Abstract
Objectives:
The prostate cancer gene 3 (PCA3), human kallikrein 2, and miRNA-141 are promising prostate cancer (Pca) specific biomarkers. Our aim was to evaluate the detection of PCA3, human glandular kallikrein 2 (hk2), and miRNA-141 mRNA in peripheral blood of patients received prostate biopsy. What's more, we want to detect the value of combination of PSA (prostate specific antigen) in the early diagnosis of PCa.
Materials and methods:
Hundred patients were divided into 2 groups according to the results of pathologic diagnosis. Quantitative real-time PCR (qRT-PCR) was used to evaluate the mRNA of PCA3, hk2, and miRNA-141 in peripheral blood. At the same time, analyze those clinical outcomes used in the patients. We compared these different outcomes to evaluate the value of new molecular markers.
Results:
The level of mRNA of PCA3, hK2, and miR-141 in Pca group were significantly higher than that in BPH. PSA had the highest sensitivity in predicting Pca diagnosis (76.7%); PCA3 had the highest specificity (82.5%). And the combination of PCA3, PSA, and hK2 improved area under the curve (AUC)-receiver operating characteristic (ROC) curve largely, especially those with PSA 4-10ng/mL.
Conclusions:
PCA3, hK2, and miRNA-141 were biomarkers of Pca with potential clinical application value, especially in patients with PSA gray area. Combining PCA3, PSA, and hK2 performed better than individual biomarkers alone in predicting Pca.
Keywords: human kallikrein 2, prostate cancer, RT-PCR, the prostate cancer gene 3
1. Introduction
Prostate cancer (Pca) is one of the most common seen cancers all over the world.[1] Suspicion of Pca is usually on the basis of digital rectal examination (DRE),[2] elevated level of prostate specific antigen (PSA), and transrectal ultrasound (TRUS) guided prostate biopsy. Until now, PSA is the only widely used biomarker for Pca. Serum PSA levels have been utilized as a Pca biomarker for over 20 years.[3] However, PSA has limitations, including lack of specificity and leading to over diagnosis of Pca.[4] Especially for the so-called gray area of PSA levels 4.0 to 10.0 ng/mL, resulting in a high negative biopsy rate. In view of this, to maximizing the specificity and sensitivity of the diagnosis of Pca, more Pca biomarkers should be discovered.
With the advent of genomic and proteomic technologies, several promising alternative biomarkers have being discovered.[4] Compared to PSA, prostate cancer gene 3 (PCA3) is more specific for Pca. Because PCA3 is over-expressed in prostate tumor tissue and is not observed in benign prostate disorders. Human glandular kallikrein 2 (hK2) is a serine protease, and its aminoacid sequence is 79% homologous to that for PSA. Past studies suggest that hK2 is expressed higher in Pca than that in normal prostate epithelium. While those individual biomarkers still have disadvantaged, until now, no individual biomarker has been proved to be more valuable in Pca than PSA. Our study detected a number of molecular markers of patients’ peripheral blood that accept prostate biopsy through real-time fluorescent quantitative PCR method to semi-quantitative. Screening molecular markers can increase diagnostic sensitivity and specificity of Pca. What's more, we sought to investigate how these biomarkers can be combined with PSA levels to develop clinically practical algorithms for Pca diagnosis.
2. Material and methods
2.1. Ethical statement
All protocols involving human subjects were reviewed and approved by the ethical committee of the First Affiliated Hospital of Zhejiang University in accordance with the Declaration of Helsinki (Association, 2000). Informed written consents from the human subjects were obtained in this study.
2.2. Data collection
All the clinical data were collected in the Department of Urology of the First Affiliated Hospital of Zhejiang University. The following data were extracted from the electronic medical record system of the First Affiliated Hospital of Zhejiang University: age, serum PSA, DRE, and TRUS results, prostate volume, magnetic resonance imaging results, biopsy results (current and history), radiologic results, and radical prostatectomy results (if applicable).
