Abstract
Pentatricopeptide repeat (PPR) proteins are a large family of helical repeat proteins that bind RNA in mitochondria and chloroplasts. Sites of PPR action have been inferred primarily from genetic data, which have led to the view that most PPR proteins act at a very small number of sites in vivo. Here, we report new functions for three chloroplast PPR proteins that had already been studied in depth. Maize PPR5, previously shown to promote trnG splicing, is also required for rpl16 splicing. Maize PPR10, previously shown to bind the atpI-atpH and psaJ-rpl33 intercistronic regions, also stabilizes a 3′-end downstream from psaI. Arabidopsis PGR3, shown previously to bind upstream of petL, also binds the rpl14-rps8 intercistronic region where it stabilizes a 3′-end and stimulates rps8 translation. These functions of PGR3 are conserved in maize. The discovery of new functions for three proteins that were already among the best characterized members of the PPR family implies that functional repertoires of PPR proteins are more complex than have been appreciated. The diversity of sequences bound by PPR10 and PGR3 in vivo highlights challenges of predicting binding sites of native PPR proteins based on the amino acid code for nucleotide recognition by PPR motifs.
INTRODUCTION
Chloroplast genomes bear a strong signature of their bacterial ancestry (reviewed in 1). Most chloroplast genes are grouped into polycistronic transcription units (reviewed in 2) that are transcribed by a bacterial-type RNA polymerase (reviewed in 3) and translated by 70S ribosomes (reviewed in 4). Superimposed on this ancient infrastructure are numerous features that arose post-endosymbiosis, such as RNA editing and protein-mediated stabilization of RNA termini (5). These acquired processes are mediated by several families of nucleus-encoded RNA-binding proteins that coevolved with the organelle genomes whose expression they impact. The paradigm for this phenomenon is the pentatricopeptide repeat (PPR) family (6), a large family of helical repeat proteins that influence gene expression in mitochondria and chloroplasts (reviewed in 7). PPR proteins harbor tandem degenerate repeats of ∼35 amino acids, each of which adopts a helix-turn-helix fold. Consecutive repeats stack to form an elongated surface that binds single-stranded RNA. PPR tracts often bind RNA in a modular 1-repeat, 1-nt fashion with nucleotide specificity strongly influenced by the identity of the amino acids at two positions in each repeat, the so-called ‘PPR code’ (8,9,10,11). Long PPR tracts bind RNA with high affinity and specificity, and they can affect gene expression by interfering with ribonuclease access or by altering local RNA folding (reviewed in 7). Many PPR proteins in plant organelles specify sites of RNA editing; these harbor variant repeat tracts and an accessory domain at the C-terminus that is closely tied to this editing function (reviewed in 12).
The identification of organelle gene expression defects in loss-of-function mutants has been the primary means for assigning functions to PPR proteins (reviewed in 7). Such studies have typically detected defects in the metabolism of one or several RNAs, implying that PPR–RNA interactions are highly specific in vivo. However, the technical challenge of comprehensively analyzing gene expression may have led to an exaggerated view of the sequence specificity of PPR proteins. In this work, we describe additional sites of action for three chloroplast PPR proteins that were already among the most thoroughly studied members of the PPR family: PPR10 (13,14,15), PGR3 (16,17) and PPR5 (18,19). These are canonical ‘P-type’ PPR proteins lacking any accessory domains (20). Comparison of the multiple RNA sequences bound by PPR10 and PGR3 in vivo reveals considerable flexibility in how each protein selects physiologically meaningful RNA ligands. These findings suggest that additional functions remain to be discovered for many PPR proteins that have been characterized previously, and highlight the importance of genome-wide transcriptome, translatome and RNA–protein interactome surveys for the functional analysis of organelle RNA-binding proteins.
MATERIALS AND METHODS
Maize mutants and growth
Maize cps1 encodes the chloroplast localized cysteinyl transfer RNA (tRNA) synthetase (21). The cps1-1/2 allele is a hypomorph that is heteroallelic for Mu transposon insertions in the 5′-untranslated region (5′-UTR) and in an exon (21,22). The ppr5 mutant and PPR5 RIP-chip data were described in (19). The ppr10 mutant and PPR10 RIP-chip data were described in (13). The ppr4-1 and ppr103-2/-3 mutants were described in (23) and (24), respectively. Maize plants were sown in soil and grown in diurnal cycles (16-h light/8-h dark) at a temperature of 28 and 26°C for the light and dark periods, respectively. Plants were illuminated using a light intensity of ∼300 μmol m−2 s−1. The second and third leaves to emerge were harvested 8 or 9 days after planting and flash frozen in liquid nitrogen prior to processing for RNA extraction or ribosome profiling.
Arabidopsis mutants and growth
Arabidopsis was grown on soil at 23°C, using a light intensity of ∼120 μmol m−2 s−1 with 16-h light/8-h dark cycles. Plants used for ribosome profiling were 2 weeks old, whereas those used for RNA extraction were 3 weeks old. The svr7-3 allele was described in (25). Seeds for the pgr3-4 allele were obtained from the Versailles Arabidopsis thaliana Stock Center (line FLAG_086D06) and also as a generous gift from Toshiharu Shikanai (Kyoto University). The position of the T-DNA insertion was determined by DNA sequencing to map 588 bp downstream from the PGR3 start codon, which encodes PGR3’s third PPR motif.
Ribosome profiling
Ribosome profiling of the maize pgr3 mutant was performed by Ribo-Seq as described in (26). Ribosome profiling of the Arabidopsis pgr3 mutant was performed using high-resolution microarrays to map ribosome footprints, similar to the method used for maize in (15). This method was modified as follows. Two-week-old wild-type (Ws) and pgr3 seedlings (1.6 g each) were homogenized in liquid nitrogen. Samples were thawed in 10.6 ml of ribosome extraction buffer, incubated with 100 units of micrococcal nuclease (Roche) per ml of extract for 1 h at 23°C, layered onto a 1.5 ml sucrose cushion and centrifuged for 3 h at 170 000 g at 4°C. RNA was extracted as described in (27), except that RNA was precipitated by addition of three volumes of ethanol to the aqueous phase. RNA between 20 and 50 nt was gel purified as described in (15). Two micrograms of the RNA recovered was labeled and purified using the Label IT® miRNA Labeling Kit (Mirus). Cy5 and Cy3 labeled RNA was hybridized in 1× hybridization solution (Mirus) to custom microarrays covering the whole Arabidopsis chloroplast genome (NCBI: NC_000932) in a tiling fashion (MYcroarray). The array consists of overlapping 50-mer oligonucleotide probes (20-nt overlap) in triplicate spots covering both strands of the chloroplast genome. Arrays were hybridized at 40°C overnight. Arrays were then washed four times in 1× SSPE for 3 min and one time in 0.5× SSPE for 1 min at room temperature. Arrays were scanned with a ScanArray Gx Plus microarray scanner (Perkin Elmer). Data were analyzed with GenePix Pro 7.0 software (Molecular Devices) using the default local background subtraction method. Only probes with at least two (of three) spots passing the background filter were used to calculate the median of Cy3/Cy5 ratios. These medians were used to calculate the median of ratios for all probes corresponding to each chloroplast open reading frame (ORF).
cRT-PCR and 3′RACE
The 3′-end in the rpl14-rps8 intergenic region in maize was mapped by cRT-PCR as described previously (13) using the following primer pair: rpl16 PE 5′ ctaacagtcacacactaagcat; rpl14-207for2 5′ attcaaaggcgacgacggta. The orthologous 3′-end in Arabidopsis was mapped by 3′-RACE as follows. Leaf RNA (2 μg) was ligated to 10 pmol of a 3′-adapter using T4 RNA ligase I (Epicenter) at 37°C for 2 h. RNA was purified by phenol/chloroform extraction and reverse-transcribed using ProtoScript II reverse transcriptase (NEB) using a primer complementary to the 3′-adapter. Polymerase chain reaction (PCR) amplification was performed using a gene-specific and an adapter-specific primer (see Supplementary Table S1). PCR products were gel-eluted using the JETSORB Gel Extraction Kit (Genomed) and cloned in the pJET1.2/blunt vector (Thermo Scientific).
