Abstract
Hotspots of disease transmission can strongly influence pathogen spread. Bee pathogens may be transmitted via shared floral use, but the role of plant species and floral trait variation in shaping transmission dynamics is almost entirely unexplored. Given the importance of pathogens for the decline of several bee species, understanding whether and how plant species and floral traits affect transmission could give us important tools for predicting which plant species may be hotspots for disease spread. We assessed variation in transmission via susceptibility (probability of infection) and mean intensity (cell count of infected bees) of the trypanosomatid gut pathogen Crithidia bombi to uninfected Bombus impatiens workers foraging on 14 plant species, and assessed the role of floral traits, bee size and foraging behavior on transmission. We also conducted a manipulative experiment to determine how the number of open flowers affected transmission on three plant species, Penstemon digitalis, Monarda didyma, and Lythrum salicaria. Plant species differed fourfold in the overall mean abundance of Crithidia in foraging bumble bees (mean including infected and uninfected bees). Across plant species, bee susceptibility and mean intensity increased with the number of reproductive structures per inflorescence (buds, flowers and fruits); smaller bees and those that foraged longer were also more susceptible. Trait-based models were as good or better than species-based models at predicting susceptibility and mean intensity based on AIC values. Surprisingly, floral size and morphology did not significantly predict transmission across species. In the manipulative experiment, more open flowers increased mean pathogen abundance fourfold in Monarda, but had no effect in the other two plant species. Our results suggest that variation among plant species, through their influence on pathogen transmission, may shape bee disease dynamics. Given widespread investment in pollinator-friendly plantings to support pollinators, understanding how plant species affect disease transmission is important for recommending plant species that optimize pollinator health.
Keywords: bee decline, bee parasites, Bombus impatiens, Crithidia, environmental reservoir, floral traits, foraging behavior, trait-based, transmission hotspots
Introduction
Pathogen transmission is mediated by environmental heterogeneity (reviewed in Paull et al. 2012) and can be influenced by features of the transmission site. ‘Hotspots’ are regions characterized by particularly high pathogen prevalence or incidence, and can be sources of transmission to less infected areas (Paull et al. 2012). For example, the bacteria that cause cholera can concentrate on water hyacinth, which prolongs pathogen longevity (Spira et al. 1981). Thus, the presence of water hyacinth at sites may create a ‘hotspot’ that results in increased transmission across the landscape. Similarly, species may vary in their ability to transmit pathogens, even given similar levels of pathogen in the environment. For example, six grass species varied in host susceptibility, competence, and vector population sizes when exposed to Barley Yellow Dwarf virus, and several traits associated with life history were associated with this variation (Cronin et al. 2010). The goal of our study was to assess the extent to which plant species vary as hotspots for bee disease transmission, and, if so, the potential for floral traits to explain this variation.
Populations of many wild bee species are in decline, and pathogens have been implicated as one of the likely causal factors (e. g., Goulson et al. 2015). There is increasing evidence that bees share pathogens within and across species (e. g., Gamboa et al. 2015, McMahon et al. 2015), including transmission from widespread managed species such as Apis mellifera to wild Bombus hosts (Fürst et al. 2014). Crithidia bombi (Zoomastigophora:Trypanosomatidae) is a gut trypanosome that infects a wide range of bumble bee species with infection rates up to 80% (Shykoff and Schmid-Hempel 1991a, Gillespie 2010). Crithidia can impair learning, reduce colony reproduction under food limitation, reduce a queen’s ability to found new colonies, and is associated with decreased reproduction in wild colonies (e. g., Shykoff and Schmid-Hempel 1991b, Brown et al. 2003, Gegear et al. 2006, Goulson et al. 2017). This pathogen is transmitted when feces from an infected individual are consumed by an uninfected bee (Durrer and Schmid-Hempel 1994). While there are obvious routes for transmission within colonies, the environmental factors that contribute to horizontal transmission are largely unknown.
Flowers can be visited by a wide range of pollinators and other species (McArt et al. 2014) and are logical suspects as hotspots of pathogen transmission among bees, but very little empirical work has addressed this (reviewed in Koch et al. 2017). More than 20 years ago, a landmark study showed that Bombus terrestris could become infected with Crithidia by foraging on inflorescences previously foraged on by infected bees (Durrer and Schmid-Hempel 1994). More recent work has demonstrated that Crithidia was shared among three Bombus species, and that potential to transmit the pathogen varied among plant species (Ruiz-Gonzalez et al. 2012). Furthermore, multiple pathogens, including Crithidia spp., Nosema spp. and viruses, can be transmitted among bee species via shared flower use (Ruiz-Gonzalez and Brown 2006, Singh et al. 2010, Graystock et al. 2015).
Plant species may differ in their potential to transmit pathogens, and such variation could be mediated by floral traits. Pathogen transmission among bees via shared flower use was different on two different plant species in each of two studies (Durrer and Schmid-Hempel 1994, Graystock et al. 2015). This suggests that plant community composition can affect transmission, but consideration of so few plant species limits our ability to generalize. Furthermore, since each study used only two plant species that differ in many ways, it is not possible to determine which traits might be responsible for differences in transmission. Only one study has manipulated floral traits to assess their role in transmission; Durrer and Schmid-Hempel (1994) manipulated inflorescence architecture in a single plant species, and found that B. terrestris were more likely to become infected after foraging on inflorescences with a ‘linear’ rather than ‘spiral’ arrangement of flowers. This suggests that floral traits can affect disease transmission in foraging bees, but more work is needed across a broader range of plant species to evaluate this hypothesis. More generally, both floral and pollinator morphology can be important for efficient pollen transfer (e. g., Montgomery and Rathcke 2012). Thus, it is a logical extension to hypothesize that floral morphology could also influence pathogen transmission. Given that pathogen transmission among bees may be widespread (Fürst et al. 2014, Graystock et al. 2016), it is important to understand whether and how plant species mediate transmission.
While documenting plant species variation in transmission would provide an important first step to understanding how plant community composition influences pollinator-disease dynamics, a trait-based approach (Westoby and Wright 2006, Webb et al. 2010) to understanding disease transmission has several potential advantages over species-by-species approaches. If traits alone can predict transmission as well as models incorporating species identity, the effort required to parameterize transmission rate models for complex communities may be greatly reduced, because many relevant traits (e.g., measures of individual size and life history) are easily obtained from publicly available databases. For example, the probability that rodent species were zoonotic reservoirs could be predicted with approximately 75% accuracy based on only five host traits; considering 11 host traits improved predictions to >90% (Han et al. 2015). Trait-based analyses are also potentially generalizable between taxonomically distinct communities, while a species-based approach requires a new study for each new species.