2.3. Sample collection
Blood samples were collected at the Department of Urology in the First Affiliated Hospital of Zhejiang University from April 2014 to June 2015. When an abnormality is felt during a DRE, levels of PSA >4ng/mL, or abnormal findings in MRI, the patients who need prostate biopsy were included in this study. Patients considered with prostatitis, severe illness cannot withstand biopsy, were excluded. For each participant included in the study, 5 mL peripheral blood with anticoagulant was collected right when he was hospitalized. Then the blood samples were used for peripheral blood mononuclear cells (PBMCs) and serum isolation in 2 hours. PBMCs were isolated by Ficoll-PaqueTM Plus (Amersham Pharmacia Biotech, Shanghai, China) according to the facture's instructions. Collected PBMCs were stored at −80°C till used.
2.4. Total RNA preparation and cDNA synthesis
Total RNA from PBMCs and serum was extracted with Trizol reagent (Invitrogen, Waltham, MA), according to the manufacturer's instruction. PrimeScript RT regent Kit (Takara) was used to synthesize cDNA according to manufacturer's instructions.
2.5. Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) for PCA3, hK2, and miRNA-141 was performed using a SYBR Premix Ex Taq Kit (Takara) on a 7500 real-time PCR system (Applied Biosystems, Foster City, CA). Data were analyzed using the SPSS (version 23.0, IBM). Primers for miR-141 amplification were: CCACTGCCTTAACCCCTTC (revise), and AGGGGTGAGACTAGGCAGGT (forward); U6 amplification were: GGAACGCTTCACGAATTTG (revise), and ATTGGAACGATACAGAGAAGATT (forward); PCA3 amplification were: ACGTTCTGGGATACATGTGC (revise) and GAGAACAGGGGAGGGAGAG (forward); hK2 amplification were: CGGTAATGCACCACCTTGGTGT (revise) and GGCTCTGGACAGGTGGTAAAGA (forward).
2.6. Detection of PSA
Both total PSA and free PSA were detected by ARCHITECT PSA Reagent Kit upon patients were admitted in the hospital.
2.7. Statistical analysis
All analyses were performed using IBM Statistical Package for the Social Sciences 22.0 (SPSS, Inc., Chicago, IL). The differences of RNA levels between Pca group and control group were analyzed by the Mann–Whitney test. Receiver operating characteristic (ROC) curve was established to determine the diagnostic value (namely, sensitivity, and specificity) of mRNA in Pca. During the ROC curve analysis, a training dataset was first adopted to determine the cut-off values and was then applied to the remaining patient population. Correlation statistics were analyzed by the Spearman correlation test. For all determinations, a P < .05 was considered statistically significant.
3. Results
3.1. Patient characteristics
Hundred patients included in the study underwent prostate biopsy. According to the final pathologic results, they were divided to Pca group and benign prostatic hyperplasia group (BPH), there are 43 cases of Pca, aged 53 to 81, the average age is 69. And 57 cases of BPH, aged 49 to 77, and average age is 65. There were 47 patients’ PSA between 4 to 10 ng/mL. In addition, there are 6 patients detected by emission computed tomography (ECT) 99 tc – MDP bone imaging in the diagnosis of consideration in bone metastatic lesions. All the 100 patients were follow-up 2 years, and 9 of them got repeated biopsy, only 1 was proved to be Pca.
To investigate the diagnostic value of biomarkers we used today, we retrospective study 831 patients took prostate biopsy in our hospital from April 2013 to June 2015. Aged 44 to 91, the average age is 69. There are 287 cases of Pca.
3.2. Validation of qRT-PCR system
The levels of biomarkers in PBMCs samples were determined by qRT-PCR using GAPDH (or U6) as a normalization control. To validate the specificity of the qRT-PCR system adopted in the study, the amplification curves and melting curves of biomarkers were analyzed. The results indicated that non-specific products were amplified in the reaction, implying that the current qRT-PCR system was validated to successfully detect hK2, PCA3, miR-141, and GAPDH in PBMCs samples.
