Abstract
The development of biocompatible nanomaterials has become a new frontier in the detection, treatment and prevention of human amyloid diseases. Here we demonstrated the use of graphene quantum dots (GQDs) as a potent inhibitor against the in vivo aggregation and toxicity of human islet amyloid polypeptide (IAPP), a hallmark of type 2 diabetes. GQDs initiated contact with IAPP through electrostatic and hydrophobic interactions as well as hydrogen bonding, which subsequently drove the peptide fibrillization off-pathway to eliminate the toxic intermediates. Such interactions, probed in vitro by a thioflavin T kinetic assay, fluorescence quenching, circular dichroism spectroscopy, a cell viability assay and in silico by discrete molecular dynamics simulations, translated to a significant recovery of embryonic zebrafish from the damage elicited by IAPP in vivo, as indicated by improved hatching as well as alleviated reactive oxygen species production, abnormality and mortality of the organism. This study points to the potential of using zero-dimensional nanomaterials for in vivo mitigation of a range of amyloidosis.
Keywords: GQDs, IAPP, amyloidosis inhibition, zebrafish, toxicity
Graphical Abstract
Graphene quantum dots are biocompatible zero-dimensional nanostructures, which displayed a potency in rescuing zebrafish embryos from the toxicity of human islet amyloid polypeptide.
Introduction
Protein aggregation into cross-beta amyloid fibrils represents a general phenomenon characteristic to more than 50 human diseases, including neurodegenerative disorders such as Alzheimer’s and Parkinson’s diseases, metabolic disorders such as type 2 diabetes (T2D), as well as cardiac arrhythmias.[1, 2] The sigmoidal fibrillization kinetics of amyloid proteins, from disordered monomers to toxic oligomers (i.e. nucleation), and from oligomers and protofibrils to amyloid fibrils (i.e. elongation and saturation), are consequential to aberrant environmental triggers such as cellular pH, concentrations of physiological metals and chaperones, and the presence of lipid membranes.[3]
Human islet amyloid polypeptide (IAPP) is a 37-residue peptide co-synthesized, co-stored, and co-secreted with insulin in pancreatic beta islets.[4] Aside from its functional role in glycemic control, IAPP is one of the most amyloidogenic peptides known, and its aggregation pathway is triggered by compromised protection of insulin, zinc and C-peptide, insufficient processing of proIAPP (the precursor of IAPP), accumulation of intermediately processed peptides, as well as conversion from disordered monomers to alpha helices and then beta sheets initiated by contact with cell membranes.[5–10] Common strategies against IAPP aggregation and toxicity often involve small molecules (e.g. biomimetics, curcumin, resveratrol and epigallocatechin gallate),[11–13] chaperone proteins (e.g. serum albumin and casein)[14] and nanoparticles (e.g. graphene, gold nanoparticles, dendrimers, star polymers, and carbon nanotubes),[15–20] utilizing their capacities in establishing hydrogen bonding, hydrophobic interaction and π-stacking with the amyloid peptide to disrupt the latter’s amyloid aggregation.
Graphene quantum dots (GQDs) are single- or few-layered graphene sheets of 10 nm or less in size, and are often referred to as zero-dimensional nanomaterials due to their extremely small dimensions.[21] Compared with other carbon-based nanomaterials such as carbon nanotubes, fullerenes and graphene, GQDs possess size-dependent auto-fluorescence originating from quantum confinement and the edge effects.[21] As a result of their physical, chemical and biological properties, GQDs have found a range of applications in drug delivery, biosensing, bioimaging, stem cell technology and photothermal therapy.[22–24] Within the field of amyloidosis, GQDs have been recently employed for the detection of monomeric amyloid-beta, and their efficacies for dose-dependent aggregation inhibition of hen-egg white lysozyme, amyloid beta and alpha synuclein have been explored.[25–27] Inspired by their molecular-like size, minimal toxicity, aromatic structure and amphiphilic properties, here we first demonstrated the use of GQDs as inhibitors against the aggregation and toxicity of IAPP in an embryonic zebrafish model, which has recently been validated as a high-throughput in vivo model for investigating amyloid-induced toxicity.[20, 28–30] Specifically, GQDs were synthesized using the top-down approach via electrochemical oxidation of a graphite electrode. The size, morphology and surface charge of the nanostructures were characterized with transmission electron microscopy (TEM) and a Zetasizer. Thioflavin T (ThT) kinetics, fluorescence quenching and pancreatic β-cell viability assays, as well as circular dichroism (CD) spectroscopy and discrete molecular dynamics (DMD) simulations were performed to probe the binding between IAPP and GQDs. Additionally, embryonic hatching, malformation, mortality and reactive oxygen species (ROS) production were examined, demonstrating the potency of GQDs in mitigating the aggregation and toxicity of IAPP in vivo. Exploiting the capacity of GQDs as a biocompatible nano-inhibitor may prove beneficial to the treatment and prevention of a range of amyloid diseases.
Methods and Materials
Materials
37-residue human islet amyloid polypeptide (IAPP) (KCNTATCATQRLANFLVHSSNNFGAILSSTNVGSNTY; disulfide bridge: 2–7; MW: 3,906; purity>95%) was purchased from AnaSpec, Inc. as lyophilized powder. All chemicals used were acquired from Sigma-Aldrich, if not specified otherwise.
Dynamic light scattering and zeta potentials
The hydrodynamic diameters and zeta potentials of GQDs in Milli-Q water and Holtfreter’s buffer were determined using a Zetasizer Nano-ZS (Malvern Instruments, UK). The measurement was conducted in triplicate using disposable folder capillary cells and analyzed by Zetasizer Software 7.02. Samples were sonicated prior to measurements.
Synthesis of GQDs
GQDs were synthesized following the protocol described in reference 31. Briefly, the GQDs were prepared by a three-electrode system consisting of a graphite working electrode, a platinum foil counter electrode and an Ag/AgCl reference electrode. Ethanol (70 mL), water (16 mL), and NaOH (0.06 g) were mixed to obtain the electrolyte. A potentiostatic voltage of 5 V was applied to the working electrode for 3 h under nitrogen ambient. Excess NaOH precipitates and other impurities were removed by centrifugation. The homogeneous GQD suspension was dialyzed for several days to fully remove the Na ions and kept at room temperature in dark. The concentration of GQDs was obtained by measuring weight loss of the graphite electrode and the final suspension volume. The size distribution of GQDs was measured by ImageJ software using a total of 200 individual particles observed in TEM images.
X-ray photoelectron spectroscopy (XPS)
X-ray photoelectron spectroscopy (XPS) data were obtained with an ESCALab220i-XL electron spectrometer (VG Scientific, West Sussex, UK) using 300 W Al Kα radiation.
Transmission electron microscopy and energy-dispersive X-ray spectroscopy
High-resolution TEM imaging and energy-dispersive X-ray spectroscopy (EDX) elemental analysis were performed on a Tecnai G2 F20 Transmission Electron Microscope (FEI, Eindhoven, The Netherlands) operated at 200 kV. Briefly, 400 mesh copper grids (Formvar film, ProSciTech) were glow discharged and IAPP (25 μM), GQDs (500 μg/mL) or GQDs pre-incubated with IAPP, which were dissolved/suspended in Milli-Q water for 24 h, were placed onto the grids. Negative staining by 1 % uranyl acetate was performed before imaging. Images were recorded using an UltraScan 1000 P 2k CCD camera (Gatan, California, USA) and Gatand Digital Micrograph 3.9.5 software.
Statistical analysis of IAPP fibrils
Fibrils tracking and their statistical analysis were performed by open-access software FiberApp, to measure the morphology and related parameters (contour and persistence length) of IAPP fibrils in the presence and absence of GQDs. Details of the software and methods employed to acquire the values of contour and persistence length (approximately 100 fibrils per sample condition analyzed) were described in previous publications.[32, 33]
Atomic force microscopy
A droplet of 20 μL of GQD suspension was deposited on freshly cleaved mica, incubated for 2 min, rinsed with Milli-Q water and dried with air. Atomic force microscopy (AFM) imaging was performed using a Cypher AFM (Asylum Research) and an AC240TS cantilever (Asylum Research; radius: 7 nm) operating in the tapping mode in air, at a scan rate of 2 Hz.