RNA gel blot hybridization
RNA gel blot analysis in maize was performed using radiolabeled probes generated by PCR and random-hexamer labeling, or radiolabeled synthetic oligonucleotides as described in (28) and (29), respectively. RNA gel blot analysis in Arabidopsis was performed as described in (30), using radiolabeled RNA probes. Oligonucleotides and primers used to generate probes are described in Supplementary Table S1.
Generation of antibody to ZmPGR3
A PCR product encoding ZmPGR3 (starting at amino acid 61 and ending at the natural stop codon) was generated using complementary DNA from the inbred line B73 and cloned into the pMAL-TEV vector (New England Biolabs). The MBP–PGR3 fusion protein was expressed, cleaved with TEV and purified by size-exclusion chromatography as described for PPR10 (13). Four milligrams of the purified protein was used for generating polyclonal antisera in rabbits at Alpha Diagnostic Inc. Antisera were affinity purified against the antigen coupled to HiTrap NHS-activated column (GE Healthcare Life Science).
RIP-seq analysis
RIP-seq was performed as previously described for RIP-CHIP (31) except that the coimmunoprecipitated RNA was identified by high-throughput sequencing. Briefly, 75 μl of maize chloroplast stromal extract (∼15 mg/ml protein) was incubated with affinity purified antibodies to either AtpB or ZmPGR3. Antibody complexes were collected using magnetic beads coupled with Protein A/G (Pierce) and washed extensively as described previously (31). RNA was purified from the beads using Trizol/Chloroform and was concentrated by ethanol precipitation using glycoblue as carrier. The RNA was sheared using the method described in the NEBNext® Magnesium RNA Fragmentation protocol (94°C for 5 min in a buffer containing 40 mM Tris-OAc, 100 mM KOAc, 30 mM Mg(OAc)2, pH 8.3 at 25°C). The 5′-ends were phosphorylated by treatment with adenosine triphosphate (ATP) and T4 polynucleotide kinase (NEB), and 20 ng of the RNA was used to prepare an RNA-seq library with the Bioo small RNA Kit v3.
RESULTS
PPR10 stabilizes a 3′-end downstream of psaI
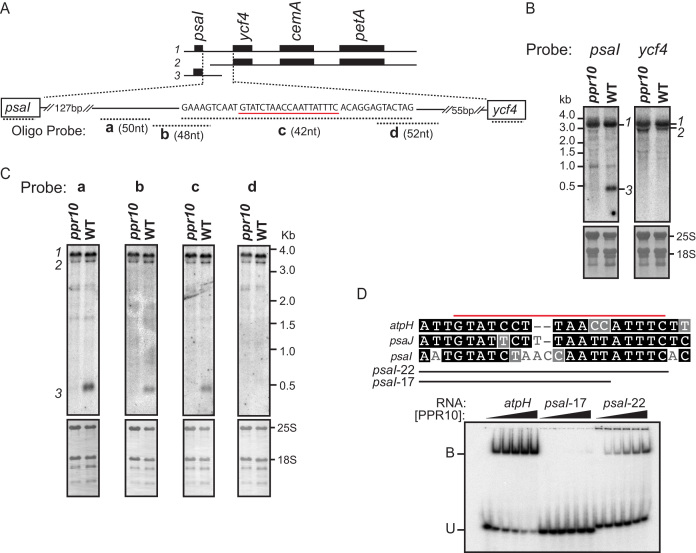
PPR10 consists of 19 P-type PPR motifs and binds to similar sequences in the chloroplast atpI-atpH and psaJ-rpl33 intergenic regions in maize (13,14). These interactions impede 5′→3′ and 3′→5′ exoribonucleases, and thereby stabilize the adjacent RNA and define positions of processed RNA termini (13,14). Additionally, PPR10’s interaction upstream of atpH increases atpH translational efficiency (14,15). Three plastome-wide assays had previously been employed to explore PPR10’s roles: an RNA-coimmunoprecipitation-and-microarray (RIP-chip) assay identified RNAs bound to PPR10 in vivo (13), microarray hybridizations quantified chloroplast RNAs in ppr10 mutants and ribosome profiling reported defects in chloroplast protein synthesis in ppr10 mutants (15). Despite the comprehensive nature of these prior studies, we detected a third site of PPR10 action serendipitously: RNA gel blots generated for a different purpose showed that ppr10 mutants lack monocistronic psaI transcripts (Figure 1, transcript 3). The tetracistronic precursor (psaI-ycf4-cemA-petA, transcript 1) and a minor tricistronic transcript (ycf4-cemA-petA, transcript 2) accumulate independently of PPR10.
Figure 1.
PPR10 defines and stabilizes the 3′-end of monocistronic psaI RNA. (A) The psaI-ycf4-cemA-petA transcription unit. The three transcripts detected by RNA gel blot hybridization are diagrammed at top. The oligonucleotide probes used in panel (C) are diagrammed below in relation to the expansion of the psaI-ycf4 intergenic region. Probe lengths are not drawn to scale, but their overlap with one another is precisely shown by the nucleotide sequence above. The sequence underlined in red resembles known PPR10-binding sites (see panel (D)), and ends 430 nt downstream from the psaI transcription start site (53). (B) RNA gel blot hybridization showing the loss of monocistronic psaI RNA in ppr10 mutants. The two panels come from the same gel and were hybridized with gene-specific probes for either psaI or ycf4 RNA. Excerpts of the same blots stained with methylene blue are shown below to illustrate rRNA abundance. The transcripts are numbered to the right, according to the scheme shown in panel (A). (C) Mapping the 3′-end of monocistronic psaI mRNA by high-resolution RNA gel blot hybridization. Replicate panels from the same gel were hybridized to the oligonucleotide probes diagrammed in panel (A). (D) Gel mobility shift assay demonstrating that PPR10 can bind to the sequence at the 3′-end of the PPR10-dependent psaI transcript. A comparison of the psaI site with the known atpH and psaJ sites is shown at top; the red line marks the minimal binding site established for atpH (14). PPR10 was used at 10, 5, 2.5, 1.25 or 0 nM.
We noticed a sequence in the psaI-ycf4 intergenic region that resembles PPR10’s known binding sites (Figure 1A–D); 3′-exonucleolytic processing to the 3′-boundary of that site would generate a 430-nt RNA, consistent with the size of the monocistronic psaI RNA whose accumulation relies on PPR10. RNA gel blot hybridizations using closely spaced oligonucleotide probes confirmed that this sequence maps very near the 3′-end of the PPR10-dependent RNA (Figure 1C). Gel mobility shift assays with recombinant PPR10 showed further that PPR10 can bind this sequence in vitro, albeit with lower affinity than it binds the atpH site (Figure 1D). Interestingly, the sequences at the 5′- and 3′-ends of this binding site are conserved with those in the atpH and psaJ sites, but the spacing between these conserved sequences is different in each case (Figure 1D).