3.3. The diagnosis efficiency of tPSA and PSAD as biomarkers for Pca
The retrospective study of 831 cases showed that tPSA and PSAD had significant difference. ROC curve cure analysis showed that tPSA at the cut-off of 12.25, the sensitivity was 61.7%, and the specificity was 69.7%, with an area under the curve (AUC) of 0.698 (95%CI: 0.661–0.735); the cut-off of PSAD was 0.30, the sensitivity was 71.7% and the specificity was 74.8%, with an AUC of 0.780 (95%CI: 0.746–0.814) (Fig. 1A).
Figure 1.
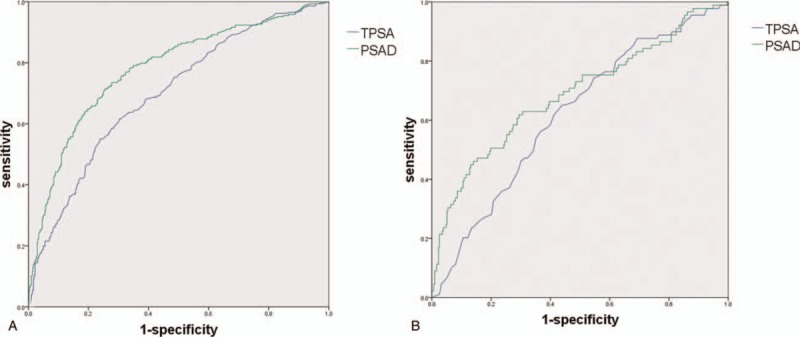
A. ROC curve analysis of PSA and PSAD in all 831 patients. B. ROC curve analysis of PSA and PSAD in patients with PSA 4 to 10ng/mL. PSA = prostate specific antigen, ROC = receiver operating characteristic.
When restricted PSA level to 4 to 10ng/mL, the cut-off of tPSA was 6.85, the sensitivity was 65.2%, and the specificity was 55.9%, with an AUC of 0.619 (95%CI: 0.555–0.683); the cut-off of PSAD was 0.19, the sensitivity was 61.8%, and the specificity was 70.6%, with an AUC of 0.780 (95%CI: 0.746–0.814) (Fig. 1B).
3.4. Comparison of hK2, PSA, PCA3, and miR-141 between Pca group and BPH group
The level of biomarkers in PBMCs was evaluated by qRT-PCR. They were significantly higher (P < .05) in Pca group of PSA, PCA3, hK2, and miR-141 (Fig. 2).
Figure 2.
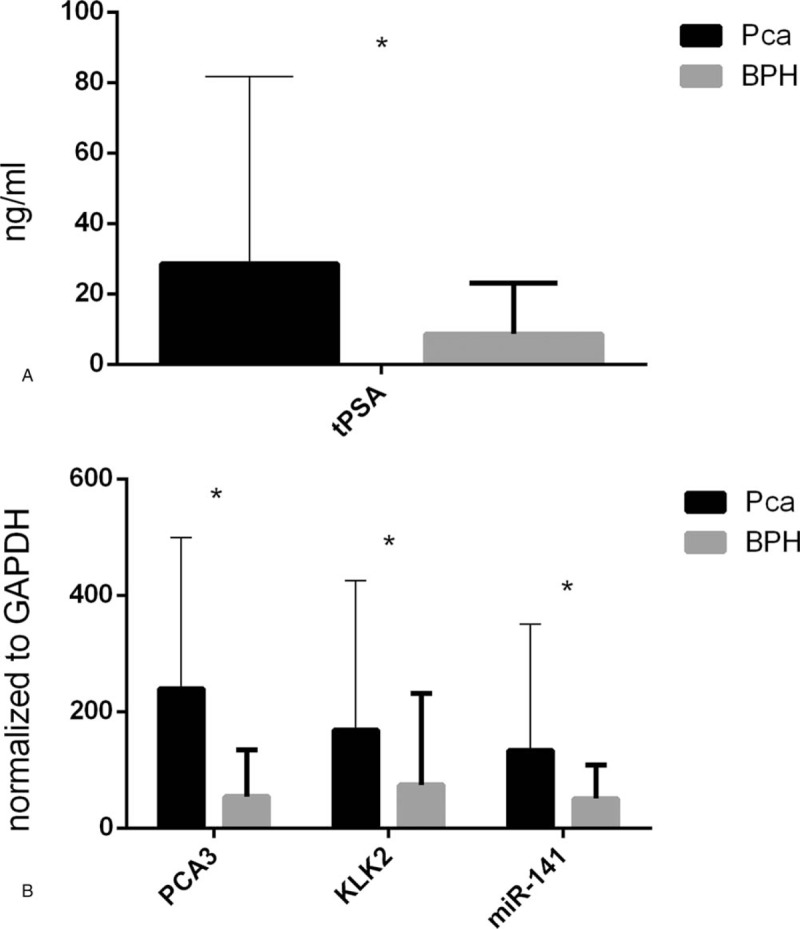
