Abstract
The KIF14 locus is gained and overexpressed in various malignancies, with prognostic relevance. Its protein product, a mitotic kinesin, accelerates growth of normal mammary epithelial cells in vitro and retinoblastoma tumours in a mouse model, while KIF14 knockdown blocks growth of brain, liver, ovarian, breast, prostate, and other tumour cells and xenografts. However, the tumour-initiating effects of Kif14 overexpression have not been studied. We aged a cohort of Kif14-overexpressing transgenic mice and wild-type littermates and documented survival, cause of death, and tumour burden. The Kif14 transgene was expressed in all tissues examined, and was associated with increased proliferation marker expression. Neither mouse weights nor overall survival differed between genotypes. However, Kif14 transgenic mice showed a higher incidence of fatal lymphomas (73 vs. 50%, p = 0.03, Fisher’s exact test), primarily follicular and diffuse B-cell lymphomas. Non-tumour findings included a bilateral ballooning degeneration of lens in 12% of Kif14 transgenic mice but no wild-type mice (p = 0.02). Overall, this work reveals a novel association of Kif14 overexpression with lymphoma but suggests that Kif14 does not have as prominent a role in initiating cancer in other cell types as it does in accelerating tumour development in response to other oncogenic insults.
Introduction
The ATP- and microtubule dependent molecular motors known as kinesins have garnered considerable interest as cancer targets. In particular, the mitotic kinesins Eg5 (KIF11) and CENP-E (KIF10) have been inhibited in clinical trials, with some success1. However, several other members of the 45-strong kinesin superfamily have been implicated in cancer as well2. Prominent amongst these is KIF14, which has an oncogenic role supported by genomic, functional, and clinical evidence3. We first identified KIF14 in a region of somatic genomic gain in retinoblastoma4, and subsequently showed low-level amplification of the KIF14 locus in this cancer5, and increasing KIF14 copy number in the transition from benign retinoma to malignant retinoblastoma6. Moreover, the KIF14 locus is gained in breast carcinoma5, hepatocellular carcinoma5,7, ovarian carcinoma8, papillary renal cell carcinoma9, and medulloblastoma10. Complementing this genomic gain, KIF14 is overexpressed in multiple tumour types, including retinoblastoma (both human and murine)4,11,12 and cancers of the breast13, cervix14, liver15, lung16, ovary8, larynx17, brain18,19, and prostate20. Importantly, this overexpression has clinical relevance: high KIF14 expression is associated with poor prognosis in breast13, cervix14, liver15, lung16, ovary8, brain18, prostate20, and other cancers. In breast cancer, KIF14 expression correlates with proliferation13. Consistent with this, knockdown of KIF14 decreases tumour cell and xenograft growth, while overexpression promotes growth both of cancer cells, and also normal mammary epithelial cells21.
The molecular functions of KIF14 are only partially known. It is essential for cytokinesis, and interacts with and helps localize cytokinesis regulators protein regulator of cytokinesis 1 (PRC1) and citron kinase (CIT)22,23. In turn, its localization is regulated by Nek7-catalysed phosphorylation24. It also may have a cytokinesis-independent role; it can regulate Rap1a-Radil signaling at the cell membrane25. At the whole organism level, a spontaneous mouse Kif14 mutant, laggard, revealed that Kif14 is essential for myelination. Laggard mice have microcephaly, markedly reduced brain size, and a severe ataxia phenotype that is lethal within three weeks of birth26. Interestingly, KIF14 loss-of-function mutations in humans have been associated with an embryonic lethal phenotype of microcephaly, renal cystic dysplasia, and genitourinary and brain malformations, traits characteristic of a ciliopathy27. More recently, causative KIF14 mutations have been documented in patients with less severe, non-lethal microcephaly phenotypes28,29. This role in microcephaly has been touted as a potential reason to consider KIF14 as a therapeutic target for CNS malignancies30.
In characterizing the laggard mutant, the Sakisaka group generated a Kif14 overexpressing transgenic (Tg) mouse, which could rescue the phenotype of laggard and Kif14 knock-out animals26. The advent of this model raised the appealing possibility of exploring Kif14’s cancer-promoting effects in animals. We recently crossed this Kif14 Tg mouse into a transgenic retinoblastoma mouse model. The Simian Virus 40 T-antigen-driven retinoblastoma model is known to upregulate endogenous Kif14 during tumourigenesis12. Transgenic Kif14 overexpression further accelerated tumour initiation, progression, and total tumour burden in these mice, thus confirming for the first time that Kif14 promotes tumour growth in vivo31. We subsequently hypothesized that Kif14 overexpression might similarly predispose mice to spontaneous tumours. Here, we report the effects of Kif14 overexpression on tumour formation in a population of otherwise normal mice allowed to live out their lifespans.
