Abstract
Cationic intrinsically disordered antimicrobial peptides (CIDAMPs) belong to a novel class of epithelial peptide antibiotics with microbicidal activity against various pathogens, including Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus and Candida albicans. Here we show that treatment of distinct bacteria with different hornerin (HRNR)-derived CIDAMPs cause formation of unique cytoplasmic protein aggregates, suggesting a common intracellular mode of action. We further found that, unlike most amphipathic antimicrobial peptides, HRNR traverses bacterial membranes energy-dependently and accumulates within the cytoplasm. Strikingly, certain structurally different, HRNR-based CIDAMPs were found to bind to an identical panel of distinct bacterial ribosomal proteins, thereby manifesting features of several known classes of antibiotics. This may cause the formation of aberrant proteins and toxic protein aggregates in HRNR-treated pathogens which eventually may induce its death. Our study reveals evidence that structurally distinct CIDAMPs of an abundant body surface protein simultaneously target multiple sites of the bacterial protein synthesis machinery.
Introduction
By unknown reasons, healthy human skin is remarkably resistant towards infection by Pseudomonas (P.) aeruginosa, an environmental opportunistic pathogen widespread in water and soil. In an effort to get insight into this unusual natural resistance, we recently identified in heel stratum corneum-extracts peptide fragments of the epidermal intrinsically disordered S100-fused-type 254 kDa protein hornerin (HRNR) as potent, at skin-relevant environmental conditions P. aeruginosa-cidal antimicrobial peptides (AMPs) (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision). HRNR is highly expressed in healthy skin1 where it can form nanofiber scaffolds in stratum corneum2. It consists of 95% of glycine- and serine-rich highly cationic repeat domains and it is present in skin as a complex mixture of multimeric polypeptide fragments1,3. These repeat domains were identified as linked series of “cationic intrinsically disordered antimicrobial peptides, CIDAMPs”, microbicidal peptides rich in disorder-promoting polar amino acids like Gly/Ser/Thr/Gln/His and low in order-promoting, hydrophobic AA like Leu/Ile/Val/Asp/Tyr/Phe/Trp (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision). Strikingly, quantitative proteome analyses identified HRNR as highly abundant protein in humans4. Here, mainly epithelial cell types revealed highest HRNR-abundance in barrier organs (kidney, skin, lung, vulva, colon, rectum, urinary bladder, uterine cervix, placenta). Further, immune privileged organs and organs where vital structures need to be protected from the potentially damaging effects of an inflammatory immune response (eye, brain, central nervous system, female gonads, teeth as well as the heart) revealed a similar HRNR abundance4. This suggests that HRNR-derived CIDAMPs are important innate defense effector peptides, acting at the outermost surface of healthy barrier organs as disinfectants, possibly helping to keep the surface of skin and mucosa free of infection by commensals as well as environmental microbes (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision).
A panel of recombinantly and by chemical synthesis generated HRNR-polypeptide-fragments revealed to be microbicidal AMPs with higher potency than most amphipathic AMPs, targeting P. aeruginosa, Escherichia (E.) coli, Candida (C.) albicans and, to a lesser degree, also Staphylococcus (S.) aureus (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision). It is therefore aimed to explore, why and how HRNR-derived CIDAMPs are able to exert its potent microbicidal activity.
Ultrastructural Analyses Reveal a Unique Morphology of CIDAMP-Treated Micro-organisms
Most known antimicrobial proteins and peptides possess diverse secondary structures with an amphipathic surface in hydrophobic environments5. These AMPs are largely targeting the bacterial membrane, but can have also multiple other modes of action that differ from those of conventional antibiotics5. Disturbance of bacterial membrane integrity can directly or indirectly cause metabolic dysfunction and cell death, besides pore formation per se6.
Observing alterations in bacterial membrane integrity by transmission electron microscopy (TEM) can give first hints on the detailed mechanisms of cell death at lethal AMP concentrations.
Having demonstrated that HRNR-polypeptide-fragments are bactericidal antimicrobials, killing also the yeast C. albicans, we wondered whether these peptides actively destroy microbial cells by direct membrane effects or whether they rather act indirectly via an intracellular target. To achieve this, selected strains of E. coli, P. aeruginosa, S. aureus and C. albicans were challenged with distinct HRNR-derived CIDAMPs, rHRNR2591–2684, rSUMO3-HRNR2591–2684, HRNR1132–1143 (HR2-8), HRNR2606–2628 (HR1-11) and HRNR2656–2677 (HR1-18), respectively, and then imaged by TEM.
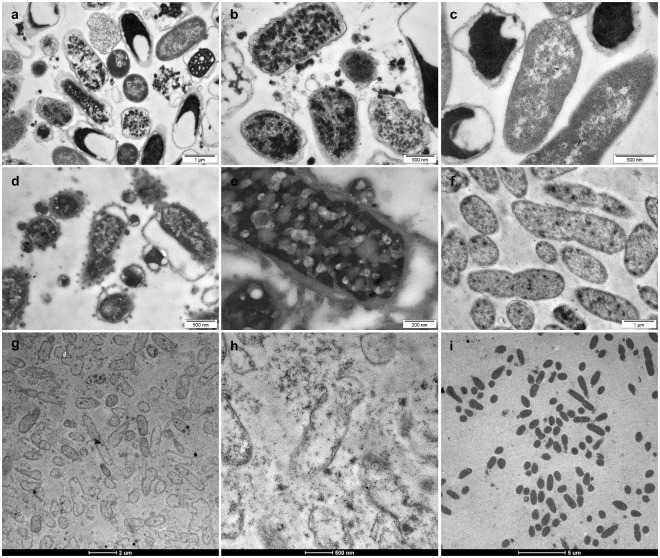
Treatment of E. coli ATCC 11775 with rHRNR2591–2684 for 2 h caused condensation of electron-dense cytoplasmic material, forming large aggregates, and in some cells apparent cytological lysis with liberation of electron dense material upon treatment at pH 5.5, in 10 mM Na- phosphate (NaP) (Fig. 1a). Incubation of control E. coli ATCC 11775 cells at pH 5.5, in 10 mM NaP, revealed in some bacteria an increased electron density of the cytosol. Here the periplasmic space of many cells looked hyperhydrated, very similar as previously reported7, but the inner and outer membranes remained intact (Fig. 1b). When P. aeruginosa ATCC 10145 was exposed to rHRNR2591–2684 at identical conditions, condensation of electron-dense cytoplasmic material and blebs of the outer membrane with an occasional blebbing were seen (Fig. 1c,d), showing evidence of inner membrane breakage. In the controls, P. aeruginosa cells did not show the signs of hyperhydration (Fig. 1e,h) seen in E. coli cells (Fig. 1b). Exposure of P. aeruginosa to rSUMO3-HRNR2591–2684 resulted in widespread peeling of the outer membrane. Many cells underwent extensive lysis and, as a result, lost most of the cytoplasmic electron-dense material. Nearly all cells exposed to this CIDAMP became ghost cells with complete extraction of cytoplasmic contents (Fig. 1f,g). Short HRNR-peptides like the duodecapeptide HRNR1132–1143 (GSGSRQSPSYGR) - the only P. aeruginosa-cidal CIDAMP with a positive net charge of +2 - and those of distinct HRNR-repeat domains like HR1-11 (HRNR2606–2628), HR2-11 (HRNR1132–1157) and HR1-18 (HRNR2656–2677), caused morphological changes (Supplementary Figs S1 and S2) very similar as seen for the long HRNR fragments (Fig. 1a,c,d). We then studied the kinetics of CIDAMP-dependent ultrastructural changes. Exposure of E. coli ATCC 11775 to rSUMO3-HRNR2591–2684 for 5 min caused a few blebs of the outer membrane, which was more prominent after 20 min exposure, showing morphological evidence of a bacterial stress response. Leakage with liberation of electron dense cytoplasmic material was not observed (Supplementary Fig. S3). Interestingly, at higher magnification nanofiber-like structures upon rSUMO3-HRNR2591–2684-treatment of E. coli were seen (Supplementary Fig. S3b). Since disordered proteins are prone to form nanofibrils and amyloid-like structures8 we surmised that rSUMO3-HRNR2591–2684 may form nanofibers. To test this, rSUMO3-HRNR2591–2684 was treated with ultrasound and then analyzed for amyloid-formation, confirming our hypothesis (Supplementary Fig. S4). Thus, nanostructures seen in samples of rSUMO3-HRNR2591–2684-treated E. coli (Fig. 1a and Supplementary Fig. S4) might have originated from rSUMO3-HRNR2591–2684, an observation supported by scaffolds of HRNR-nanofibers seen in vivo in the eye lid2.
Figure 1.
TEM analyses of HRNR-treated E. coli and P. aeruginosa. (a,b) Transmission electron microscopy (TEM) analyses of 6.25 × 107/ml E. coli ATCC 11775 in 10 mM NaP, pH 5.5, 1 h treatment with 312.5 µg/ml rHRNR2591–2684. (c) E. coli control. Note the absence of membrane perturbation and the presence of intracytoplasmic electron-dense aggregates in rHRNR2591–2684-treated bacteria (a,b). The hyperhydrated looking periplasmic space of many cells in the control (a), is similar as seen for E. coli treated with low ion strength and acidic buffers7. TEM of 6.25 × 107/ml P. aeruginosa ATCC 10145, in 10 mM NaP, pH 5.5, 1 h treatment with 312.5 µg/ml rHRNR2591–2684 (d,e). (f) P. aeruginosa control. Note condensation of electron-dense cytoplasmic material and blebs of the outer membrane with an occasional ballooning (d,e). 1 h treatment of 6.25 × 107/ml P. aeruginosa ATCC 10145 with 469 µg/ml rSUMO3-HRNR2591–2684 in 10 mM NaP, pH 5.5 revealed widespread peeling of the outer membrane (g,h). (i) P. aeruginosa control. Images are representative of two independent experiments, sampling on average 10 images per condition and species in each experiment.