Level of biomarkers in PBMCs evaluated by qRT-PCR. PBMCs = peripheral blood mononuclear cells, qRT-PCR = quantitative real-time PCR.
3.5. Evaluation of diagnostic accuracy of hK2, PCA3, and miR-141
We tested the diagnostic value of hK2, PCA3, and miR-141 in PBMCs. They were evaluated using ROC curve analysis. At the cut-off of 10.64 of tPSA, the sensitivity was 76.7% and the specificity was 66.7%, with an AUC of 0.735 (95% CI: 0.625–0.835). At the cut-off of 104.45 of miR-141, the sensitivity was 34.9% and the specificity was 89.5%, with an AUC of 0.625 (95% CI: 0.511–0.739). For PCA3, the cut-off is 121.2, the sensitivity was 55.8%, and the specificity was 82.5%, with an AUC of 0.667 (95% CI: 0.558–0.775). For hK2, the cut-off is 268.33, the sensitivity was 25.6%, and the specificity was 96.5%, with an AUC of 0.607 (95% CI: 0.493–0.721) (Fig. 3A).
Figure 3.
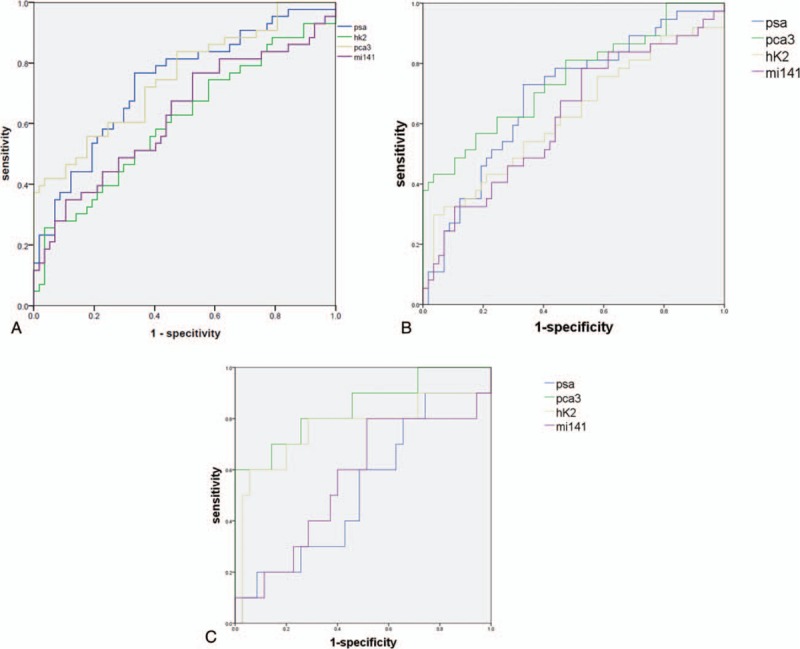
A. ROC curve analysis of PSA, PCA3, hK2, and miR-141 in 100 patients. B. ROC curve analysis of PSA, PCA3, hK2, and miR-141 in patients without bone metastases. C. ROC curve analysis of PSA, PCA3, hK2, and miR-141 in patients with PSA between 4 to 10 ng/mL. hK2 = human glandular kallikrein 2, PCA3 = prostate cancer gene 3, PSA = prostate specific antigen, ROC = receiver operating characteristic.
