Abstract
The transcriptional activator RbpA associates with Mycobacterium tuberculosis RNA polymerase (MtbRNAP) during transcription initiation, and stimulates formation of the MtbRNAP-promoter open complex (RPo). Here, we explored the influence of promoter motifs on RbpA-mediated activation of MtbRNAP containing the stress-response σB subunit. We show that both the ‘extended −10’ promoter motif (T-17G-16T-15G-14) and RbpA stabilized RPo and allowed promoter opening at suboptimal temperatures. Furthermore, in the presence of the T-17G-16T-15G-14 motif, RbpA was dispensable for RNA synthesis initiation, while exerting a stabilization effect on RPo. On the other hand, RbpA compensated for the lack of sequence-specific interactions of domains 3 and 4 of σB with the extended −10 and the −35 motifs, respectively. Mutations of the positively charged residues K73, K74 and R79 in RbpA basic linker (BL) had little effect on RPo formation, but affected MtbRNAP capacity for de novo transcription initiation. We propose that RbpA stimulates transcription by strengthening the non-specific interaction of the σ subunit with promoter DNA upstream of the −10 element, and by indirectly optimizing MtbRNAP interaction with initiation substrates. Consequently, RbpA renders MtbRNAP promiscuous in promoter selection, thus compensating for the weak conservation of the −35 motif in mycobacteria.
INTRODUCTION
In bacteria, transcription is performed by the multi-subunit DNA-dependent RNA polymerase (RNAP) that is composed of the catalytic core (E, subunits 2αββ'ω) and the σ subunit, required for promoter-specific initiation of RNA synthesis (reviewed in (1,2)). During exponential growth, expression of most genes is controlled by the housekeeping (principal) σ subunit (σ70 in Escherichia coli, and σA in Mycobacterium tuberculosis) that belongs to the Group 1 σ subunits. Alternative Group 2 σ subunits (σS in E. coli, and σB in M. tuberculosis) are the most similar to the principal σ subunit, and are responsible for the expression of specialized genes in response to stress, during the stationary growth phase and dormancy (3,4).
Most bacterial promoters recognized by Group 1 and 2 σ subunits belong to the −10/−35 class and contain two consensus elements: the −10 element (E. coli consensus motif: T-12A-11T-10A-9A-8T-7) and the −35 element (E. coli consensus motif: T-35T-34G-33A-32C-31A-30). These motifs are recognized by domain 2 (σ2) and 4 (σ4) of the σ subunit, respectively. The ‘extended −10’ class of promoters contains the extended −10 motif (T-17R-16T-15G-14; R = purine) that is located one nucleotide upstream of the −10 element (5–7) and is recognized by domain 3 of the σ subunit (σ3). It has been shown that the extended −10 motif bypasses the requirement of the σ4/-35 element interaction (8,9). The percentage of promoters containing at least the downstream part of the extended −10 motif (T-15G-14) varies among bacteria, from ∼18% in E. coli to ∼45% in Bacillus subtilis (6,7,10).
During transcription initiation, RNAP binds to the promoter and forms an unstable ‘closed complex’ (RPc) that isomerizes spontaneously into a transcriptionally competent ‘open complex’ (RPo) through the formation of several intermediate complexes (RPi) (11–13). The concerted action of the RNAP core and σ subunit triggers the opening of ∼13 bp of the promoter DNA around the transcription start site, and makes the single-stranded DNA template available for initiation of RNA synthesis (14–16).
M. tuberculosis RNAP (MtbRNAP) differs from the extensively studied E. coli RNAP because it requires auxiliary factors (CarD and RbpA) to form stable RPo on housekeeping gene promoters (17,18). RbpA is a global transcriptional activator essential for M. tuberculosis growth, and could be implicated in the control of its physiological state (19–22). RbpA selectively binds to the σA and σB subunits of MtbRNAP and stimulates RPo formation (19,23,24). It has been shown that the stress-response σB-MtbRNAP displays stronger dependence on RbpA than σA-MtbRNAP (24)
Structural studies demonstrated that RbpA C-terminal domain interacts with σ2 via its σ-interacting domain (SID), whereas RbpA basic linker (BL) interacts with promoter sequences upstream of the −10 element (25,26). RbpA seems not to recognize any DNA motif, although its requirement for transcription has been shown to be promoter sequence-dependent (18,24). Indeed, RbpA is required for the stable binding of σB-MtbRNAP at promoters of the −10/-35 (rrnAP3, sigAP, lacUV5) and extended −10 class (galP1cons) (24). However, it is dispensable for RPo formation at the extended −10 class sinP3 promoter of B. subtilis (24). Recently, we demonstrated that RbpA stabilizes the ‘open’ conformation of the σB subunit in MtbRNAP. This is optimal for recognition of the −10/-35 promoters, but is dispensable for recognition of the extended −10 promoters (27). Here, to better understand the molecular basis of this promoter specificity, we explored the effect of mutations in σB and RbpA on MtbRNAP activity at promoter variants that harbor different combinations of the extended −10 and −35 motifs. We found that interaction between domain 3 of σB and the extended −10 motif strongly influences MtbRNAP activity, but has no effect on its ability to respond to RbpA activation. Furthermore, we found that RbpA modulates MtbRNAP selectivity for nucleotide substrates.
