Abstract
Objective
The pathogenetic underpinnings of extra-pulmonary small cell carcinomas (EPSCCs) of the head and neck are poorly understood. We sought to describe the clinical case and whole-exome DNA sequencing data of a patient with sinonasal Schneiderian carcinoma in-situ whose tumor progressed to small cell carcinoma (SCC).
Study Design
Case report and whole exome sequencing of tumor DNA.
Setting
Academic medical center.
Subjects and Methods
A 52-year-old male with sinonasal Schneiderian carcinoma in-situ whose tumor progressed to small cell carcinoma. We performed whole exome genetic sequencing and copy-number variation (CNV) analysis of tumor and normal DNA extracted from flash-frozen, paraffin-embedded (FFPE) samples.
Results
A total of 93 high-confidence, non-synonymous somatic mutation events were identified in sinonasal SCC, including loss-of-function mutations in TP53, MAML3, a transcriptional coactivator of the Notch pathway, and GAS6, an activating ligand of the TAM family of tyrosine kinase receptors. Focal amplifications of chromosomal regions 6p25-11.1, containing SOX4 and VEGFA, and 14q32.1-32.3, containing AKT1 and the Notch inhibitory ligand DLK1 were additionally seen. Further CNV analysis revealed deletions in the critical cell-cycle regulators CDKN2A, RB1, RBL1, and RBL2 and the chromatin modifier EP300.
Conclusions
Small cell carcinoma may rarely arise from sinonasal Schneiderian carcinoma in-situ and exhibits similar genomic aberrations (e.g. SOX amplification, Notch pathway inactivation) to pulmonary small cell carcinoma.
Keywords: Sinonasal SCC, NOTCH, SOX4
Introduction
Extrapulmonary small cell carcinomas (EPSCCs) are exceptionally rare neoplasms that arise from primary sites of the genitourinary tract, gastrointestinal tract, or head and neck (e.g. oral cavity, salivary glands, larynx, nasal cavity and paranasal sinuses)1–3 EPSCCs of the head and neck are clinically aggressive; patients typically present with distant metastatic disease and exhibit dismal 5-year overall- and disease-specific survival rates of less than 25 %.4,5 In contrast to the genetic understanding of many other cancers of the head and neck region that have driven advances in precision medicine,6–8 the pathogenetic basis of head and neck EPSCCs is currently unknown, limiting the utility of advancing targeted therapy for this disease.
EPSCCs appear to share some common genomic aberrations with pulmonary small cell carcinoma (SCLC), a disease characterized by an extraordinary mutational burden and frequent inactivation of RB1 and TP53.9,10 These cancers are thought to either clonally proliferate from a multipotent stem cell progenitor, as in SCLC, or arise as a late-stage evolution in the genetic progression of more organ-typical cancers.11 The latter hypothesis is supported by genetic and histologic evidence of tumor evolution into EPSCC. EPSCCs of the breast demonstrate loss of heterozygosity at BRCA-1, BRCA-2, 8p21-24, 11q23.3, and 12q25 typical of invasive ductal carcinoma.12 Histologically, EPSCCs of the appendix and urinary bladder often arise focally in a background of adenocarcinoma and urothelial carcinoma, respectively.13,14 However, these posited molecular associations are derived from limited genomic sequencing data of genitourinary and gastrointestinal EPSCCs. Little is known about the pathogenetic underpinnings of EPSCCs of the head and neck.
Inverted papillomas (IP) of the paranasal sinuses are pathologically benign tumors that tend to recur after surgical resection.15 Synchronous or metachronous development of in situ or invasive carcinoma (e.g. Schneiderian carcinoma) develops in 10 – 25 % of IP cases.16,17 Most often, these cancers are of squamous differentiation (i.e. sinonasal squamous cell carcinoma) though isolated cases of other malignant pathologies associated with IP have been reported.18,19
Herein, we report a case of a patient with a multiply-recurrent papillary Schneiderian carcinoma in-situ of the sphenoethmoidal sinus who developed a subsequent small cell carcinoma in the surgical resection bed that was subjected to whole exome sequencing. We posit clinical evidence that this patient’s original tumor devolved into an EPSCC and present the first comprehensive sequencing data on EPSCCs of the head and neck.