When we removed the patients whose ECT showed bone metastases, PCA3 showed the highest diagnostic performance (AUC = 0.747, 95% CI: 0.643–0.852), at the cut-off of 237.8, the sensitivity was 43.2% and the specificity was 96.5%. For tPSA, the cut-off is 10.64, the sensitivity was 73.0%, and the specificity was 66.7%, with an AUC of 0.692 (95% CI: 0.582–0.802). For hK2, the cut-off is 268.3, the sensitivity was 29.7%, and the specificity was 96.5%, with an AUC of 0.623 (95% CI: 0.502–0.744). For miR-141, the cut-off is 23.4, the sensitivity was 78.4%, and the specificity was 47.4%, with an AUC of 0.621 (95% CI: 0.503–0.739) (Fig. 3B).
When restricted PSA level to 4 to 10ng/mL, PCA3 showed the highest diagnostic performance (AUC = 0.843, 95% CI: 0.687–0.998), at the cut-off of 349.2, the sensitivity was 60.0% and the specificity was 100.0%. For tPSA, the cut-off is 5.88, the sensitivity was 60.0%, and the specificity was 51.4%, with an AUC of 0.520 (95% CI: 0.316–0.724). For hK2, the cut-off is 248.3, the sensitivity was 60.0%, and the specificity was 94.3%, with an AUC of 0.760 (95% CI: 0.549–0.971). For miR-141, the cut-off is 20.6, the sensitivity was 80.0%, and the specificity was 48.6%, with an AUC of 0.621 (95% CI: 0.503–0.739) (Fig. 3C).
3.6. Logistic regression analysis
Binary logistic regression analysis was carried out to assess tPSA, hK2, PCA3, and miR-141 associated with Pca diagnosis. tPSA, hK2, and PCA3 had statistical significance. And they composed logistic regression model. ROC curve of the model was evaluated compared with other biomarkers. The model showed the highest diagnostic performance (AUC = 0.841, 95% CI: 0.761–0.920), at the cut-off of 0.437, the sensitivity was 69.8% and the specificity was 84.2% (Fig. 4A). Especially when restricted PSA level to 4 to –10ng/mL, the logistic model showed great advantage compared to PSA (AUC = 0.853, 95% CI: 0.659–1), at the cut-off of 0.494, the sensitivity was 72.7% and the specificity was 89.2% (Fig. 4B).
Figure 4.

Multivariable logistic regression resulted in a model combining PSA, PCA3, and hK2. A. ROC curve analysis of the model, PSA, PCA3, hK2, and miR-141 in 100 patients B. ROC curve analysis of the model and PSA in patients with PSA between 4 to 10 ng/mL. hK2 = human glandular kallikrein 2, PCA3 = prostate cancer gene 3, PSA = prostate specific antigen, ROC = receiver operating characteristic.