When PPR10 binds the atpI-atpH and psaJ-rpl33 intergenic regions, it stabilizes both 5′- and 3′-RNA termini; the locations of the processed 5′-ends are defined by the position of PPR10’s N-terminus on the RNA, whereas the positions of the processed 3′-ends are defined by PPR10′s C-terminus. If PPR10 were to act in this way at this newly discovered site, it would stabilize a 5′-end upstream of ycf4. However, the (minor) tricistronic transcript spanning ycf4-cemA-petA accumulates independently of PPR10 (transcript 2 in Figure 1B). Furthermore, this tricistronic RNA was detected by hybridization to two oligonucleotides mapping upstream of the PPR10-binding site (probes ‘a' and ‘b' in Figure 1C), indicating that its 5′ end is not defined by PPR10. These results imply that PPR10 blocks 3′→5′ but not 5′→3′ RNA decay when bound to the site in the psaI-ycf4 intergenic region.
PGR3 stabilizes rpl16-rpl14 dicistronic RNA and stimulates rps8 translation
PGR3 has 28 P-type PPR motifs and is among the largest and best characterized PPR proteins in Arabidopsis. PGR3 binds to a site in the petL 5′-UTR, where it stabilizes the downstream messenger RNA (mRNA) and increases petL translational efficiency (16). PGR3 also promotes the accumulation of the NADH Dehydrogenase-like (NDH) complex, which has been proposed to result from its binding to the ndhA 5′-UTR and activation of ndhA translation (16,17); however, the published evidence for PGR3′s effects on ndhA RNA is inconclusive. We revisited the functions of PGR3 for two reasons. First, we hoped to clarify the basis for the NDH deficiency in pgr3 mutants. Second, the functions described for Arabidopsis PGR3 are not sufficient to account for the phenotype of a pgr3 mutant in maize (Zm-pgr3): the petL RNA stabilization function is conserved in maize, but Zm-pgr3 mutants also have a reduction in plastid ribosomes whose basis is unknown (21).
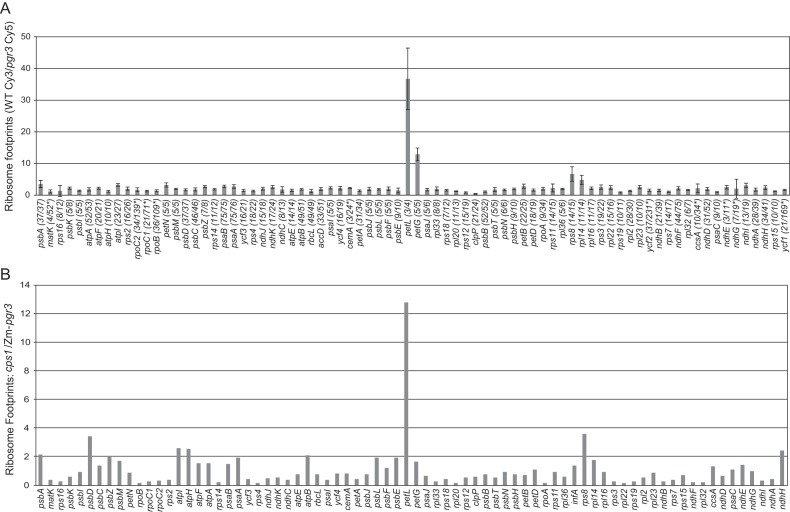
To gain a more comprehensive view of the effects of PGR3, we used ribosome profiling to analyze chloroplast gene expression in Arabidopsis and maize pgr3 mutants. Ribosome profiling provides a genome-wide accounting of ribosome footprints, the short mRNA segments that are protected by bound ribosomes from ribonuclease attack. Changes in ribosome footprint abundance can result from changes in mRNA abundance and/or changes in translational efficiency. Chloroplast ribosome footprints from wild-type and pgr3 Arabidopsis seedling leaves were mapped by hybridization to high-resolution microarrays covering the entire chloroplast genome. These experiments used a previously unreported allele, pgr3-4, which has an insertion that disrupts the ORF and is likely to be a null allele. pgr3-4 mutants were slightly chlorophyll deficient and grew more slowly than the wild-type under moderate light conditions (Supplementary Figure S1). The ribosome profiling data are summarized in Figure 2A as the ratio of ribosome footprints in the wild-type relative to the mutant for each chloroplast gene. The results revealed defects in petL and petG expression, as reported previously (16,32). However, in contradiction with the proposed role for PGR3 in ndhA translation, the abundance of ribosome footprints on the ndhA ORF was not reduced in the mutant. Furthermore, all of the genes encoding NDH subunits appeared to be expressed normally, although the signals for ndhE and ndhG were too weak to make firm conclusions. Unexpectedly, the results revealed a decrease in ribosome footprints on the rpl14 and rps8 genes in the pgr3 mutant (Figure 2A). This finding was explored in experiments described below.
Figure 2.
Analysis of chloroplast gene expression in Arabidopsis and maize pgr3 mutants by ribosome profiling. (A) Ribosome footprints from seedling leaves of the Arabidopsis pgr3 mutants and their normal siblings were detected by hybridization to high-resolution microarrays spanning the whole chloroplast genome. The values shown are the median ratio (wild-type:mutant) of the median normalized signal intensities among all 50-nt array probes mapping within each ORF. Error bars represent the standard deviation calculated from all probes covering the ORF. Each ORF is annotated with the number of probes whose signal was above background as a fraction of the total number of probes. Genes for which fewer than half of the probes were above background are marked with an asterisk; their values could not be assessed with confidence. (B) Ribosome footprints from seedling leaves of the maize pgr3 mutant (Zm-pgr3) were mapped by deep sequencing. The values shown are the ratio of normalized read counts for each gene in cps1-1/2, a mutant with a plastid ribosome deficiency similar in magnitude to that of Zm-pgr3. Read counts were normalized to the total number of reads mapping to chloroplast ORFs. A comparison to the wild-type is shown in Supplementary Figure S2A.
Ribosome footprints from the maize pgr3 mutant were mapped by deep sequencing. Zm-pgr3 mutants have a reduced content of plastid ribosomes (21), and defects of this nature cause characteristic changes in the chloroplast transcriptome (e.g. 33,34). Therefore, we compared the ribosome profiling data for Zm-pgr3 to that from a cps1-1/2 mutant, which has a ribosome deficiency of similar magnitude due to mutation of a gene encoding a chloroplast tRNA synthetase (21,22). The loss of petL expression in Zm-pgr3 stood out clearly in the data (Figure 2B). As in Arabidopsis, this assay did not reveal a defect in ndhA; together, these results provide strong evidence that PGR3 is not needed for ndhA expression. Also as in Arabidopsis, the data suggested a role for Zm-PGR3 in rps8 expression. Minor defects in the expression of several other genes were suggested by the data (e.g. psbD, atpI and ndhH), most of which were further examined by RNA gel blot hybridization (Supplementary Figure S2B). The abundance and pattern of transcripts from these genes in Zm-pgr3 mutants were similar to that in other mutants that are deficient for plastid ribosomes (Supplementary Figure S2B). Furthermore, RNA coimmunoprecipitation did not detect interactions between Zm-PGR3 and these RNAs (see below).