Methods
Whole Exome Sequencing
All patient material was collected under an IRB-approved protocol from the University of Michigan pathology archive. Regions of tumor or adjacent normal were identified using an H&E slide, and punch cores were taken from the block as described.20 In-situ hybridization for Epstein-Barr Virus was performed and negative. Genomic DNA was isolated from the sinonasal small cell carcinoma and adjacent normal mucosa using the Qiagen Allprep DNA/RNA FFPE kit as previously described21,22 and quantified using a Qubit and Bioanalyzer to determine the quality DNA yields using previously defined thresholds for molecular analysis.23 An Illumina gDNA library was prepared from 300 ng of DNA and enriched for exomes using the Roche Nimblegen SeqCap EZ v3.0 kit (Roche Nimblegen, Indianapolis, IN) as described.24 Exome-enriched libraries were then sequenced on an Illumina HiSeq 2500 System at the University of Michigan sequencing core, resulting in 86-million (mean of 50.3 reads/base) and 76-million (mean of 34.7 reads/base) mapped reads for tumor DNA and normal DNA, respectively.
Variant Calling and Copy Number analysis
FastQC (v.0.11.5) was used to assess quality of the exome sequencing data. The sequencing reads were mapped to the hg19 version of the human genome using BWA (v.0.7.15). GATK best practices were followed to prepare BAM files for variant calling. VarScan2 (v.2.4.1) was employed for variant calling.25 VarSeq was used to annotate and filter variants of interest. All variants in introns and intergenic regions were eliminated from the analysis. Only variants with 5 or more alternate reads were considered as true positives. ADTEx26 was used to estimate the copy number variations (CNV) seen in the sinonasal small cell carcinoma.
Results
Case Description
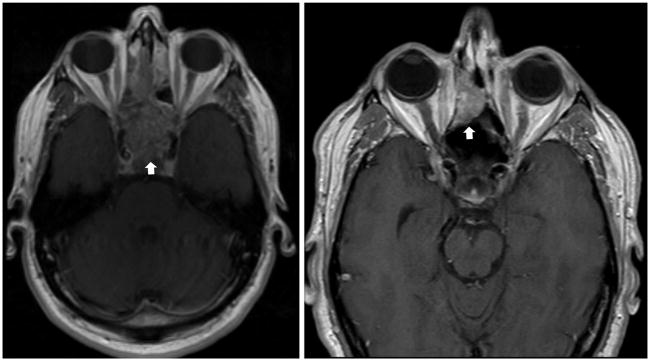
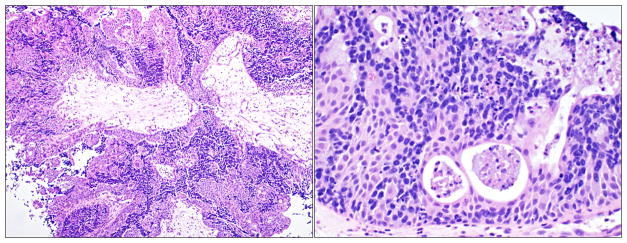
A 52-year-old male presented with a six-month history of nasal congestion, rhinorrhea, and dysosmia; MRI revealed an inhomogeneous, enhancing mass in the sphenoethmoidal sinus (Figure 1). Tissue biopsy was consistent with a poorly-differentiated, papillary Schneiderian carcinoma in-situ, negative for human papillomavirus on in-situ hybridization (Figure 2). The tumor had a dimorphic population of typical Schneiderian papilloma epithelial cells and a subpopulation with a more discohesive morphology reminiscent of small cell carcinoma. A diagnosis of small cell carcinoma was entertained, though was ultimately excluded on the basis of large cell size, minimal mitotic activity, lack of invasive growth and absence of staining for neuroendocrine markers and thyroid transcription factor-1 (TTF-1). The tumor was resected via an anterior sub-cranial approach with negative margins. Over the ensuing five years of clinical surveillance, the patient developed two consecutive recurrences of sphenoethmoidal masses, both resected endoscopically with pathologic confirmation of recurrent papillary Schneiderian carcinoma in-situ.
FIGURE 1.
Left: Schneiderian carcinoma in-situ (arrow) shown as a 6.5-cm enhancing, ethmoid sinus mass (T1, post-contrast image).