Materials and Methods
Animals
All animal experiments were approved by the Indiana University School of Medicine Institutional Animal Care and Use Committee and were in accordance with all standards set forth in the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Visual Research. Hemizygous mice constitutively overexpressing Kif14 (BDF1-Tg(pCAGGS-Kif14)#28; Accession Number CDB0476T, http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html)26 or their wild-type BDF1 littermates of both sexes were used for the tumour formation and aging study. Mice were genotyped as described26. All pups from consecutive litters entered the study until enrolment numbers were met. In total, 125 mice entered the study (n = 61 Kif14 Tg, 64 wild-type; 2 animals per genotype were subsequently censored due to accidental death). Animals were maintained under standard housing conditions32 until natural death or veterinarian-mandated euthanasia by CO2 asphyxiation followed by cervical dislocation. Veterinary staff were masked to animal genotype in making euthanasia decisions. Mice were checked daily and were weighed monthly starting at five weeks of age. For gene expression analyses, 5–6-week-old female Kif14 Tg or BDF1 mice (Charles River Laboratories) were used.
Necropsy & Histopathology
Necropsies were performed at the Melvin and Bren Simon Cancer Center In Vivo Therapeutics Core facility by researchers masked to animal genotype. Tissues collected for histopathology included liver, lung, brain, pituitary, eyes, mammary gland, ovary, seminal vesicle, kidney, spleen, and any abnormal masses. These tissues were fixed in 10% neutral buffered formalin overnight, and transferred to ethanol prior to embedding, sectioning, and H&E staining at the Indiana University School of Medicine Histology Core. Representative sections were read by a board-certified pathologist and two pathology residents masked to genotype. Lymphomas were classified according to the Mouse Models for Human Cancers Consortium “Bethesda” criteria33. Histopathological images were taken with an Aperio Slide Scanner.
Gene expression analysis
We devised a custom qRT-PCR assay to monitor Kif14 transgene expression without the influence of endogenous Kif14 expression. The vector used for transgene construction, pCAGGS, contains an intron in the promoter region34, so we designed TaqMan primer/probe sequences spanning this intron with the reverse primer within the Kif14 cDNA sequence to ensure specificity for the transgene without amplification from genomic DNA. Forward primer = 5′-TCTGACTGACCGCGTTACTC-3′, Reverse primer = 5′-CTTCCGA-TGTTGTGTCTGCTATG-3′, and probe = 5′-ACAGCACAATAACCAGCACGTTGC-3′. PCR efficiency for this assay was >90%, and no amplification from Kif14 Tg or wild-type gDNA, or wild-type cDNA was observed. We used an Applied Biosystems TaqMan assay to monitor endogenous expression of Kif14 (Mm01291408_m1) in wild-type mice. In order to assess proliferative potential, we utilized a TaqMan assay for Mki67 (Mm01278617_m1); this gene encodes the proliferation marker Ki-67. Trizol (Invitrogen) was used to isolate RNA from unfixed tissues dissected from healthy Kif14 Tg and wild-type mice and stored at −80 °C in RNAlater. First-strand cDNA was synthesized from 250 or 500 ng RNA using iScript Select (Bio-Rad) and random primers. PCR reactions included TaqMan Fast Advanced Master mix (Applied Biosystems), and TaqMan primer/probe mixes. For the Kif14 transgene custom assay, we used 500 nM each primer, and 250 nM probe labeled with FAM and ZEN quencher (IDT). qPCR was performed on a ViiA7 system, and results normalized to housekeeping gene TaqMan assays (Applied Biosystems) for Tbp (Mm00446973_m1) and Hprt (Mm01545399_m1). Results are reported relative to a pool of all tissues from one mouse.
Statistical analysis
Mouse survival was compared by the log-rank test. Differential sex ratios and pathologies were assessed by Fisher’s exact test. For a priori power calculations, mouse survival was the primary endpoint. Assuming 75% of the transgenic mice and 50% of the non-transgenic mice would develop tumours by 130 weeks and totally 65 mice with tumours (event) would be observed, we had 80% power to detect such difference with n = 57 per group at p = 0.05 using the log-rank test. Correlation between Mki67 and Kif14 expression was assessed by Spearman’s ρ. Statistical analyses were performed with GraphPad Prism v. 7.0. Two-sided p-values < 0.05 were considered statistically significant in all tests.