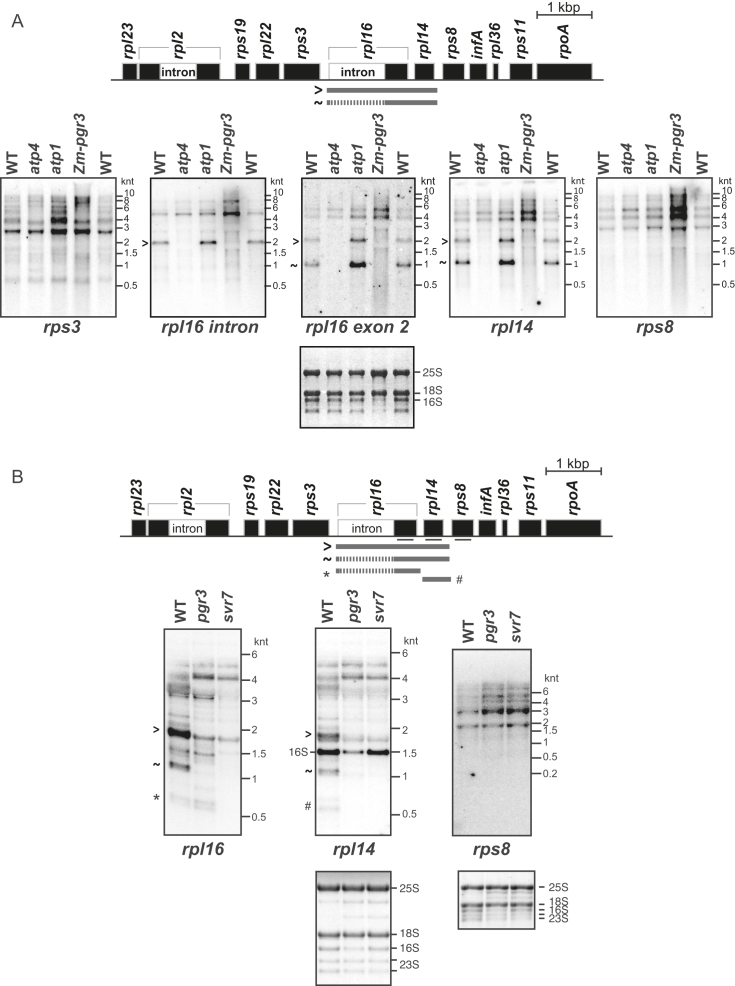
The ribosome profiling data suggested that PGR3 stimulates the expression of rps8 and possibly the adjacent rpl14 gene in both maize and Arabidopsis. To determine whether these effects arise from a defect in mRNA metabolism, we analyzed transcripts from these genes in maize and Arabidopsis pgr3 mutants by RNA gel blot hybridization (Figure 3). The rpl16 and rpl14 genes in maize are represented on dicistronic transcripts whose accumulation requires the PLS-type PPR protein PPR103, and the PPR-SMR protein ATP4 (15,24). Interestingly, the transcript patterns from Zm-pgr3 and atp4 mutants were similar (Figure 3A): dicistronic rpl16-rpl14 transcripts that either include or lack the rpl16 intron are absent in both mutants, whereas transcripts from the adjacent genes (rps3 and rps8) accumulate to normal or slightly elevated levels.
Figure 3.
RNA gel blot hybridizations demonstrating loss of dicistronic rpl16-rpl14 transcripts in maize and Arabidopsis pgr3 mutants. Seedling leaf RNA from maize (A) or Arabidopsis (B) plants of the indicated genotype was analyzed by RNA gel blot hybridization, using probes specific for the indicated regions. The maize atp1 mutant lacks the chloroplast ATP synthase (54) and was included to control for effects resulting from the loss of this complex. The first three lanes of the blots shown in (A) (WT, atp4 and atp1) were published previously (15) and are reproduced here with permission from: Zoschke, R., Watkins, K.P. and Barkan, A. (2013) A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell,25, 2265–2275; DOI:10.1105/tpc.113.111567, www.plantcell.org, Copyright American Society of Plant Biologists (2013). Arabidopsis svr7 is orthologous to maize atp4. The rpl16 and rpl14 blots in panel (B) came from the same gel, whereas the rps8 blot came from a different gel. The probe for rpl14 in Arabidopsis cross-hybridized with the 16S rRNA, as marked in (B).
In Arabidopsis, transcripts from the pgr3 mutant were compared with those from an svr7 mutant, which is orthologous to maize atp4 (25,35). The major dicistronic spliced rpl16-rpl14 transcript was missing in the pgr3 and svr7 mutants, indicating that this function of PGR3 and ATP4/SVR7 is conserved between maize and Arabidopsis. However, unlike the situation in maize, there were minor differences in transcript patterns between svr7 and pgr3 mutants: a low-abundance monocistronic rpl16 transcript accumulates in pgr3 mutants but is absent in svr7 mutants (asterisk in Figure 3B). Therefore, both SVR7 and PGR3 are required to stabilize the dicistronic rpl16-rpl14 transcript, whereas this monocistronic rpl16 RNA is dependent only on SVR7. Note that the abundance of chloroplast ribosomal RNA (rRNA) is reduced in Arabidopsis pgr3 mutants as in maize, albeit less severely (see 16S rRNA on methylene-blue stained blots in Figure 3B and RNA gel blot hybridizations in Supplementary Figure S2C), indicating a previously undetected role for Arabidopsis PGR3 in promoting chloroplast ribosome accumulation.
The loss of rpl16 and rpl14 ribosome footprints in pgr3 and Zm-pgr3 mutants seems modest in comparison with the loss of the corresponding mRNA (compare Figures 2 and 3). The rpl16 and rpl14 ribosome footprints in the mutants must derive from translation of the large transcript isoforms, which accumulate to somewhat higher levels than the corresponding transcripts in wild-type plants (Figure 3). It is also possible that the translational efficiency of these transcripts is elevated in the mutants, as might occur if autogenous control mechanisms regulate ribosomal protein synthesis in chloroplasts, as in Escherichia coli (36).