Right: Invasive small cell carcinoma (arrow) shown as a 2-cm polypoid, enhancing mass within the sinonasal surgical defect.
FIGURE 2.
Representative H & E images of papillary Schneiderian carcinoma in situ at 10x (left) and 20x (right) magnification.
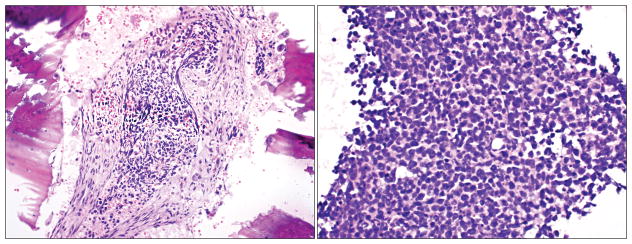
Six years after original diagnosis, surveillance MRI noted a new polypoid, enhancing mass within the sphenoethmoidal surgical defect with infiltration into the cribriform plate and right medial orbital wall (Figure 1). Small cell carcinoma with bony invasion and extensive in-situ component was diagnosed on pathology (Figure 3). The patient underwent repeat sub-cranial resection with calvarial bone graft inset of medial orbital wall and radial forearm free flap reconstruction. Post-operatively, three cycles of adjuvant cisplatin and etoposide and skull base radiation (54 Gy) were administered. At time of manuscript preparation, the patient was five years out from treatment completion with no evidence of small cell carcinoma recurrence or metastases.
FIGURE 3.
Representative H & E images of small cell carcinoma depicting bony invasion at 10x (left) and 20x (right) magnification.
Exome Sequencing Analysis of Sinonasal Small Cell Carcinoma (SCC)
Deep whole exome sequencing identified 93 high-confidence, non-synonymous somatic mutation events in the sinonasal SCC (supplemental figure 1, supplemental table 1). Sixty-three (67.8 %) were single-nucleotide substitutions and 30 (32.2 %) were disruptive insertion-deletion (indel) events. Only 18 (19.4 %) of these mutational events have been previously documented in the Catalogue of Somatic Mutations in Cancer (COSMIC).27
A heterozygous missense mutation in TP53 (c.528C>G; p.C176W) was the most significant mutational event confirmed in the small cell carcinoma sample. This TP53 variant is pathogenic and frequently present across multiple human cancers.28,29 Importantly, no single-nucleotide substitution or indel events were identified in genes involved in cell cycle regulation (RB1, RBL1, RBL2, TP73), chromatin modification (EP300, CREBBP), or kinase signaling (KIT, PIK3CA, BRAF) pathways recurrently implicated in SCLC.9,10,30 However, we identified a disruptive in-frame indel mutation in MAML3 (c.2302_2304delCAG), which may play a role in regulating downstream NOTCH signaling.31 We also noted a missense mutation in GAS6 (c.629C>T,p.A210V), a secreted peptide that binds to and activates the TAM family of tyrosine kinase receptors.32
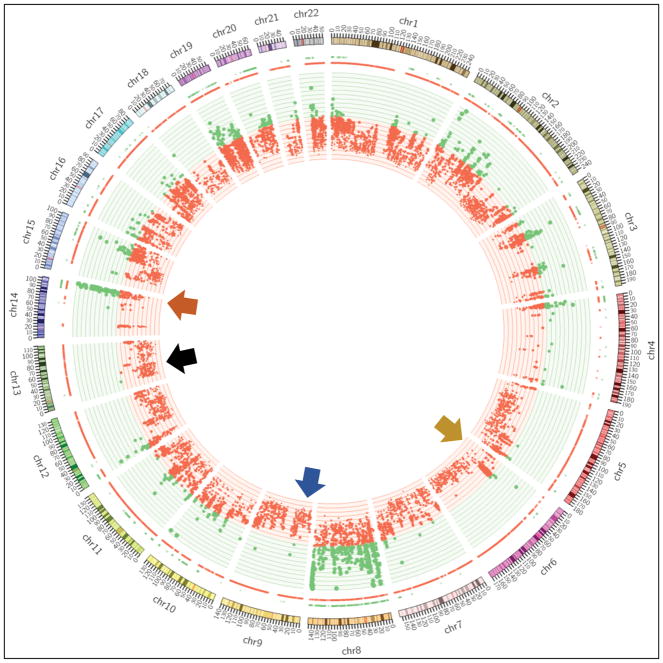
Whole exome CNV analysis identified a focal amplification of the chromosomal region 6p25-11.1 containing SOX4, a transcription factor essential to embryologic differentiation, and VEGFA (vascular endothelial growth factor-alpha). Focal amplification of 14q32.1-32.3 containing AKT1, a serine/threonine protein-kinase and known oncogene, was additionally present. Genomic losses at 3p14.3-14.2 (harboring FHIT) and 3p12.3-12.2 (harboring ROBO1), typical of SCLC,33 were not identified in the sinonasal SCC, but we did identify an amplification of the Notch pathway inhibitor DLK1 further suggesting that inhibition of NOTCH signaling is an important step in the pathogenesis of small cell carcinoma (Figure 4).