To evaluate whether variation among plant species can shape pathogen transmission to foraging bees and to assess the role of floral traits in mediating these dynamics, we used 14 bee-pollinated plant species from eight different families, encompassing a range of floral trait variation. We allowed uninfected, individual Bombus impatiens workers to forage on inflorescences provided with Crithidia inoculum, and then reared these bees and compared transmission across species, measured as susceptibility (probability of becoming infected) and mean intensity (cell counts of infected bees). We also measured floral traits for each species, and evaluated the effect of these traits, bee foraging behavior, and bee size on susceptibility and mean intensity across plant species. We then compared how well trait-based models and species-based models explained variation in susceptibility and mean intensity. We hypothesized that traits that increase encounter rate with pathogens, such as small flowers and wide or nonexistent corolla tubes, and bee behavior, such as number of flowers visited or total time foraging, would increase susceptibility or intensity of infection. Floral traits could also affect how much pathogen inoculum is consumed per encounter, which positively relates to infection intensity (Otterstatter and Thomson 2006). Finally, nectar production or floral morphology could affect desiccation, which is important for viability of some pathogens such as Crithidia (Figueroa et. al., unpublished manuscript). Because our results suggested that reproductive structures per inflorescence predicted transmission, we then conducted transmission trials on three plant species in which we experimentally manipulated open flowers per inflorescence. Ultimately, our goal was to elucidate the role of flowering species and floral traits in bee disease dynamics.
Materials and Methods
Transmission trials across 14 plant species
Study site and species.
This research was conducted at the University of Massachusetts Center for Agriculture (South Deerfield, MA, U.S.A., 42° 28.6’ N, 72° 34.8’ W) in 2014. The 14 plant species included in the study were Antirrhinum majus (Plantaginaceae), Asclepias incarnata (Asclepiadaceae), Digitalis purpurea (Plantaginaceae), Eupatorium perfoliatum (Asteraceae), Helianthus annuus (Asteraceae), Impatiens capensis (Balsaminaceae), Linaria vulgaris (Plantaginaceae), Lobelia siphilitica (Campanulaceae), Lythrum salicaria (Lythraceae), Monarda didyma (Lamiaceae), Penstemon digitalis (Plantaginaceae), Solidago canadensis (Asteraceae), Thymus vulgaris (German variety; Lamiaceae), and Verbascum thapsus (Scrophulariaceae); for simplicity we refer to all species by genus hereafter. Many of these species were selected from ‘bee friendly’ suggested planting lists (e.g. Xerces society; http://www.xerces.org/pollinator-conservation/plant-lists/), and others were chosen for particular interest as invasive, common horticultural species. Overall, we chose species representing a wide range of variation in traits including flower size, number, and morphology. Some species were obtained from local nurseries or grown from seed and transplanted to the field site; others were collected from naturally-growing areas nearby (Appendix S1).
We used commercial colonies of Bombus impatiens (Biobest, Leamington, Ontario, Canada), the common eastern bumble bee, which is the most prevalent wild bumble bee species in our region (Gillespie 2010). Because B. impatiens is widely distributed commercially, understanding how plants mediate transmission in this species is particularly important due to the potential for spread from commercial to wild bees (Colla et al. 2006). Colonies were confirmed to be Crithidia-free with weekly dissections of five bees per colony. Crithidia was maintained in a ‘source’ colony that was originally infected from wild B. impatiens workers collected from two sites in Amherst, MA, U.S.A. (42°24′32.47″N 72°31′39.57″W; 42°23′20″N 72°31′21″W) and then transferred to new source colonies as needed. Five source colonies were used over the course of the experiment; usually only one source was used per day, but on five dates two sources were used to produce enough inoculum. Colonies were provided ad libitum with 30% sucrose solution replaced weekly, and approximately 10 g pollen loaves made of 30% sucrose mixed with multifloral honey bee-collected pollen (Koppert Biological Systems, Howell, Michigan) added every other day.
Inoculum preparation.
To create Crithidia inoculum for use in transmission trials, we dissected up to 10 bees per day from a single source colony. Guts were ground in 300 μL deionized water in microcentrifuge tubes and left to sit for 4 h. Moving Crithidia cells were counted on a Neubauer hemacytometer in a 0.02 μL subsample from a 10 μL sample per bee using a light microscope at 40x magnification. Because one of the three life stages of Crithidia is non-motile, we note that by counting only moving Crithidia cells we introduced some variation in the number of infective cells in inoculum made each day; this value also varies daily because we estimate concentration from a small subsample of gut solution. Thus, random variation in daily inoculum concentration makes our results a more conservative test of plant species differences. After counting Crithidia cells, we then combined 150 μL of gut solution from up to five bees each day, diluted this with deionized water, and then mixed with an equal volume of 50% sucrose to create a final solution of 25% sucrose with 600 cells μL−1.. Thus, our inoculum had a Crithidia cell concentration within the natural concentration occurring in feces (Otterstatter and Thomson 2006) and also a sugar concentration within the range of nectar; the average concentration from species we were able to measure in this study was 30% (range: 11.5–55%; data not shown). We recorded the time at which inoculum preparation was completed each day, and transported inoculum to the field site in a cooler with ice packs to minimize loss of viability.
Transmission trials.
During natural foraging to wild plants, floral traits could influence transmission of bee pathogens at flowers by altering 1) the likelihood of depositing pathogens on flowers, 2) pathogen viability in flowers, 3) the likelihood of encountering flowers that contain pathogens, and 4) pathogen acquisition and establishment in hosts upon visiting flowers that contain pathogens (reviewed in McArt et al. 2014). Although we would have ideally assessed all four of these mechanisms by allowing uninfected bees to forage on plants previously visited by infected bees (as in Durrer and Schmid Hempel 1994, which used B. terrestris), we were unable to replicate results of that study, suggesting that natural transmission rates in B. impatiens are too low to be detected by this approach. Instead, we compared transmission potential between plant species by adding controlled amounts of inoculum to Crithidia-free inflorescences, allowing a single uninfected bee to forage, rearing the bee for 7 days and then assessing susceptibility (presence/absence of pathogens following exposure) and mean intensity (cell counts in infected bees). This methodology evaluates processes three and four – likelihood of encountering flowers that contain pathogens (via foraging behavior upon encountering an infected plant), and pathogen acquisition and establishment in hosts upon visiting flowers that contain pathogens. Each of these are major unexplored components of pathogen transmission by bees at flowers (McArt et al. 2014) that could be affected by floral number, size, shape, or nectar production, as well as (or in addition to) bee foraging behavior. This methodology does not account for likelihood of depositing pathogen cells on flowers (process one), or variation in viability in flowers (process two), which are beyond the scope of this paper but are being explored in a forthcoming manuscript (Figueroa et al., unpublished manuscript). In a subsequent large-scale experiment, we ranked plant species as ‘high’ or ‘low’ transmission based on the trials reported here, and conducted an experiment with infected bee microcolonies foraging on uninoculated ‘high’ or ‘low’ transmission plants. Average colony-level infection after two weeks was approximately twice as high when foraging on ‘high’ compared to ‘low’ transmission plants (Adler et al., unpublished data), suggesting that processes of transmission we tested in this study explain substantial variation in longer-term transmission dynamics.