Results
Kif14 transgene expression and proliferation
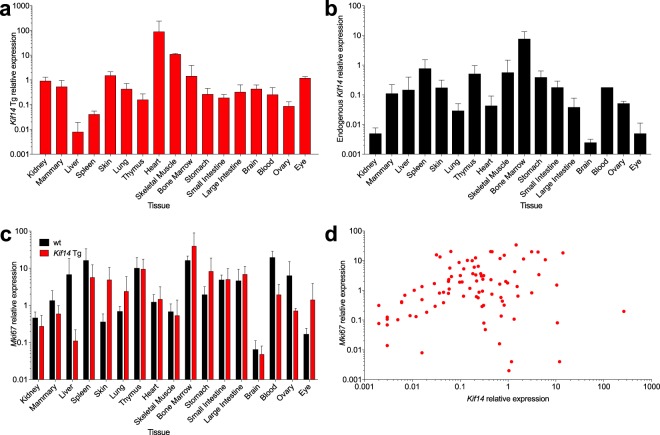
To assess the Kif14 transgene expression pattern across tissues, we developed a qRT-PCR assay that would specifically amplify the transgene cDNA. As expected, given the strong “AG” promoter and cytomegalovirus immediate early enhancer used in constructing this strain26, we observed Kif14 transgene expression in all normal tissue types analysed (Fig. 1a). There was considerable variability in expression, however, with highest expression in heart and skeletal muscle, and lowest expression in liver and spleen. In wild-type mice, endogenous Kif14 was likewise expressed in all tissues examined, with highest expression in bone marrow and spleen, and lowest expression in kidney and brain (Fig. 1b).
Figure 1.
Kif14 gene expression and proliferation in Kif14 transgenic (Tg) and wild-type (wt) mice. (a) The Kif14 transgene is expressed in all tissues analyzed in Tg mice. (b) Endogenous Kif14 is expressed in all tissues analyzed in wt mice. (c) Mki67 proliferation marker is increased in some Kif14 Tg tissues compared with wt tissues. (d) Mki67 expression positively correlates with Kif14 expression (Spearman’s ρ = 0.21, p = 0.04). All individual samples from all tissue types analyzed are shown (both wt and Tg mice, endogenous Kif14 and Kif14 Tg, respectively). qRT-PCR for the indicated genes, normalized to Hprt and Tbp and reported relative to a pool of all tissues. Mean ± s.d. of results from 3 individual mice shown. Note differences in y-axis scales between panels.
Since Kif14 expression is associated with enhanced proliferation13, we assessed expression of the proliferation marker Mki67 (encoding Ki-67) in all tissues. In some cases, Mki67 was increased in Kif14 Tg tissues compared with wild-type counterparts, notably in bone marrow, skin, stomach, lung, and eye (Fig. 1c). Across all tissues, there was a significant positive correlation between Kif14 levels and Mki67 levels (Fig. 1d; Spearman’s ρ = 0.21, p = 0.04).
Mouse characteristics and survival
We monitored the weights and survival of the Kif14 Tg and wild-type mice to assess Kif14’s effects on these parameters. Kif14 Tg mice were grossly indistinguishable from their wild-type littermates; there was no difference in sex ratio between the two genotypes (Kif14 Tg: 55% female, wild-type: 45% female; p = 0.28) and both genotypes showed an identical pattern of weight gain throughout early life (Fig. 2a). During late life, female Kif14 Tg mice lost weight somewhat more rapidly than female wild-type mice, but this difference was not observed for their male counterparts. Overall survival also was not influenced by Kif14 overexpression (Fig. 2b; 781 vs. 748 days median survival for Kif14 Tg and wild-type mice, respectively, log-rank p = 0.68). Age at death ranged from 120–1234 days for wild-type and 308–973 days for Kif14 Tg mice. Moreover, no survival difference was observed when analysing females and males separately (Females: 777 vs. 722 days median survival, log-rank p = 0.64; Males: 788 vs. 749 days median survival, log-rank p = 0.92).
Figure 2.