Fluorescence spectra of GQDs
The excitation and emission spectra from GQD suspension were acquired by a Xe-lamp (Oriel Instrument, 300W) as the tuneable excitation source. A monochromator (Newport, Oriel Cornerstone 130) was used to alter the excitation wavelength. QE65 Pro (Ocean Optics) with spectral correction was used as the detector. For emission spectra scanning, calibration of the spectral irradiance of excitation light was made using a PTI QuantaMaster fluorometer as the reference system.
GQD fluorescence quenching by IAPP
GQDs at 500 μg/mL were pre-incubated with IAPP at progressively increasing concentrations (10, 20, 30, 40 and 50 μM) in a black/clear bottom Costar 96 well plate. The total volume of each sample was kept constant. The fluorescence intensity of GQDs was read after 10 min of incubation using a Clariostar (BMG LABTECH, Germany) microplate reader. The excitation was fixed at 355 nm, while the emission was scanned from 400 to 600 nm. The fluorescence intensities of Milli-Q water and IAPP were used as control. The experiment was done in triplicate to ensure reproducibility.
Thioflavin T (ThT) assay of IAPP w/o GQDs
A thioflavin T (ThT) kinetic assay was used to measure IAPP fibrillization in the presence of GQDs. IAPP was weighed on a microbalance and dissolved in Milli-Q water. In a black/clear bottom Costar 96 well plate, 25 μM aqueous solution of IAPP, ThT fluorescent dye (50 μM) with and without GQDs (250, 375 and 500 μg/mL) were incubated for 13 h at 28.5 °C. ThT fluorescence was recorded with excitation at 440 nm and emission at 485 nm, at a 1 h interval (PerkinElmer, EnSpire 2300). ThT and GQDs alone were measured as controls. ThT fluorescence was normalized based on the fluorescence of IAPP alone and background fluorescence was deducted. The experiment was done in triplicate to ensure reproducibility.
Circular dichroism (CD) spectroscopy
Changes in the secondary structure of IAPP, induced by different concentrations of GQDs, were probed by circular dichroism (CD) spectroscopy. IAPP was incubated with 500, 750 and 1000 μg/mL of GQDs before CD spectra scanning. IAPP dissolved in water and incubated for 0 h and 12 h and GQDs solution alone at 750 μg/mL were used as controls. CD spectra were recorded by Aviv Biomedical Model 410 from 190 to 280 nm, with a step size of 1 nm, a reading time of 4 sec per step, and 3 readings per step at room temperature. IAPP concentration in all samples was adjusted at 50 μM for good signal intensities. The data were analyzed via Dichroweb, and Contin/reference set 4 was used to estimate the percentage secondary structure.[34]
Discrete molecular dynamics (DMD) simulations
The GQD used in the simulations possessed a diameter of ~3.0 nm. The initial IAPP coordinate was taken from the NMR structure solved in detergent micelles at neutral pH (model 1, PDBID: 2L86).[5] In order to investigate the GQD effect on IAPP aggregation, both IAPP monomer and dimer were simulated in the absence or presence of the GQD. For each system, 20 independent simulations were performed, each of which lasted 400 ns at 300 K starting with different initial configurations (i.e., coordinates and velocities). The IAPP peptides and the GQD were initially randomly placed in the simulation box with different orientations and a minimum inter-molecular distance of 1.5 nm. All simulations were performed with all-atom DMD,[35, 36] a molecular dynamics algorithm with enhanced conformational sampling efficiency, widely used for studying protein folding,[37] amyloid aggregation,[38] and protein-nanoparticle interactions.[16] The detailed description of the DMD algorithm can be found in previous studies.[16, 37] The forcefield used in the simulations was based on the Medusa forcefield,[39] which included van der Waals (VDW), solvation, hydrogen bond and electrostatic interactions.
DMD analysis methods
Secondary structure analyses were performed using the dictionary secondary structure of protein (DSSP) method.[40] The two-dimensional (2D) potential of mean force (PMF) was computed as -kBTlnP(Rg, NH-bonds), where P(Rg, NH-bonds) denotes the probability of a conformation having a given value of radius of gyration, Rg, and the total number of backbone hydrogen bonds (Nh-bonds). A backbone hydrogen bond was considered to be formed when the distance between the backbone N and O atoms was within 3.5 Å and the NH--O angle was greater than 150°. Two residues in contact had more than one inter-atomic contact, with a cutoff distance of 0.55 nm between any two heavy atoms.
In vitro viability assay
GQDs dissolved in water/ethanol solution were first distilled to remove ethanol and replaced with Milli-Q water with pH adjusted to 7.4. A black/clear bottom Costar 96 well plate was coated with poly-L-lysine for 30 min at 37 °C and then washed 3× in DPBS to enhance cell adhesion. Pancreatic βTC-6 (purchased from ATCC) beta cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal bovine serum (FBS) prior to experiment. Cells were seeded at a density of ~50,000 per well in 200 μL media and incubated for 48 h at 37 °C and 5% CO2 to reach ~80% confluency. The medium was then refreshed and samples were added into the wells to make up the final tested concentrations (GQDs: 200, 300 and 400 μg/mL; IAPP: 20 μM). After 24 h of treatment, the medium was aspirated and wells were washed 3× in DPBS and then 100 μL pre-diluted (1 in 10 dilution) alamarBlue dye in medium was added to each well under dark. The plate was returned to incubator for 2 h before endpoint fluorescence was read on a Flexstation 3 plate reader (Molecular Devices), with excitation at 544 nm and emission at 590 nm. Percentage cell viability was calculated as relative fluorescence intensity to untreated cells after deduction of background fluorescence. Experiments were performed in triplicate. Statistical analysis was conducted by student’s t-test.
In vivo toxicity assay using zebrafish embryos by microinjection
Wild-type zebrafish circulatory breeding system (Haisheng, Shanghai, China) was maintained at 28.5 °C based on a 14 h:10 h light/dark cycle. Embryos were generated by mating of female and male adult fish (1:1 ratio), of which two genders were separated manually the night before and the spawning was triggered by removing the divider in the following morning. Embryos were collected 0.5 h afterwards, washed with 0.5 ppm methylene blue solution, and then transferred to Holtfreter’s medium in a Petri-dish. Healthy and fertilized embryos were selected for further microinjection under a stereomicroscope (Olympus-SZ61, Olympus Ltd., Japan). GQDs dissolved in water/ethanol solution were first distilled to remove ethanol and replaced with Milli-Q water with pH adjusted to 7. Microinjection was performed with a fixed injection pressure using a pneumatic microinjection system (PV830 Pneumatic Picopump, WPI). 1 nL of each sample (GQDs at 100, 150 or 200 μg/mL and IAPP at 10, 15 or 20 μM, as well as GQDs pre-incubated with IAPP) diluted in Holtfreter’s medium was microinjected into the yolk of the embryos at 1 h post fertilization (hpf). Embryos were examined under a bright-field optical microscope (Olympus-SZ61, Olympus Ltd., Japan) and physically damaged embryos (caused by microinjection) were removed. Embryos were placed on U-bottom transparent 96 well plates (Costar-3599, Corning, US) with one single embryo in each well containing 200 μL of Holtfreter’s medium. The treated and control embryos were incubated at 28.5 °C and the development of embryos was observed over time by the same optical microscope and bright-field images were taken by a built-in camera. The toxicities of IAPP and GQDs induced on embryos were quantified by the hatching and malformation rates (mainly tail deformation and yolk sac edema) at 72 hpf, and mortality rate at 120 hpf. All experiments were performed in triplicate to ensure reproducibility. Statistical analysis was conducted by student’s t-test. All experiments related to zebrafish were carried out in accordance with the Animal Ethics Committee at Tongji University, with the protocol approved by the Animal Center of Tongji University (Protocol #TJLAC-018–020).
