic mutation identification by recovering circulating tumor DNA mutations. Cancer Res 2016;76:5954–61. [DOI] [PubMed] [Google Scholar]
- [21].Garcia JM, Garcia V, Silva J, et al. Extracellular tumor DNA in plasma and overall survival in breast cancer patients. Genes Chromosomes Cancer 2006;45:692–701. [DOI] [PubMed] [Google Scholar]
- [22].Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [DOI] [PubMed] [Google Scholar]
- [23].Gasch C, Oldopp T, Mauermann O, et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Mol Oncol 2016;10:1330–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gyanchandani R, Kota KJ, Jonnalagadda AR, et al. Detection of ESR1 mutations in circulating cell-free DNA from patients with metastatic breast cancer treated with palbociclib and letrozole. Oncotarget 2017;8:66901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hrebien S, O’Leary B, Beaney M, et al. Reproducibility of digital PCR assays for circulating tumor DNA analysis in advanced breast cancer. PLoS One 2016;11:e0165023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang ZH, Li LH, Hua D. Quantitative analysis of plasma circulating DNA at diagnosis and during follow-up of breast cancer patients. Cancer Lett 2006;243:64–70. [DOI] [PubMed] [Google Scholar]
- [27].Ibrahim IH, Kamel MM, Ghareeb M. Circulating DNA in Egyptian women with breast cancer. Asian Pac J Cancer Prev 2016;17:2989–93. [PubMed] [Google Scholar]
- [28].Iqbal S, Vishnubhatla S, Raina V, et al. Circulating cell-free DNA and its integrity as a prognostic marker for breast cancer. Springerplus 2015;4:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jansen MP, Martens JW, Helmijr JC, et al. Cell-free DNA mutations as biomarkers in breast cancer patients receiving tamoxifen. Oncotarget 2016;7:43412–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kanwar N, Hu P, Bedard P, et al. Identification of genomic signatures in circulating tumor cells from breast cancer. Int J Cancer 2015;137:332–44. [DOI] [PubMed] [Google Scholar]
- [31].Kawasaki H, Igawa E, Kohosozawa R, et al. Detection of aberrant methylation of tumor suppressor genes in plasma from cancer patients. Pers Med Univers 2013;2:20–4. [Google Scholar]
- [32].Kirkizlar E, Zimmermann B, Constantin T, et al. Detection of clonal and subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl Oncol 2015;8:407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kristiansen S, Nielsen D, Soletormos G. Detection and monitoring of hypermethylated RASSF1A in serum from patients with metastatic breast cancer. Clin Epigenetics 2016;8:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lee JJ, Ko E, Cho J, et al. Methylation and Immunoexpression of p16(INK4a) tumor suppressor gene in primary breast cancer tissue and their quantitative p16(INK4a) hypermethylation in plasma by real-time PCR. Korean J Pathol 2012;46:554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liang DH, Ensor JE, Liu ZB, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat 2016;155:139–49. [DOI] [PubMed] [Google Scholar]
- [36].Ma F, Zhu W, Guan Y, et al. ctDNA dynamics: a novel indicator to track resistance in metastatic breast cancer treated with anti-HER2 therapy. Oncotarget 2016;7:66020–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Madic J, Kiialainen A, Bidard FC, et al. Circulating tumor DNA and circulating tumor cells in metastatic triple negative breast cancer patients. Int J Cancer 2015;136:2158–65. [DOI] [PubMed] [Google Scholar]
- [38].Markou A, Farkona S, Schiza C, et al. PIK3CA mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin Cancer Res 2014;20:5823–34. [DOI] [PubMed] [Google Scholar]
- [39].Matsui S, Kagara N, Mishima C, et al. Methylation of the SEPT9_v2 promoter as a novel marker for the detection of circulating tumor DNA in breast cancer patients. Oncol Rep 2016;36:2225–35. [DOI] [PubMed] [Google Scholar]
- [40].Matuschek C, Bölke E, Lammering G, et al. Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. Eur J Med Res 2010;15:277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mirza S, Sharma G, Parshad R, et al. Clinical significance of Stratifin, ERalpha and PR promoter methylation in tumor and serum DNA in Indian breast cancer patients. Clin Biochem 2010;43:380–6. [DOI] [PubMed] [Google Scholar]
- [42].Mirza S, Sharma G, Parshad R, et al. Clinical significance of promoter hypermethylation of ERbeta and RARbeta2 in tumor and serum DNA in Indian breast cancer patients. Ann Surg Oncol 2012;19:3107–15. [DOI] [PubMed] [Google Scholar]
- [43].Mishima C, Kagara N, Matsui S, et al. Promoter methylation of TRIM9 as a marker for detection of circulating tumor DNA in breast cancer patients. Springerplus 2015;4:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nakauchi C, Kagara N, Shimazu K, et al. Detection of TP53/PIK3CA mutations in cell-free plasma DNA from metastatic breast cancer patients using next generation sequencing. Clin Breast Cancer 2016;16:418–23. [DOI] [PubMed] [Google Scholar]
- [45].Oshiro C, Kagara N, Naoi Y, et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat 2015;150:299–307. [DOI] [PubMed] [Google Scholar]
- [46].Page K, Guttery DS, Fernandez-Garcia D, et al. Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin Chem 2017;63:532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Page K, Hava N, Ward B, et al. Detection of HER2 amplification in circulating free DNA in patients with breast cancer. Br J Cancer 2011;104:1342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Papadopoulou E, Davilas E, Sotiriou V, et al. Cell-free DNA and RNA in plasma as a new molecular marker for prostate and breast cancer. Ann N Y Acad Sci 2006;1075:235–43. [DOI] [PubMed] [Google Scholar]