Exposure of S. aureus towards HRNR2591–2684, at pH 5.5, elicited small blebs and - as seen in P. aeruginosa and E. coli - condensation of electron dense cytoplasmic material (Fig. 2). HRNR2591–2684 caused S. aureus aggregation, similar as observed in E. coli (Supplementary Fig. S3). Aggregated cells are connected via electron-dense contacts (Fig. 2d) - resembling features seen for the sweet water polyp Hydra vulgaris-derived antimicrobial peptide hydramacin-1 upon exposure towards E. coli9.
Figure 2.
Ultrastructural analyses of rHRNR2591–2684-treated S. aureus. TEM analyses of 6.25 × 107/ml S. aureus ATCC 6538, in 10 mM NaP, pH 5.5, treated with 312.5 µg/ml rHRNR2591–2684 for 2 h. Note the condensation of electron-dense cytoplasmic material and formation of membrane blebs (a–c). Occasionally ballooning (c) and aggregated cells (d), connected via electron-dense contacts, were found upon rHRNR2591–2684-treatment. (e,f) Control. Images are representative of two independent experiments, sampling on average 10 images.
Also treatment of the yeast C. albicans with rHRNR2591HRNR2591–2684 led to characteristic ultrastructural patterns with the release of electron-dense membrane vesicles and changes mainly of the nucleus, cytoplasmic structures and condensation and alteration of the chromatin (Fig. 3). Chromatin margination and condensation along the nucleus and blebs from the nucleus are hallmark ultrastructural signs of apoptosis in fungi10, indicating that rHRNR2591–2684 might kill C. albicans similar as AMPs like lactoferrin, human ß-defensins or plant defensins by apoptosis-like cell death11.
Figure 3.
Ultrastructural analyses of HRNR-treated C. albicans. TEM analyses of 6.25 × 107/ml C. albicans ATCC 244433, treated for 2 h with 312.5 µg/ml rHRNR2591–2684 in 10 mM NaP, pH 5.5 (a–d). (e,f) Control. Note the release of electron dense vesicles (a–c) and marked changes of the intracytoplasmic morphology. Images are representative of two independent experiments, sampling on average 10 images in each experiment.
In summary, ultrastructural analyses of rHRNR2591–2684-treated bacteria like P. aeruginosa, E. coli and S. aureus reveal for various different HRNR-fragments no signs of immediate membrane alteration – typical features of pore-forming amphipathic antimicrobial peptides5. Instead a unique cytosolic aggregation of electron-dense particles, indicative for protein misfolding or assembly of misfolded polypeptides into cytotoxic aggregates12, was observed. In contrast, rHRNR2591–2684 causes in C. albicans ultrastructural changes reminiscent to fungal apoptosis-like programmed cell death.
The Uptake of HRNR-Derived CIDAMPs is an Active Process
Although the action principle of the majority of yet known amphipathic AMPs is based on membrane permeabilization5,13, there is ample evidence now that some, or even most of these AMPs affect microbial viability also by other mechanisms, in addition or alternative to their membrane-permeabilizing/disrupting properties5,14.
The main characteristics of amphipathic AMPs for high binding and selectivity toward microbial membranes are the amino acid composition and sequence15. These determine the physicochemical properties of the peptide in respect to charge, amphipathicity, hydrophobicity, flexibility and H-bonding capacity as key factors for their mode of action and selectivity toward microbial cells16. The antimicrobial mechanism of classical amphipathic and cationic AMPs mostly relates to targeting the microbial cytoplasmic membrane by creating transmembrane pores or channels that cause leakage of intracellular molecules, which finally leads to cell death17,18. For AMPs having intracellular targets, the precise mechanism how bacterial cells are entered is not clear. Due to their small size, AMPs seem to diffuse rapidly inside and outside of the cell membrane19.
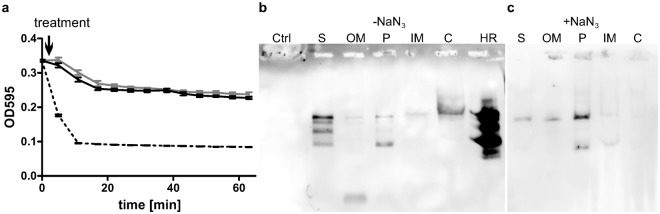
To get insight into the mechanism how HRNR-based CIDAMPs may traverse bacterial membranes, we analyzed the permeabilizing properties of rHRNR2591–2684 and its partition in subcellular compartments in P. aeruginosa PAO1. Confirming our TEM data, no membrane permeabilizing properties of this CIDAMP were found (Fig. 4a). Using fractionation techniques, we detected immunoreactive rHRNR2591–2684 in treated bacteria within the cytosol (Fig. 4b) upon Western blot analyses. Interestingly, pretreatment of the bacteria with the respiratory chain blocker sodium azide caused accumulation of rHRNR2591–2684 mainly in the outer membrane and periplasmic fraction (Fig. 4c) - indications for a passive interaction of this CIDAMP with the bacterial envelope and an active uptake mechanism of rHRNR2591–2684 into bacterial cytosol. Intriguingly, this observation appears to be similar to the uptake of certain colicin bacteriocins into E. coli. Here, an intrinsically disordered N-terminal domain facilitates the translocation across the outer membrane and is involved in an energy-dependent, TonB-mediated uptake20.
Figure 4.
rHRNR2591–2684 is a non-permeabilizing, energy-dependently translocating CIDAMP. (a) rHRNR2591–2684 does not permeabilize the bacterial membrane. Lysis by 50 μg/mL lysozyme (expressed as OD595 against time ± s. e. m., n = 3) of chloramphenicol-treated P. aeruginosa PAO1 cells (gray line) in the presence of polymyxin B (PMB, 10 μg/mL; dotted line) or rHRNR2591–2684 (5 μg/mL; black line). (b) rHRNR2591–2684 is translocated into bacterial cytosol. HRNR-Western blot of fractionated, rhHRNR2591–2684-treated PAO1, Ctrl: control (untreated PAO1). S: sample supernatant, OM: outer membrane-, P: periplasmic-, IM: inner membrane-, C: cytoplasmic fraction, HR: Hornerin fragment rHRNR2591–2684. (c) rHRNR2591–2684 translocation is energy-dependent. PAO1 was treated with rHRNR2591–2684 in the presence of NaN3, fractionated and analyzed by HRNR-Western blot.
There are three proposed mechanisms by which cationic AMPs enter the cytoplasm of microbes: A spontaneous lipid-assisted translocation, a stereospecific receptor-mediated membrane translocation21 and a spontaneous translocation without pore formation. The main mechanism is most likely a spontaneous translocation with pore formation, as contemplated by the Shai-Matsuzaki-Huang model13. A second mechanism, based on translocation of the inner membrane peptide transporter SbmA, has been reported for several Pro-rich AMPs22,23. Uptake of the Pro-rich cathelicidin Bac7, which has an extended secondary structure14, occurs in a stereospecificity-dependent manner since the all-D-enantiomers were inactive as antimicrobials and were excluded from uptake into bacterial cells24. Since all-(D)-CIDAMPs we had investigated are similarly active as its all-(L)-enantiomers (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision), a stereospecific cellular uptake of CIDAMPs seems to be less likely.
HRNR-Derived CIDAMPs are Targeting Bacterial Ribosomal Proteins
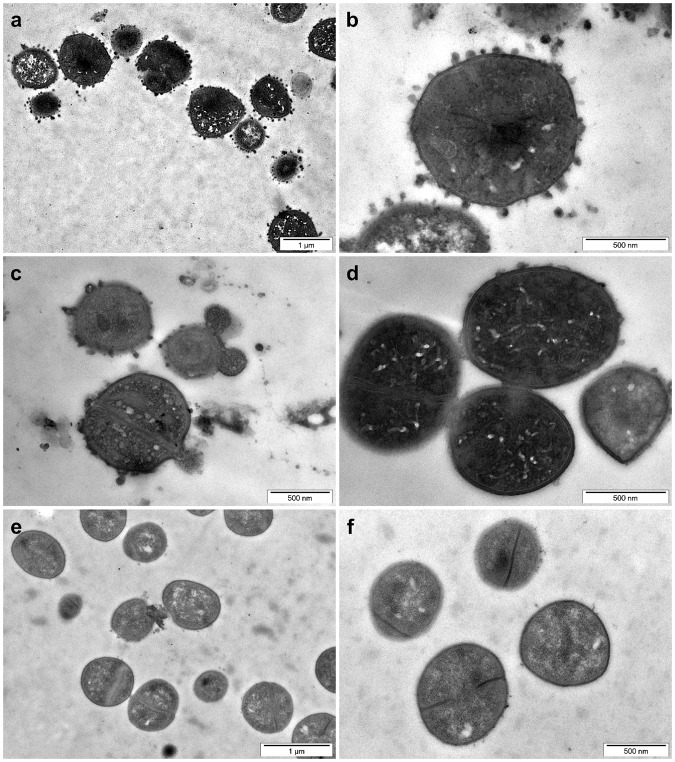
Ultrastructural investigations of distinct bacteria treated with different HRNR-based CIDAMPs revealed electron dense cytoplasmic aggregates as a common characteristic (Fig. 4), assuming a unique killing mechanism for HRNR-derived CIDAMPs. To identify the intracellular target of rHRNR2591–2684, we performed immunogold-electron microscopy with rHRNR2591–2684-treated P. aeruginosa ATCC 10145 using post-embedding immunogold-staining techniques with an affinity-purified, rHRNR2591–2684-directed polyclonal antibody1 to localize cellular CIDAMP-binding sites. Exposure of P. aeruginosa ATCC 10145 to rHRNR2591–2684 caused gold particle accumulation at electron-dense aggregates within the cytoplasm, but not at the membrane, nourishing the hypothesis that rHRNR2591–2684 possibly binds to bacterial ribosomes (Fig. 5) and contrasting to the membrane interactions of Pro-rich AMPs, where the peptides localize uniformly around the E. coli membrane25. To delineate the mechanisms of the antimicrobial activity of CIDAMPs further, and following the hypothesis that bacterial ribosomes are the target of CIDAMPs, E. coli ribosomes were separated by SulfoLink®– coupling resin-chromatography, a method allowing its efficient isolation from bacterial lysates without harsh conditions and is rapidly enough to limit degradation, resulting in highly active ribosomes26. To explore whether selected CIDAMPs bind to ribosomal proteins, we analyzed SulfoLink®–chromatography fractions by a Far-Western blot analysis27, where protein-coated nitrocellulose membranes were incubated with rSUMO3-HRNR2591–2684 as “bait” protein and HRNR2591–2684-antibodies for visualization of HRNR-ribosomal protein interaction. Far-Western blot analyses of SulfoLink®–column-bound E. coli-proteins revealed multiple proteins interacting with HRNR2591–2684 (Supplementary Fig. S5).