FIGURE 4.
Circos Plot showing amplification (green dots) of SOX4 on chr 6 (yellow arrow), DLK1 on chr 14 (orange) and deletion (red dots) of CDKN2A on chr 9 (blue), RB1 on chr 13 (black).
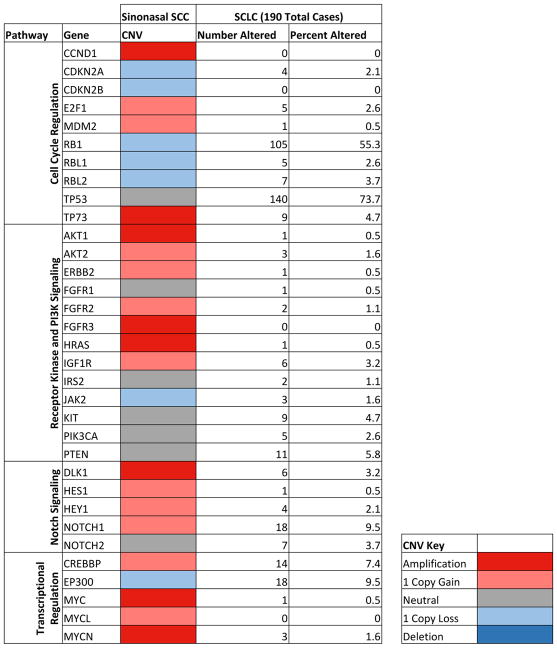
Significant CNVs were identified in several genes central to SCLC pathogenesis (Figure 5). Recurrent amplification of the pro-growth genes CCND1, E2F1, and MDM2 and hemizygous deletion of tumor suppressors CDKN2A, RB1, RBL1, and RBL2 contributed to aberrant cell cycle regulation and cellular proliferation. Amplification of Notch pathway members, MYC family genes, and CNVs of chromatin modifiers CREBBP and EP300 were similarly important genomic events in sinonasal SCC pathogenesis.
FIGURE 5.
Whole Exome CNV Analysis of Sinonasal SCC Reveals Significant Amplification and Deletion Events in Genes Recurrently Implicated in SCLC Pathogenesis9,10,30 (Frequency of individual gene alterations in SCLC calculated using cBioPortal)34,35
Discussion
Benign IP, Schneiderian carcinoma, and more lethal sinonasal carcinomas (e.g. squamous cell carcinoma, small cell carcinoma) exist on a spectrum of tumor evolution, though the molecular drivers and clinical prediction of tumor progression remain uncertain. The patient described herein had a multiply-recurrent Schneiderian carcinoma in-situ of the paranasal sinuses with subsequent tumor devolution into sinonasal SCC. The development of SCC within the previously-resected sinonasal surgical defect and absence of distant metastatic disease and potential primary SCC foci support a direct progression from Schneiderian carcinoma in-situ to sinonasal SCC. Additionally, the patient’s continued survival with no evidence of disease recurrence concurs with superior prognosis of sinonasal SCC compared to SCLC and EPSCCs as reported in the literature.4
A growing body of evidence supports the versatility and unpredictability of tumor progression in IP and Schneiderian carcinoma. Recent case series have posited associations between these tumors and synchronous adenocarcinoma and esthesioneuroblastoma of the paranasal sinuses.13,36 Our study similarly suggests that IP and Schneiderian carcinoma have the potential to devolve into sinonasal SCC, though certainly robust investigations of genetic associations between these tumor types are essential to confirm direct tumor progression.