Plants were grown and trials were conducted in the field at the University of Massachusetts Center for Agriculture (South Deerfield, MA, U.S.A., 42° 28.6’ N, 72° 34.8’ W) from June 24 through August 28, 2014. Whenever possible, we used multiple plant species on each trial date, but each species was only used on a subset of all possible dates due to phenology. To conduct transmission trials, inflorescences of all plant species were covered with organza bags (ULine, Pleasant Prairie, Wisconsin) before flowers opened to prevent wild bee visitation and potential pathogen deposition. To conduct a trial, an inflorescence with at least five open flowers was clipped with scissors and immediately placed in a florist’s water tube. We counted open flowers, placed four 10 μL inoculum drops within four separate flowers (one drop per flower) using a pipette, and marked these flowers at the outside base, calyx or stem with paint pens (Craftsmart® Fine Line 6 Count, Basic, Michaels Stores, Inc., Irving, Texas). For Eupatorium and Solidago, capitula were considered ‘flowers,’ while in Helianthus we used a single capitulum and individual florets were counted as flowers. We chose 10 μL to simulate the volume of feces from a single defecation event. Four drops were used to facilitate encounters during foraging; by having a minimum of five open flowers we ensured there was at least one un-inoculated flower to visit. Bees almost always consumed inoculum upon first contact. We placed inoculum in contact with reproductive structures whenever possible; this was typically inside tubular flowers (e.g., Lobelia, Penstemon) or on top of open flowers (e.g., Lythrum). In some cases, flowers were so small that the drop rested on top of the corolla (e.g., Solidago, Thymus, Eupatorium) and may not have contacted nectar. We placed drops inside flowers due to initial findings of Crithidia in nectar and bees defecating on flowers (Durrer and Schmid-Hempel 1994, Otterstatter and Thomson 2006). More recent work suggests that feces are more likely to be deposited on outer floral surfaces and not in nectar (Cisarovsky and Schmid-Hempel 2014), although data show that bees deposit up to 47% of their feces within flowers on some plant species (Figueroa et al., unpublished manuscript). Our goal was to standardize the amount and presentation of inoculum across species so we could assess variation in susceptibility and intensity of infection given the same starting conditions, after controlling for foraging-induced differences in exposure.
Each inflorescence was individually placed into a small cage (45.7 cm x 71.0 cm x 55.6 cm) constructed of a wood frame with plexiglass or cloth sides with a chilled, uninfected experimental B. impatiens worker initially placed on the inflorescence. For each trial, we recorded the plant species, experimental bee colony of origin, start and end time, time spent foraging (i.e., actively probing flowers), and the number of open flowers probed and number of inoculum drops probed. For the latter two measures, every new entry into a flower in which reproductive parts were contacted was considered a new flower probe (and if the flower was inoculated, it was also a new drop probed) because we could not ascertain whether bees consumed all the inoculum drop in a single probe. A trial was concluded after the bee ceased foraging, if at least one inoculated flower was probed. We did not limit trials to a specified time period because the rate at which bees probed individual flowers varied widely with species, and so limiting trial time period would create a de facto difference in the number of flowers probed per species. Bees that did not forage on an inoculated flower after 20 min were excluded. After each trial, inflorescences were disposed of and experimental bees were returned to a cooler on ice until transport to the laboratory at the end of the field day. We used 6–10 experimental colonies per plant species and had 11–36 bees with successful trials and pathogen counts per plant species.
Assessing pathogen infection.
Upon returning to the laboratory, each bee was placed in a 20 mL plastic scintillation vial with a nectar feeder with 500 μL of 30% sucrose solution and a 0.1–0.2 g portion of a pollen loaf; all pollen loaves were made from the same pollen source used to maintain colonies. Bees were housed in a growth chamber at 27°C in darkness, and placed in new vials with fresh nectar and pollen daily. After 7 days, bees were dissected and Crithidia cells were counted as in ‘Inoculum preparation’ above, except that guts were left for 5 h instead of 4 h before counting (the shorter time for inoculation preparation allowed us to begin field trials sooner). We collected the right forewing of each bee to measure radial cell length as an estimate of bee size (Harder 1982); we refer to this as ‘bee size’ in analysis.
Measuring floral traits.
To understand the role of floral traits in mediating bee disease transmission, we measured reproductive structures per inflorescence, floral size and morphology, nectar production, and nectar secondary chemistry. We measured these floral traits on single inflorescences from 22–38 (median 30) individuals of each plant species that were not used in transmission trials (sample sizes are provided in Appendix S1, Table S1). While it would have been ideal to measure floral traits on the inflorescences used in trials, this would have been prohibitively time consuming and, in the case of nectar measurements, potentially damaging to flowers. However, we included the number of open flowers for each trial in analysis. For all other floral traits, we used separate plants to measure species-level values for use in analyses relating traits to transmission. We measured corolla length and width using digital calipers to the nearest 0.01 mm (Appendix S2), and used these traits in a principal components analysis to generate a first component that reflected floral size (PC1 = 0.87*corolla length + 0.50*corolla width, accounting for 91% of total variance), and a second component representing floral shape (PC2 = 0.5*corolla length - 0.87*corolla width, accounting for 9% of total variance), which correlated strongly (r = 0.88) with the ratio of corolla length:width. We counted reproductive structures per inflorescence (including buds, flowers and fruits), and measured the height of the tallest flower. We measured nectar volume after 24 h of bagging using glass microcapillary tubes; we did not remove nectar prior to bagging flowers to avoid damaging nectaries. We did not include sugar content in this study since nectar production was too low to measure sugar on several species. For a subset of species, in 2015 we measured floral longevity by marking buds and noting the date of first opening and senescence. We present means, sample sizes, and standard deviation for all predictor traits used in analyses in Data S1 and Metadata S1.
Statistical analysis.
Statistical analyses were conducted in R (R Core Team 2017) version 3.3 or higher. We analyzed susceptibility (presence/absence of Crithidia) and mean intensity (mean raw Crithidia cell count per 0.02 μl gut sample in infected bees) as two separate components of pathogen transmission to bees. Although using a single response with negative binomial regression should be more powerful, this was inappropriate for our data (see Appendix S3 for justification). However, we summarize patterns using a combined response variable for the purposes of comparing species only. Susceptibility is a binary (0–1) response and was therefore analyzed by logistic regression. Mean intensity (values of all non-zero counts) was highly right-skewed, so our analyses used log-transformed counts; these had a symmetric and approximately Gaussian distribution, and were therefore analyzed by linear regression assuming Gaussian error distributions. Models with only fixed effects were fitted using glm (for susceptibility) and lm (for intensity) functions in R. Models that included any random effects were fitted using the gam function in the mgcv package (Wood 2006). Note that the fitted models were logistic or linear mixed regression models, not generalized additive models. We used the gam function because, for the models we consider here (which do not include multiple random effects with correlations), gam reports the statistical significance (P value) for random effect terms specified through the random effect (“re”) basis.
Our statistical analyses assessed whether susceptibility and mean intensity were predicted by plant species and by floral traits, and whether these responses were better predicted by species identity or by floral traits. We initially fitted generalized additive models for nonlinear effects of day of year (Julian date), the elapsed time between inoculum preparation and each trial, or both, on susceptibility and mean intensity using gam in the mgcv package, but these covariates did not affect susceptibility or mean intensity (P > 0.12 from anova.gam in all cases) and were omitted from subsequent analyses. We could not include the effect of experimental colony in our analyses because this was confounded with plant species, since we used different colonies over the course of the summer and plant species bloomed at different times. However, we have no a priori reason to think that experimental colonies, sourced commercially and reared in the lab, would vary systematically in susceptibility to infection over a three-month period.