Figure 5.
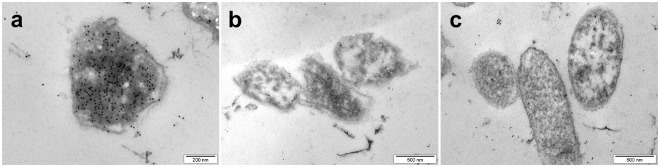
HRNR binds to intracytoplasmic aggregates in P. aeruginosa. P. aeruginosa ATCC 10145 was treated for 5 min with rHRNR2591–2684 (45 µg/ml 10 mM NaP, pH 5.5), and then cellular localization of this CIDAMP was analyzed by immunocytochemistry with a HRNR2591–2684-specific polyclonal antibody, followed by incubation with a gold-conjugated secondary antibody (a). Bacteria, treated with rHRNR2591–2684 (b) or buffer (c), followed by incubation with the gold-conjugated secondary antibody, served as controls. Note accumulation of intracytoplasmic immuno-gold (a), which is corresponding to electron dense cytoplasmic aggregates seen upon TEM-analyses of CIDAMP-treated P. aeruginosa (Fig. 1c,d). Images are representative of two independent experiments, sampling on average 10 images in each experiment.
56 ribosomal protein subunits, which to a high extent are posttranslationally modified28, have been identified in E. coli K1228, and its biochemical separation is a technically challenging task. Direct ion exchange HPLC allows the purification of only nine proteins29. RP-HPLC, however, was successfully utilized for separation of E. coli 30S ribosomal proteins and was shown to yield much higher numbers and greater resolution of these proteins than other HPLC-methods30 do. Also 50S ribosomal proteins could be separated by RP-HPLC31. Therefore we used RP-HPLC methods to yield both, E. coli 30S- and 50S-ribosomal subunits. Due to a limited solubility of distinct ribosomal proteins in different organic solvents, it was found to be impossible to separate all 50S and 30S ribosomal subunits at a single RP-HPLC-column and a single organic solvent as eluent31. We identified two different RP-HPLC columns (Jupiter® C18 widepore RP-HPLC column and an Aeris® C18 widepore RP-HPLC column) and gradients of two different eluents (either acetonitrile (ACN) in aqueous 0.1% trifluoroacetic acid (TFA) or 2-propanol (Prp) in aqueous 0.1% TFA) to be useful in this study.
Aliquots of each HPLC-fraction were applied either to a nitrocellulose membrane for a modified Far-dot blot analysis or to an SDS-PAGE gel for a Far-Western blot analysis27. Here, protein-coated nitrocellulose membranes were incubated with rHRNR2591–2684, biotinylated rSUMO3-HRNR2591–2684, biotinylated rHRNR2591–2684, biotinylated HR1-18, or rHRNR1075–1172. Thereafter, Strep-Tactin® or polyclonal antibodies1 against HRNR2591–2684 and HRNR1075–1172, respectively, together with biotinylated secondary antibodies, were used for visualization of HRNR-ribosomal protein-interaction.
Results indicate a specific binding of the recombinant biotinylated HRNR-derived CIDAMP rHRNR2591–2684 or its SUMO3-fusion protein to a panel of ribosomal proteins. We identified the E. coli 50S ribosomal proteins L2, L18, L22, L28 as well as the E. coli 30S ribosomal proteins S11, S13, S18, S19 and S20 when proteins of HPLC fractions were sequenced (Table 1, Supplementary Figs S6–10). Using a more sensitive LC-MS/MS analyses of HRNR-interacting ribosomal proteins, we identified, apart from the E. coli 50S ribosomal proteins L2, L18, L22, L28 and the 30S ribosomal proteins S11, S18, S19 and S20, in addition the 30S ribosomal proteins S3, S4, S6, S10, S12 as well as the 50S proteins L13 and L14 in HRNR-active HPLC fractions (Table 1, Supplementary Table S1, Figs S10–S12). Thus, the investigated CIDAMPs, rHRNR2591–2684 and biotinylated rSUMO3-HRNR2591–2684, may bind to a panel of distinct ribosomal protein subunits of E. coli (Table 1), each having unique affinity to HRNR polypeptides. Since resolution of SDS-PAGE analyses and RP-HPLC is not high enough for a complete separation of all different subunits, including truncated and posttranslationally modified forms, further studies are necessary to identify all HRNR-binding E. coli-ribosomal proteins and its specific affinity to distinct HRNR peptides and possibly other CIDAMPs.
Table 1.
E. coli ribosomal proteins interacting with Hornerin*.
| 30S ribosomal proteins: S3, S4, S5, S6, S7, S10, S11, S12, S13, S18, S19, S20, S21 |
| 50S ribosomal proteins: L1, L2, L3, L4, L5, L6, L9, L13, L14, L16, L18, L19, L21, L22, L23, L28, L31 |
*Ribosomal proteins known to be phosphorylated62 are in italic, and those found at apparent relative high abundance in RP-HPLC-fractions (estimated via total ion current estimation upon ESI-MS analyses) are shown in bold.
One of the most prominent Far-Western blot bands of HRNR-binding ribosomal proteins corresponded to a 37 kDa protein (Supplementary Figs S5 and S6). Sequence analyses revealed the E. coli ribosomal 50S protein L2, a 29,729 Da protein28 migrating upon SDS-PAGE like a 38 kDa protein32. Thus, the E. coli ribosomal 50S protein L2 may be a major target of the investigated HRNR fragments, its biotin-derivatives and SUMO3-fusion protein.
Another major HRNR-binding protein seems to be the E. coli ribosomal 50S protein L22, which gave a strong 15 kDa band (Supplementary Fig. S8). This protein eluted as principle ribosomal protein upon HPLC-analyses with ACN as eluent (Supplementary Fig. S8b), but did not elute with Prp (Supplementary Fig. S6) – confirming previous findings31. We also noticed that L22, a 12,226 Da protein, migrates upon SDS-PAGE like a 15 kDa protein (Supplementary Fig. S8b), confirming a previous study33. Thus, two of the HRNR-stained bands (37 kDa and 15 kDa) correspond to the E. coli 50S ribosomal proteins L2 and L22. Due to close similarities of ribosomal proteins in its physicochemical properties28 and unknown affinities toward HRNR polypeptide fragments of the other, yet uncharacterized HRNR-binding ribosomal proteins, its structural analysis deserves further detailed investigations.
Distinct HRNR-Derived CIDAMPs Reveal Similar Ribosomal Protein Binding Patterns
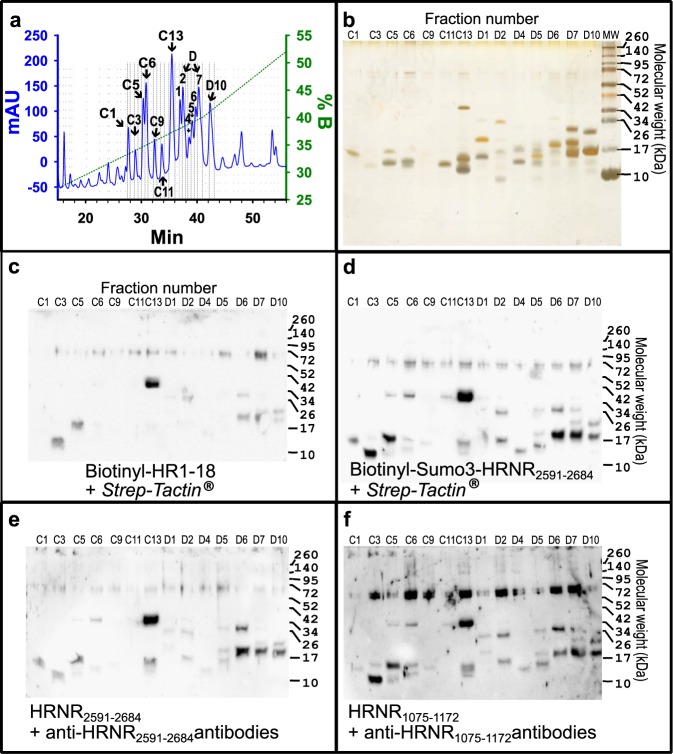
Next, we asked whether the structure of the CIDAMP and/or the method to detect CIDAMP-binding to bacterial ribosomal protein sub-units defines the outcome. We compared the binding patterns of four structurally different, HRNR-based CIDAMPs: biotinylated rSUMO3-HRNR2591–2684, biotinylated HR1-18 (HRNR2656–2677, which represents a 22-mer fragment of HRNR2591–2684), rHRNR2591–2684 and rHRNR1075–1172, each detected using the HRNR-Far-Western blot technique. Either Strep-Tactin® or antibodies against rHRNR2591–2684 and rHRNR1075–1172, respectively, were utilized (Fig. 6): SulfoLink®–column-bound proteins of an E. coli-extract were separated on a Jupiter® C18 widepore RP-HPLC column with a gradient of 2-propanol in aqueous 0.1% TFA and aliquots of fractions containing UV-absorbing peaks (C1–D10) (Fig. 6a) were adjusted in parallel to five PAGE-gels and analyzed for silver-stained proteins (Fig. 6b), for binding of biotinylated HR1-18 (Fig. 6c), biotinylated rSUMO3-HRNR2591–2684 (Fig. 6d), rHRNR2591–2684 (Fig. 6e), and for binding of rHRNR1075–1172 (Fig. 6f). We observed marked similarities of the staining patterns, irrespective whether a biotin-labeled or an antibody-detectable CIDAMP was used to probe and detect the target protein on the membrane. Biotinylated HR1-18 showed far less intensive bands, but an almost identical pattern with some bands missing (Fig. 6), which were weakly stained in experiments with long HRNR-peptides (Fig. 6). It is tempting to speculate that the affinity of the biotinylated 22-mer peptide HR1-18 towards distinct ribosomal proteins may be lower than that of the long HRNR-fragments. Most likely not only the peptide chain length, but also biotinylation of the CIDAMP affects its affinity to ribosomal proteins, a hypothesis supported by a comparatively much lower antimicrobial potency and efficacy of the biotinylated HRNR-peptide-fragments Biotin-rHRNR2591–2684 and rHRNR2591–2684 (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision).