In vivo reactive oxygen species (ROS) generation by IAPP w/o GQDs
An ROS assay was performed with the 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA) dye (Purchased from Aladdin, China). Upon cleavage of the acetate groups by esterases and oxidation, the non-fluorescent H2DCFDA was converted to the highly fluorescent 2’,7’-dichlorofluorescein (DCF). Briefly, 1 nL of buffer, containing 10 μM of IAPP with and without GQDs (100, 150 and 200 μg/mL), and 10 μM of DCF dye were microinjected inside the yolk of zebrafish embryos (1 hpf). The embryos were incubated in 200 μL of buffer in a black/clear bottom Costar 96 well plate for 24 h at 28.5 °C and fluorescence was recorded by a microplate reader (Varioskan™ LUX, Thermo Fisher Scientific, US). The fluorescence intensity of DCF at 530 nm, excited at 485 nm, was used to determine the level of ROS generation. DCF fluorescence was normalized based on the control and background fluorescence was deducted. 0.3% H2O2 was used as positive control and Holtfreter’s buffer as negative control. Statistical analysis was conducted by student’s t-test.
In vivo fluorescence imaging of ThT-labelled IAPP w/o GQDs
Embryos or larvae at different developmental stages (0.5, 4, 24, 48 and 72 hpf) were washed, anaesthetized in a 0.01% tricaine solution, and embedded in 1% low-melt agarose for positioning. Fluorescence images of the samples were taken using a FITC filter set (Ex: 488 nm, Em: 540 nm) under a fluorescence microscope (Olympus-SZ2-ILA, Olympus Ltd., Japan). The exposure time was fixed at 150 ms for all samples and the brightness and contrast of images were kept constant. ThT dye was microinjected together with IAPP (2:1 molar ratio) with and without GQDs (150 μg/mL). Images of embryos injected with ThT alone were used as control.
Results and Discussion
X-ray photoelectron spectroscopy (XPS) was used to measure the surface composition of as-prepared GQDs. The high-resolution spectrum exhibited a C 1s peak at 285 eV and an O 1s peak at 532 eV (Figure S1a, Electronic Supplementary Information) and the C 1s peak confirmed the enrichment of hydroxyl, carbonyl and carboxylic acid groups on the surface of GQDs (Figure S1b).[41] TEM images of as-synthesized GQDs revealed spherical and monodispersed nanostructures (Figure S2a) with an average diameter of 3.6 ± 0.3 nm (n = 200) (Figure S2b). The GQDs were further characterized with atomic force microscopy (AFM) and energy-dispersive X-ray spectroscopy (EDX), showing relatively uniform sizes and good purities of the nanomaterial (Figure S2c&d). Blue fluorescence was observed based on scanning the emission spectra of GQDs, with the maximum fluorescence intensity obtained at an excitation wavelength of 355 nm (Figure S2e). The zeta potential and hydrodynamic size of GQDs in Milli-Q water were measured to determine their surface charge and suspendibility. A net negative charge of −20.6 mV and a hydrodynamic diameter of 18.3 nm were obtained for GQDs in aqueous solution (Table 1). In comparison, IAPP carried a net charge of +36.8 mV at neutral pH, according to our previous publication.[33]
Table 1.
Zeta potentials and hydrodynamic sizes of GQDs in Milli-Q water and H buffer.
| Nanomaterials | Zeta potential (mV) | Hydrodynamic size (nm) | PDI |
|---|---|---|---|
| GQDs in Milli-Q water | −20.6 ± 0.4 | 18.3 ± 2.2 | 0.3 ± 0.0 |
| GQDs in H buffer | −10.4 ± 0.4 | 23.3 ± 3.1 | 0.2 ± 0.0 |
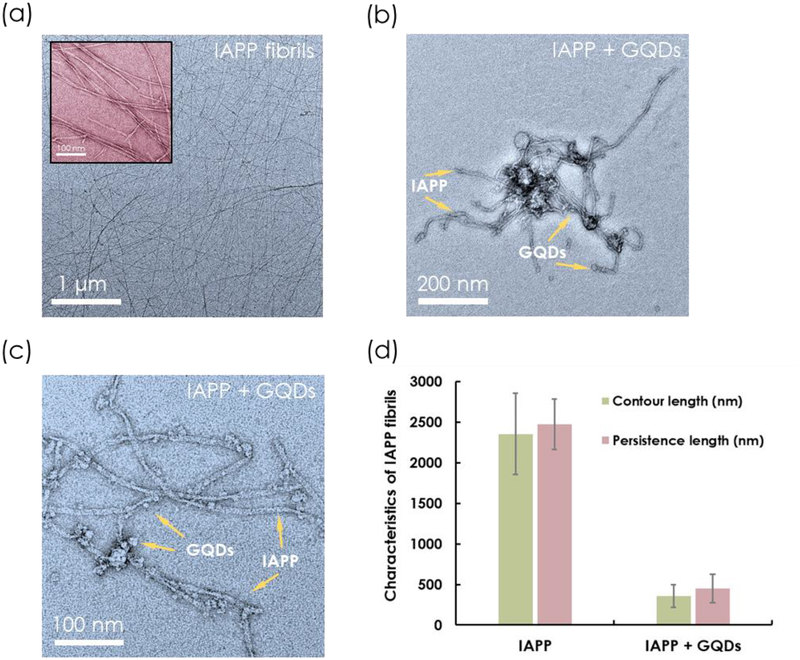
For all experiments involving the interactions between IAPP and GQDs, the weight ratios between the two species were fixed at 1:5, 1:3.75 and 1:2.5, to accommodate the varying instrument resolution used for this study. TEM imaging was utilized for direct visualization of the mesoscopic properties of IAPP fibrillization in the presence and absence of GQDs. For IAPP control, straight and long fibrils were clearly visible after 24 h of incubation, as shown in Figure 1. Statistical analysis by FiberApp[32] revealed an average contour length of 2,355 ± 503 nm and a persistence length of 2,471 ± 310 nm for the IAPP fibrils. To elicit maximal inhibition, GQDs were added during the initial stage of IAPP nucleation when oligomers and protofibrils were actively forming.[42] Here, fresh IAPP monomers were pre-incubated with GQDs, and considerably shorter and softer fibrils than those in the control were visible under TEM after 24 h. An average contour length of 359 ± 141 nm and a persistence length of 447 ± 175 nm for IAPP were obtained, based on FiberApp analysis (Figure 1d). IAPP was bundled in clusters and adsorbed by GQDs (Figure 1b&c). No such phenomenon occurred when IAPP was pre-incubated with large graphene oxide sheets (GO), wherein only the physical adsorption of IAPP on the GO surfaces was observed.[43]
Figure 1.
(a-c) TEM images of IAPP and IAPP pre-incubated with GQDs for 24 h and (d) their respective contour length and persistence length. IAPP concentration: 25 μM. GQD concentration: 500 μg/mL.