Plant species and pathogen transmission.
To assess covariates for inclusion in models that used plant species as a predictor of susceptibility and mean intensity, we first fitted a series of linear (intensity) and generalized linear (susceptibility) models with single predictors, including plant species, inoculum source colony, bee size (estimated as wing radial cell length), and bee foraging behavior (number of flowers probed, number of inoculum drops probed, and total time foraging) as fixed covariates, using both susceptibility and mean intensity as responses in separate analyses. Species identity and inoculum source colony were factor variables; species identity was fitted as a random effect but inoculum source colony was fitted as fixed since there were only six levels (four sources plus two combinations used on some days); all other covariates are numerical and were fitted as fixed effects. Only covariates that were significantly or marginally significantly related to susceptibility or mean intensity were retained for model selection (described in Results and Appendix S3).
Traits and transmission.
For models using floral traits rather than species identity to predict susceptibility and mean intensity, we again assessed potential covariates by fitting a set of linear or generalized linear models with potential covariates as single fixed effects. Potential covariates were floral traits (corolla size, corolla shape, number of open flowers, reproductive structures per inflorescence, nectar volume and floral longevity), bee size (estimated as wing radial cell length), and bee foraging behavior (number of flowers probed, number of inoculum drops probed, and total time foraging). Traits that were significant predictors in these analyses were used in model selection (described in Results) to produce final trait-based models for susceptibility and mean intensity. Species identity was not included in trait-based models because it is confounded with floral traits, which were measured at the species level. Helianthus was an outlier for several floral traits and foraging behavior measures (see Appendix S3), and so was omitted from analyses of trait-dependent transmission but included in analyses that assessed species differences in susceptibility or mean intensity without considering floral traits.
Species vs. traits as predictors of susceptibility and mean intensity.
Trait-based and species-based models are non-nested, so model comparison was done using AIC omitting Helianthus, because comparison is only possible when all models are fitted to the same data. We selected the lowest-AIC models for both species (with bee size and behavior as potential covariates, but not floral traits) and traits (with bee size, behavior and floral traits as potential covariates, but not species) to determine which most effectively predicted susceptibility and mean intensity.
Transmission trials manipulating flower number
Experimental trials.
In our observational transmission trials, reproductive structures per inflorescence was the most consistent predictor of susceptibility and mean intensity (see Results). This was a surprising result, since the number of open flowers did not predict responses as strongly. This suggests that some unmeasured trait correlated with reproductive structures per inflorescence affects transmission. Alternatively, because number of open flowers and reproductive structures per inflorescence were tightly correlated across species (Spearman’s ρ = 0.79, n = 13, P < 0.001 for all species but Helianthus), this observational approach may not be able to distinguish whether number of open flowers or some other correlated unmeasured trait is the underlying cause of altered transmission.
To determine whether the number of open flowers or some other correlated trait underlies the relationship between number of reproductive structures per inflorescence and transmission, we conducted transmission trials manipulating flower number and comparing transmission within three plant species. Although intraspecific variation in number of open flowers may affect susceptibility or mean intensity differently than interspecific variation, manipulating this trait within species allows us to assess its importance in the absence of confounding species-level differences in other traits, and using three plant species provides some generality to this assessment. Trials were conducted in 2016 on Penstemon (June 13–29), Monarda (June 30 – July 15) and Lythrum (July 18 – Aug 9) using plants and protocols from the same site and trials described previously, except that each inflorescence was assigned to a high or low flower number treatment in alternating sequence, and inoculum was made using ¼ strength Ringer’s solution ((Sigma-Aldrich, St. Louis, MO, USA) instead of deionized water. The number of open flowers for each treatment varied with species; the low and high range of open flowers was 5–7 and 11–13 for Penstemon, 10–15 and 25–30 for Monarda, and 6–10 and 16–20 for Lythrum. These numbers were based on typical flower production for each species, and such that the upper bound of the low treatment was half of the upper bound of the high treatment, with constant range of values within treatment. We only selected inflorescences with at least one more open flower than the maximum ‘high’ treatment value (e.g., 14 for Penstemon). We then randomly assigned inflorescences to treatments and removed open flowers using dissecting scissors or forceps to achieve the appropriate number; at least one open flower was removed from every inflorescence to control for damage effects. The number of open flowers removed per inflorescence and the final number of open flowers were recorded for each trial, along with the bee behavior variables described for the previous transmission trials. Bees came from six experimental colonies.
Statistical analysis.
Prior to analysis, we discarded six bees due to unusual foraging (e. g, spending several minutes inside a single flower), death or missing data, and two extreme outliers (Appendix S3), resulting in final sample sizes of 63, 49 and 65 bees in Penstemon, Monarda and Lythrum trials respectively. Because this experiment focused on the within-species effect of number of open flowers, rather than comparing species differences, we analyzed each species separately. This allowed us to analyze counts for each species as a single response, including both infected and uninfected bees, using negative binomial regression, because the treatment with higher mean count was also the treatment with a higher frequency of nonzero counts. Using mean abundance of Crithidia as the response (including uninfected bees) combines susceptibility and mean intensity into one response variable. We used R functions glm.nb for models with only fixed effects and gam with family=nb for models including random effects, in both cases using the default log link function.
As in the multi-species infection trials, we first assessed whether other potential covariates (bee size, trial time, time foraging, number of inoculum drops probed, number of flowers probed, minutes between inoculum preparation and trial, bee dissection time) should be included in subsequent analyses. Because we analyzed species separately and inoculum strength can vary daily, we also included trial date as an unordered, categorical random effect. We fitted negative binomial regression models (Appendix S3) that always included treatment as a fixed effect, and trial date and bee colony of origin as random effects. In each model, only one additional covariate was included, whose significance was tested by anova.gam. Additional covariates that were significant predictors in these analyses were used in model selection (described in Appendix S3) to produce the final model for testing treatment effects.
Results
Transmission trials across 14 plant species
Plant species and pathogen transmission
Tabulating mean pathogen counts per foraging bee showed that plant species varied fourfold in mean Crithidia abundance (mean count including uninfected bees; Fig. 1A). Mean abundance was highest in Asclepias, and high in Monarda, Lythrum, and Lobelia, and lowest in Digitalis, Antirrhinum, Linaria, and Thymus.
Fig. 1.