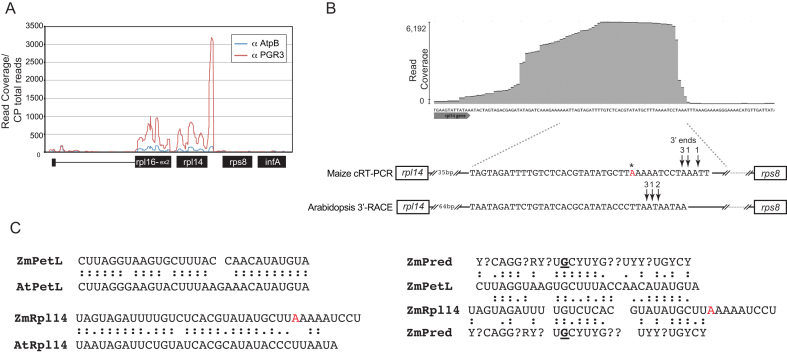
The absence of spliced and unspliced dicistronic rpl16-rpl14 RNA in maize and Arabidopsis pgr3 mutants suggested that PGR3 stabilizes either the 5′-end upstream of rpl16 or the 3′-end downstream of rpl14. A different PPR protein, PPR103, has been shown to stabilize the 5′-end of these transcripts by binding the rpl16 5′-UTR (24). As the 5′-stabilization function is accounted for, it seemed most likely that PGR3 blocks 3′→5′ degradation by binding downstream of rpl14. This view was confirmed by sequencing RNAs that coimmunoprecipitate with Zm-PGR3 from stromal extract (RIP-seq). In comparison with a control assay using a different antibody, immunoprecipitation with anti-PGR3 enriched RNA from across the rpl16-rpl14 region, with peak enrichment mapping to a short segment in the rpl14-rps8 intergenic region (Figure 4A and B; Supplementary Figure S3). We mapped the 3′-end of the PGR3-dependent rpl16-rpl14 RNA in maize and found that it coincides with the peak of RNA enrichment in the RIP-seq data (Figure 4B). We showed further that the PGR3-dependent rpl14 3′-end in Arabidopsis maps a short distance downstream from a sequence with strong similarity to the maize PGR3-binding site (Figure 4B). Taken together, these data provide strong evidence that PGR3 in Arabidopsis and maize binds within the rpl14-rps8 intergenic region and, in so doing, stabilizes the RNA upstream of its binding site. This function is consistent with the canonical barrier mechanism for PPR-mediated RNA stabilization (13,14,29). Our data suggest, in addition, that the binding of PGR3 to the rpl14-rps8 intergenic region stimulates rps8 translation in both maize and Arabidopsis: rps8 transcripts are of normal size and somewhat elevated abundance in pgr3 mutants (Figure 3), yet the abundance of ribosome footprints on rps8 is reduced at least as much as that on rpl14 (Figure 2). We conclude, therefore, that the binding of PGR3 in the rpl14-rps8 intergenic region simultaneously stabilizes a processed 3′-RNA terminus and stimulates rps8 translation. PGR3’s stimulatory effect on the expression of two genes that encode ribosomal proteins (rpl14 and rps8) likely contributes to the loss of plastid ribosomes in maize and Arabidopsis pgr3 mutants.
Figure 4.
RIP-seq analysis showing that Zm-PGR3 associates with RNA from the rpl14-rps8 intergenic region in vivo. Antibody to Zm-PGR3 was used for immunoprecipitation from maize chloroplast stroma. RNA purified from the immunoprecipitate was analyzed by deep sequencing. An immunoprecipitation with antibody to AtpB (a subunit of the chloroplast ATP synthase) served as a negative control. (A) Sequence read counts in the rpl14 region. Read counts were normalized to the total number of reads mapping to the chloroplast genome. A genome-wide view of the data is shown in Supplementary Figure S3. (B) Position of the PGR3-binding site and PGR3-dependent 3′-ends in the rpl14-rps8 intergenic region. A screen capture of an excerpt of the PGR3 RIP-seq data displayed with the Geneious browser is shown at top. The sequence is expanded below, where the positions of the PGR3-dependent 3′-ends as mapped by circular RT-PCR (maize) or 3′-RACE (Arabidopsis) are marked. The number of clones representing each 3′-end is indicated. The asterisk marks a difference in the sequences we obtained from that in the reference maize chloroplast sequence (GENBANK Accession NC_001666). (C) Comparison of PGR3’s binding sites near petL and rpl14. Alignments between the orthologous sites in maize (Zm) and Arabidopsis (At) are shown at left. An alignment between the maize petL and rpl14 sites and the Zm-PGR3 binding site as predicted by the PPR code (ZmPred) is shown to the right. The code prediction was based on the nucleotide specificities according to the published PPR code (8). PPR motifs that lacked canonical amino acids at the specificity-determining positions are marked with question marks. The binding site prediction for Arabidopsis PGR3 is similar, with the exception of the position in bold underline near the center of the protein, which is predicted to be a pyrimidine in Arabidopsis.
A comparison of the sequences bound by Zm-PGR3 near rpl14 and petL and the orthologous regions in Arabidopsis is shown in Figure 4C. The sequence in each region is highly conserved between maize and Arabidopsis (Figure 4C, left). However, the similarity between the petL and rpl14 sites is patchy, and their optimal alignment requires incorporation of two gaps (Figure 4C, right). The sequence that is predicted to bind Zm-PGR3 based on the PPR code is aligned to each binding site in Figure 4C (right). The predicted binding site includes seven contiguous matches with the central segment of both the petL and rpl14 sites. However, the petL site shows greater similarity to the predicted site in the 5′-region, whereas the rpl14 site is more similar to the predicted site in the 3′-region. Thus, the predicted ‘tightness’ of the interaction at each end of the RNA site correlates with the ability to block either 5′- or 3′-exonucleolytic degradation (at petL and rpl14, respectively). However, these alignments are speculative, and experimental data would be required to define the register between the protein and RNAs.
A genome-wide view of the Zm-PGR3 RIP-seq data revealed, in addition to the interaction near rpl14, the known interaction at petL (Supplementary Figure S3). A small peak of enrichment was also observed near atpF. RNA gel blot hybridizations showed a small reduction in the abundance of a transcript with a 3′-end mapping to the atpF-atpA intergenic region, similar to what is observed in atp4 mutants (Supplementary Figure S2B) (37). This suggests that PGR3 and ATP4 cooperate to stabilize that 3′-end, just as they cooperate to stabilize the 3′-end in the rpl14-rps8 intergenic region. The RIP-seq data did not detect interactions with mRNAs encoding NDH subunits, suggesting that the minor defects in ndh expression detected by ribosome profiling (Figure 2B) are secondary effects that do not result from direct interactions with Zm-PGR3. Likewise, the other minor gene expression defects suggested by the Zm-pgr3 ribosome profiling and RNA gel blot data were not reflected in the RIP-seq data, suggesting that these are also secondary effects. However, false-negatives are possible in RNA coimmunoprecipitation assays, so we cannot rule out the possibility of additional sites of Zm-PGR3–RNA interaction. In addition, a role for Arabidopsis PGR3 in translation of ndhE or ndhG cannot be excluded from our ribosome profiling data due to the low signal for these genes even in the wild-type (Figure 2A).
PPR5 promotes rpl16 splicing
PPR5 is a P-type PPR protein in maize with ten PPR motifs. PPR5 binds the group II intron in the chloroplast trnG-UCC precursor, where it prevents endonucleolytic cleavage of the intron while also stimulating splicing (18,19). We discovered a second site of PPR5 action serendipitously when RNA from a ppr5 mutant was included as a control on an RNA gel blot that had been prepared for other purposes (Figure 5). The results showed that spliced rpl16-rpl14 RNA fails to accumulate in the ppr5 mutant, whereas the unspliced transcript accumulates normally. This effect contrasts with the loss of both spliced and unspliced isoforms in ppr103 (24) (Figure 5) and pgr3 mutants (Figure 3). These results indicate that PPR5 is required for the removal of the group II intron in rpl16. In fact, RIP-chip data reported previously for PPR5 had suggested an interaction with the rpl16 intron and had prompted examination of rpl16 transcripts in ppr5 mutants (19). However, only hypomorphic ppr5 alleles had been examined and their effect on rpl16 splicing was incomplete (19). The current data in conjunction with the prior RIP-chip data (19) provide strong evidence that PPR5 interacts with the rpl16 intron, and that this interaction stimulates splicing.
Figure 5.