4. Discussion
The early diagnosis of Pca depends mainly on DRE, PSA, and prostate magnetic resonance imaging. However, PSA is a prostate-specific but not a Pca -specific marker.[5] The human body can synthesize PSA not only confined to the Pca tissue, but also including normal prostate tissue, many studies have pointed out that all disease destroy prostate epithelial cells can cause elevated PSA in the blood, but PSA rise more apparent in patients with Pca, which leads to PSA as markers for diagnosis of Pca has a high misdiagnosis rate, especially in the PSA ”grey area” (4–10 ng/mL) patients. When PSA is 4 to 10 ng/mL, specificity rate is only 48.6%, with 50% of the rate of misdiagnosis. Owing to significant limitations of PSA as a biomarker, recent studies were focus on discovering new biomarkers. Many new biomarkers were discovered over the past years. However, there still no biomarker can replace PSA. A large inventory of candidate biomarkers that might prove to be more useful than individual biomarker when combined in a multiplex model.[6–8]
PCA3 gene located in human chromosome 9(q21–22), consist of 4 exons and 3 introns, there are 3 different positions have selective poly adenosine acidification site on the fourth exon (include 4a, 4b, 4c as 3 subunits).[9,10] PCA3 expressed in Pca organizations with high specificity, many studies indicate that PCA3 is strongly overexpressed in malignant prostate tissue compared to benign or normal adjacent one.[9] Besides, in other organizations or tumor PCA3 is not detected, especially in normal prostate tissue. So, PCA3 usually showed higher specificity than PSA. We found PCA3 express was more stable in PBMCs than post-DRE urine, so we detect PCA3 in PBMCs instead of post-DRE urine in our study. At the cut-off of 121.2, the sensitivity was 55.8% and the specificity was 82.5% of PCA3. When we rule out patients with bone metastases, PCA3 showed great diagnosis value than PSA. The specificity was much high than that of PSA (82.5% vs 66.7%).
Human kallikrein 2 is a kind of serine protease, which possesses 79% of the amino acid sequence identical with PSA, is also a novel biomarker for Pca.[11,12] hK2 is mostly produced in the prostate, secreting with the form of proenzyme in the body and outside the cell is activated into active enzymes. hK2 exists in blood, semen and saliva, and other bodily fluids, furthermore, 80% to 95% of hK2 in the blood exists in the free form. Many experiments have confirmed that the serum hK2 is helpful to the detection of Pca and prognosis.[12] We found when the cut-off is 268.33, the sensitivity was 25.6% and the specificity was 96.5% of hK2, with an AUC of 0.607 (95% CI: 0.493–0.721), the results was familiar with other studies. What's more, for those patients with PSA “grey area,” the sensitivity was up to 60.0% and the specificity to 94.3%.
In this study, the mRNA of the hK2, PCA3, and miR-141 in Pca group is significantly higher than that in BPH group. Combination of PSA, PCA3, and hK2 can greatly improve the sensitivity and specificity of the early diagnosis of Pca, especially in the removal of the patients with significantly bone metastases. The advantages of joint application of molecular markers become more obvious. At the same time, compared to tPSA, free PSA ratio, PSAD and other low diagnostic sensitivity, and specificity PSA ”grey area” (4–10 ng/mL) biomarkers, combined diagnosis shows great advantages.
Prior studies had reported PCA3 and hK2 as biomarker in early diagnosis of Pca,[13,14] however, they had not evaluated the utility of combining serum PSA with PCA3 and hK2 in developing models in early diagnosis of Pca as we have done in the study. Combine PSA, PCA3, and hK2 can both increase sensitivity and specific degree, improve the accuracy of diagnosis of Pca, reduce the present situation of PSA screening high false positive rate, excessive care, and burden on patients.
There are, however, several limitations to our study: (1) the sample size needed to further expand, and we still need multicenter studies to confirm the diagnosis value of the model. (2) Although we have followed these patients for 2 years, there still may be false negative. Because pathological findings were according to prostate biopsy instead of total prostate resection. So we think further studies with more patients and longer follow-up times would be useful.
5. Conclusion
PCA3, hK2, and miR-141 are early diagnosis of Pca markers which have potential clinical application values, especially in the patients of PSA gray area. They have great diagnosis value, especially in combination with serum PSA, are very attractive targets with respect to early detection of Pca.