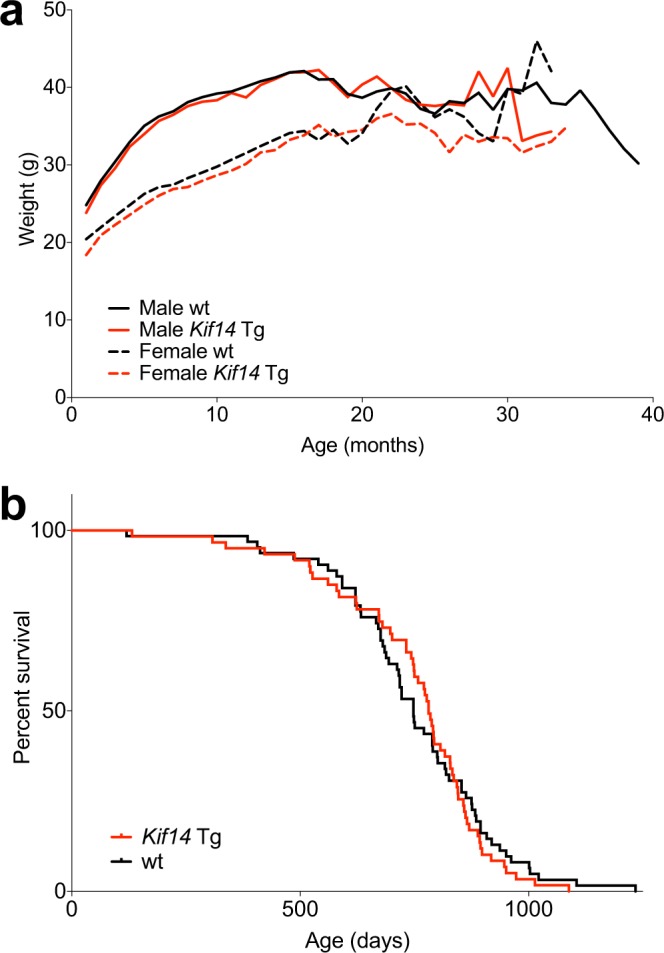
Characteristics of Kif14 transgenic (Tg) mice. (a) Mean weights of Kif14 Tg mice and their wild-type (wt) littermates, measured monthly through the lifespan. (b) Survival of Kif14 Tg mice and their wt littermates. No survival difference was observed between genotypes (p = 0.68, log-rank test).
Spectrum of tumours
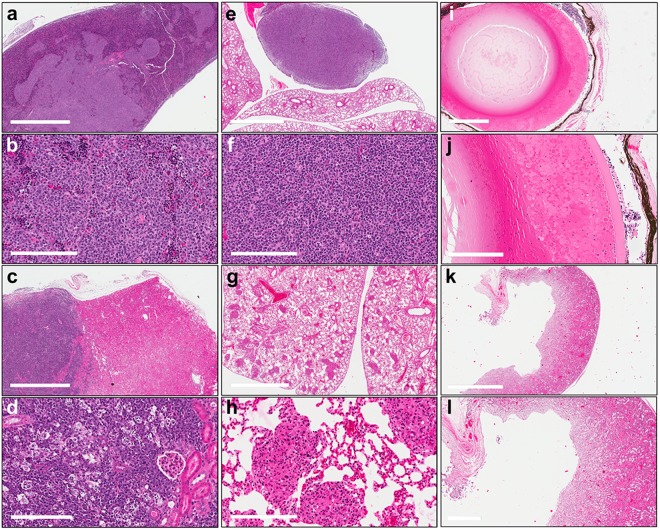
Both Kif14 Tg and wild-type mice developed a wide variety of tumours (Table 1), and there was a non-significantly increased frequency of overall cancer deaths in the Kif14 Tg group: the presumed cause of death was cancer-related in 90% of the Kif14 Tg and 78% of wild-type mice for which cause of death could be inferred (p = 0.15). Lymphoid malignancies were most common. Follicular lymphomas were the most commonly observed tumours (Fig. 3a,b), followed by diffuse large B cell lymphomas (Fig. 3c,d). Thymic lymphoma was also seen (Fig. 3e,f), as well as varied sarcomas (Fig. 3g,h), carcinomas, myeloid dysplasia, and pituitary tumours. Although total incidence of lymphoma did not differ between genotypes (85 vs. 74%, p = 0.29, Table 1), lymphoma as the cause of death was more common in Kif14 Tg mice than in their wild-type littermates (73 vs. 50%, p = 0.03, Table 1). No other tumour type significantly differed between the two groups in incidence or as cause of death (Table 1). Multiple animals bore more than one tumour type at time of death: seven Kif14 Tg mice had two types of tumours and one Kif14 Tg mouse had three distinct types of tumours. There were nine wild-type mice with two types of tumours and two wild-type mice with three distinct types of tumours.
Table 1.