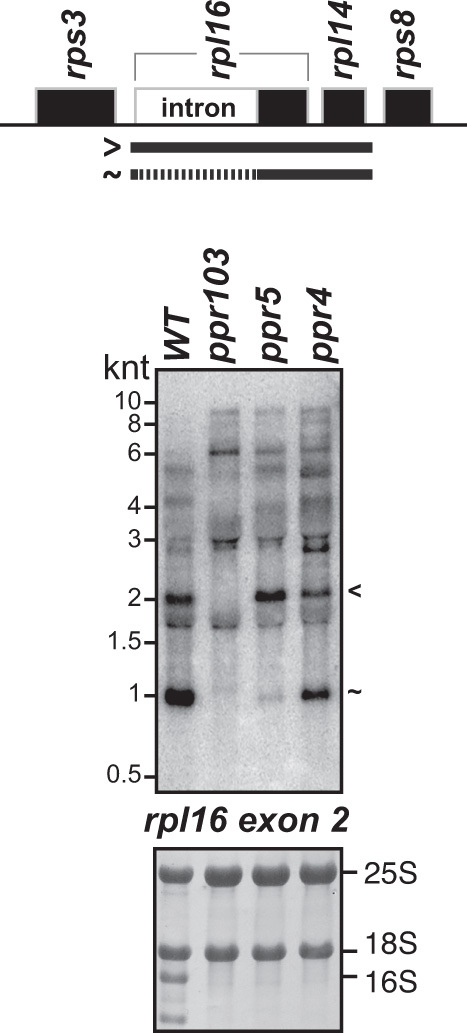
RNA gel blot hybridization demonstrating a defect in rpl16 splicing in ppr5 mutants. Seedling leaf RNA from ppr5-1 mutants was compared to that in ppr103-2/-3 and ppr4-1 mutants. The ppr103 mutants lack both spliced and unspliced rpl16 RNAs due to a defect in stabilizing the processed 5′-end in the rpl16 5′-UTR (24). The ppr4-1 mutants are deficient for plastid ribosomes due to a defect in rps12 splicing (23). rRNA abundance is shown on the methylene blue stained blots below. The blot was probed to detect the second exon of rpl16. Transcripts were identified based on the data in Figure 3 and in (15).
DISCUSSION
Genetic and biochemical data support the view that many PPR proteins bind RNA with a high degree of sequence specificity (reviewed in 7). However, the assays used to detect sites of PPR action have typically been limited in scope, so the degree to which PPR proteins bind RNA selectively in vivo remains unclear. Results presented here show that three PPR proteins that were already among the best characterized members of the PPR family have functions that previously escaped detection (summarized in Figure 6): PPR10 stabilizes a 3′-RNA terminus in the psaI-ycf4 intergenic region, PGR3 acts in the rpl14-rps8 intergenic region to stabilize a 3′-RNA terminus and stimulate rps8 translation, and PPR5 promotes the splicing of the group II intron in rpl16. In vitro RNA binding and/or in vivo RNA coimmunoprecipitation data strongly suggest that these functions involve direct interactions between each protein and the affected RNAs. In addition, our results show that PGR3 does not activate ndhA translation, as had previously been suggested. Implications of these findings with regard to PPR functions, nuclear-organellar coevolution and the engineering of PPR proteins to bind specified RNAs are discussed below.
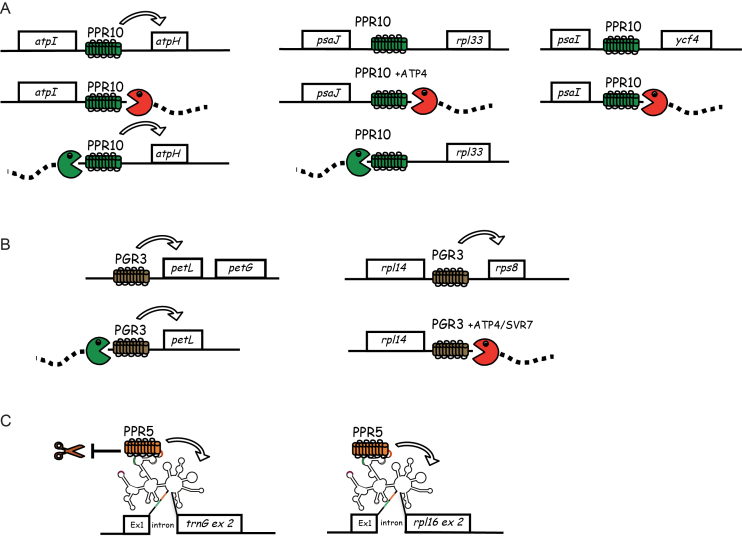
Figure 6.
Summary of the known functions of PPR10 (A), PGR3 (B) and PPR5 (C). See text for details. Arrows indicate an enhancement of translational efficiency (PPR10, PGR3) or RNA splicing (PPR5). RIP-seq data suggest that Zm-PGR3 may also interact weakly in the atpF-atpA intergenic region and possibly with a transcript related to the 23S rRNA. The diagrammed binding sites for PPR10, PGR3 and PPR5 are supported by RNA coimmunoprecipitation and/or in vitro RNA-binding assays. However, the effects of ATP4/SVR7 have not been shown to result from direct interaction with RNA. The fact that Zm-PGR3 mutants have a much more severe plastid ribosome deficiency than does Arabidopsis pgr3 implies that Zm-PGR3 has an as yet unidentified target involved in ribosome biogenesis. The RIP-seq data for Zm-PGR3 (Supplementary Figure S3) suggest that this might involve an interaction with 23S rRNA or a precursor thereof.
Need for multiple genome-wide assays to reveal functional repertoires of organelle RNA-binding proteins
A variety of genome-wide assays have been used to detect gene expression defects in mutants lacking PPR and other organellar RNA-binding proteins. Changes in RNA abundance, editing, splicing and end-processing have been detected by qRT-PCR, RNA-seq, microarray hybridization and cRT-PCR (e.g. 15,38,39,40), defects in translation have been detected by ribosome profiling (15,41) and the loss of in vivo RNA ‘footprints’ has been detected by small RNA sequencing (e.g. 42,43). Genome-wide RNA coimmunoprecipitations (RIP-chip and RIP-seq) have complemented genetic data by identifying RNAs that associate with a particular protein in vivo (e.g. 31,44). However, each of these assays has limitations. For example, false-negatives can occur in RNA coimmunoprecipitation assays due to low affinity RNA–protein interactions, qRT-PCR assays do not report defects in transcript end-processing and ribosome profiling does not capture defects in rRNA or tRNA metabolism. Thus, although we had previously assayed RNA abundance and translation in ppr10 mutants by microarray hybridization and ribosome profiling (15), and we had also assayed PPR10–RNA interactions by RIP-chip (13), we had nonetheless missed PPR10’s site of action downstream of psaI.
In this context, it seems likely that additional functions remain to be discovered for numerous PPR proteins that have already been characterized to some extent. In particular, roles of PPR proteins as translational activators have almost certainly been underestimated due to the technical challenge of identifying translation defects. Ribosome profiling now makes it possible to uncover effects on translation that would otherwise have escaped detection (e.g. PGR3’s effect on rps8 translation), and to rule out putative translational effects that had been difficult to assess (e.g. the proposed role for PGR3 in ndhA translation). The use of ribosome profiling and RNA-seq to detect gene expression defects in ppr mutants, in conjunction with RIP-seq to identify sites of PPR-RNA interaction, can be expected to reveal the majority of sites of PPR action.