Figure 6.
Distinct HRNR-derived CIDAMPs show similar ribosomal protein binding patterns. (a) SulfoLink®–column-bound proteins of an E. coli-extract were separated on a Jupiter® C18 RP-HPLC column with a Prp-gradient. HPLC fractions containing UV-absorbing peaks (C1–D10) were divided into five aliquots and adjusted in parallel to five PAGE-gels and separated. (b) Silver-stained proteins. (c) HRNR-Far-Western blot for probing with biotinylated HR1-18 (HRNR2556–2677) using Strep-Tactin®, (d) HRNR-Far-Western blot for probing with biotinylated rSumo3-HRNR2591–2684 using Strep-Tactin®, (e) HRNR-Far-Western blot for probing with rHRNR2591–2684 using anti-HRNR2591–2684 antibodies, (f) HRNR-Far-Western blot for probing with rHRNR1075–1172 using anti-HRNR1075–1172 antibodies. Note similarities of the staining patterns, irrespective the CIDAMP AA-sequence or biotin-labeling and irrespective whether a Strep-Tactin®- or antibody-detectable CIDAMP was used to probe and detect the target protein on the membrane. Note the presence of 70 kDa bands upon HRNR-Far-Western blot analyses in most of the investigated HPLC fractions with highest intensity for rHRNR1075–1172 binding (f). The most intensive band, corresponding to a 37 kDa protein in fraction number C13, was identified as E. coli ribosomal protein L2.
Discussion
Our findings clearly show that distinct HRNR-derived CIDAMPs exert bactericidal activity in E. coli by targeting the ribosome. HRNR-Far-Western blot analyses identified several distinct HRNR-binding ribosomal proteins (Table 1 and Supplementary Figs S6–S12), among them the 50S ribosomal proteins L2, L22 and L28 as well as yet, for technical reasons, not be clearly assigned 14 additional 50S and 13 30S ribosomal subunits. Thus, distinct HRNR-derived CIDAMPs simultaneously target different sites of the bacterial protein synthesis machinery, suggesting a common mechanism of action.
The most intensive 37 kDa band seen upon HRNR-Far-Western blot analyses (Fig. 6) originated from E. coli 50S ribosomal protein L2. L2 is the second largest 50S ribosomal protein in E. coli and is one of the most highly conserved ribosomal proteins34 with numerous functions in protein synthesis35,36. It is absolutely required for subunit association as a primary rRNA binding protein and important for peptidyl transferase activity36. Thus, HRNR-derived CIDAMPs may interfere with these essential cellular processes.
The second clearly identified HRNR-interacting ribosomal protein is the E. coli 50S ribosomal protein L22, which is a component of the binding site for erythromycin on the ribosome33. L22 is important during the early stages of 50S assembly. It is one of the proteins that surrounds the polypeptide exit tunnel, where it can interact with nascent translation products in the exit tunnel, providing there one of the earliest contacts with a nascent peptide chain past the peptidyl transferase center37.
The third clearly identified HRNR-interacting protein is the E. coli 50S ribosomal protein L28 (Supplementary Fig. S7a,b), which is required for ribosome assembly38.
Therefore we surmise that CIDAMPs, by manifesting the features of several known classes of ribosome inhibiting antibiotics by simultaneously blocking the ribosome assembly, the peptidyl transferase center and the peptide-exit tunnel of the ribosome, and by targeting simultaneously multiple other ribosomal proteins, may cause the synthesis of aberrant and toxic proteins forming large disordered aggregates12 in CIDAMP-treated bacteria (Fig. 1 and Supplementary Figs S1–S3).
CIDAMPs show marked similarities to proline-rich antimicrobial peptides (PrAMPs) found in insects and some mammals. Like most CIDAMPs (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision), PrAMPs are predominantly active against many Gram-negative bacteria39. Both, insect-derived PrAMPs and mammalian PrAMPs kill bacteria in a non-lytic mode of action by inhibiting bacterial protein translation at the 70S ribosome40. Crystal structure analyses of the PrAMPs revealed binding to the Thermus thermophilus 70S ribosome41. Each of the PrAMPs blocks the peptide exit tunnel of the ribosome by simultaneously occupying three well characterized antibiotic-binding sites and interferes with the initiation step of translation, thereby revealing a common mechanism of action used by these PrAMPs to inactivate protein synthesis41.
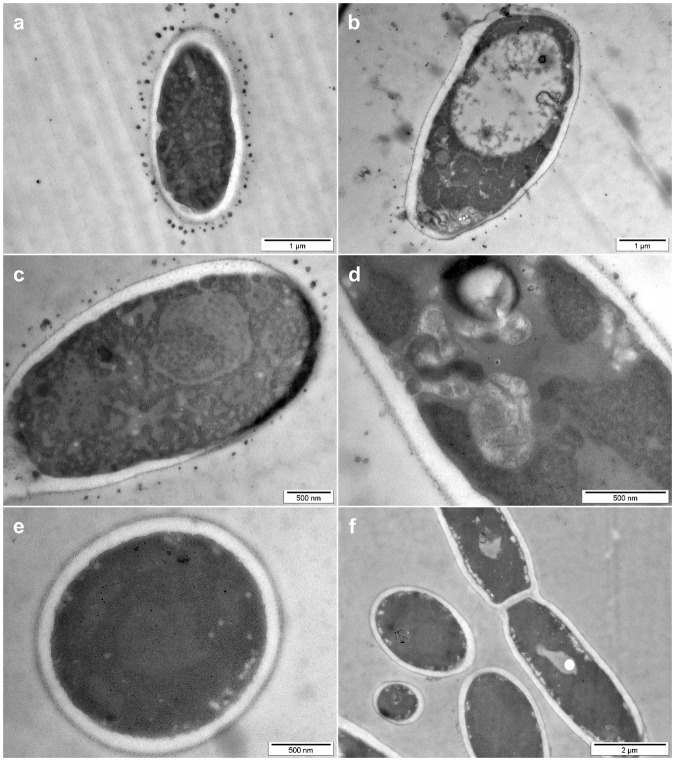
The key role of the bacterial ribosome makes it an important target for antimicrobial agents and it is not surprising that a large number of clinically relevant antibiotics target this protein synthesis machinery of bacteria. The majority of antibiotics bind to one of three key sites in the ribosome: the decoding site (or A-site) on the 30S ribosome, the peptidyl-transferase center (PTC) on the 50S ribosome, and the peptide exit tunnel on the 50S ribosome42. Whereas aminoglycosides bind to the A-site and interfere with codon recognition and specificity causing synthesis of aberrant proteins, antibiotics such as chloramphenicol, clindamycin, and the oxazolidinone linezolid bind at the PTC and inhibit peptide bond formation. Macrolides such as erythromycin block elongation of the growing peptide chain at the peptide exit tunnel43. In support with our findings, ultrastructural analyses of aminoglycoside-treated P. aeruginosa shows marked electron-dense cytosolic aggregates (Fig. 7), very similar as seen upon challenge with almost all CIDAMPs we had studied (Fig. 1 and Supplementary Figs S1–S3). Since these aggregates are indicative for protein misfolding or assembly of misfolded polypeptides into insoluble and cytotoxic aggregates, known to be able to induce the bacterial death12, it is tempting to speculate that CIDAMPs, at least in part, are killing microbes in a self-assembly nanostructure-dependent manner. Several short cationic peptides, conjugated with fatty acids, mediate their antimicrobial activity from the formation of nanostructures44. Thus, it might be possible that at least some CIDAMPs may also express their antimicrobial activity via nanofibrils. May be, this is the case for the nanofiber-forming, HRNR-based CIDAMP rSUMO3-HRNR2591–2684 (Supplementary Fig. S4), which resembles several amphiphilic peptides, where the molecular self-assembly affects antibacterial properties45.
Figure 7.
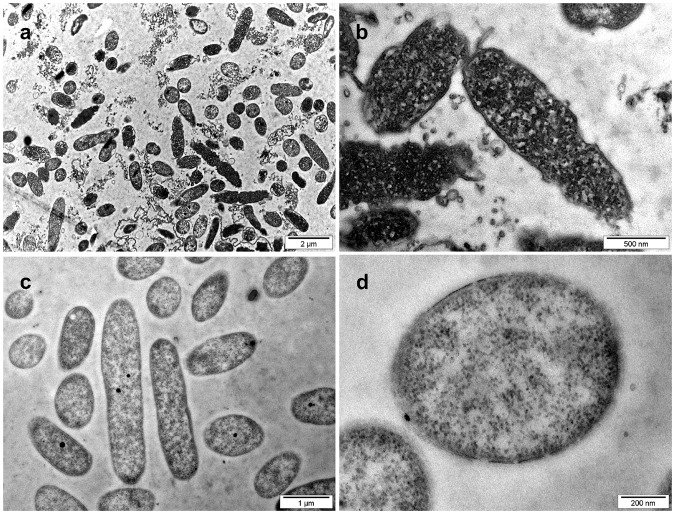
TEM of Gentamicin-treated P. aeruginosa at pH 5.5. (a,b) TEM of P. aeruginosa ATCC 11145, treated with 30 µg/mL Gentamicin in 10 mM NaP, pH 5.5, for 2 h at 37 °C. (c,d) Buffer control (also Supplementary Fig. S1g,h). Note condensation of electron-dense cytoplasmic material in Gentamicin-treated bacteria (a,b). Images are representative of two independent experiments, sampling on average 10 images.