Spectrum of pathological observations in Kif14 transgenic (Tg) mice versus wild-type (wt) littermates.
| Pathology | Observed | Cause of death | ||||
|---|---|---|---|---|---|---|
| wt | Kif14 Tg | p | wt | Kif14 Tg | p | |
| lymphoma | 34 (74) | 35 (85) | 0.29 | 23 (50) | 30 (73) | 0.03 |
| follicular lymphoma | 14 (30) | 19 (46) | 10 (22) | 16 (39) | ||
| diffuse B cell lymphoma | 12 (26) | 12 (29) | 9 (20) | 11 (27) | ||
| thymic lymphoma | 5 (11) | 3 (7) | 4 (9) | 3 (7) | ||
| lacrimal gland lymphoma | 2 (4) | 0 (0) | 0 (0) | 0 (0) | ||
| unspecified lymphoma | 1 (2) | 1 (2) | 0 (0) | 0 (0) | ||
| sarcoma | 4 (9) | 7 (17) | 0.34 | 4 (9) | 6 (15) | 0.51 |
| undifferentiated sarcoma | 1 (2) | 2 (5) | 1 (2) | 1 (2) | ||
| histiocytic sarcoma | 1 (2) | 3 (7) | 1 (2) | 3 (7) | ||
| hemangiosarcoma | 1 (2) | 1 (2) | 1 (2) | 1 (2) | ||
| fibrosarcoma | 1 (2) | 0 (0) | 1 (2) | 0 (0) | ||
| lung sarcoma | 0 (0) | 1 (2) | 0 (0) | 1 (2) | ||
| carcinoma | 8 (17) | 2 (5) | 0.09 | 6 (13) | 1 (2) | 0.11 |
| hepatocellular carcinoma | 2 (4) | 1 (2) | 2 (4) | 0 (0) | ||
| adenocarcinoma | 4 (9) | 0 (0) | 2 (4) | 0 (0) | ||
| spindle cell carcinoma | 1 (2) | 0 (0) | 1 (2) | 0 (0) | ||
| abdominal carcinoma | 1 (2) | 0 (0) | 1 (2) | 0 (0) | ||
| ductal carcinoma | 0 (0) | 1 (2) | 0 (0) | 1 (2) | ||
| myeloid dysplasia | 5 (11) | 7 (17) | 0.54 | 1 (2) | 1 (2) | 1.0 |
| pituitary tumour | 4 (9) | 2 (5) | 0.41 | 4 (9) | 1 (2) | 0.18 |
| hydronephrosis | 15 (33) | 16 (39) | 0.65 | 3 (7) | 2 (5) | |
| ballooning lens degeneration | 0 (0) | 5 (12) | 0.02 | 0 (0) | 0 (0) | |
| hemangioma | 2 (4) | 1 (2) | 1.0 | 1 (2) | 0 (0) | |
| adenoma | 3 (7) | 5 (12) | 0.47 | 1 (2) | 0 (0) | |
Data are stratified into animals in which a pathology was observed, and the subset of animals in which that pathology was the cause of death. Note that some animals exhibited multiple pathologies. Number of animals and (% of total necropsied) per genotype indicated (N = 46 for wt, 41 for Kif14 Tg). Fisher’s exact test p-values for comparing genotypes shown; significant differences are bold and italicized.
Figure 3.
Histology of Kif14 transgenic mouse tumour and eye phenotypes. Low and high magnification histological H&E images of (a,b) follicular lymphoma in spleen; (c,d) diffuse large B cell lymphoma in kidney; (e,f) thymic lymphoma in lung; (g,h) sarcoma in lung; (i,j) ballooning degeneration of lens; and (k,l) hydronephrosis. Scale bars in (a,c,e,g,k) = 2 mm; in (b,d,f,h,j) = 200 µm; in (i,l) = 600 µm.
Non-tumour pathology
Despite the preponderance of tumours, other non-malignant pathologies were evident in these mice as well. Intriguingly, a bilateral, ballooning degeneration of the lens was seen in five Kif14 Tg mice but no wild-type mice (p = 0.02) (Fig. 3i,j). Other pathologies included hydronephrosis in 30–40% of animals (Fig. 3k,l), adenomas, and hemangiomas (Table 1), plus isolated cases of sepsis and pneumonia. However, the incidence of other pathologies did not differ between genotypes.
Discussion
KIF14 is a cancer gene with broad relevance to epithelial and neuronal cancers. Clinical, in vitro, xenograft, and transgenic animal studies have indicated that when gained or overexpressed, this gene promotes tumour growth (reviewed in ref.3). Beyond the correlative clinical studies, functional examples include accelerated proliferation after overexpression of KIF14 in ovarian cancer8 or hepatocellular carcinoma35 cell lines and increased tumor formation in response to Kif14 overexpression in a mouse model of retinoblastoma31. However, KIF14’s role as a tumour initiator has not previously been assessed.