Author contributions
A.L.J. and Q.Z. conceived the initial planning, Z.J.M, X.W, W.H and S.W. collected the blood, K.B.Y, Y.F.H. and Z.J.M. completed the PCR of the biomarkers, D.H.Z. and L.P.X analyzed the results. All authors wrote and reviewed the manuscript.
Data curation: Alin Ji, Yingfang Hu.
Formal analysis: Alin Ji.
Funding acquisition: Dahong Zhang, Liping Xie.
Investigation: Kebing Yang, Wei He.
Methodology: Kebing Yang.
Resources: Dahong Zhang.
Software: Wei He.
Writing – original draft: Zujie Mao.
Writing – review & editing: Qi Zhang, Liping Xie.
Footnotes
Abbreviations: AUC = area under the curve, DRE = digital rectal examination, ECT = emission computed tomography, hK2 = human glandular kallikrein 2, PBMCs = peripheral blood mononuclear cells, Pca = prostate cancer, PCA3 = prostate cancer gene 3, PSA = prostate specific antigen, qRT-PCR = quantitative real-time PCR, ROC = receiver operating characteristic, TRUS = transrectal ultrasound.
This study was supported by Natural Science Foundation of China (81502541), Huzhou public welfare technology research project (2016GY68) and Zhejiang provincial natural science foundation of China (LY16H160015).
The authors declare that they have no competing interests.
The authors report no conflicts of interest.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Wu J, Ji A, Xie B, et al. Is magnetic resonance/ultrasound fusion prostate biopsy better than systematic prostate biopsy? An updated meta- and trial sequential analysis. Oncotarget 2015;6:43571–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhang K, Bangma CH, Roobol MJ. Prostate cancer screening in Europe and Asia. Asian J Urol 2017;4:86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Saini S. PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016;39:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhu Y, Han CT, Zhang GM, et al. Effect of body mass index on the performance characteristics of PSA-related markers to detect prostate cancer. Sci Rep 2016;6:19034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 2005;5:845–56. [DOI] [PubMed] [Google Scholar]
- [7].Goodison S, Ogawa O, Matsui Y, et al. A multiplex urinary immunoassay for bladder cancer detection: analysis of a Japanese cohort. J Transl Med 2016;14:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van der Heijden AG, Mengual L, Lozano JJ, et al. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur J Cancer 2016;64:127–36. [DOI] [PubMed] [Google Scholar]
- [9].Roberts MJ, Chow CW, Schirra HJ, et al. Diagnostic performance of expression of PCA3, Hepsin and miR biomarkers inejaculate in combination with serum PSA for the detection of prostate cancer. Prostate 2015;75:539–49. [DOI] [PubMed] [Google Scholar]
- [10].Filella X, Foj L. Prostate cancer detection and prognosis: from prostate specific antigen (PSA) to exosomal biomarkers. Int J Mol Sci 2016;17: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Timmermand OV, Ulmert D, Evans-Axelsson S, et al. Preclinical imaging of kallikrein-related peptidase 2 (hK2) in prostate cancer with a (111)In-radiolabelled monoclonal antibody, 11B6. EJNMMI Res 2014;4:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Satkunasivam R, Zhang W, Trachtenberg J, et al. Human kallikrein-2 gene and protein expression predicts prostate cancer at repeat biopsy. Springerplus 2014;3:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bachour DM, Chahin E, Al-Fahoum S. Human kallikrein-2, prostate specific antigen and free- prostate specific antigen in combination to discriminate prostate cancer from benign diseases in Syrian patients. Asian Pac J Cancer Prev 2015;16:7085–8. [DOI] [PubMed] [Google Scholar]
- [14].Fenner A. Prostate cancer: PCA3 as a grade reclassification predictor. Nat Rev Urol 2017;14:390. [DOI] [PubMed] [Google Scholar]