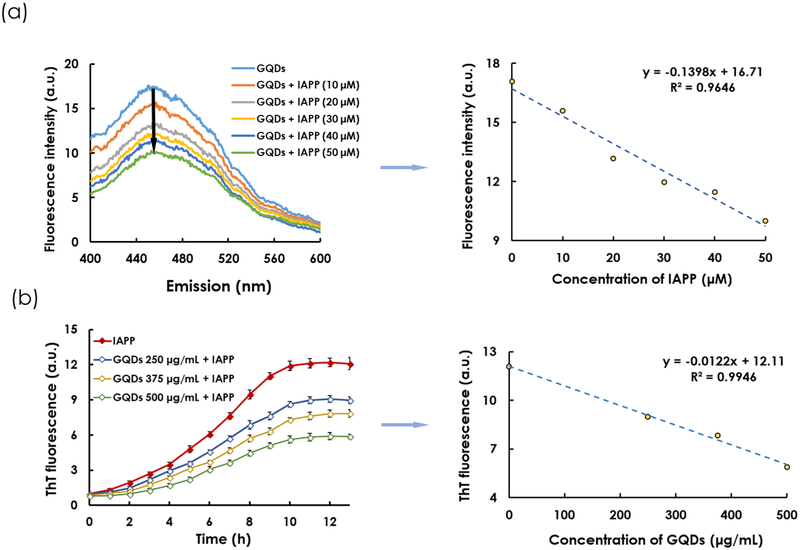
To further evaluate the association between GQDs and IAPP, the fluorescence intensities of GQDs pre-incubated with IAPP at progressively increasing concentrations were quantified. At fixed excitation (355 nm) and GQD concentration (500 μg/mL), the peak emission fluorescence intensity of GQDs at 450 nm decreased linearly (R2 = 0.9646) with increasing IAPP concentrations (Figure 2a; IAPP control in Figure S3), similarly to the observation made for the detection of monomeric amyloid beta by GQDs.[44] This phenomenon can be explained by the intercalation of GQDs into IAPP protofibrils and fibrils during aggregation (Figure 1b&c), and therefore a perturbed local environment of the nanostructure for fluorescence emission, via hydrogen bonding, electrostatic and hydrophobic interactions between the GQDs and the amyloidogenic region of the cationic IAPP.
Figure 2.
(a) Quenching of GQD fluorescence at increasing IAPP concentrations. GQD concentration: 500 μg/mL. Excitation wavelength: 355 nm. Right panel: A linear (R2 = 0.9646) reduction of GQD fluorescence with increased IAPP concentration at emission wavelength of 450 nm. (b) ThT kinetic assay of IAPP fibrillization in the presence and absence of GQDs. IAPP concentration: 25 μM. Right panel: A linear (R2 = 0.9946) reduction of ThT fluorescence in the saturation phase (points selected at 13 h) with increased GQD concentration is evident.
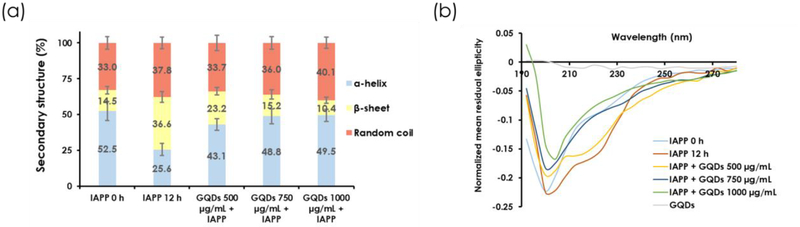
A ThT fluorescence assay was performed to examine how GQDs interfered with the kinetics of IAPP aggregation. ThT is a fluorescence dye which has high affinity for the β sheets of amyloid proteins, including IAPP. As IAPP fibrillated in an aqueous environment, ThT bound to IAPP and its fluorescence intensity excited at 440 nm exhibited a sigmoidal curve to indicate the processes of nucleation, elongation and saturation. Upon addition of GQDs of different concentrations, IAPP fibrillization was drastically suppressed, evidently from a prolonged elongation phase coupled with much reduced ThT fluorescence in the nucleation phase up to the saturation phase (Figure 2b; controls of IAPP, ThT dye and GQDs see Figures S4&S5), with a downward linear relationship between ThT fluorescence intensity and GQD concentration (R2 = 0.9946) observed in the saturation phase. In accordance with the ThT assay, CD spectroscopy and analysis revealed a high proportion of a β-sheet content in IAPP after 12 h. IAPP pre-incubated with GQDs significantly reduced the β-sheet content, shifting that to a α-helix- and random coil-rich conformation (Figure 3a&3b). The β-sheet composition of IAPP decreased with increasing GQD concentration.
Figure 3.
(a) Secondary structure (α-helix, β-sheet and random coil) compositions of IAPP (50 μM) at 0 and 12 h and GQDs (500, 750 and 1000 μg/mL) pre-incubated IAPP at 12 h. (b) CD spectra of IAPP at 0 and 12 h as well as GQDs pre-incubated with IAPP at 12 h.
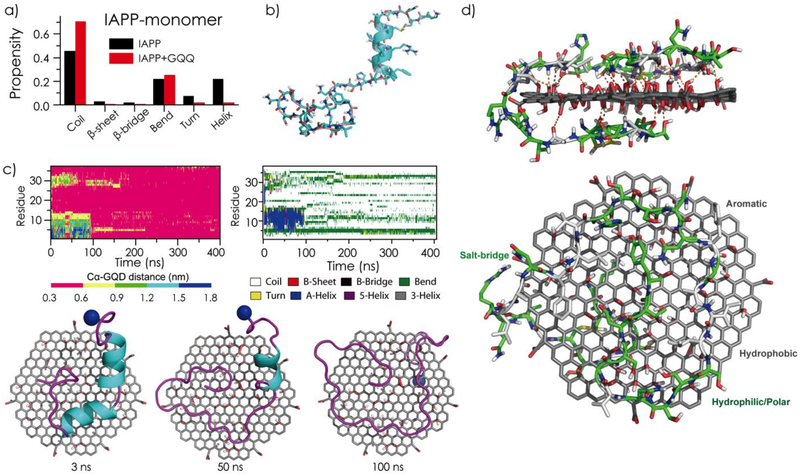
To investigate the inhibition effect of GQDs on IAPP aggregation at the molecular level, all-atom DMD simulations[35] were performed. Specifically, the structure and dynamics of IAPP monomer and dimer were characterized in the absence or presence of GQDs (structure configurations of GQD and IAPP see Figure S6). DMD simulations have been widely used to study protein folding and aggregation[38, 45–47] due to their high computational efficiency and predictive power. An isolated IAPP monomer mainly adopted random coil and helical conformations (Figure 4a–b and Figure S7), where residues 8–15 assumed helices (Figure 4b and Figure S7) as previously observed by solution NMR[48] and 2D IR[49]. A transient β-hairpin-like structure was also observed around residues 16–20 and 24–28, in agreement with previous computational studies.[50, 51] In the presence of a GQD, the IAPP monomer was absorbed onto the nanosheet surface, and the nano-bio interaction rendered the ordered helical and β-sheet structures of IAPP into random coils (Figure S7). As illustrated by snapshots and binding dynamics of a typical simulation trajectory (Figure 4c), the peptide first partially bound to the GQD surface with the N-terminal helix remained intact. Due to thermodynamic fluctuations, the helix underwent transient partial-unfolding (e.g., during the first 100 ns of the particular trajectory), and became completely unfolded and fully absorbed onto the GQD surface with the Cα atoms of all residues located within 6 Å to the nanosheet after 100 ns. As exemplified by an equilibrium structure of the GQD-IAPP monomer complex in Figure 4d, the hydrophilic and hydrophobic residues of IAPP were partitioned onto oxidated and non-oxidated regions of the GQD, respectively. The positively charged residues (e.g. Arg11) in IAPP formed salt-bridges with the carboxyl acids, and polar residues made extensive hydrogen bond interactions with the epoxide, carbonyl and hydroxyl groups of the GQD.[52] Hence, the strong binding between IAPP and GQD was driven by a combination of hydrophobic interactions, aromatic stacking (e.g. Phe15, Phe23 and Tyr37), hydrogen bonds and electrostatics, subsequently reducing ordered protein secondary structures.
Figure 4.
Effects of GQDs on the structure and dynamics of IAPP monomer. (a) Secondary structure contents of IAPP monomer in the absence and presence of a GQD. (b) A representative IAPP monomer solution structure in DMD simulations. (c) Time evolution of each residue’s distance to the GQD (Cα atom was used in calculation) and secondary structure from a typical DMD simulation trajectory. Snapshots of an IAPP-GQD complex at 3, 50, and 100 ns are shown for the given simulation trajectory. (d) A typical GQD-IAPP monomer binding structure, randomly selected from equilibrated DMD simulations, in side (upper) and top (lower) views. The hydrogen bonds between the IAPP monomer and the GQD are shown as orange dots, while the hydrophobic and polar/charged residues are colored in grey and green, respectively.