HRNR-derived CIDAMPs are also targeting C. albicans (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision; Fig. 1). The mode of fungicidal action of these CIDAMPs is currently speculative. Ultrastructural analyses of HRNR-treated C. albicans (Fig. 3) revealed morphological signs reminiscent of apoptosis-like cell death (ALD) in fungi10. If fungicidal activity of CIDAMPs should be based on ALD, one may hypothesize that at least some CIDAMPs, due to their amyloidogenic properties might cause homologous seeding of toxic CIDAMP-nanostructures or seeding of heterologous, toxic protein aggregates.
Amyloidogenic peptides are toxic to bacterial cells46, and it was even proposed that the Alzheimer β-peptide may itself be an overlooked AMP, for which C. albicans was identified as the most sensitive microbial target47. Further, introduction of positively charged amino acids into other amyloidogenic peptides lead to the identification of novel potent AMPs with broad spectrum antimicrobial activity48, which would support the hypothesis that supramolecular structures of CIDAMPs may also have microbicidal properties. Formation of intracellular protein aggregates induces oxidative stress, including production of free radicals49. This would result in damage to other cellular proteins and induction of apoptosis in eukaryotic cells50.
Although the precise chain of events that leads to CIDAMP-dependent cell death in bacteria and C. albicans remains to be elucidated, the ability of CIDAMPs to elicit antimicrobial activity via protein aggregate-formation in bacteria represents a rich and unexplored chemical space to be mined in search of novel therapeutic strategies to fight infectious diseases12.
Methods
Synthetic Peptides
Peptides were purchased as TFA-salts from Genecust Europe (Luxembourg). When necessary, peptides were further purified by RP-HPLC, adopting conditions successfully used for purification of antimicrobial peptides51. The identity of the peptides was confirmed by ESI-MS analyses and its purity was better than 95% as determined by RP-HPLC and mass spectrometry.
Peptides were stored as a stock at 3 mg/mL in 0.01% (v/v) aqueous acetic acid at −20 °C until further use and dilutions were always freshly prepared.
Recombinant expression of hornerin polypeptide fragments
Two recombinant hornerin polypeptides (rHRNR1075–1172, rHRNR2591–2684) were expressed in different expression systems (Supplementary Table S1). First, we attempted to generate HRNR repeat-domain peptides using a thioredoxin-reductase-(His)6-HRNR fusion protein, which had to be cleaved by enterokinase to liberate the full length HRNR-peptide. To subclone into the expression vector pET-32a (Novagen, North Ryde, Australia), PCR with Pfu DNA polymerase (Promega, Mannhein, Germany) was performed under the following conditions: 45 s at 98 °C; 5 cycles (45 s at 98 °C; 45 s at 55 °C; 1 min at 72 °C); 25 cycles (45 s at 98 °C; 1 min at 72 °C). The inserts were cut with Bgl II and Not I, gel purified and inserted into the pET-32a vector that had been double-digested with Bgl II and Not I. Although the fusion protein could be generated, all of our attempts to generate full length HRNR peptides failed. In particular the rHRNR2591–2684-fusion protein was found to be extremely sensitive towards treatment with enterokinase resulting in excessive degradation. Using a different strategy, we could generate defined HRNR polypeptides from SUMO3-His-tag-fusion proteins. To subclone the rHRNR1075–1172 into the expression vector pET-SUMO (Invitrogen), PCR was performed with Pfu DNA polymerase for 30 cycles (45 s at 98 °C; 45 s at Tm-5 °C; 1 min at 72 °C). The inserts were gel purified and inserted into the linerazied pET-SUMO vector. To subclone the rHRNR2591–2684 into the expression vector pSumo3 (LifeSensors, Malvern, USA), PCR was performed with PfuTurbo Hotstart Polymerase (Stratagene/Agilent, Waldbronn, Germany) under the following conditions: 1 min at 98 °C; 10 cycles (30 s at 95 °C; 30 s at 53 °C; 1 min at 72 °C); 23 cycles (30 s at 95 °C; 30 s at 64 °C; 1 min at 72 °C). The Insert were cut with Bsa I and Bam HI, gel purified and inserted into the linearized pSumo3 vector. Specific primer pairs used in this study are listed in (Supplementary Table S2). All positive clones were identified and verified by sequencing. The plasmids were introduced into the E. coli host strain BL21(DE3)pLysS or Rosetta-gami 2(DE3)pLysS (Novagen). Subsequently, these were grown at 37 °C in TSB containing appropriate antibiotics. Expression of the recombinant protein was induced with 1 mM isopropyl thio-β-D-galactoside (IPTG) for 3 h at 37 °C. Bacteria were harvested by centrifugation at 5,000 × g for 5 min at 4 °C, lysed by sonication, centrifugated at 15,500 × g for 60 min at 4 °C (Beckman Coulter, Krefeld, Germany) and 0,45 µm filtered. Recombinant proteins were trapped with Ni2+ prepared columns (Macherey-Nagel, Dueren, Germany) and Ni2+-affinity column-bound proteins were subjected to preparative reversed-phase high-performance liquid chromatography (RP-HPLC) with a column (SP250/10 Nucleosil 300-7 C8; Macherey-Nagel) that was previously equilibrated with 0.1% (v/v) TFA in HPLC-grade water containing 10% acetonitrile. The polyhistidine-tagged fusion proteins were eluted with a gradient of increasing concentrations of acetonitrile containing 0.1% (v/v) TFA (at a flow rate of 3 mL/min). Fractions containing UV (215 nm)-absorbing material were collected, lyophilized and analysed by ESI-QTOF-mass spectrometry (Micromass, Manchester, U.K.). Purified histidine-tagged Sumo- and SUMO3-fusion proteins were then digested with SUMO protease 1 or 2 (Lifesensors) according to the manufacturer’s suggestion. The target polypeptide was purified by RP-HPLC on a Jupiter-5 µ-C4-300 Å HPLC column (Phenomenex, Aschaffenburg, Germany) equilibrated with 0.1% TFA in 10% acetonitrile. Peptides were eluted with a gradient of increasing concentrations of acetonitrile containing 0.1% (v/v) TFA (at a flow rate of 0.5 mL/min). Fractions of each peak were collected. Purity of recombinant fusion-proteins was determined by SDS-PAGE. Briefly, proteins were separated on NuPAGE® Novex 10% Bis-Tris gels with MES SDS buffer (Invitrogen). Fusion proteins in Bis-Tris gels were stained with either silver nitrate (Sigma) or Coomassie blue R-250 (Sigma). The SeeBlue® Plus 2 Pre-stained Standard marker (Invitrogen) was used as molecular weight markers. The polypeptide purity and molecular masses were assessed using QTOF-ESI-MS. Further details (in german language) are available from (http://macau.uni-kiel.de/receive/dissertation_diss_00018004).
Biotin- and fluorescein-labeling of recombinant HRNR polypeptides
N-terminal biotinylation or fluorescein-labeling of recombinant HRNR polypeptides was performed with commercial biotinylation or fluorescein-labeling kits (Thermo Fisher ScientificTM) according to the manufacturer. The HPLC-purified biotinylated and fluorescein-labeled products were monitored by ESI-MS and were found to contain >90% of labeled polypeptides.
Formation of rSUMO3-HRNR2591–2684 amyloid-like nanostructures
For the formation of amyloid-like nanostructures52, 1–2 µg lyophilized rSUMO3-HRNR2591–2684 or fluorescein-labeled rSUMO3-HRNR2591–2684 was dissolved in 100 µl distilled water. Samples were sonicated 5 times for 15 sec. at a power of 70% (Sonopuls GM70/Bandelin Elektronik) with intermissions of 15 sec, while cooling on ice. Samples were further analyzed for amyloid-formation using thioflavin T fluorescence monitoring53, for DAPI-staining and monitored by light microscopy (Supplementary Fig. S4).
Microbes used in this study
Candida albicans ATCC 2443, Escherichia coli ATCC11775, Pseudomonas aeruginosa ATCC 10145, Pseudomonas aeruginosa ATCC 11446, Pseudomonas aeruginosa PA01, Staphylococcus aureus ATCC 6538.
Microbial growth conditions
Bacteria were cultivated in either brain heart infusion medium (BHI), lysogeny broth (LB) or tryptic soy broth (TSB)54. If not otherwise stated, microbes were incubated under shaking conditions (37 °C at 170 rpm) or as recommended by ATCC or DSM. Candida albicans was cultured for 3 days on Yeast Extract-Peptone Dextrose (YPD) agar at 30 °C and yeast suspensions at appropriate density were treated with CIDAMPs as indicated.
Colony-Forming Unit (CFU) assay of antimicrobial activity
All purified peptides and recombinant proteins were applied using a colony forming unit (CFU) assay in different media as described elsewhere (TL, UG, Zhihong Wu, JB, AB, AT and JMS, Sci. Rep., in revision).
Transmission Electron Microscopy
All Transmission electron microscopy (TEM) imaging was performed by the Christian-Albrechts-University (CAU) Kiel Center of Biologic Imaging Core at the microscopy core facility. Logarithmic grown microorganisms were concentrated at ambient temperature to an OD600nm of 4 in 10 mM NaP, pH 7.4/ 1% TSB or 10 mM NaP, pH 5.5/0.25% glucose, washed with the respective medium and then suspended to an OD600nm of 2.0, depending on the species, corresponding to 109–1010 microorganisms. The amount of the CIDAMPs used was about 2 × 107 molecules per colony forming unit (CFU). 150 µL microorganism suspension (6.25 × 107/mL) were then incubated with 10 µL CIDAMPs either at ambient temperature or at 37 °C for defined time periods (5 min, 30 min, 90 min, 120 min or 180 min) in the respective media (10 mM NaP, pH 5.5 with or without 0.25% (w/v) glucose or 1% TSB), which also served as controls to identify medium effects. Microorganisms were then fixed in 2.5% glutaraldehyde at 4 °C overnight. Bacteria were centrifuged at 3,220 × g for 10 min, supernatants discarded and then the pelleted samples were suspended at 44 °C (in a thermoblock) in a vial containing 2% Noble agar in distilled water. Samples in the vial were centrifuged, cooled to 4 °C and the bacteria-containing agar-block taken from the tip of the vial. This was then dehydrated in an ascending graded ethanol (EtOH) series. For embedding, the EtOH was replaced stepwise by a polyhydroxy-aromatic acrylic resin (LR White), starting at a resin:EtOH ratio of 1:2, followed by 1:1, 2:1, and three times in resin only, each for 30 min. Finally, samples were embedded in resin at 60 °C. The hardened resin was then cut into 5 nm sections and transferred onto a grid. All samples were analyzed with a transmission electron microscope (Philips TEM 208 or FEI Tecnai G2 Spirit BioTwin).