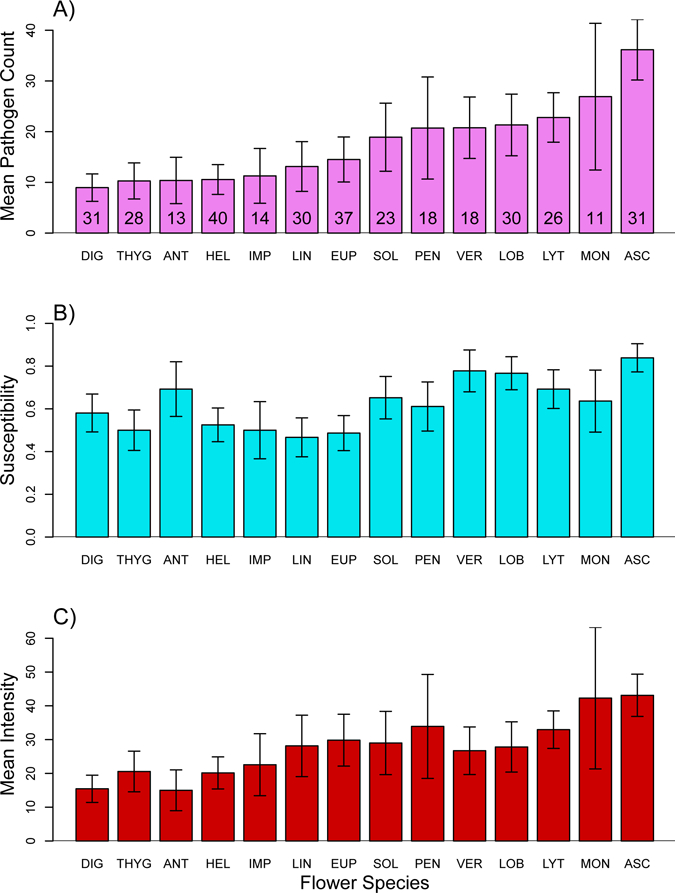
A) Mean Crithidia cell count (in a 2 μl sample) of bees foraging at different plant species provided with the same inoculum, including both zero and positive counts (mean abundance). Error bars are ±1 s.e.m.; numbers in bars are the sample size. Species acronyms begin with the first three letters of the genus. B) Susceptibility, the fraction of trials on each plant species for which the pathogen count was positive. Error bars are binomial standard errors on the fraction of positive counts. C) Mean intensity, the mean of positive cell counts. Error bars are ±1 s.e.m. Figure generated by R script SusceptibilityAndIntensityPlots.R; Dryad repository.
Plant species and bee size were significant predictors of variation in susceptibility in single-variable analyses, with larger bees having lower susceptibility (Table 1). Both remained significant in a generalized linear mixed model including both variables with species as a random effect (species: n = 292, df = 4.66, χ2 = 9.04, P = 0.029; bee size: n = 292, z = −2.015, P = 0.044; Fig. 2B). Total time foraging (marginally significant in the single-variable analysis; Table 1) was not significant in a generalized linear mixed model that also included species as a random effect (n = 303, z = 1.754, P = 0.078). Plant species also predicted mean intensity (Table 2, Fig. 1C). No other bee behavior covariates were significant predictors of mean intensity in the single-variable analyses. Patterns in mean abundance were largely reflected in the patterns for mean intensity (Figs 1A vs. 1C). Some species, such as Antirrhinum, had high susceptibility but low mean intensity, leading to low overall mean abundance.
Table 1.
Analysis of bee susceptibility to infection as a function of species identity, inoculum source colony, floral traits, bee traits, and bee foraging behavior, using generalized linear models with each focal variable as the one covariate (see text for details). Only the Species model includes data on Helianthus. Source file: SpeciesTraitsAndSusceptibility.R and scripts that it sources; Dryad repository.
| Variable | p value1 | χ2 | n | Coefficient |
|---|---|---|---|---|
| Species | 0.029 | 10.146 | 350 | - - - |
| Inoculum Source | 0.443 | 4.779 | 281 | - - - |
| Nectar Volume | 0.550 | 0.357 | 297 | −0.058 |
| Number of Open Flowers | 0.323 | 0.978 | 304 | 0.004 |
| Corolla Size | 0.501 | 0.453 | 310 | −0.006 |
| Corolla Shape | 0.152 | 2.057 | 310 | −0.050 |
| Repro. Structures per Infl. | 0.009 | 6.751 | 310 | 0.004 |
| Floral Longevity | 0.274 | 1.119 | 215 | 0.066 |
| Bee Size | 0.042 | 4.155 | 292 | −0.871 |
| Total Time Foraging (min) | 0.057 | 3.616 | 303 | 0.054 |
| Number of Flowers Probed | 0.993 | 0 | 304 | 0 |
| Number of Inoc. Drops Probed | 0.789 | 0.071 | 303 | 0.007 |
p-values were obtained from summary.gam (for Species, fitted as a random effect) or drop1 with test= “Chisq” (all others, fitted as fixed effects), χ2 is the value of the test statistic which has an approximately chi-square distribution, n is the sample size and coefficient is the coefficient of the focal variable in the linear predictor.
Fig. 2.
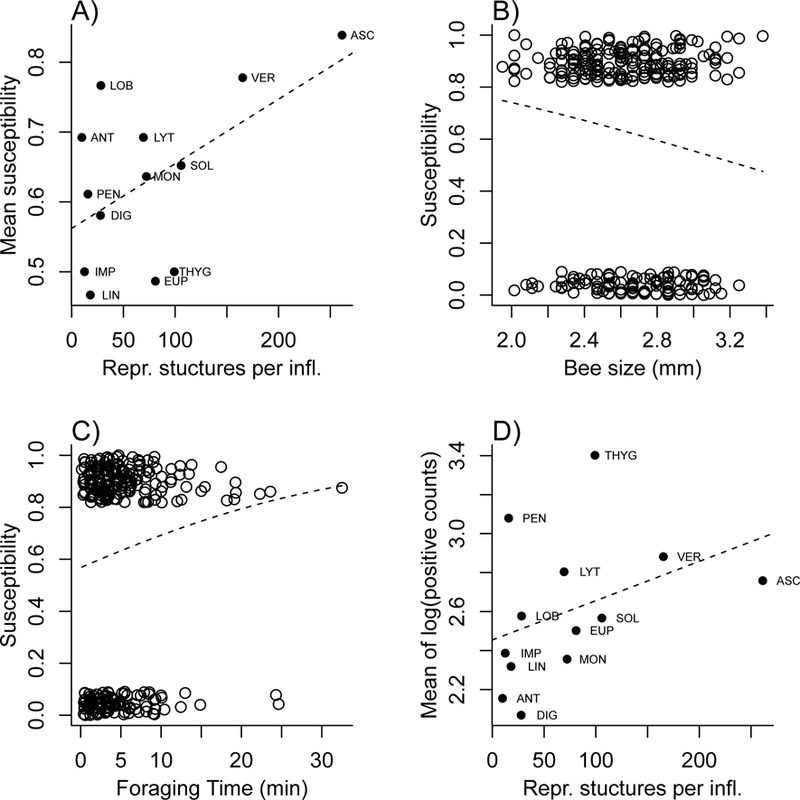
Relationships between traits and components of pathogen transmission that were statistically significant in the transmission trials using 14 plant species. A) Mean susceptibility (over all trials using a particular species) versus reproductive structures per inflorescence (estimated mean for the species). B), C) Susceptibility in each trial irrespective of flower species (0=not infected, 1=infected; values jittered to separate points) as a function of B) bee size, estimated by the length of the wing radial cell, and C) total time foraging by the bee. D) Mean intensity (mean of all log-transformed positive pathogen cell counts for each species) as a function of reproductive structures per inflorescence (estimated mean for the species). The dashed lines in each panel are regressions fitted to the plotted points (linear regression in panels A and D, logistic regression in B and C). Figure generated by R script SusceptibilityAndIntensityPlots.R; Dryad repository.
Table 2.