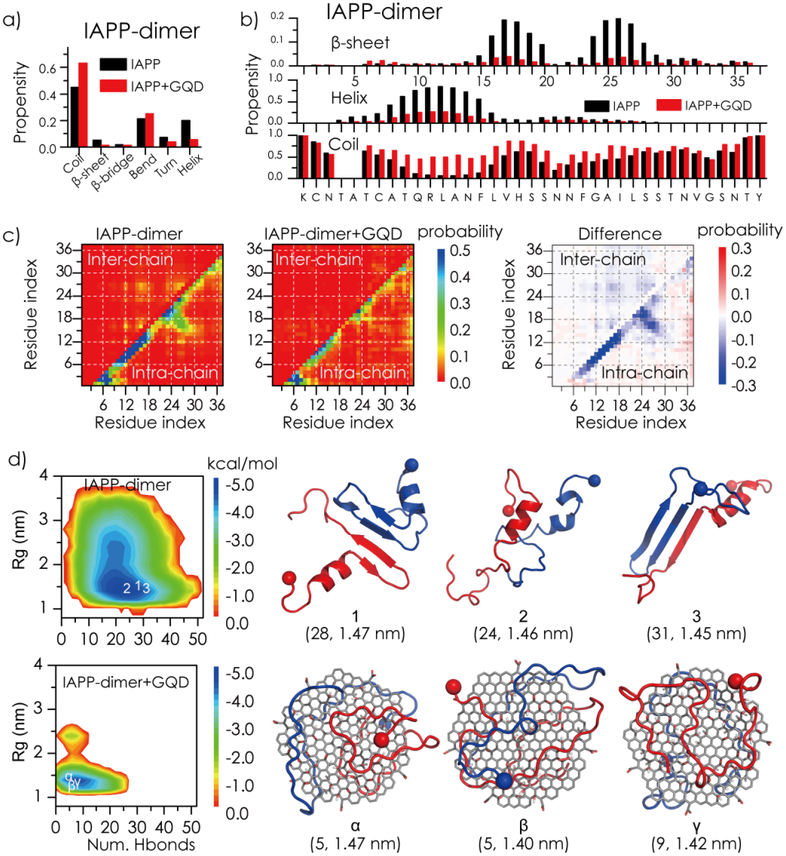
To investigate the effect of GQDs on IAPP self-association, an IAPP dimer in the absence or presence of a GQD was simulated. Compared to IAPP monomer, the β-sheet content of an IAPP dimer in the absence of the GQD was doubled from ~2.5% to ~5.1% (Figure 5a). Similar to the case of an IAPP monomer, binding with GQD effectively induced unfolding of the ordered helix and β-sheet structures in the IAPP dimer. We also computed the residue-wise contact frequency maps for both intra- and inter-peptides (lower and upper diagonals in Figure 5c). The intra-chain contact pattern along the diagonal in the N-termini of the IAPP dimer in the absence of GQD denotes formation of helices (Figure 5a), and the intra-chain contact pattern perpendicular to the diagonal between residues 16–20 and 24–28 reflects formation of a β-hairpin like structure (Figure 5c). Inter-peptide interactions were mainly located around residues 8–18 and 22–28, indicating that these regions were hot-spots for IAPP self-association, consistent with previous experimental[53] and computational studies.[54] In the presence of the GQD, both intra- and inter-chain interactions were significantly reduced (Figure 5c), since IAPP peptides preferred to interact with the GQD instead of with themselves (intra-chain) and each other (inter-chain). As shown by coarse-grained simulations,[55] strong interactions between proteins and nanoparticles inhibited amyloid aggregation.
Figure 5.
Effects of GQDs on the structure and dynamics of an IAPP dimer. Average secondary structure contents (a) as well as secondary structure propensities per residue (b) were computed for the IAPP dimer in the absence and presence of GQD. (c) The intra- and inter-chain contact frequency maps of the IAPP dimer. The changes of IAPP contact frequencies upon binding of GQD are presented. (d) The potential mean force (PMF) of the IAPP dimer (top left) or IAPP dimer + GQD (bottom left) is shown on the left as a function of the total number of hydrogen bonds (Num. Hbond) and radius of gyration (Rg, nm). Three snapshots of the IAPP dimer (top right) or IAPP dimer + GQD (bottom right) near the corresponding free energy basins (the coordinates labelled in the PMF on the left are given in the parentheses on the right).
To probe the conformational properties of IAPP dimer in the absence and presence of the GQD, we computed the potential of mean force (PMF, the effect free energy landscape) as a function of the total number of hydrogen bonds formed among peptide backbones and the radius of gyration (Rg) of the IAPP dimer (Figure 5d). In the absence of the GQD, the PMF of the IAPP dimer featured a single basin with the number of backbone hydrogen bonds featured at 15~25 and Rg at 1.3~1.8 nm. Typical snapshots near the free energy surface basin indicate that the IAPP dimer in the absence of the GQD contained significant helical and β-sheet structures. Previous experimental studies suggested that helical intermediates might be important for IAPP aggregation,[48, 53] while ion mobility mass spectrometry (IMS-MS) combined with molecular simulations showed that an ordered β-hairpin structure was a possible amyloidogenic precursor of IAPP aggregation.[56] Our observation of the co-existence of both helix and β-hairpin conformations in the IAPP dimer was consistent with these seemingly contradicting experiments, and revealed that both types of conformations were present in the IAPP aggregation intermediates. With GQD, the free energy basin of the IAPP dimer became narrower possessing a significantly smaller number of backbone hydrogen bonds at ~5–10, corresponding to IAPP mainly adopting unstructured conformation on the GQD surface (Figure 5d). Taken together, the DMD simulations revealed that strong interactions between the amphiphilic GQD and IAPP rendered unstructured coils and impeded the self-association of IAPP, in agreement with our biophysical characterizations.
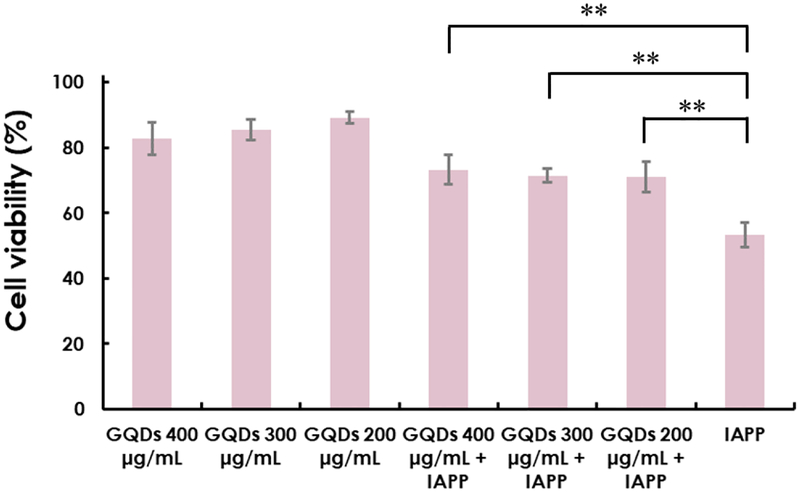
To enable amyloidosis inhibition of GQDs, a viability assay was performed using a βTC 6 pancreatic beta cell line (Figure 6). GQDs at concentrations up to 400 μg/mL elicited a low rate of cell death (>80% cell viability), while at 200 μg/mL, a minimal rate of cell death (~90% cell viability). In comparison, IAPP induced significant cytotoxicity (~50% cell death), and IAPP pre-incubated with GQDs elicited much lower toxicity (<30%) in comparison, indicating the capability of GQDs in mitigating IAPP toxicity in vitro.
Figure 6.