Post-embedding immunogold electron microscopy
Logarithmic grown P. aeruginosa ATCC 10145 were concentrated at ambient temperature to an OD600nm of 4 in 10 mM NaP, pH 5.5/ 0.25% glucose, washed with 10 mM NaP, pH 5.5/0.25% glucose and then suspended to an OD600nm of 2. To 100 µL bacteria suspension, 10 µL rHRNR2591–2684 (500 µg/mL) or 10 mM NaP, pH 5.5/0.25% glucose for control were added and then incubated at ambient temperature for 5 min. Bacteria were fixed in 4% paraformaldehyde in 0.1 M NaP, pH 7.4, for 1 h, dehydrated and embedded in LR White resin. Ultrathin sections were prepared and mounted on nickel grids. After incubation with 1% BSA for 30 min, the sections were incubated with a HRNR2591–2684-specific polyclonal antibody (0.1 mg/ml in 0.1 M NaP, pH 7.4) at 4 °C overnight. After washing with 0.1 M NaP, pH 7.4, the sections were incubated with a rabbit anti-goat IgG, conjugated to 5 nm gold particles, (dilution 1:10) at ambient temperature for 3 h. The sections were then washed with water and stained with uranyl acetate. Transmission electron microscopy (TEM) imaging was performed by the Christian-Albrechts-University (CAU) Kiel Center of Biologic Imaging Core at the microscopy core facility with a Philips TEM 208 or FEI Tecnai G2 Spirit BioTwin.
Studies on lytic properties of rSUMO3-HRNR2591–2684 in P. aeruginosa
P. aeruginosa ATCC11446 was grown over night at 37 °C and 170 rpm in LB medium without salt. 10 ml BHI medium was inoculated with an aliquot (1/50) of the overnight culture and incubated at 37 °C with shaking until an OD600 of 0.4. Bacteria were centrifuged (5 min 2,000 × g) and the pellet was washed once in 10 mM NaP, pH 7.4, and resuspended in 10 mM NaP, pH 7.4, supplemented with 15 µg/ml Chloramphenicol. The OD600 was adjusted to 0.8–1.0.
200 µl of the bacterial suspension was transferred into a round bottom microtiter plate. If appropriate, combinations of lysozyme (50 µg/ml) and Polymyxin B (1 mg/ml) or rSUMO3-HRNR2591–2684 (5 µg/ml) were added and bacterial lysis was followed by monitoring the OD620 over 1 h (measurements all 5 min) in a TECAN reader.
Studies on the localization of rSUMO3-HRNR2591–2684 in bacterial compartments
2 ml of a P. aeruginosa PaO1 suspension was incubated at 37 °C/170 rpm overnight. A starter culture, prepared with 12 ml BHI/100 ml flask and 200 µl of an overnight culture, was incubated at 37 °C/170 rpm up to an OD620 of 0.4. The bacterial pellet was washed with 10 mM NaP, pH 5.5, containing 0.25% glucose, and re-suspended to an OD620 of 0.6 in the same buffer. If appropriate, 0.2% (w/v) NaN3, was added. Bacteria (100 µl) were treated with 100 µl of rSUMO3-HRNR2591–2684 (1 µg/ml) in 10 mM NaP, pH 5.5, containing 0.25% glucose or buffer (control) for 30 min at 37 °C statically. Thereafter bacteria were centrifuged (13,000 × g, 10 min, 4 °C) and supernatants were harvested. To isolate the periplasm, bacteria were resuspended in 100 µl 20% sucrose and 25 µl 5 mM EDTA, pH 8, followed by incubation for 10 min at 180 rpm and room temperature. The bacterial pellet was then spun down (13,000 × g for 10 min at 4 °C) and the supernatant removed. Thereafter, ice-cold water (100 µl) was added and the test mixture incubated for 10 min on ice at 180 rpm followed by 10 min storage on ice without shaking. Then the suspension was spun down (13,000 × g for 10 min at 4 °C) and the periplasm-containing supernatant was harvested.
To isolate the cytosolic content, the bacterial pellet was suspended in 100 µl ice-cold water, bacterial cells were broken up by heating (5 min at 95 °C) and, after centrifugation (13,000 × g for 10 min at 4 °C), the cytoplasm-enriched supernatant was harvested.
To isolate the inner membrane fraction, bacteria were washed once with 100 µl water. The bacterial cell pellet was suspended in 100 µl 1% Sarkosyl in 30 mM TRIS, pH 8, and incubated for 30 min at 37 °C under shaking conditions and subsequently spun down at 20,000 × g for 1 h at 4 °C. Whereas the inner membrane fraction was enriched in the supernatant, the remaining pellet contained the outer membrane fraction. The harvested bacterial compartments were separated by SDS-PAGE and rSUMO3-HRNR2591–2684 was detected by Western blotting.
Ribosome isolation by SulfoLinkTM affinity chromatography
E. coli ATCC 11775 was inoculated into 10 mL TSB and incubated slantwise overnight and then cultured in 500 mL TSB at 37 °C/175 rpm until an OD600 (max.: 0.6–0.8) was reached. The culture was spun down at 13,000 × g for 10 min and the pellet was suspended in 10 mL 10 mM NaP, pH 7.3. After centrifugation (13,000 × g for 10 min, 4 °C), the sediment was re-suspended in 5 mL E. coli lysis buffer (20 mM Tris-HCl (pH 7.5), 10.5 mM Mg-acetate, 60 mM NH4Cl, 0.5 mM EDTA, and 3 mM β–mercaptoethanol), 5 × treated with ultrasound (70 sec, 15 sec., 30 sec. brake) and finally spun down at 20,000 × g for 30 min at 4 °C. The supernatant was sterile filtered and immediately applied to a SulfoLinkTM-affinity column (Thermo Fisher ScientificTM). Bound material was eluted with elution buffer according to the protocol provided by the manufacturer. Eluted material (“crude ribosomes and ribosomal proteins”) were stored at 4 °C until further use.
Generation of a crude ribosomal subunit-preparation for HPLC-separation
For isolation of ribosomal subunits, SulfoLink TM- affinity column-enriched ribosomes were treated according to a protocol recently described55. Briefly, the affinity column eluate (2.7 mL) was supplemented with 300 µl 1 M MgCl2. Then 6 mL glacial acetic acid was added and the sample was stirred for 45 min on ice. Precipitated RNA was removed by centrifugation (10,000 × g for 10 min, 4 °C). Proteins were precipitated by adding 5 vol. ice-cold acetone to the supernatant and stored at −20 °C overnight. Unlike the original protocol, which recommended washing the sediment with cold acetone, we omitted this step due to dramatic losses of material and problems to precipitate proteins again. The pellet was dried in vacuum for 5 min., dissolved in 0.1% (v/v) TFA in water containing 4 M urea (37 °C, 1 h shaking).
Analytical RP-HPLC of ribosomal proteins
Separation of E. coli ribosomal subunits was performed by reversed-phase (RP) high performance liquid chromatography (HPLC), using separation strategies successfully performed for separation and isolation of cationic antimicrobial peptides and chemokines56–58. Prior to injection into the HPLC-column, crude ribosomal subunit preparations in TFA-water/urea were spun down and the supernatant was separated by RP-HPLC. We either used a Jupiter® 300 Å, 250 × 12.6 mm, C18-RP-HPLC column (Phenomenex) or an Aeris® widepore XB C18, 250 × 12.6 mm, RP-HPLC column (Phenomenex) and either a gradient of acetonitrile (ACN) in aqueous 0.1% TFA, or a gradient of 2-propanol (Prp) in aqueous 0.1% TFA – as indicated for separation of ribosomal proteins. The presence of proteins was monitored at 215 nm, 254 nm and 280 nm. Fractions were collected according to the presence of UV (215 nm) absorbing peaks.
It is important to note that samples to be eluted either with an ACN- or Prp-gradient have been tested for possible precipitate formation by these solvents (80%) prior to application. Precipitate-depleted application samples were then lyophilized, dissolved in 0.1% TFA and then injected into the RP-HPLC column. Nevertheless, we often faced high pressure problems, possibly due to precipitation of some of the ribosomal subunits as a result of stationary phase interaction. This also caused an apparently more or less ribosomal subunit-selective depletion of at least some of the ribosomal subunits, which were missing in the eluates. The precipitation/binding of ribosomal subunits to the column material caused different UV-absorbance profiles of the eluates when repeating the HPLC-separation with additional crude ribosomal protein preparations.
SDS-PAGE-analyses and protein staining
Electrophoretic mobility of ribosomal proteins was investigated using 12% SDS-polyacrylamide gels (SDS-PAGE) in the presence of 8 M urea and tricine59, as described for chemokines and antimicrobial peptides51,60. Peptides were visualized by silver staining or by Far-Western blot analysis (see below).
SDS-PAGE separation of selected RP-HPLC fractions was performed simultaneously with five different gels, where one gel was silver-stained and the other four were used for Far-Western blot-analysis. To achieve this, all HPLC-fractions were lyophilized within the vials and then the residues in each vial were dissolved in 55 µl 0.01% (v/v) acetic acid, followed by blending with 20 µl of 4 × SDS-PAGE loading buffer, which contained 1 mg/mL DTT. The mixtures, corresponding to the selected fractions, were heated to 95 °C for 5 min, centrifuged and then 14 µl of each were applied to the five different gels and separated.