Analysis of mean intensity (log of positive pathogen counts) as a function of species identity, inoculum source colony, floral traits, bee traits, and bee foraging behavior, using linear models with each focal variable as the one covariate (see text for details). Only the Species model includes data on Helianthus. Source file: SpeciesTraitsAndIntensity.R and scripts that it sources; Dryad repository.
| Variable | p value1 | F | n | Coefficient |
|---|---|---|---|---|
| Species | 0.048 | 0.574 | 215 | - - - |
| Inoculum source | 0.352 | 1.120 | 175 | - - - |
| Nectar Volume | 0.021 | 5.432 | 185 | −0.181 |
| Number of Open Flowers | 0.218 | 1.527 | 192 | 0.003 |
| Corolla Size | 0.013 | 6.270 | 194 | −0.017 |
| Corolla Shape | 0.031 | 4.739 | 194 | −0.056 |
| Repro. Structures per Infl. | 0.001 | 11.709 | 194 | 0.004 |
| Floral Longevity | 0.476 | 0.510 | 134 | −0.032 |
| Bee size | 0.701 | 0.148 | 183 | −0.121 |
| Total Time Foraging (min) | 0.160 | 1.922 | 191 | −0.027 |
| Number of Flowers Probed | 0.459 | 0.550 | 192 | 0.003 |
| Number of Inoc. Drops Probed | 0.262 | 1.267 | 192 | 0.021 |
Table entries are as in Table 1, except that p-values are based on an F statistic.
Traits and transmission
In our initial analysis testing each potential predictor one at a time on susceptibility, reproductive structures per inflorescence, bee size, and total time foraging were significant or marginally significant (Table 1). Bee size and total time foraging were correlated (n = 283, r = −0.26, P < 0.001), so we fitted two GLMs including each of these separately as a covariate in addition to reproductive structures per inflorescence. In the first model, bees visiting plant species with more reproductive structures per inflorescence were more likely to acquire Crithidia (n = 291, χ2 = 6.366, P = 0.011; Fig. 2A), as were smaller bees (n = 291, χ2 = 4.75, P = 0.029; Fig. 2B). In the second model, bees visiting plant species with more reproductive structures per inflorescence were again more likely to acquire Crithidia (n = 302, χ2 = 9.24, P = 0.002), as were bees with greater total time foraging (n = 302, χ2 = 6.08, P = 0.014; Fig. 2C). Of these two models, the one with total time foraging has the lower AIC (ΔAIC=3.93), and in a GLM with both time foraging and bee size as covariates, bee size is not significant (P = 0.16) while both time foraging (n = 285, χ2 = 5.88, P = 0.015) and reproductive structures per inflorescence were (n = 285, χ2 = 8.26, P = 0.004). The relationship between susceptibility and reproductive structures per inflorescence (for which we have only one value per species) remained even if we used a grouped response with one susceptibility value per species (t = 2.22, P = 0.048).
Using a similar approach for mean intensity as the response, four variables (reproductive structures per inflorescence, nectar production, corolla size, corolla shape) were significant as individual predictors in separate models (Table 2). In models that included reproductive structures per inflorescence as a predictor of mean intensity, none of the other variables was significant as an additional predictor (P > 0.3 for all three), while reproductive structures per inflorescence was significant in all cases (F > 5.2, P < 0.025 for all). The final trait-based model for mean intensity thus had reproductive structures per inflorescence as the only covariate; bees that became infected after visiting plant species with more reproductive structures per inflorescence had higher mean Crithidia loads (n=194, F=11.71, P<0.001; Fig. 2D). The relationship between intensity and reproductive structures per inflorescence remained even if we used a grouped response with one intensity value per species (t = 3.7, P = 0.0035).
Species vs. traits as predictors of susceptibility and intensity
For susceptibility, the lowest AIC species-based model was a GLMM including species as a random effect, and bee size and total time foraging as fixed effects. The lowest AIC trait-based model was a GLM including reproductive structures per inflorescence, bee size and total time foraging as fixed effects. AIC for the trait-based model was somewhat lower (ΔAIC = 3.03). The two models made similar predictions (r = 0.84 between the two models’ fitted values), but the species-based model required more parameters (df = 6.97 for the species-based model, and 4 for the trait-based model). For prediction of mean intensity, the best species-based model included only species as a random effect, and the best trait-based model included only reproductive structures per inflorescence as a fixed effect. Comparing these models, AIC for the trait-based model was substantially lower (ΔAIC = 5.85), because the predictions were very similar (r = 0.83 between the two models’ fitted values) but the trait-based model had fewer parameters (df = 5.93 for the species-based model, and 3 for the trait-based model). Thus, for both susceptibility and intensity, traits and species identity had similar predictive power, and so a trait-based model is preferred due to greater simplicity. We also found no evidence of bias in the trait-based predictions (Appendix S3).
Transmission trials manipulating flower number
For Penstemon, there were no significant covariates in model selection, and no significant treatment effect on mean Crithidia abundance (mean counts including zero values; n = 66, χ2 = 0.867, P = 0.352). In Lythrum, there was no significant treatment effect in a model with (n = 68, χ2 = 0.005, P = 0.943) or without significant covariates (n = 71, χ2 = 0.042, P = 0.847). In Monarda, the effect of treatment was tested in a model including flowers probed as a fixed effect; both treatment and number of flowers probed were significant (treatment: n = 51, χ2 = 5.374, P = 0.02; number of flowers probed: n = 51, χ2 = 6.24, P = 0.01). The estimated coefficient for the lower flower number treatment (−1.375) corresponds to a roughly 4-fold reduction in mean pathogen abundance in the low compared to high flower treatments, aligning closely with raw mean abundance per bee in each treatment (mean ± se: low: 7.07 ± 2.00; high: 30.91 ± 12.13 cells per 0.02 μl).
Discussion
Overall, plant species differed fourfold in the mean abundance of pathogen cells established after a single bee foraging bout (Fig. 1A), with species explaining significant variation in both susceptibility and mean intensity (Figs 1B, 1C). These results complement earlier work which reported that the probability of Crithidia infection in B. terrestris workers differed on two plant species (Durrer and Schmid-Hempel 1994). Research more than twenty years later showed that B. terrestris and Apis mellifera can vector pathogens of both bee species via shared floral foraging, and the extent of vectoring differed between two plant species (Graystock et al. 2015). To our knowledge, these are the only previous studies asking whether plant species modulate pathogen acquisition among bees. Here we greatly extend the evidence of earlier work and, based upon the considerable variation in the effectiveness of different plant species to act as transmission hubs, suggest that plant community composition is likely to mediate bee-pathogen transmission dynamics. Future work should manipulate plant community composition in structured microcosms including bees and pathogen to assess longer-term effects.