βTC 6 pancreatic beta cell viability determined by alamarBlue after treatment of GQDs (200, 300 and 400 μg/mL) and IAPP alone (20 μM) as well as GQDs pre-incubated IAPP. (ns: P ≥ 0.05, *: P < 0.05 and **: P < 0.01)
Research on amyloidosis mitigation has so far been conducted mostly in vitro or ex vivo in the literature.[1] In this study, the efficacy of GQDs in rescuing the toxicity of IAPP was assessed using an embryonic zebrafish model. The zebrafish model is commonly employed as a facile, high-throughput in vivo model in the fields of nanotoxicology and pharmaceutical sciences,[28, 57, 58] and has recently been validated for amyloidosis research.[20, 59, 60] However, the conventional method of testing the toxicity of materials by direct exposure to zebrafish embryos in buffer imposes limitations for the study of amyloidosis with nanomaterial inhibitors. For example, the protective chorionic membranes of fresh zebrafish embryos can hinder the penetration of large proportion of external nanomaterials, which may render an overestimate of toxic concentrations. In addition, consumption of large quantities of expensive amyloid proteins is required in direct exposure. Here, we employed a microinjection method to circumvent the above drawbacks. Upon microinjection into the yolk, GQDs or IAPP directly interacted with the yolk and diffused into the cells of embryos to render, if any, toxic effects on the development of embryos into larvae.
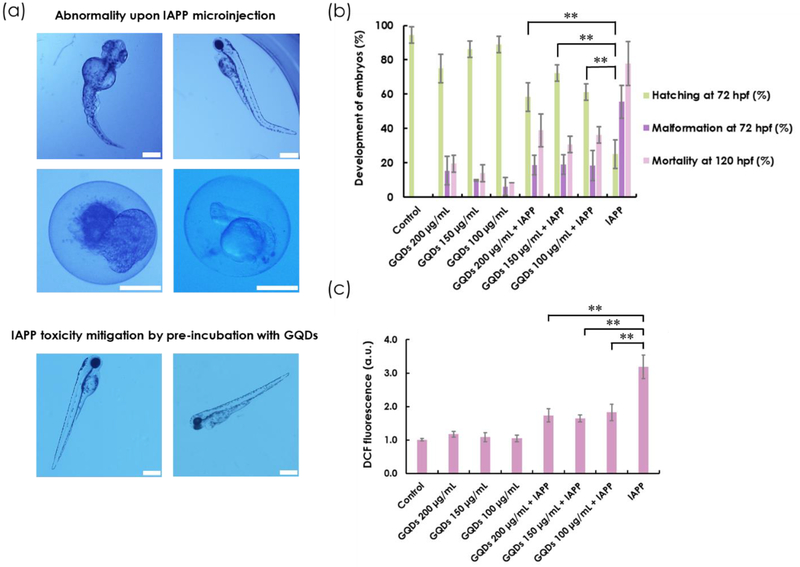
The hatching rate of zebrafish embryos is a major indicator of toxicity. Normally, zebrafish embryos can hatch into larvae by 72 h post fertilization (hpf). The embryos that are hatched later than 72 hpf are considered delayed. Here the development of embryos was observed under a bright-field microscope (Figure S8). The toxic effect of IAPP caused significantly delayed hatching of the embryos even at 10 μM of IAPP at 72 hpf (Figure S9). In addition, low concentrations of GQDs (≤ 150 μg/mL) elicited a minor effect on zebrafish hatching (hatching rate > 80%). Next, IAPP (10 μM) was pre-incubated with GQDs at 200, 150 and 100 μg/mL (weight ratios same as with in vitro experiments), respectively, and it was observed that the hatching rate more than doubled for those pre-incubated with GQDs. Specifically, GQDs at 150 μg/mL resulted in the highest improvement in hatching rate when pre-incubated with IAPP (Figure 7b). Concordantly, the malformation rate of larvae induced by IAPP preincubated with GQDs at 72 hpf was significantly lower in comparison with IAPP alone. Malformation was observed during different stages of embryonic development and images were taken under a bright-field microscope to record the morphology and health status of embryos and larvae. Most embryos failed to hatch upon IAPP microinjection. For those that did develop into larvae, the most common abnormalities were tail deformity and swollen yolk sac (yolk sac edema) caused by the toxicity originated from IAPP fibrillization. Interestingly, GQDs significantly reduced deformity in larvae caused by IAPP (Figure 7a–b). To further characterize the toxicity profiles of IAPP and GQDs, the mortality rate of zebrafish larvae at 120 hpf was determined and the result was consistent with that of hatching and malformation, wherein IAPP toxicity was greatly alleviated by the addition of GQDs (Figure 7b). An ROS assay was performed to measure oxidative stress occurring inside zebrafish embryos. H2DCFDA dye, which can be converted to highly fluorescent DCF upon oxidation, was mixed with IAPP monomers immediately before microinjection into the chorionic fluids of embryos. Consistent with the in vivo toxicity assay, a significant reduction of DCF fluorescence was observed, indicating a considerable decrease of ROS production when IAPP was pre-incubated with GQDs (Figure 7c).
Figure 7.
(a) Bright-field imaging of abnormality upon IAPP microinjection into the yolk of embryos and mitigation of toxicity by pre-incubating IAPP with GQDs. (b) Percentage hatching, malformation and mortality of zebrafish embryos upon microinjection of GQDs (100, 150 and 200 μg/mL) and IAPP (10 μM) alone as well as IAPP pre-incubated with GQDs. Holtfreter’s buffer solution was used as control. (c) ROS generation characterized by DCF fluorescence. (ns: P ≥ 0.05, *: P < 0.05 and **: P < 0.01). Scale bar: 500 μm.
Fluorescence imaging was performed to visualize IAPP fibrillization inside zebrafish embryos and in early developing larvae. ThT was employed as a β-sheet content indicator, and was injected together with IAPP, GQDs or IAPP pre-incubated with GQDs, into the yolk of embryos. After binding onto IAPP during the fibrillization process, ThT fluorescence was observed in the GFP channel at 0.5, 4, 24, 48 and 72 hpf, respectively. The fluorescence of ThT resulting from binding to IAPP fibrils was clearly visible inside the embryos from 24 hpf. Minimal fluorescence was observed when ThT was injected alone or together with GQDs. In addition, when IAPP was pre-incubated with GQDs, ThT fluorescence was not observed under the same settings mainly due to the inhibitory effect of IAPP fibrillization by GQDs, indicating that considerably less fibrils and other β sheet-rich structures were present due to the effective mitigation of IAPP aggregation by the nanostructures (Figure S10).
Conclusion
As-prepared GQDs, as well as GQDs doped with nitrogen and fluorine[25–27, 42] for biological and sensing purposes, have recently found applications in the field of amyloidosis, demonstrating various degrees of efficacy in inhibiting the aggregation and toxicity of functional and pathogenic proteins.[15, 26, 27] For these applications, electrostatic and hydrophobic interactions with the GQDs have been mainly attributed to the prevention or retardation of protein structural conversions from functional monomeric states to toxic intermediates and β-sheet rich amyloid fibrils. In the present study, strong binding between IAPP and the amphiphilic GQDs - governed by the combination of hydrophobic interactions, aromatic stacking, hydrogen bonds and salt-bridges - converted co-existing helix and β-hairpin conformations in the peptides to random coils, impeding their self-assembly as manifested by a reduced number of H-bonds between the peptide backbones, thermodynamic shrinkages of their Rg values, and loss of conformational flexibility with narrower PMF basins. It is interesting to note that strong association between GQDs and IAPP induced both quenching of the nanostructure autofluorescence and reduction of the β-sheet content in the peptide in a dose-dependent manner, while inhibited IAPP toxicity in zebrafish embryos which also contained zebrafish hatching enzyme 1 (ZHE1), glycoprotein subunits, and different forms of metabolites during embryonic development.[61, 62] This GQD-IAPP association in the embryonic environment may be attributed to their high affinity at psychological pH, which prohibited IAPP toxicity in vivo and rescued zebrafish from impaired hatching, malformation, mortality, as well as ROS production elicited by IAPP from the embryonic to the larval stage. This robust inhibiting potential of GQDs against peptide aggregation and toxicity, coupled with their known biocompatibility and small sizes, points to their potential as a nanomedicine for the in vivo mitigation of a range of amyloidosis.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by ARC Project No. CE140100036 (Davis), the Recruitment Program of “1000plan Youth” and Startup Funds from Tongji University (Lin), NSF CBET-1553945 (Ding) and NIH R35GM119691 (Ding). TEM imaging was performed at Bio21 Advanced Microscopy Facility, University of Melbourne.