HRNR-Far-Western-blot-analyses
For HRNR-Far-Western-blot analysis, ribosomal proteins-containing samples (14 µl each, see above) were separated on a 12% SDS-tricine polyacrylamide gel containing 8 M urea. Proteins were then transferred to a nitrocellulose membrane (pore size: 0.2 µm, Schleicher & Schuell BioScience, Dassel, Germany) or polyvinylidenfluoride membrane (pore size: 0.2 µm, GE Healthcare) using an alkaline transfer buffer (48 mM Tris, 39 mM glycine, 0.0375% (w/v) SDS and 20% EtOH (pH 9.2)). An alkaline transfer buffer is essential to get cationic proteins efficiently transferred to a membrane1. Thereafter, the membrane was blocked for 1 h in blocking buffer (5% (w/v) bovine serum albumin (BSA) in PBS/Tris, pH 7.4 + 0.05% Tween) and, after washing with PBS/Tris, used for further Far-Western blot analyses: Membranes were incubated with biotinylated HRNR2591–2684 (4 µg/mL) in PBS/Tris, biotinylated HR1-18 in PBS/Tris (4 µg/mL), recombinant HRNR2591–2684 (4 µg/mL) and recombinant HRNR1075–1172 (4 µg/mL), respectively, at 4 °C overnight. This was followed by a 4 × wash with PBS/Tris, pH 7.4. Thereafter, membranes were incubated on a rotator overnight, with either Streptactin®-HRP (iba-lifesciences) in PBS/Tris, pH 7.4 (1:20,000) for 1 h (for biotinylated HRNR-peptides), or with goat-anti-HRNR2591–2684 (1 µg/mL)1 in PBS/Tris, pH 7.4, and with goat-anti-HRNR1075–1172 (1 µg/mL)1 in PBS/Tris, pH 7.4, for recombinant HRNR-peptides, respectively. Membranes were then washed 6-fold with PBS/Tris, and incubated with peroxidase substrate (Roche Lumilight Western Blotting Substrate No. 12015196001) in the case of biotinylated HRNR peptides. The other membranes were then incubated with mouse anti goat IgG-HRP (Jackson, No. 205-035-108) 1:10,000 at 4 °C overnight. After an additional washing step (6×) with PBS/Tris, pH 7.4, membranes were incubated with a peroxidase substrate (Roche Lumilight Western Blotting Substrate No. 12015196001) at ambient temperature and documented with a “Diana III Digital CCD Imaging System” or “FUSION FX7”.
HRNR-Far-Immuno-Dot-blot-analysis
10–30 µL-aliquots of RP-18-HPLC-fractions were lyophilized in a microtiter plate, each residue dissolved in 5 µL of water, and 2 µL applied to a nitrocellulose membrane (0.2 µm-pore size; Bio-Rad) in a DotBlot System (Manifold I). After blotting, the membrane was saturated with PBS/M [PBS, pH 7.4, containing 5% (wt/vol) freeze-dried low-fat milk or BSA] at ambient temperature for 1 h and then washed three times with PBS/T [1 × PBS containing 0.05% (vol/vol) Tween 20]. Membranes were then incubated overnight with biotinylated rSUMO3-HRNR2591–2684 (4 µg/mL) in PBS/T or recombinant HRNR2591–2684 (4 µg/mL) in PBS/T at 4 °C. This was followed by a 4x wash with PBS/T. Thereafter, membranes were incubated on a rotator, either with Streptactin®-HRP (iba-lifesciences) in PBS/T (1:20,000) for 1 h to detect biotinylated rSUMO3-HRNR2591–2684, or overnight with goat-anti-HRNR2591–2684 (1 µg/mL)1 in PBS/T to detect rHRNR2591–2684. Membranes were then washed 6-times with PBS/T and incubated with peroxidase substrate (Roche Lumilight Western Blotting Substrate No. 12015196001) in the case of biotinylated HRNR peptides. Other membranes were then incubated overnight with mouse-anti-goat HRP 1:20,000 (Invitrogen-thermofisher) in PBS/T at 4 °C. After an additional 4x wash with PBS/T and twice with PBS, membranes were incubated with a peroxidase substrate (Roche LumiLight or SuperSignal West Dura; Pierce) at ambient temperature for 5 min and the chemiluminescence was monitored with a “Diana III Digital CCD Imaging System” (Raytest) or “FUSION FX7” (Filber). Biotinylated rSUMO3-HRNR2591–2684 (50 ng/dot) or rHRNR2591–2684 (50 ng/dot) served as positive controls.
Amino acid sequencing of ribosomal proteins
For amino acid sequencing of proteins, RP-HPLC fractions were lyophilized, and the residues dissolved in 0.01% acetic acid (v/v), reduced with DTT, alkylated with iodoacetamide followed by digestion of the alkylated peptides with a mixture of sequencing grade LysC and modified trypsin as described51. Tryptic digests were then subjected to MS/MS-analyses on a QTOF-2 instrument utilizing electrospray ionization and data subjected to Mascot database search (http://www.matrixscience.com/)61. MS/MS sequence evaluation of peptide fragments was done using MassLynx PepSeq software.
Tryptic digestion of HPLC-fractions for nano-LC-ESI-MS of ribosomal proteins
Samples have been filled up with ammonium bicarbonate buffer (50 mM) to a final volume of approximately 50 µL. To reduce disulfide linkages, 1 µL TCEP solution (100 mM) was added and the samples heated to 60 °C for 20 minutes. Cysteine residues were blocked through alkylation by adding 1 µL acrylamide solution (200 mM) and incubated at room temperature for 20 min. For digestion, 50 ng trypsin was added and incubated at 37 °C for 4 h. The digest was stopped by adding 1 µL TFA.
10 µL of the digested samples were injected into a Dionex U3000 nano-LC system (Dionex, Idstein, Germany) coupled online to a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptides were desalted on a trap column (Acclaim Pepmap C-18, 300 µm × 5 mm, 5 µm, 100 Å, Dionex) at a flow-rate of 30 µL/min with loading buffer for 2 min before being eluted onto an analytical column (Acclaim Pepmap C-18, 75 µm × 500 mm, 3 µm, 100 Å, Dionex) at a flow-rate of 300 nL/min. For peptide elution and separation, a linear gradient with eluent A (0.05% FA) and eluent B (0.04% FA in 80% ACN) was employed: 4–50% B in 28 min, 50–90% B in 5 min, 90% B for 10 min, 90–4% B in 0.1 min and 4% B for 15 min. MS data were recorded from 5 to 60 min. MS full scans at a resolution of 70,000 were acquired between 300 and 2,000 m/z. The 10 most intense precursors with a charge state of at least 2+ and at most 7+ were isolated and fragmented using HCD and a normalized collision energy of 25% applied (isolation width was set to 3 m/z). The resolution for MS/MS acquisition was set to 17,500. After fragmentation, precursors were excluded from further isolation for 15 sec.
Data interpretation was performed using the Proteome Discoverer software (version 1.4, Thermo Fisher) and the search engine Mascot (version 2.2.07, Matrix Science, London, UK). All precursors between 350 and 5,000 Da with a signal-to-noise ratio of at least 1.5 were considered. The search was performed against a FASTA database of the combined human and E. coli proteomes (76377 sequences, source: www.uniprot.org, last updated in 04/2016) with tryptic enzyme specificity selected.
Precursor and fragment mass tolerances were set to 10 ppm and 0.02 Da, respectively. Oxidation of methionine, acetylation of N-termini and deamidation of asparagine and glutamine residues were set as dynamic modification. Propionamidation of cysteine residues was set as only static modification. All sequences were also searched against a decoy list of peptides with a strict (0.01) and relaxed (0.05) false discovery rate (FDR). For identification, only high confident peptide spectrum matches were allowed.
Electronic supplementary material
Acknowledgements
This work was supported by Deutsche Forschungsgemeinschaft as a high risk grant “Reinhart-Koselleck-Project” (SCHR 305/5-1) provided to JMS. AB and AT received support from the excellence cluster “Inflammation at Interfaces” and the SFB 877, project Z2. We thank Silvia Voss, Dept. of Infection Medicine, UKSH, for performing antimicrobial assays, Jutta Quitzau, Anke Rose and Claudia Mehrens for performing HPLC-analyses, SDS-PAGE-analyses, Far-Westernblot analyses and Far-immuno-dot-analyses. Maria Mulisch, Marita Beese and Kay Kruse, Central Microscopy Facility of Kiel University CAU, are acknowledged for performing transmission electron microscopy analyses.
Author Contributions
J.M.S. designed and coordinated the study, performed experiments, analyzed data and wrote the manuscript. U.G. performed antimicrobial assays, performed biochemical experiments, generated figures and wrote the manuscript. T.L. generated recombinant proteins, performed TEM analyzes, analyzed data and generated figures. J.B. performed ESI-MS analyses, interpreted MS-data and prepared figures. A.B. and A.T. performed LC-MS/MS analyses, interpreted data and wrote the manuscript.
Competing Interests
Kiel University has filed a provisional patent application with JMS as inventor. It covers cationic intrinsically disordered antimicrobial peptides (CIDAMPs) as designer peptide antibiotics, as well as the application of CIDAMPs as disinfectants and for the prevention and treatment of bacterial and fungal infections (European patent application number EP16199780.4 “Cationic intrinsically disordered antimicrobial peptides”). All other authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ulrich Gerstel and Ties Latendorf contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34467-8.