In the transmission trials with 14 plant species, we found that models predicting susceptibility and mean intensity based on floral traits made similar in-sample predictions to models based on species identity. However, the trait-based models had lower AIC, and are therefore expected to have better out-of-sample predictive accuracy (i.e., more accurate forecasts of new observations), because the trait-based models required fewer parameters to fit the data. Moreover, only the trait-based models have any predictive power for species not represented in the data set. These gains in parsimony and generalization are the potential benefits of trait-based models, which has inspired trait-based approaches to many different aspects of community ecology (e.g., Westoby and Wright 2006, Webb et al. 2010). Given enough data on all species in a community the situation would be reversed, because species always have idiosyncratic differences that cannot be fully captured by a list of traits. But in species-rich communities, getting “enough data” on ecological processes in each species may require prohibitive time or expense. Measuring relevant traits on all species, and using a subset to estimate trait-transmission relationships, may be far more feasible. If we can identify specific floral traits that shape pathogen transmission, these could be used to guide recommendations for pollinator-friendly habitat, within the context of other constraints such as phenology and providing diverse resources to support specialist as well as generalist pollinator species.
No measure of floral morphology significantly predicted transmission, which was surprising given the importance of floral morphology for pollen transfer by bees (e. g., Costa et al. 2017). However, we note that whenever possible we added inoculum within the corolla tube. Naturally foraging infected bees are likely to deposit feces on the corolla lip or outside the flower, and floral traits may shape risk or exposure by affecting where and how much bees defecate as well as trypanosome survival, although we note that in our study the number of inoculum drops probed had surprisingly little relationship with susceptibility or mean intensity of infection. The ultimate effect of floral traits on transmission will depend on whether their effects on risk amplify or counter their effects on susceptibility and intensity.
In our observational trials, species with more reproductive structures per inflorescence had greater transmission, measured as both susceptibility and mean intensity (Figure 2A, D). This was the most consistent floral trait that predicted transmission, more than the number of flowers probed per trial or the amount of nectar each species produced. Reproductive structures per inflorescence even explained transmission more than the number of open flowers, which was surprising for two reasons. First, reproductive structures per inflorescence was measured at the species level (i.e., one value per plant species) while number of open flowers was counted for each trial. We would expect that a variable that was evaluated specifically for each trial would have more predictive power than a similar variable at the species level. Secondly, it is difficult to explain how reproductive structures per inflorescence (including buds and fruits) could mediate transmission more than the number of open flowers, given that bees only foraged on open flowers in our trials. If transmission occurs through spreading inoculum across all floral surfaces, then increased reproductive structures per inflorescence could provide more surface for spread via contact. Similarly, if more reproductive structures create more inflorescence complexity, this may affect micro-climates conducive to pathogen viability (such as increased humidity) or alter bee foraging behavior in ways that increase exposure. It is also possible that some underlying trait we did not consider is correlated with species-level variation in reproductive structures per inflorescence. For example, if plants that produced fewer flowers also produced longer-lasting flowers with more effective antimicrobial defense through nectar proteins or volatiles (e. g., Thornburg et al. 2003), this could help explain our result. We measured floral longevity on a subset of our species and found no relationship with transmission (Tables 1 and 2), but have not exhaustively tested this hypothesis.
Because reproductive structures per inflorescence and number of open flowers are often tightly correlated, we manipulated the number of open flowers to determine whether this trait influences variation in bee host susceptibility and infection intensity within species of plants, although we acknowledge that the same trait may affect pathogen dynamics differently within versus across species. We found only partial support for the hypothesis that the relationship between reproductive structures per inflorescence and transmission was due to an underlying correlation with number of open flowers. Flowers increased trypanosome pathogen abundance in bees nearly fourfold in Monarda, but there was no effect in Lythrum or Penstemon. Thus, the number of open flowers is unlikely to be the only mechanism explaining the relationship between reproductive structures per inflorescence and transmission.
Bee size and total time foraging were correlated with each other and with susceptibility in the observational transmission trials; smaller bees foraged for longer total time in the trials, and were more likely to become infected than larger bees. It is interesting that smaller bees foraged for longer periods but did not probe more flowers or inoculum drops in that time, suggesting that other mechanisms underlie the relationship between bee size and susceptibility. Small bees may be able to access more of the inoculum drops, particularly in plant species with narrow corollas. Furthermore, consuming the same amount of inoculum provides proportionally more pathogen cells per gram of bee tissue in a smaller bee, perhaps resulting in higher probability of infection. Finally, smaller bees could have less ability to resist infection. Although reduced food availability can decrease bee size (Schmid-Hempel and Schmid-Hempel 1998, Rotheray et al. 2017) and affect immune function (Schmid-Hempel and Schmid-Hempel 1998, Brown et al. 2003, Alaux et al. 2010), our bees were commercially reared and should not have been food stressed. Regardless of the mechanism, greater infection in smaller bees could have consequences for within-hive transmission since smaller bees are more often nurse bees while larger bees are foragers (Goulson 2010).
In conclusion, plant species varied widely in the transmission of Crithidia to B. impatiens, suggesting that plant species, through their influence on pathogen transmission, may play an important role in shaping bee disease dynamics. Surprisingly, reproductive structures per inflorescence best predicted variation in transmission; floral size and morphology did not play significant roles. Across species, trait-based models were as good or better at predicting susceptibility and mean intensity based on AIC values, indicating the potential to use traits to select plant species that minimize pathogen spread, rather than requiring an evaluation of every plant species. Our manipulative experiment suggested that, within species, open flowers play a partial role explaining variation in transmission; such intraspecific variation may play important roles in plant-pollinator-pathogen dynamics. Given widespread investment in pollinator-friendly plantings to support pollinators, determining how plant species affect disease transmission is critical for recommending plant species that optimize pollinator health.
Supplementary Material
Acknowledgements.
We thank two anonymous reviewers for feedback that improved the manuscript, Biobest for generously donating bee colonies, M. Ha as project manager and P. Anderson, O. Biller, S. J. Connon, D. Delaney, E. Duvall, S. Fogel, G. Killough-Hill, J. Giacomini, A. Hogeboom, A. Lee, G. LoCascio, E. Mann, L. Metz, N. Milano, R. Pasquale, K. Rothchild, D. Shaheen, T. Shaya, C. Shuja, T. Truong, A. Turkle, I. Weston and A. Zhao for field and lab assistance, C. Joyner for greenhouse support, and N. Woodard for field site management. This project was supported by NSF-DEB-1258096 (LSA and REI), by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM122062 (LSA, SHM, REI, and SPE), by the National Research Initiative (NRI) Arthropod and Nematode Biology and Management Program of the USDA Cooperative State Research, Education, and Extension Service (CSREES) Grant no. USDA-AFRI 2013–02536 (LSA and REI), and by UMass Amherst Biology Department Summer Research Fellowship and the Summer Scholars program at the UMass Amherst Center for Agriculture, Food and the Environment (LSA and KMM). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation or the official views of the National Institutes of Health. LSA and REI conceived the ideas and designed methodology for the transmission trials; SPE and SHM conceived the comparison of trait-based and species-based models; LSA collected transmission and floral trait data for the observational study; LSA and KMM designed methodology for the manipulative experiment and KMM collected those data; PCS collected chemical data that were ultimately omitted from the manuscript, and contributed related ideas and content; SPE and LSA analysed the data; LSA led the writing of the manuscript with contributions from all authors. All authors contributed critically to the drafts and gave final approval for publication.