Footnotes
Electronic Supplementary Information (ESI): Figures S1–S10 on XPS, TEM, AFM, EDX and auto-fluorescence characterizations of GQDs, fluorescence controls, ThT controls, raw CD spectra, GQD and IAPP configurations, monomeric IAPP secondary structure w/o a GQD, examples of microinjection, zebrafish development and IAPP-impaired hatching, as well as fluorescence imaging of ThT-labeled IAPP w/o GQDs.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ke PC; Sani M-A; Ding F; Kakinen A; Javed I; Separovic F; Davis TP; Mezzenga R, Chemical Society Reviews 2017, 46 (21), 6492–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenberg D; Jucker M, Cell 2012, 148 (6), 1188–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowles TPJ; Vendruscolo M; Dobson CM, Nature Reviews Molecular Cell Biology 2014, 15, 384. [DOI] [PubMed] [Google Scholar]
- 4.Zraika S; Hull RL; Verchere CB; Clark A; Potter KJ; Fraser PE; Raleigh DP; Kahn SE, Diabetologia 2010, 53 (6), 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanga RPR; Brender JR; Vivekanandan S; Ramamoorthy A, Biochimica et Biophysica Acta (BBA) - Biomembranes 2011, 1808 (10), 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haataja L; Gurlo T; Huang CJ; Butler PC, Endocrine Reviews 2008, 29 (3), 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brender JR; Hartman K; Nanga RPR; Popovych N; de la Salud Bea R; Vivekanandan S; Marsh ENG; Ramamoorthy A, Journal of the American Chemical Society 2010, 132 (26), 8973–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larson JL; Miranker AD, Journal of Molecular Biology 2004, 335 (1), 221–231. [DOI] [PubMed] [Google Scholar]
- 9.Sciacca Michele F.; Milardi D; Messina Grazia M.; Marletta G; Brender Jeffrey R.; Ramamoorthy A; La Rosa C, Biophysical Journal 2013, 104 (1), 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge X; Kakinen A; Gurzov EN; Yang W; Pang L; Pilkington EH; Govindan-Nedumpully P; Chen P; Separovic F; Davis TP; Ke PC; Ding F, Chemical Communications 2017, 53 (68), 9394–9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abedini A; Meng F; Raleigh DP, Journal of the American Chemical Society 2007, 129 (37), 11300–11301. [DOI] [PubMed] [Google Scholar]
- 12.Yan L-M; Tatarek-Nossol M; Velkova A; Kazantzis A; Kapurniotu A, Proceedings of the National Academy of Sciences of the United States of America 2006, 103 (7), 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng F; Abedini A; Plesner A; Verchere CB; Raleigh DP, Biochemistry 2010, 49 (37), 8127–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorn DC; Meehan S; Sunde M; Rekas A; Gras SL; MacPhee CE; Dobson CM; Wilson MR; Carver JA, Biochemistry 2005, 44 (51), 17027–17036. [DOI] [PubMed] [Google Scholar]
- 15.Mahmoudi M; Akhavan O; Ghavami M; Rezaee F; Ghiasi SMA, Nanoscale 2012, 4 (23), 7322–7325. [DOI] [PubMed] [Google Scholar]
- 16.Javed I; Sun Y; Adamcik J; Wang B; Kakinen A; Pilkington EH; Ding F; Mezzenga R; Davis TP; Ke PC, Biomacromolecules 2017, 18 (12), 4316–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anika G; Bernd A; Jelger RH, Angewandte Chemie International Edition 2016, 55 (37), 11242–11246.27513605 [Google Scholar]
- 18.Pilkington EH; Lai M; Ge X; Stanley WJ; Wang B; Wang M; Kakinen A; Sani M-A; Whittaker MR; Gurzov EN; Ding F; Quinn JF; Davis TP; Ke PC, Biomacromolecules 2017, 18 (12), 4249–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.N. GE; Bo W; H. PE; Pengyu C; Aleksandr K; J. SW; A. LS; G. HE; P. DT; Feng D; Chun KP, Small 2016, 12 (12), 1615–1626.26808649 [Google Scholar]
- 20.Javed I; Yu T; Peng G; Sánchez-Ferrer A; Faridi A; Kakinen A; Zhao M; Mezzenga R; Davis TP; Lin S; Ke PC, Nano Letters 2018, 18 (9), 5797–5804. [DOI] [PubMed] [Google Scholar]
- 21.Iannazzo D; Ziccarelli I; Pistone A, Journal of Materials Chemistry B 2017, 5 (32), 6471–6489. [DOI] [PubMed] [Google Scholar]
- 22.Volarevic V; Paunovic V; Markovic Z; Simovic Markovic B; Misirkic-Marjanovic M; Todorovic-Markovic B; Bojic S; Vucicevic L; Jovanovic S; Arsenijevic N; Holclajtner-Antunovic I; Milosavljevic M; Dramicanin M; Kravic-Stevovic T; Ciric D; Lukic ML; Trajkovic V, ACS Nano 2014, 8 (12), 12098–12109. [DOI] [PubMed] [Google Scholar]
- 23.Markovic ZM; Ristic BZ; Arsikin KM; Klisic DG; Harhaji-Trajkovic LM; Todorovic-Markovic BM; Kepic DP; Kravic-Stevovic TK; Jovanovic SP; Milenkovic MM; Milivojevic DD; Bumbasirevic VZ; Dramicanin MD; Trajkovic VS, Biomaterials 2012, 33 (29), 7084–7092. [DOI] [PubMed] [Google Scholar]
- 24.Ristic BZ; Milenkovic MM; Dakic IR; Todorovic-Markovic BM; Milosavljevic MS; Budimir MD; Paunovic VG; Dramicanin MD; Markovic ZM; Trajkovic VS, Biomaterials 2014, 35 (15), 4428–4435. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y; Xu L-P; Dai W; Dong H; Wen Y; Zhang X, Nanoscale 2015, 7 (45), 19060–19065. [DOI] [PubMed] [Google Scholar]
- 26.Zeng H.-j.; Miao M; Liu Z; Yang R; Qu L.-b., International Journal of Biological Macromolecules 2017, 95, 856–861. [DOI] [PubMed] [Google Scholar]
- 27.Kim D; Yoo JM; Hwang H; Lee J; Lee SH; Yun SP; Park MJ; Lee M; Choi S; Kwon SH; Lee S; Kwon S-H; Kim S; Park YJ; Kinoshita M; Lee Y-H; Shin S; Paik SR; Lee SJ; Lee S; Hong BH; Ko HS, Nature Nanotechnology 2018. DOI 10.1038/s41565-018-0179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howe K; Clark MD; Torroja CF; Torrance J; Berthelot C; Muffato M; Collins JE; Humphray S; McLaren K; Matthews L; McLaren S; Sealy I; Caccamo M; Churcher C; Scott C; Barrett JC; Koch R; Rauch G-J; White S; Chow W; Kilian B; Quintais LT; Guerra-Assunção JA; Zhou Y; Gu Y; Yen J; Vogel J-H; Eyre T; Redmond S; Banerjee R; Chi J; Fu B; Langley E; Maguire SF; Laird GK; Lloyd D; Kenyon E; Donaldson S; Sehra H; Almeida-King J; Loveland J; Trevanion S; Jones M; Quail M; Willey D; Hunt A; Burton J; Sims S; McLay K; Plumb B; Davis J; Clee C; Oliver K; Clark R; Riddle C; Elliott D; Threadgold G; Harden G; Ware D; Begum S; Mortimore B; Kerry G; Heath P; Phillimore B; Tracey A; Corby N; Dunn M; Johnson C; Wood J; Clark S; Pelan S; Griffiths G; Smith M; Glithero R; Howden P; Barker N; Lloyd C; Stevens C; Harley J; Holt K; Panagiotidis G; Lovell