References
- 1.Wu Z, et al. Highly complex peptide aggregates of the S100 fused-type protein hornerin are present in human skin. J. Invest. Dermatol. 2009;129:1446–1458. doi: 10.1038/jid.2008.370. [DOI] [PubMed] [Google Scholar]
- 2.Garreis F, et al. Expression and Regulation of S100 Fused-Type Protein Hornerin at the Ocular Surface and Lacrimal Apparatus. Invest. Ophthalmol. Vis. Sci. 2017;58:5968–5977. doi: 10.1167/iovs.17-22637. [DOI] [PubMed] [Google Scholar]
- 3.Henry J, et al. Hornerin is a component of the epidermal cornified cell envelopes. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2011;25:1567–1576. doi: 10.1096/fj.10-168658. [DOI] [PubMed] [Google Scholar]
- 4.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 6.Pag U, et al. Analysis of in vitro activities and modes of action of synthetic antimicrobial peptides derived from an alpha-helical ‘sequence template’. J. Antimicrob. Chemother. 2008;61:341–352. doi: 10.1093/jac/dkm479. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann M, et al. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 2010;54:3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front. Biosci. Landmark Ed. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 9.Jung S, et al. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan Hydra. J. Biol. Chem. 2009;284:1896–1905. doi: 10.1074/jbc.M804713200. [DOI] [PubMed] [Google Scholar]
- 10.Ramsdale M. Programmed cell death in pathogenic fungi. Biochim. Biophys. Acta. 2008;1783:1369–1380. doi: 10.1016/j.bbamcr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Andrés MT, Viejo-Díaz M, Fierro JF. Human lactoferrin induces apoptosis-like cell death in Candida albicans: critical role of K+ -channel-mediated K+ efflux. Antimicrob. Agents Chemother. 2008;52:4081–4088. doi: 10.1128/AAC.01597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bednarska NG, et al. Protein aggregation as an antibiotic design strategy. Mol. Microbiol. 2016;99:849–865. doi: 10.1111/mmi.13269. [DOI] [PubMed] [Google Scholar]
- 13.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 14.Scocchi M, Mardirossian M, Runti G, Benincasa M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016;16:76–88. doi: 10.2174/1568026615666150703121009. [DOI] [PubMed] [Google Scholar]
- 15.Melo MN, Ferre R, Castanho MARB. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009;7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- 16.Findlay B, Zhanel GG, Schweizer F. Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold. Antimicrob. Agents Chemother. 2010;54:4049–4058. doi: 10.1128/AAC.00530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1999;1462:11–28. doi: 10.1016/S0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 18.Dathe M, et al. General aspects of peptide selectivity towards lipid bilayers and cell membranes studied by variation of the structural parameters of amphipathic helical model peptides. Biochim. Biophys. Acta. 2002;1558:171–186. doi: 10.1016/S0005-2736(01)00429-1. [DOI] [PubMed] [Google Scholar]
- 19.Nissen-Meyer J, Nes IF. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch. Microbiol. 1997;167:67–77. doi: 10.1007/s002030050418. [DOI] [PubMed] [Google Scholar]
- 20.Kim YC, Tarr AW, Penfold CN. Colicin import into E. coli cells: a model system for insights into the import mechanisms of bacteriocins. Biochim. Biophys. Acta. 2014;1843:1717–1731. doi: 10.1016/j.bbamcr.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas P. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 2009;276:6483–6496. doi: 10.1111/j.1742-4658.2009.07359.x. [DOI] [PubMed] [Google Scholar]
- 22.Scocchi M, Tossi A, Gennaro R. Proline-rich antimicrobial peptides: converging to a non-lytic mechanism of action. Cell. Mol. Life Sci. CMLS. 2011;68:2317–2330. doi: 10.1007/s00018-011-0721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Runti G, et al. Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide Bac7 (1–35) J. Bacteriol. 2013;195:5343–5351. doi: 10.1128/JB.00818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podda E, et al. Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim. Biophys. Acta. 2006;1760:1732–1740. doi: 10.1016/j.bbagen.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Li W, et al. Multimerization of a Proline-Rich Antimicrobial Peptide, Chex-Arg 20, Alters Its Mechanism of Interaction with the Escherichia coli Membrane. Chem. Biol. 2015;22:1250–1258. doi: 10.1016/j.chembiol.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Maguire BA, Wondrack LM, Contillo LG, Xu Z. A novel chromatography system to isolate active ribosomes from pathogenic bacteria. RNA N. Y. N. 2008;14:188–195. doi: 10.1261/rna.692408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall RA. Studying protein-protein interactions via blot overlay or Far Western blot. Methods Mol. Biol. Clifton NJ. 2004;261:167–174. doi: 10.1385/1-59259-762-9:167. [DOI] [PubMed] [Google Scholar]
- 28.Arnold RJ, Reilly JP. Observation of Escherichia coli ribosomal proteins and their posttranslational modifications by mass spectrometry. Anal. Biochem. 1999;269:105–112. doi: 10.1006/abio.1998.3077. [DOI] [PubMed] [Google Scholar]
- 29.Cachia C, Flamion PJ, Schreiber JP. Purification of E. coli 30S ribosomal proteins by high-performance liquid chromatography under non-denaturing conditions. J. Chromatogr. 1991;539:343–353. doi: 10.1016/S0021-9673(01)83943-1. [DOI] [PubMed] [Google Scholar]
- 30.Kerlavage AR, Weitzmann CJ, Hasan T, Cooperman BS. Reversed-phase high-performance liquid chromatography of Escherichia coli ribosomal proteins. Characteristics of the separation of a complex protein mixture. J. Chromatogr. 1983;266:225–237. doi: 10.1016/S0021-9673(01)90896-9. [DOI] [PubMed] [Google Scholar]
- 31.Kamp RM, Wittmann-Liebold B. Purification of Escherichia coli 50S ribosomal proteins by high performance liquid chromatography. FEBS Lett. 1984;167:59–63. doi: 10.1016/0014-5793(84)80832-7. [DOI] [PubMed] [Google Scholar]
- 32.Okada S, Okada T, Aimi T, Morinaga T, Itoh T. HSP70 and ribosomal protein L2: novel 5S rRNA binding proteins in Escherichia coli. FEBS Lett. 2000;485:153–156. doi: 10.1016/S0014-5793(00)02184-0. [DOI] [PubMed] [Google Scholar]
- 33.Arévalo MA, Tejedor F, Polo F, Ballesta JP. Protein components of the erythromycin binding site in bacterial ribosomes. J. Biol. Chem. 1988;263:58–63. [PubMed] [Google Scholar]
- 34.Chandler, J. R. et al. Bactobolin resistance is conferred by mutations in the L2 ribosomal protein. mBio3 (2012). [DOI] [PMC free article] [PubMed]
- 35.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 36.Willumeit R, et al. Localization of the protein L2 in the 50 S subunit and the 70 S E. coli ribosome. J. Mol. Biol. 2001;305:167–177. doi: 10.1006/jmbi.2000.4289. [DOI] [PubMed] [Google Scholar]
- 37.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/S0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 38.Maguire BA, Wild DG. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol. Microbiol. 1997;23:237–245. doi: 10.1046/j.1365-2958.1997.2131578.x. [DOI] [PubMed] [Google Scholar]
- 39.Yi H-Y, Chowdhury M, Huang Y-D, Yu X-Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014;98:5807–5822. doi: 10.1007/s00253-014-5792-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mardirossian M, et al. The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem. Biol. 2014;21:1639–1647. doi: 10.1016/j.chembiol.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Gagnon MG, et al. Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition. Nucleic Acids Res. 2016;44:2439–2450. doi: 10.1093/nar/gkw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCoy LS, Xie Y, Tor Y. Antibiotics that target protein synthesis. Wiley Interdiscip. Rev. RNA. 2011;2:209–232. doi: 10.1002/wrna.60. [DOI] [PubMed] [Google Scholar]
- 43.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 44.Makovitzki A, Baram J, Shai Y. Antimicrobial lipopolypeptides composed of palmitoyl Di- and tricationic peptides: in vitro and in vivo activities, self-assembly to nanostructures, and a plausible mode of action. Biochemistry (Mosc.) 2008;47:10630–10636. doi: 10.1021/bi8011675. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. Self-assembled cationic amphiphiles as antimicrobial peptides mimics: Role of hydrophobicity, linkage type, and assembly state. Nanomedicine Nanotechnol. Biol. Med. 2017;13:343–352. doi: 10.1016/j.nano.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Last NB, Miranker AD. Common mechanism unites membrane poration by amyloid and antimicrobial peptides. Proc. Natl. Acad. Sci. USA. 2013;110:6382–6387. doi: 10.1073/pnas.1219059110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soscia SJ, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. Plos One. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torrent M, Valle J, Nogués MV, Boix E, Andreu D. The generation of antimicrobial peptide activity: a trade-off between charge and aggregation? Angew. Chem. Int. Ed Engl. 2011;50:10686–10689. doi: 10.1002/anie.201103589. [DOI] [PubMed] [Google Scholar]
- 49.Bednarska NG, Schymkowitz J, Rousseau F, Van Eldere J. Protein aggregation in bacteria: the thin boundary between functionality and toxicity. Microbiol. Read. Engl. 2013;159:1795–1806. doi: 10.1099/mic.0.069575-0. [DOI] [PubMed] [Google Scholar]
- 50.Sharma SK, Christen P, Goloubinoff P. Disaggregating chaperones: an unfolding story. Curr. Protein Pept. Sci. 2009;10:432–446. doi: 10.2174/138920309789351930. [DOI] [PubMed] [Google Scholar]
- 51.Schröder J-M. Purification of antimicrobial peptides from human skin. Methods Mol. Biol. Clifton NJ. 2010;618:15–30. doi: 10.1007/978-1-60761-594-1_2. [DOI] [PubMed] [Google Scholar]
- 52.Bang JH, Suslick KS. Applications of ultrasound to the synthesis of nanostructured materials. Adv. Mater. Deerfield Beach Fla. 2010;22:1039–1059. doi: 10.1002/adma.200904093. [DOI] [PubMed] [Google Scholar]
- 53.Meisl G, et al. Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 2016;11:252–272. doi: 10.1038/nprot.2016.010. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual. (CSHL Press, 2001).
- 55.Hardy SJ, Kurland CG, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry (Mosc.) 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 56.Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 57.Harder J, Schroder J-M. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 58.Gläser R, et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat. Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 59.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 60.Schröder JM. Purification of chemokines from natural sources. Methods Mol. Biol. Clifton NJ. 2000;138:1–10. doi: 10.1385/1-59259-058-6:1. [DOI] [PubMed] [Google Scholar]
- 61.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 62.Soung GY, Miller JL, Koc H, Koc EC. Comprehensive analysis of phosphorylated proteins of Escherichia coli ribosomes. J. Proteome Res. 2009;8:3390–3402. doi: 10.1021/pr900042e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.