EPSCCs of the head and neck and other anatomic sites share identical histopathological features with SCLC and are treated with similar chemo-radiation protocols. However, recent evidence suggests that EPSCCs are a heterogeneous group of neuroendocrine neoplasms harboring unique genetic drivers of oncogenesis, making comparative genomic analyses with SCLC imperative.37 Whole exome sequencing and CNV analysis of this rare case of sinonasal SCC revealed several unique genetic aberrations but also important shared mutational signatures with SCLC.
We identified a total of 93 high-confidence, non-synonymous somatic alterations in exomes of sinonasal SCC, a relatively modest mutational burden compared to that typically seen in SCLC.9,10,30 The 75 (80.6 %) somatic alterations not presently listed in COSMIC are potential targets for future elucidation of their role in EPSCC initiation and progression. We did not identify any single-nucleotide substitution or indel events in the majority of genes recurrently altered in SCLC, though significant CNVs in these genes were almost uniformly present (Table 1). While a heterozygous, loss-of-function mutation in TP53 (c.528C>G; p.C176W) was confirmed in our sample, one allele remained intact with no loss-of-heterozygosity seen for this critical tumor suppressor.
Focal amplification of chromosomal regions 6p25-11.1 and 14q32.1-32.3 were seen in our sinonasal SCC sample. The former is a known genomic fragile site and amplification hotspot containing the SOX4 gene.38 Notably, amplification of SOX family genes is a recurrent driver of SCLC initiation and proliferation.30 Amplification of 14q31.2-32.3 resulted in activation of DLK1, an inhibitory ligand of the Notch signaling pathway. Overexpression of DLK1 and recurrent inactivating mutations in NOTCH1 are typical of SCLC, supporting an important tumor suppressor function of the Notch pathway in these neuroendocrine neoplasms. Homozygous deletions of FHIT at 3p14.3-14.2 and ROBO1 at 3p12.3-12.2, genes posited to be critical tumor suppressors in SCLC and other human malignancies, were not present in sinonasal SCC.39,40
Whole exome CNV analysis of sinonasal SCC revealed numerous amplification and deletion events in genes critical to SCLC pathogenesis (Table 1). Hemizygous deletion of CDKN2A, RB1, RBL1, and RBL2 were present in sinonasal SCC. Therefore, inactivating alterations of these critical cell cycle regulators is likely essential for both EPSCC and SCLC pathogenesis. In the Notch signaling pathway, DLK1 amplification and inactivation of the downstream transcriptional co-activator MAML3 likely resulted in net inactivation of Notch signaling in sinonasal SCC. Notch signaling has been posited to have a critical tumor suppressor function in SCLC and other human malignancies, thus this pathway is an attractive target for future mechanistic studies in both EPSCC and SCLC.9,10,41 Finally, copy gain of the chromatin modifier CREBBP in sinonasal SCC diverged from the predominant pattern of inactivation of chromatin modifiers in SCLC.
Conclusions
EPSCCs of the head and neck may rarely arise as a late-stage evolution in the genetic progression of sinonasal IP and Schneiderian carcinoma. Whole exome sequencing of sinonasal SCC revealed inactivating alterations in TP53, CDKN2A, and RB1 similar to SCLC. Likewise, the probable inactivation of NOTCH signaling through an indel in MAML3 and amplification of the pathway inhibitor DLK1 is consistent with the inactivation of NOTCH signaling commonly seen in SCLC. Future comprehensive genomic studies on EPSCCs of the head and neck and other anatomic sites are needed to guide therapeutic strategies in these rare neoplasms.
Supplementary Material
Frequency of mutational events in sinonasal SCC.
List of 93 high-confidence, non-synonymous somatic mutation events in sinonasal SCC with predicted amino acid change, when known. (Mutation events in bolded genes are currently documented in COSMIC).
Acknowledgments
Grant Support: J.C.B. received funding from NIH Grants U01-DE025184, P30-CA046592 and R01-CA194536. M.L.L. was funded by F31 CA20634 and J.E.M. by F31 DE027600. J.D.S. received funding from NIH T32 DC 5356-15.