- [49].Parsons HA, Beaver JA, Cimino-Mathews A, et al. Individualized molecular analyses guide efforts (IMAGE): a prospective study of molecular profiling of tissue and blood in metastatic triple-negative breast cancer. Clin Cancer Res 2017;23:379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Payne RE, Hava NL, Page K, et al. The presence of disseminated tumour cells in the bone marrow is inversely related to circulating free DNA in plasma in breast cancer dormancy. Br J Cancer 2012;106:375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pestrin M, Salvianti F, Galardi F, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol 2015;9:749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Roth C, Pantel K, Muller V, et al. Apoptosis-related deregulation of proteolytic activities and high serum levels of circulating nucleosomes and DNA in blood correlate with breast cancer progression. BMC Cancer 2011;11:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rothe F, Laes JF, Lambrechts D, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol 2014;25:1959–65. [DOI] [PubMed] [Google Scholar]
- [54].Sawada T, Watanabe M, Fujimura Y, et al. Sensitive cytometry based system for enumeration, capture and analysis of gene mutations of circulating tumor cells. Cancer Sci 2016;107:307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schwaederle M, Husain H, Fanta PT, et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget 2016;7:9707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sefrioui D, Perdrix A, Sarafan-Vasseur N, et al. Short report: monitoring ESR1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int J Cancer 2015;137:2513–9. [DOI] [PubMed] [Google Scholar]
- [57].Shan M, Yin H, Li J, et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget 2016;7:18485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sharma G, Mirza S, Parshad R, et al. CpG hypomethylation of MDR1 gene in tumor and serum of invasive ductal breast carcinoma patients. Clin Biochem 2010;43:373–9. [DOI] [PubMed] [Google Scholar]
- [59].Sharma G, Mirza S, Parshad R, et al. Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci 2010;87:83–91. [DOI] [PubMed] [Google Scholar]
- [60].Shaw JA, Guttery DS, Hills A, et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin Cancer Res 2017;23:88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Silva JM, Silva J, Sanchez A, et al. Tumor DNA in plasma at diagnosis of breast cancer patients is a valuable predictor of disease-free survival. Clin Cancer Res 2002;8:3761–6. [PubMed] [Google Scholar]
- [62].Skvortsova TE, Rykova EY, Tamkovich SN, et al. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br J Cancer 2006;94:1492–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Spoerke JM, Gendreau S, Walter K, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 2016;7:11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Strauss WM, Carter C, Simmons J, et al. Analysis of tumor template from multiple compartments in a blood sample provides complementary access to peripheral tumor biomarkers. Oncotarget 2016;7:26724–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Takahashi H, Kagara N, Tanei T, et al. Correlation of methylated circulating tumor DNA with response to neoadjuvant chemotherapy in breast cancer patients. Clin Breast Cancer 2017;17:61.e3–9.e3. [DOI] [PubMed] [Google Scholar]
- [66].Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Prognostic role of PIK3CA mutations of cell-free DNA in early-stage triple negative breast cancer. Cancer Sci 2015;106:1582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Clinical significance of monitoring ESR1 mutations in circulating cell-free DNA in estrogen receptor positive breast cancer patients. Oncotarget 2016;7:32504–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Van der Auwera I, Elst HJ, Van Laere SJ, et al. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. Br J Cancer 2009;100:1277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang P, Bahreini A, Gyanchandani R, et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin Cancer Res 2016;22:1130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zaher ER, Anwar MM, Kohail HMA, et al. Value of circulating DNA concentration and integrity as a screening test for detection of cancer in an Egyptian cohort. Alex J Med 2012;48:187–96. [Google Scholar]
- [71].Zhang JJ, Ouyang T, Wan WH, et al. Detection and significance of APC gene promoter hypermethylation in serum of breast cancer patients [in Chinese]. Ai Zheng 2007;26:44–7. [PubMed] [Google Scholar]
- [72].Lee JH, Jeong H, Choi JW, et al. Clinicopathologic significance of MYD88 L265P mutation in diffuse large B-cell lymphoma: a meta-analysis. Sci Rep 2017;7:1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Borenstein M, Hedges LV, Higgins JP, et al. A basic introduction to fixed-effect and random-effects models for meta-analysis. Research synthesis methods 2010;1:97–111. [DOI] [PubMed] [Google Scholar]
- [74].Crowther M, Lim W, Crowther MA. Systematic review and meta-analysis methodology. Blood 2010;116:3140–6. [DOI] [PubMed] [Google Scholar]
- [75].Angus L, Beije N, Jager A, et al. ESR1 mutations: moving towards guiding treatment decision-making in metastatic breast cancer patients. Cancer Treat Rev 2017;52:33–40. [DOI] [PubMed] [Google Scholar]
- [76].Han X, Wang J, Sun Y. Circulating tumor DNA as biomarkers for cancer detection. Genomics Proteomics Bioinformatics 2017;15:59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