J; Beasley H; Henderson C; Gordon D; Auger K; Wright D; Collins J; Raisen C; Dyer L; Leung K; Robertson L; Ambridge K; Leongamornlert D; McGuire S; Gilderthorp R; Griffiths C; Manthravadi D; Nichol S; Barker G; Whitehead S; Kay M; Brown J; Murnane C; Gray E; Humphries M; Sycamore N; Barker D; Saunders D; Wallis J; Babbage A; Hammond S; Mashreghi-Mohammadi M; Barr L; Martin S; Wray P; Ellington A; Matthews N; Ellwood M; Woodmansey R; Clark G; Cooper JD; Tromans A; Grafham D; Skuce C; Pandian R; Andrews R; Harrison E; Kimberley A; Garnett J; Fosker N; Hall R; Garner P; Kelly D; Bird C; Palmer S; Gehring I; Berger A; Dooley CM; Ersan-Ürün Z; Eser C; Geiger H; Geisler M; Karotki L; Kirn A; Konantz J; Konantz M; Oberländer M; Rudolph-Geiger S; Teucke M; Lanz C; Raddatz G; Osoegawa K; Zhu B; Rapp A; Widaa S; Langford C; Yang F; Schuster SC; Carter NP; Harrow J; Ning Z; Herrero J; Searle SMJ; Enright A; Geisler R; Plasterk RHA; Lee C; Westerfield M; de Jong PJ; Zon LI; Postlethwait JH; Nüsslein-Volhard C; Hubbard TJP; Crollius HR; Rogers J; Stemple DL, Nature 2013, 496, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S; Zhao Y; Nel AE; Lin S, Small 2013, 9 (9–10), 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalueff AV; Stewart AM; Gerlai R, Trends in Pharmacological Sciences 2014, 35 (2), 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu M; Xu Y; Niu F; Gooding JJ; Liu J, Analyst 2016, 141 (9), 2657–2664. [DOI] [PubMed] [Google Scholar]
- 32.Usov I; Mezzenga R, Macromolecules 2015, 48 (5), 1269–1280. [Google Scholar]
- 33.Wang M; Kakinen A; Pilkington EH; Davis TP; Ke PC, Biomaterials Science 2017, 5 (3), 485–493. [DOI] [PubMed] [Google Scholar]
- 34.Whitmore L; Wallace BA, Biopolymers 2008, 89 (5), 392–400. [DOI] [PubMed] [Google Scholar]
- 35.Ding F; Tsao D; Nie H; Dokholyan NV, Structure (London, England: 1993) 2008, 16 (7), 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Žganec M; Žerovnik E; Urbanc B, Journal of Chemical Theory and Computation 2015, 11 (5), 2355–2366. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y; Ding F; Nie H; Serohijos AW; Sharma S; Wilcox KC; Yin S; Dokholyan NV, Archives of biochemistry and biophysics 2008, 469 (1), 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y; Ge X; Xing Y; Wang B; Ding F, Scientific Reports 2018, 8 (1), 10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin S; Biedermannova L; Vondrasek J; Dokholyan NV, Journal of chemical information and modeling 2008, 48 (8), 1656–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W; Sander C, Biopolymers 1983, 22 (12), 2577–2637. [DOI] [PubMed] [Google Scholar]
- 41.Li Y; Hu Y; Zhao Y; Shi G; Deng L; Hou Y; Qu L, Advanced Materials 2011, 23 (6), 776–780. [DOI] [PubMed] [Google Scholar]
- 42.Yousaf M; Huang H; Li P; Wang C; Yang Y, ACS Chemical Neuroscience 2017, 8 (6), 1368–1377. [DOI] [PubMed] [Google Scholar]
- 43.Nedumpully-Govindan P; Gurzov EN; Chen P; Pilkington EH; Stanley WJ; Litwak SA; Davis TP; Ke PC; Ding F, Physical Chemistry Chemical Physics 2016, 18 (1), 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H; Li P; Zhang M; Yu Y; Huang Y; Gu H; Wang C; Yang Y, Nanoscale 2017, 9 (16), 5044–5048. [DOI] [PubMed] [Google Scholar]
- 45.Sun Y; Wang B; Ge X; Ding F, Physical Chemistry Chemical Physics 2017, 19 (41), 28414–28423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dokholyan NV; Buldyrev SV; Stanley HE; Shakhnovich EI, Folding and Design 1998, 3 (6), 577–587. [DOI] [PubMed] [Google Scholar]
- 47.Urbanc B; Betnel M; Cruz L; Bitan G; Teplow DB, Journal of the American Chemical Society 2010, 132 (12), 4266–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez Camargo DC; Tripsianes K; Buday K; Franko A; Göbl C; Hartlmüller C; Sarkar R; Aichler M; Mettenleiter G; Schulz M; Böddrich A; Erck C; Martens H; Walch AK; Madl T; Wanker EE; Conrad M; de Angelis MH; Reif B, Scientific Reports 2017, 7, 44041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maj M; Lomont JP; Rich KL; Alperstein AM; Zanni MT, Chemical Science 2018, 9 (2), 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mo Y; Lei J; Sun Y; Zhang Q; Wei G, Scientific Reports 2016, 6, 33076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu C; Shea J-E, PLOS Computational Biology 2013, 9 (8), e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stankovich S; Dikin DA; Piner RD; Kohlhaas KA; Kleinhammes A; Jia Y; Wu Y; Nguyen ST; Ruoff RS, Carbon 2007, 45 (7), 1558–1565. [Google Scholar]
- 53.Andreetto E; Yan L-M; Tatarek-Nossol M; Velkova A; Frank R; Kapurniotu A, Angewandte Chemie International Edition 2010, 49 (17), 3081–3085. [DOI] [PubMed] [Google Scholar]
- 54.Ge X; Yang Y; Sun Y; Cao W; Ding F, ACS Chemical Neuroscience 2018, 9 (5), 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radic S; Davis TP; Ke PC; Ding F, RSC advances 2015, 5 (127), 105498–105498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupuis NF; Wu C; Shea J-E; Bowers MT, Journal of the American Chemical Society 2009, 131 (51), 18283–18292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzo LY; Golombek SK; Mertens ME; Pan Y; Laaf D; Broda J; Jayapaul J; Möckel D; Subr V; Hennink WE; Storm G; Simon U; Jahnen-Dechent W; Kiessling F; Lammers T, Journal of Materials Chemistry B 2013, 1 (32), 3918–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleming A; Alderton WK, Drug Discovery Today: Disease Models 2013, 10 (1), e43–e50. [Google Scholar]
- 59.Fontana BD; Mezzomo NJ; Kalueff AV; Rosemberg DB, Experimental Neurology 2018, 299, 157–171. [DOI] [PubMed] [Google Scholar]
- 60.Stewart AM; Braubach O; Spitsbergen J; Gerlai R; Kalueff AV, Trends in Neurosciences 2014, 37 (5), 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okada A; Sano K; Nagata K; Yasumasu S; Ohtsuka J; Yamamura A; Kubota K; Iuchi I; Tanokura M, Journal of Molecular Biology 2010, 402 (5), 865–878. [DOI] [PubMed] [Google Scholar]
- 62.Lin S; Zhao Y; Ji Z; Ear J; Chang CH; Zhang H; Low-Kam C; Yamada K; Meng H; Wang X; Liu R; Pokhrel S; Mädler L; Damoiseaux R; Xia T; Godwin HA; Lin S; Nel AE, Small 2013, 9 (9‐10), 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.