References
- 1.Brennan SM, Gregory DL, Stillie A, Herschtal A, Mac Manus M, Ball DL. Should extrapulmonary small cell cancer be managed like small cell lung cancer? Cancer. 2010;116(4):888–95. doi: 10.1002/cncr.24858. [DOI] [PubMed] [Google Scholar]
- 2.Gnepp DR, Ferlito A, Hyams V. Primary anaplastic small cell (oat cell) carcinoma of the larynx. Review of the literature and report of 18 cases. Cancer. 1983;51:1731–45. doi: 10.1002/1097-0142(19830501)51:9<1731::aid-cncr2820510929>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Misawa K, Kawasaki H, Endo S, et al. Primary combined small and squamous cell carcinoma of the hypopharynx: a case report. Mol Clin Oncol. 2016;4(5):709–14. doi: 10.3892/mco.2016.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pointer KB, Ko HC, Brower JV, et al. Small cell carcinoma of the head and neck: an analysis of the National Cancer Database. Oral Oncol. 2017;69:92–98. doi: 10.1016/j.oraloncology.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Megwalu UC, Nuyen BA. Survival outcomes in oropharyngeal small-cell carcinoma compared with squamous cell carcinoma: A population-based study. JAMA Otolaryngol Head Neck Surg. doi: 10.1001/jamaoto.2017.0025. Published Online: March 16, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoesli RC, Ludwig ML, Michmerhuizen NL, et al. Genomic sequencing and precision medicine in head and neck cancers. Eur J Surg Oncol. 2017;43(5):884–92. doi: 10.1016/j.ejso.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birkeland AC, Swiecicki PL, Brenner JC, Shuman AG. A review of drugs in development for the personalized treatment of head and neck squamous cell carcinoma. Expert Rev Precis Med Drug Dev. 2016;1(4):379–85. doi: 10.1080/23808993.2016.1208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkeland AC, Ludwig ML, Meraj TS, Brenner JC, Prince ME. The tip of the iceberg: Clinical implications of genomic sequencing projects in head and neck cancer. Cancers. 2015;7(4):2094–109. doi: 10.3390/cancers7040879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small cell lung cancer. Nat Genet. 2012;44(10):1104–110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner B, Tang LH, Klimstra DS, Kelsen DP. Small-cell carcinomas of the gastrointestinal tract: A review. J Clin Oncol. 2004;22:2730–39. doi: 10.1200/JCO.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 12.Hoang MP, Maitra A, Gazdar AF, Albores-Saavedra J. Primary mammary small-cell carcinoma: A molecular analysis of 2 cases. Hum Pathol. 2001;32:753–57. doi: 10.1053/hupa.2001.25603. [DOI] [PubMed] [Google Scholar]
- 13.Rossi G, Bertolini F, Sartori G, et al. Primary mixed adenocarcinoma and small cell carcinoma of the appendix: A clinicopathologic, immunohistochemical, and molecular study of a hitherto unreported tumor. Am J Surg Pathol. 2004;28:1233–39. doi: 10.1097/01.pas.0000128666.89191.48. [DOI] [PubMed] [Google Scholar]
- 14.Terracciano L, Richter J, Tornillo L, et al. Chromosomal imbalances in small cell carcinomas of the urinary bladder. J Pathol. 1999;189:230–35. doi: 10.1002/(SICI)1096-9896(199910)189:2<230::AID-PATH407>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Kim JS, Kwon SH. Recurence of sinonasal inverted papilloma following surgical approach: A meta-analysis. Laryngoscope. 2017;127:52–58. doi: 10.1002/lary.26222. [DOI] [PubMed] [Google Scholar]
- 16.Hong SL, Kim BH, Lee JH, Cho KS, Roh HJ. Smoking and malignancy in sinonasal inverted papilloma. Laryngoscope. 2013;123:1087–91. doi: 10.1002/lary.23876. [DOI] [PubMed] [Google Scholar]
- 17.Udager AM, Rolland DCM, McHugh JB, et al. High-frequency targetable EGFR mutations in sinonasal squamous cell carcinomas arising from inverted sinonasal papilloma. Cancer Res. 2015;75(13):2600–606. doi: 10.1158/0008-5472.CAN-15-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips P, Gustafson RO, Facer GW. The clinical behavior of inverting papilloma of the nose and paranasal sinuses: Report of 112 cases and review of the literature. Laryngoscope. 