Our results suggest that Kif14 does not act broadly as an initiating oncogene, although may enhance proliferation and severity of cancer, specifically the development of fatal lymphoma, at least in mice. This result is in keeping with findings that KIF14 overexpression could enhance the proliferation of immortalized human mammary epithelial cells, but did not render them tumourigenic21, and with the correlation between Kif14 expression and Mki67 expression seen in human breast cancer samples13. Here, we found that Kif14 overexpression moderately enhanced proliferation of cells in the bone marrow, skin, stomach, lung, and eye of Kif14 Tg mice compared to wild-type mice. Moreover, Kif14 expression positively correlated with Mki67 overall. Perhaps in a compromised state (such as after loss of a tumor suppressor gene), the increased proliferation in these tissues is sufficient to enhance the incidence and severity of lymphomas in this mouse model. Thus, KIF14 appears to be an enabler, accelerating tumour formation and growth against the background of tumour suppressor gene loss and/or other genomic changes31. In contrast, overexpression of Kif11 (Eg5) in mice promoted tumour formation without other oncogenic genetic insults36. No overexpressing strains are documented (www.informatics.jax.org) for the other kinesins with known roles in mitosis (KIF2A, KIF2B, KIF2C, KIFC1, KIF4, KIF10, KIF15, KIF18A, KIF18B, KIF19, KIF20A, KIF20B, KIF23, and KIF22)37.
Of course, it remains possible that sufficiently high KIF14 overexpression, beyond the levels attained in this mouse model, or genomic gain/amplification could initiate tumour formation. It will likewise be interesting to assess any reduced incidence of tumours in mice with loss of Kif14. Based on the preponderance of evidence from in vitro and xenograft studies of KIF14 knockdown8,10,15,16,19,20,35, (partial) loss of Kif14 would be likely to reduce tumour growth. However, such loss-of-function experiments in vivo would require conditional knockouts to bypass the early postnatal lethality seen in constitutive Kif14 knockouts26 and severe CNS phenotypes seen in humans with KIF14 mutations27–29.
The spectrum of neoplasia in the parental BDF1 mouse strain used for creating the Kif14 Tg was previously described, allowing comparisons with our data38. The median survival of mice in our study (107–112 weeks) was similar to that reported (112–118 weeks), although our longest-lived mouse (a wild-type male aged 176 weeks) lived substantially longer than the longest previously reported (158 weeks). Mouse weight curves were also broadly similar. Lymphocytic tumours were most frequent in the previous study (44% of mice), consistent with our findings. However, hepatocellular carcinoma was also prevalent in that study (21% of mice)38, but rare in our cohort (4% of wild-type mice). This difference may be due to environmental factors or source of the animals studied. We also did not observe the marked sex differences in tumour incidence previously reported; we observed no difference in tumour frequency between the sexes.
As tumourigenesis was our focus in this study, the non-neoplastic changes we observed were largely incidental. The common hydronephrosis we observed (which did not differ in frequency between genotypes) is perhaps related to the previously-reported renal calculi, proteinaceous casts, and other kidney pathologies seen in BDF1 mice38. The lens phenotype, seen exclusively in Kif14 Tg mice, is more interesting. Since the ballooning degeneration was bilateral, it is likely genotype-related. However, the rarity of this phenotype (~12% of Kif14 Tg mice) suggests that other genetic or environmental factors beyond Kif14 overexpression may influence its formation.
Our findings of increased fatal lymphomas in mice overexpressing Kif14 suggest that further study of this gene in the context of hematopoietic malignancies is warranted. KIF14 has not been extensively investigated in blood cancers, although both genomic gain of the KIF14 locus and overexpression have been observed in some human genome-wide lymphoma studies3. In particular, one genomic study of human diffuse large B-cell lymphoma showed >30% gain of a genomic region containing KIF1439, while recent work indicated that the KIF14 locus was in a genomic region gained in a small cohort of relapsing B-cell precursor acute lymphoblastic leukemia patients40. It will be interesting to see if KIF14 can accelerate growth of human lymphoma cells, or if its loss can block growth of hematopoietic cancers.
In summary, we have shown that although transgenic Kif14 overexpression does not increase mouse mortality or overall tumor burden, it is associated with an increase in fatal lymphomas, plus a lens phenotype in some cases. Kif14 overexpression is also associated with proliferation in mouse tissues. Based on these data, Kif14 perhaps rarely acts as an initiating oncogene. But given its effects on proliferation, and since it has a well-established role in tumour development and progression3, it remains an appealing gene for therapeutic targeting in cancer41.
Acknowledgements
We thank Hai Liu for assistance with the power analysis, Toshiaki Sakisaka for the Kif14 Tg mice, Courtney Nicole Hemenway and Tiaishia Kia Spragins of the IUSCC In Vivo Therapeutics Core (supported by IU Simon Cancer Center Support Grant P30CA082709) for necropsy assistance, and the staff of the Indiana University School of Medicine Laboratory Animal Resource Center for husbandry. This research was supported by the American Cancer Society IRG-84–002–28, NIH/NCATS KL2TR001106, St. Baldrick’s Foundation, and Research to Prevent Blindness, Inc.