Although mutant phenotypes have been invaluable for directing experiments to potential sites of PPR action, some PPR–RNA interactions have no apparent physiological consequence (e.g. 45,46). Indeed, it is unclear whether PPR10’s RNA-stabilization effect downstream of psaI affects the rate of PsaI protein synthesis. Nonetheless, discovery of such interactions elucidates the evolutionary trajectory through which PPR proteins gain and lose functionalities, clarifies the principles that govern PPR–RNA interactions in vivo and provides a knowledge base for inferring potential binding sites of both native and engineered PPR proteins.
Complexity of the RNA–protein interactome in chloroplasts
Post-transcriptional steps in chloroplast gene expression have gained a remarkable complexity since the divergence of chloroplasts from their cyanobacterial ancestor (reviewed in 1). The expression of many chloroplast genes requires distinct RNA-binding proteins to foster RNA stabilization, RNA editing, RNA splicing and/or translation. The psbB transcription unit has served as the paradigm for this phenomenon, with 12 RNA-binding proteins required to promote specific steps in the expression of its five genes (reviewed in 1,47). With the results reported here, the known RNA–protein interactome on rpl16-rpl14-rps8 RNA has reached a similar complexity. Four PPR proteins have been shown to act on the RNA transcribed from these three genes: PPR103 stabilizes the processed 5′-end upstream of rpl16 (24), PGR3 and ATP4/SVR7 collaborate to stabilize a 3′-end downstream of rpl14, PGR3 stimulates rps8 translation and PPR5 promotes the splicing of the group II intron in rpl16. In addition, one PORR domain protein (WTF1), two CRM domain proteins (CFM3 and CAF1) and two proteins that evolved from ancient RNA metabolizing enzymes (CRS2 and RNC1) promote the splicing of the rpl16 intron (reviewed in 5). This complexity does not serve any apparent adaptive function. We favor the hypothesis that promiscuous RNA–protein interactions that were initially of no physiological consequence became cogs in an evolutionary ratchet that led to the fixation of these interactions by relaxing prior evolutionary constraints (7,48,49). A thorough cataloging of the RNAs bound in vivo by orthologous PPR proteins will facilitate progress in understanding the evolutionary transformations of chance binding interactions into essential partnerships.
Implications for predicting PPR-binding sites and effects on gene expression
Despite the elucidation of an amino acid code that influences the nucleotide specificity of PPR motifs, native PPR proteins have idiosyncratic features that complicate prediction of their sequence specificity. For example, a comprehensive analysis of PPR10’s sequence specificity in vitro revealed several nucleotides whose identities are critical for a high-affinity interaction that cannot be explained by the PPR code (50). Synthetic proteins consisting of consensus PPR motifs behaved more predictably in a similar analysis, but extension of consensus PPR tracts beyond ∼10 motifs resulted in a high tolerance for ‘mismatches’ and, thus, a diversity of high-affinity RNA ligands (51). As PPR10 and PGR3 have 19 and 28 PPR motifs, respectively, it is not surprising that they engage multiple RNA ligands in chloroplasts. Even so, the details of the newly discovered PGR3 and PPR10 binding sites highlight the flexibility in target site recognition by long PPR tracts. PPR10’s three binding sites share sequences at the 5′- and 3′-ends, including the nucleotides that are most important for high-affinity binding in vitro (Figure 1D) (50). However, the spacing between these sequences differs among the three sites, with the atpH spacing correlating with the highest affinity for PPR10. PGR3’s two binding sites are highly conserved between maize and Arabidopsis (Figure 4C, left) but show only patchy similarity with one another and with the binding site predicted by the PPR code (Figure 4C, right). This degree of flexibility in target site recognition poses a challenge for PPR-binding site prediction and the design of synthetic PPR proteins, especially when aiming to target a protein to an RNA in the nucleo-cytosolic compartment with its complex sequence space.
A previous study provided evidence that PGR3’s 5′-RNA stabilization and translational activation functions partition between its N- and C- terminus, respectively (17). That PPR10 and PGR3 stabilize a 3′-end but not a 5′-end at their newly discovered binding sites adds to the evidence that the two ends of long PPR tracts can act independently of one another. This view is supported by the hypothetical alignment between the predicted Zm-PGR3 binding site and its actual binding sites (Figure 4C), which suggests a correlation between the tightness of the binding interface at each end of the RNA–PGR3 ‘duplex’ and the ability to block exoribonucleases invading from each direction.
Despite the wealth of knowledge about PPR10, PGR3 and other PPR proteins, many questions remain about the factors that determine whether a particular PPR protein establishes physiologically meaningful interactions with particular RNAs in vivo, and how such interactions impact gene expression. For example, it is known that both PPR10 and PGR3 cooperate with the PPR-SMR protein ATP4/SVR7 for a subset of their functions: PPR10 and ATP4 are required to stabilize the 3′-end downstream of psaJ (13,37), whereas both PGR3 and ATP4 are required to stabilize the 3′-end downstream of rpl14 (this work and 15). The mechanisms underlying this cooperation are mysterious. Another unresolved question concerns mechanisms by which PPR proteins stimulate translation. PPR10 and PGR3 bind adjacent to the translation initiation region at atpH and petL, respectively, where they are well placed to activate translation by preventing RNA structures that would otherwise occlude the ribosome binding region (14,16,52). However, this mechanism seems less likely to account for the small but clear effect of PGR3 on rps8 translation, where the binding interaction is ∼80 and ∼100 nt from the start codon in maize and Arabidopsis, respectively. Also unknown are the precise PPR5-binding sites in the trnG and rpl16 introns, the structural basis for those interactions, or how they stimulate splicing. The rapidly expanding toolkit for discovering the locations and consequences of RNA–protein interactions can be anticipated to speed progress in answering these and other questions of PPR function and evolution in the coming few years.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Toshiharu Shikanai (Kyoto University) for providing the exon insertion allele of pgr3 and Ian Small (University of Western Australia) for useful discussions. We also wish to acknowledge the valuable assistance of Kenneth Watkins, Rosalind Williams-Carrier and Prakitchai Chotewutmontri (University of Oregon) with the RIP-seq and maize ribosome profiling experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Science Foundation [MCB-1243641 to A.B.]; Deutsche Forschungsgemeinschaft [SCHM 1698/5-1, TRR175-A02 to C.S.L.; ZO 302/4-1 to R.Z.]; National Institutes of Health [T32-GM007759 to R.G.M]. Funding for open access charge: University of Oregon Academic Support Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011; 155:1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bock R. Bock R. Structure, function, and inheritance of plastid genomes. Cell and Molecular Biology of Plastids. 2007; Heidelberg: Springer-Verlag; 29–63. [Google Scholar]
- 3. Borner T., Aleynikova A.Y., Zubo Y.O., Kusnetsov V.V.. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta. 2015; 1847:761–769. [DOI] [PubMed] [Google Scholar]
- 4. Zoschke R., Bock R.. Chloroplast Translation: structural and functional organization, operational control and regulation. Plant Cell. 2018; 30:745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Germain A., Hotto A.M., Barkan A., Stern D.B.. RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA. 2013; 4:295–316. [DOI] [PubMed] [Google Scholar]
- 6. Small I., Peeters N.. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000; 25:46–47. [DOI] [PubMed] [Google Scholar]
- 7. Barkan A., Small I.. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014; 65:415–442. [DOI] [PubMed] [Google Scholar]