1990;100:463–69. doi: 10.1288/00005537-199005000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Singh G, Singh M, Chandana M, Singh S, Nargotra N. Intestinal type adenocarcinoma from inverted papilloma: a rare recurrence. J Clin Diagn Res. 2016;10(11):12. doi: 10.7860/JCDR/2016/22038.8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birkeland AC, Burgin SJ, Yanik M, et al. Pathogenetic analysis of sinonasal teratocarcinosarcomas reveal actionable beta-catenin overexpression and a beta-catenin mutation. J Neurol Surg B Skull Base. 2017;78(4):346–52. doi: 10.1055/s-0037-1601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birkeland AC, Yanik M, Tillman BN, et al. Identification of targetable ERBB2 aberrations in head and neck squamous cell carcinoma. JAMA Otolaryngol Head Neck Surg. 2016;142(6):559–67. doi: 10.1001/jamaoto.2016.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tillman BN, Yanik M, Birkeland AC, et al. Targeted sequencing of an epidemiologically low risk patient defines fibroblast growth factor family aberrations as a putative driver of head and neck squamous cell carcinoma. Head Neck. 2016;38:1646–52. doi: 10.1002/hed.24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birkeland AC, Foltin SK, Michmerhuizen NL, et al. Correlation of Crtc1/3-Maml2 fusion status, grade and survival in mucoepidermoid carcinoma. Oral Oncol. 2017;68:5–8. doi: 10.1016/j.oraloncology.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Pan S, Hsieh MH, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci USA. 2013;110(50):20224–9. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koboldt D, Zhang Q, Larson D, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amarasinghe KC, Li J, Hunter SM, et al. Inferring copy number and genotype in tumour exome data. BMC Genomics. 2014;15:732. doi: 10.1186/1471-2164-15-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–83. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boora GK, Kanwar R, Kulkarni AA, et al. Exome-level comparison of primary well-differentiated neuroendocrine tumors and their cell lines. Cancer Genet. 2015;208:374–81. doi: 10.1016/j.cancergen.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–16. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa M. Notch signaling in the nucleus: roles of Mastermind-like (MAML) transcriptional coactivators. J Biochem. 2016;159(3):287–94. doi: 10.1093/jb/mvv123. [DOI] [PubMed] [Google Scholar]
- 32.Kimani SG, Kumar S, Bansal N, et al. Small molecule inhibitors block GAS6-inducible TAM activation and tumorigenicity. Sci Rep. 2017;7:43908. doi: 10.1038/srep43908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovatich A, Friedland DM, Druck T, et al. Molecular alterations to human chromosome 3p loci in neuroendocrine lung tumors. Cancer. 1998;83(6):1109–17. doi: 10.1002/(sici)1097-0142(19980915)83:6<1109::aid-cncr9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karam SD, Jay AK, Anyanwu C, et al. Pathologic collision of inverted papilloma with esthesioneuroblastoma. Front Oncol. 2014;4:44. doi: 10.3389/fonc.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng X, Liu D, Fallon JT, Zhong M. Distinct genetic alterations in small cell carcinoma from different anatomic sites. Exp Hematol Oncol. 2015;4:2. doi: 10.1186/2162-3619-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myllykangas S, Himberg J, Bohling T, Nagy B, Hollmen J, Knuutila S. DNA copy number amplification profiling of human neoplasms. Oncogene. 2006;25:7324–32. doi: 10.1038/sj.onc.1209717. [DOI] [PubMed] [Google Scholar]
- 39.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 40.Angeloni D, Elst A, Wei MH, et al. Analysis of a new homozygous deletion in the tumor suppressor region at 3p12.3 reveals two novel intronic noncoding RNA genes. Genes Chromosomes Cancer. 2006;45(7):676–91. doi: 10.1002/gcc.20332. [DOI] [PubMed] [Google Scholar]
- 41.Yap LF, Lee D, Khairuddin A, et al. The opposing roles of NOTCH signaling in head and neck cancer: a mini review. Oral Dis. 2015;21(7):850–7. doi: 10.1111/odi.12309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency of mutational events in sinonasal SCC.
List of 93 high-confidence, non-synonymous somatic mutation events in sinonasal SCC with predicted amino acid change, when known. (Mutation events in bolded genes are currently documented in COSMIC).