Author Contributions
K.S. performed all live animal work and data analysis, with the assistance of M.S., M.N.O.’H. and R.S.S.; A.L.S., K.C. and K.E.P. performed necropsies and tissue preparation; N.P. and G.E.S. performed the histopathological evaluation; T.W.C. and K.S. performed the qPCR; and T.W.C. and K.S. drafted the manuscript. All authors reviewed the manuscript.
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chandrasekaran G, Tatrai P, Gergely F. Hitting the brakes: targeting microtubule motors in cancer. Br. J. Cancer. 2015;113:693–698. doi: 10.1038/bjc.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozielski, F. (ed.) Kinesins and Cancer. (Springer 2015).
- 3.Thériault, B. L. & Corson, T. W. KIF14: a clinically relevant kinesin and potential target for cancer therapy. [F. Kozielski (ed.)] Kinesins and Cancer. 149–170 (Springer 2015).
- 4.Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–4753. doi: 10.1038/sj.onc.1208641. [DOI] [PubMed] [Google Scholar]
- 5.Bowles E, et al. Profiling genomic copy number changes in retinoblastoma beyond loss of RB1. Genes Chromosomes Cancer. 2007;46:118–129. doi: 10.1002/gcc.20383. [DOI] [PubMed] [Google Scholar]
- 6.Dimaras H, et al. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 2008;17:1363–1372. doi: 10.1093/hmg/ddn024. [DOI] [PubMed] [Google Scholar]
- 7.Kim TM, et al. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int. J. Cancer. 2008;123:2808–2815. doi: 10.1002/ijc.23901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thériault BL, Pajovic S, Bernardini MQ, Shaw PA, Gallie BL. Kinesin family member 14: An independent prognostic marker and potential therapeutic target for ovarian cancer. Int. J. Cancer. 2012;130:1844–1854. doi: 10.1002/ijc.26189. [DOI] [PubMed] [Google Scholar]
- 9.Szponar A, Zubakov D, Pawlak J, Jauch A, Kovacs G. Three genetic developmental stages of papillary renal cell tumors: duplication of chromosome 1q marks fatal progression. Int. J. Cancer. 2009;124:2071–2076. doi: 10.1002/ijc.24180. [DOI] [PubMed] [Google Scholar]
- 10.Li KK, et al. The kinesin KIF14 is overexpressed in medulloblastoma and downregulation of KIF14 suppressed tumor proliferation and induced apoptosis. Lab. Invest. 2017;97:946–961. doi: 10.1038/labinvest.2017.48. [DOI] [PubMed] [Google Scholar]
- 11.Madhavan J, et al. High expression of KIF14 in retinoblastoma: association with older age at diagnosis. Invest. Ophthalmol. Vis. Sci. 2007;48:4901–4906. doi: 10.1167/iovs.07-0063. [DOI] [PubMed] [Google Scholar]
- 12.Pajovic S, et al. The TAg-RB murine retinoblastoma cell of origin has immunohistochemical features of differentiated Müller glia with progenitor properties. Invest. Ophthalmol. Vis. Sci. 2011;52:7618–7624. doi: 10.1167/iovs.11-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corson TW, Gallie BL. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int. J. Cancer. 2006;119:1088–1094. doi: 10.1002/ijc.21954. [DOI] [PubMed] [Google Scholar]
- 14.Wang W., Shi Y., Li J., Cui W., Yang B. Up-regulation of KIF14 is a predictor of poor survival and a novel prognostic biomarker of chemoresistance to paclitaxel treatment in cervical cancer. Bioscience Reports. 2016;36(2):e00315–e00315. doi: 10.1042/BSR20150314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang T, Zhang XB, Zheng ZM. Suppression of KIF14 expression inhibits hepatocellular carcinoma progression and predicts favorable outcome. Cancer Sci. 2013;104:552–557. doi: 10.1111/cas.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corson TW, et al. KIF14 messenger RNA expression is independently prognostic for outcome in lung cancer. Clin. Cancer Res. 2007;13:3229–3234. doi: 10.1158/1078-0432.CCR-07-0393. [DOI] [PubMed] [Google Scholar]
- 17.Markowski J, et al. Metal-proteinase ADAM12, kinesin 14 and checkpoint suppressor 1 as new molecular markers of laryngeal carcinoma. Eur. Arch. Oto-rhino-laryngol. 2009;266:1501–1507. doi: 10.1007/s00405-009-1019-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q, et al. Kinesin family member 14 is a candidate prognostic marker for outcome of glioma patients. Cancer Epidemiol. 2013;37:79–84. doi: 10.1016/j.canep.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, et al. Inhibition of KIF14 suppresses tumor cell growth and promotes apoptosis in human glioblastoma. Cell. Physiol. Biochem. 2015;37:1659–1670. doi: 10.1159/000438532. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Overexpression of a novel candidate oncogene KIF14 correlates with tumor progression and poor prognosis in prostate cancer. Oncotarget. 2017;8:45459–45469. doi: 10.18632/oncotarget.17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singel SM, et al. KIF14 promotes AKT phosphorylation and contributes to chemoresistance in triple-negative breast cancer. Neoplasia. 2014;16:247–256. doi: 10.1016/j.neo.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carleton M, et al. RNA interference-mediated silencing of mitotic kinesin KIF14 disrupts cell cycle progression and induces cytokinesis failure. Mol. Cell. Biol. 2006;26:3853–3863. doi: 10.1128/MCB.26.10.3853-3863.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruneberg U, et al. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cullati SN, Kabeche L, Kettenbach AN, Gerber SA. A bifurcated signaling cascade of NIMA-related kinases controls distinct kinesins in anaphase. J. Cell Biol. 2017;216:2339–2354. doi: 10.1083/jcb.201512055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahmed SM, et al. KIF14 negatively regulates Rap1a-Radil signaling during breast cancer progression. J. Cell Biol. 2012;199:951–967. doi: 10.1083/jcb.201206051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujikura K, et al. Kif14 mutation causes severe brain malformation and hypomyelination. PLoS One. 2013;8:e53490. doi: 10.1371/journal.pone.0053490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filges I, et al. Exome sequencing identifies mutations in KIF14 as a novel cause of an autosomal recessive lethal fetal ciliopathy phenotype. Clin. Genet. 2014;86:220–228. doi: 10.1111/cge.12301. [DOI] [PubMed] [Google Scholar]
- 28.Moawia A, et al. Mutations of KIF14 cause primary microcephaly by impairing cytokinesis. Ann. Neurol. 2017;82:562–577. doi: 10.1002/ana.25044. [DOI] [PubMed] [Google Scholar]
- 29.Makrythanasis P, et al. Biallelic variants in KIF14 cause intellectual disability with microcephaly. Eur. J. Hum. Genet. 2018;26:330–339. doi: 10.1038/s41431-017-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang PY, Gershon TR. A new way to treat brain tumors: targeting proteins coded by microcephaly genes? Bioessays. 2018;40:e1700243. doi: 10.1002/bies.201700243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Hare M, et al. Kif14 overexpression accelerates murine retinoblastoma development. Int. J. Cancer. 2016;139:1752–1758. doi: 10.1002/ijc.30221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wenzel AA, O’Hare MN, Shadmand M, Corson TW. Optical coherence tomography enables imaging of tumor initiation in the TAg-RB mouse model of retinoblastoma. Mol. Vis. 2015;21:515–522. [PMC free article] [PubMed] [Google Scholar]
- 33.Morse HC, et al. & Hematopathology subcommittee of the Mouse Models of Human Cancers Consortium. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.V100.1.246. [DOI] [PubMed] [Google Scholar]
- 34.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 35.Xu H, et al. Silencing of KIF14 interferes with cell cycle progression and cytokinesis by blocking thep27(Kip1) ubiquitination pathway in hepatocellular carcinoma. Exp. Mol. Med. 2014;46:e97. doi: 10.1038/emm.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castillo A, Morse HC, 3rd, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- 37.Verhey, K. J., Cochran, J. C. & Walczak, C. E. The kinesin superfamily. Kinesins and Cancer [F. Kozielski (ed.)] Kinesins and Cancer. 1-26 (Springer 2015).
- 38.Yamate J, Tajima M, Kudow S, Sannai S. Background pathology in BDF1 mice allowed to live out their life-span. Lab. Anim. 1990;24:332–340. doi: 10.1258/002367790780865976. [DOI] [PubMed] [Google Scholar]
- 39.Yoon H, et al. Integrated copy number and gene expression profiling analysis of Epstein-Barr virus-positive diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2015;54:383–396. doi: 10.1002/gcc.22249. [DOI] [PubMed] [Google Scholar]
- 40.Ribera J, et al. Copy number profiling of adult relapsed B-cell precursor acute lymphoblastic leukemia reveals potential leukemia progression mechanisms. Genes Chromosomes Cancer. 2017;56:810–820. doi: 10.1002/gcc.22486. [DOI] [PubMed] [Google Scholar]
- 41.Basavarajappa HD, Corson TW. KIF14 as an oncogene in retinoblastoma: a target for novel therapeutics? Future Med. Chem. 2012;4:2149–2152. doi: 10.4155/fmc.12.158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.