- 8. Barkan A., Rojas M., Fujii S., Yap A., Chong Y.S., Bond C.S., Small I.. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012; 8:e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T.. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One. 2013; 8:e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takenaka M., Zehrmann A., Brennicke A., Graichen K.. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One. 2013; 8:e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H.W. et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013; 504:168–171. [DOI] [PubMed] [Google Scholar]
- 12. Shikanai T. RNA editing in plants: Machinery and flexibility of site recognition. Biochim. Biophys. Acta. 2015; 1847:779–785. [DOI] [PubMed] [Google Scholar]
- 13. Pfalz J., Bayraktar O., Prikryl J., Barkan A.. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009; 28:2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prikryl J., Rojas M., Schuster G., Barkan A.. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zoschke R., Watkins K., Barkan A.. A rapid microarray-based ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013; 25:2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cai W., Okuda K., Peng L., Shikanai T.. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 2011; 67:318–327. [DOI] [PubMed] [Google Scholar]
- 17. Fujii S., Sato N., Shikanai T.. Mutagenesis of individual pentatricopeptide repeat motifs affects RNA binding activity and reveals functional partitioning of Arabidopsis PROTON gradient regulation3. Plant Cell. 2013; 25:3079–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams-Carrier R., Kroeger T., Barkan A.. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008; 14:1930–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A.. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 2008; 28:5337–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., Bruyere C., Caboche M., Debast C., Gualberto J., Hoffmann B. et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004; 16:2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Belcher S., Williams-Carrier R., Stiffler N., Barkan A.. Large-scale genetic analysis of chloroplast biogenesis in maize. Biochim. Biophys. Acta. 2015; 1847:1004–1016. [DOI] [PubMed] [Google Scholar]
- 22. Barkan A. Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell. 1993; 5:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmitz-Linneweber C., Williams-Carrier R.E., Williams-Voelker P.M., Kroeger T.S., Vichas A., Barkan A.. A pentatricopeptide repeat protein facilitates the Trans-Splicing of the maize chloroplast rps12 Pre-mRNA. Plant Cell. 2006; 18:2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammani K., Takenaka M., Miranda R., Barkan A.. A PPR protein in the PLS subfamily stabilizes the 5′-end of processed rpl16 mRNAs in maize chloroplasts. Nucleic Acids Res. 2016; 44:4278–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zoschke R., Qu Y., Zubo Y.O., Borner T., Schmitz-Linneweber C.. Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J. Plant Res. 2013; 126:403–414. [DOI] [PubMed] [Google Scholar]
- 26. Chotewutmontri P., Barkan A.. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 2016; 12:e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weber K., Bolander M.E., Sarkar G.. PIG-B: a homemade monophasic cocktail for the extraction of RNA. Mol. Biotechnol. 1998; 9:73–77. [DOI] [PubMed] [Google Scholar]
- 28. Barkan A. Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 1998; 297:38–57. [Google Scholar]
- 29. Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suarez M., Borner T., Barkan A.. Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 2012; 40:3092–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I., Schmitz-Linneweber C.. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell. 2012; 24:4266–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmitz-Linneweber C., Williams-Carrier R., Barkan A.. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell. 2005; 17:2791–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamazaki H., Tasaka M., Shikanai T.. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004; 38:152–163. [DOI] [PubMed] [Google Scholar]
- 33. Williams P., Barkan A.. A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 2003; 36:675–686. [DOI] [PubMed] [Google Scholar]
- 34. Prikryl J., Watkins K.P., Friso G., Wijk K.J., Barkan A.. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008; 36:5152–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu X., Yu F., Rodermel S.. An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 2010; 154:1588–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sykes M.T., Sperling E., Chen S.S., Williamson J.R.. Quantitation of the ribosomal protein autoregulatory network using mass spectrometry. Anal. Chem. 2010; 82:5038–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zoschke R., Kroeger T., Belcher S., Schottler M.A., Barkan A., Schmitz-Linneweber C.. The pentatricopeptide Repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 2012; 72:547–558. [DOI] [PubMed] [Google Scholar]
- 38. Forner J., Weber B., Thuss S., Wildum S., Binder S.. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007; 35:3676–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chateigner-Boutin A.L., Ramos-Vega M., Guevara-Garcia A., Andres C., de la Luz Gutierrez-Nava M., Cantero A., Delannoy E., Jimenez L.F., Lurin C., Small I. et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 2008; 56:590–602. [DOI] [PubMed] [Google Scholar]
- 40. Bentolila S., Oh J., Hanson M.R., Bukowski R.. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013; 9:e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zoschke R., Watkins K.P., Miranda R.G., Barkan A.. The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J. 2016; 85:594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu W., Liu S., Ruwe H., Zhang D., Melonek J., Zhu Y., Hu X., Gusewski S., Yin P., Small I.D. et al. SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J. 2016; 85:607–621. [DOI] [PubMed] [Google Scholar]
- 43. Douchi D., Qu Y., Longoni P., Legendre-Lefebvre L., Johnson X., Schmitz-Linneweber C., Goldschmidt-Clermont M.. A Nucleus-Encoded chloroplast phosphoprotein governs expression of the photosystem I subunit PsaC in Chlamydomonas reinhardtii. Plant Cell. 2016; 28:1182–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meurer J., Schmid L.M., Stoppel R., Leister D., Brachmann A., Manavski N.. PALE CRESS binds to plastid RNAs and facilitates the biogenesis of the 50S ribosomal subunit. Plant J. 2017; 92:400–413. [DOI] [PubMed] [Google Scholar]
- 45. Okuda K., Hammani K., Tanz S.K., Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T.. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J. 2010; 61:339–349. [DOI] [PubMed] [Google Scholar]
- 46. Brehme N., Bayer-Csaszar E., Glass F., Takenaka M.. The DYW subgroup PPR protein MEF35 targets RNA editing sites in the mitochondrial rpl16, nad4 and cob mRNAs in Arabidopsis thaliana. PLoS One. 2015; 10:e0140680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stoppel R., Meurer J.. Complex RNA metabolism in the chloroplast: an update on the psbB operon. Planta. 2013; 237:441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lefebvre-Legendre L., Merendino L., Rivier C., Goldschmidt-Clermont M.. On the complexity of chloroplast RNA metabolism: psaA trans-splicing can be bypassed in chlamydomonas. Mol. Biol. Evol. 2014; 31:2697–2707. [DOI] [PubMed] [Google Scholar]
- 49. Lukes J., Archibald J.M., Keeling P.J., Doolittle W.F., Gray M.W.. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life. 2011; 63:528–537. [DOI] [PubMed] [Google Scholar]
- 50. Miranda R.G., Rojas M., Montgomery M.P., Gribbin K.P., Barkan A.. RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10. RNA. 2017; 23:586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miranda R.G., McDermott J.J., Barkan A.. RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions. Nucleic Acids Res. 2018; 46:2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klinkert B., Elles I., Nickelsen J.. Translation of chloroplast psbD mRNA in Chlamydomonas is controlled by a secondary RNA structure blocking the AUG start codon. Nucleic Acids Res. 2006; 34:386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhelyazkova P., Sharma C.M., Forstner K.U., Liere K., Vogel J., Borner T.. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012; 24:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McCormac D., Barkan A.. A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell. 1999; 11:1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.