Footnotes
Data Availability
Data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.0vm264s
Literature Cited
- Alaux C, Ducloz F, Crauser D, and Le Conte Y. 2010. Diet effects on honeybee immunocompetence. Biology Letters 6:562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MJF, Schmid-Hempel R, and Schmid-Hempel P. 2003. Strong context-dependent virulence in a host-parasite system: reconciling genetic evidence with theory. Journal of Animal Ecology 72:994–1002. [Google Scholar]
- Cisarovsky G, and Schmid-Hempel P. 2014. Combining laboratory and field approaches to investigate the importance of flower nectar in the horizontal transmission of a bumblebee parasite. Entomologia Experimentalis et Applicata 152:209–215. [Google Scholar]
- Colla SR, Otterstatter MC, Gegear RJ, and Thomson JD. 2006. Plight of the bumble bee: Pathogen spillover from commercial to wild populations. Biological Conservation 129:461–467. [Google Scholar]
- Costa J, Castro S, Loureiro J, and Barrett SCH. 2017. Experimental insights on Darwin’s cross-promotion hypothesis in tristylous purple loosestrife (Lythrum salicaria). American Journal of Botany 104:616–626. [DOI] [PubMed] [Google Scholar]
- Cronin J, Welsh M, Dekkers M, Abercrombie S, and Mitchell C. 2010. Host physiological phenotype explains pathogen reservoir potential. Ecology Letters 13:1221–1232. [DOI] [PubMed] [Google Scholar]
- Durrer S, and Schmid-Hempel P. 1994. Shared use of flowers leads to horizontal pathogen transmission. Proceedings of the Royal Society of London Series B 258:299–302. [Google Scholar]
- Fürst MA, McMahon DP, Osborne JL, Paxton RJ, and Brown MJF. 2014. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506:364–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa V, Ravoet J, Brunain M, Smagghe G, Meeus I, Figueroa J, Riano D, and de Graaf DC. 2015. Bee pathogens found in Bombus atratus from Colombia: A case study. Journal of Invertebrate Pathology 129:36–39. [DOI] [PubMed] [Google Scholar]
- Gegear RJ, Otterstatter MC, and Thomson JD. 2006. Bumble-bee foragers infected by a gut parasite have an impaired ability to utilize floral information. Proceedings of the Royal Society B-Biological Sciences 273:1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie S 2010. Factors affecting parasite prevalence among wild bumblebees. Ecological Entomology 35:737–747. [Google Scholar]
- Goulson D 2010. Bumble bees: behaviour, ecology and conservation Oxford University Press, New York, New York. [Google Scholar]
- Goulson D, Nicholls E, Botias C, and Rotheray EL. 2015. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347:1435–1439. [DOI] [PubMed] [Google Scholar]
- Goulson D, O’Connor S, and Park KJ. 2017. The impacts of predators and parasites on wild bumblebee colonies. Ecological Entomology
- Graystock P, Blane EJ, McFrederick QS, Goulson D, and Hughes WOH. 2016. Do managed bees drive parasite spread and emergence in wild bees? International Journal for Parasitology Parasites and Wildlife 5:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graystock P, Goulson D, and Hughes WOH. 2015. Parasites in bloom: flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proceedings of the Royal Society B-Biological Sciences 282:20151371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BA, Schmidt JP, Bowden SE, and Drake JM. 2015. Rodent reservoirs of future zoonotic diseases. Proceedings of the National Academy of Sciences, USA 112:7039–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder LD 1982. Measurement and estimation of functional proboscis length in bumblebees (Hymenoptera: Apidae). Canadian Journal Of Zoology 60:1073–1079. [Google Scholar]
- Koch H, brown MJF, and Stevenson PC. 2017. The role of disease in bee foraging ecology. Current Opinion in Insect Science 21:60–67. [DOI] [PubMed] [Google Scholar]
- McArt SH, Koch H, Irwin RE, and Adler LS. 2014. Arranging the bouquet of disease: floral traits and the transmission of plant and animal pathogens. Ecology Letters 17:624–636. [DOI] [PubMed] [Google Scholar]
- McMahon DP, Furst MA, Caspar J, Theodorou P, Brown MJF, and Paxton RJ. 2015. A sting in the spit: widespread cross-infection of multiple RNA viruses across wild and managed bees. Journal of Animal Ecology 84:615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BR, and Rathcke BJ. 2012. Effects of floral restrictiveness and stigma size on heterospecific pollen receipt in a prairie community. Oecologia 168:449–458. [DOI] [PubMed] [Google Scholar]
- Otterstatter MC, and Thomson JD. 2006. Within-host dynamics of an intestinal pathogen of bumble bees. Parasitology 133:749–761. [DOI] [PubMed] [Google Scholar]
- Paull SH, Song S, McClure KM, Sackett LC, Kilpatrick AM, and Johnson PTJ. 2012. From superspreaders to disease hotspots: linking transmission across hosts and space. Frontiers in Ecology and the Environment 10:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rotheray EL, Osborne JL, and Goulson D. 2017. Quantifying the food requirements and effects of food stress on bumble bee colony development. Journal of Apicultural Research 56:288–299. [Google Scholar]
- Ruiz-Gonzalez MX, and Brown MJF. 2006. Honey bee and bumblebee trypanosomatids: specificity and potential for transmission. Ecological Entomology 31:616–622. [Google Scholar]
- Ruiz-Gonzalez MX, Bryden J, Moret Y, Reber-Funk C, Schmid-Hempel P, and Brown MJF. 2012. Dynamic transmission, host quality, and population structure in a multihost parasite of bumblebees. Evolution 66:3053–3066. [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel R, and Schmid-Hempel P. 1998. Colony performance and immunocompetence of a social insect, Bombus terrestris, in poor and variable environments. Functional Ecology 12:22–30. [Google Scholar]
- Shykoff JA, and Schmid-Hempel P. 1991a. Incidence and effects of four parasites in natural populations of bumble bees in Switzerland Apidologie 22:117–125. [Google Scholar]
- Shykoff JA, and Schmid-Hempel P. 1991b. Parasites delay worker reproduction in bumblebees: Consequences for eusociality. Behavioral Ecology 2:242–248. [Google Scholar]
- Singh R, Levitt AL, Rajotte EG, Holmes EC, Ostiguy N, Vanengelsdorp D, Lipkin WI, Depamphilis CW, Toth AL, and Cox-Foster DL. 2010. RNA viruses in hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. Plos One 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira WM, Huq A, Ahmed QS, and Saeed YA. 1981. Uptake of Vibrio cholerae biotype feltor from contaminated water by water hyacinth (Eichornia crassipes) Applied and Environmental Microbiology 42:550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg RW, Carter C, Powell A, Mittler R, Rizhsky L, and Horner HT. 2003. A major function of the tobacco floral nectary is defense against microbial attack. Plant Systematics and Evolution 238:211–218. [Google Scholar]
- Webb CT, Hoeting JA, Ames GM, Pyne MI, and Poff NL. 2010. A structured and dynamic framework to advance traits-based theory and prediction in ecology. Ecology Letters 13:267–283. [DOI] [PubMed] [Google Scholar]
- Westoby M, and Wright IJ. 2006. Land-plant ecology on the basis of functional traits. Trends in Ecology & Evolution 21:261–268. [DOI] [PubMed] [Google Scholar]
- Wood SN 2006. Generalized additive models: An introduction with R Chapman and Hall/CRC, Boca Raton, FL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