MATERIALS AND METHODS
Proteins and DNA fragments
MtbRNAP, the σB subunit and RbpA were expressed and purified as described before (24). Mutations in σB and RbpA were introduced using the Agilent Quick Change Lightening Site-directed Mutagenesis Kit, following the manufacturer's protocol. Variants of the sigAP promoter were prepared by annealing two oligonucleotides followed by primer extension and PCR amplification with Pfu using fluorescent primers (Table S1). The amplified promoter DNA fragments were resolved by 8% native PAGE and extracted using the Nucleospin® Gel and PCR Clean-up Kit (Macherey Nagel). The sigAP-TGTG promoter labeled with Cy3 at the +2 position was purified through 6% PAGE after primer extension.
EMSA and KMnO4 probing
Core MtbRNAP (100 nM) was mixed with σB (300 nM) and RbpA (300 nM) in transcription buffer (TB, 40 mM HEPES pH 8.0, 50 mM NaCl, 5 mM MgCl2, and 5% glycerol) and incubated at 37°C for 10 min. Then, fluorescein-labeled promoter DNA (50 nM) was added and samples were incubated at 37°C for 10 min. The competitor poly(dA-dT) was added to a final concentration of 20 ng/μl and incubated at 37°C for 5 min. Samples were resolved on 6% native PAGE in 1× TBE buffer. Gels were scanned with a Amersham Imager 600 (GE Healthcare) and quantified using the ImageQuant software. For KMnO4 probing experiments, 5 mM KMnO4 was added to the reaction mixtures formed at the indicated temperatures for 30 s, and quenched by addition of 1 M β-mercaptoethanol, 1.5 M Na(CH3COO) pH 7.0. Reactions were incubated with 0.5 M piperidine at 90°C for 15 min, and DNA fragments were precipitated by adding 1/10 volume of 5 M LiCl and 4 volumes of ice-cold ethanol. Precipitated DNA fragments were washed with 80% ethanol, vacuum-dried, dissolved in 90% formamide and analyzed on 8% sequencing gels. Gels were scanned with a Typhoon 9400 Imager (GE Healthcare) and quantified using the ImageQuant software. Graphs were plotted using the Graphpad7 and Grace-5.1.23 software (http://plasma-gate.weizmann.ac.il/Grace/) software. The apparent dissociation constants (Kd) were calculated from equation: RP = A0[RNAP]/([RNAP]+ Kd, where RP is the RNAP fraction bound to DNA.
Transcription assays
Multiple-round transcription assays were performed in 10 μl of TB with 50 μM/each of ATP, GTP, CTP, 5 μM of UTP and 0.5 μM of [α32P]-UTP at 37°C for 5 min. The GpC primer (Eurogentec) was added to 100 μM, when indicated.
Single-round transcription assays, to monitor RPo formation, were performed in 10 μl of TB. First, 180 nM MtbRNAP core, 590 nM σB and 590 nM RbpA were mixed and incubated at 37°C for 5 min. After addition of 50 nM of promoter DNA, samples were incubated at 37°C for 1, 2, 3, 5 and 10 min. Transcription was initiated by adding 50 μM/each of ATP, GTP, CTP, 10 μM of UTP, 0.5 μM of [α32P]-UTP and poly(dI-dC) (0.1 mg/ml final concentration) and performed at 37°C for 3 min. Single-round transcription assays, to monitor promoter escape, were performed using the same conditions as for RPo formation. MtbRNAP-promoter complexes were incubated at 37°C for 15 min (longer incubation at 37°C resulted in MtbRNAP inactivation). Then, after addition of the NTPs/poly(dI-dC) mixture, transcription was performed for 0.5, 1, 2, 5 and 10 min. Abortive transcription assays using the lacUV5 bubble template (28) were performed in 10 μl of TB. First, 180 nM MtbRNAP core was mixed with 590 nM σB or 1 μM σBΔ4 and incubated at 37°C for 5 min. Then, 50 nM bubble DNA was added and incubated at room temperature (RT; 22°C) for 15 min. After addition of 0.5 mM ApA, 100 μM GTP, and 0.5 μM of [α32P]-UTP, samples were incubated at RT for 10 min, and then reactions were stopped by addition of an equal volume of 7M urea/100 mM EDTA solution. RNA transcripts were analyzed on denaturing 18% PAGE/7M urea gels. Gels were scanned with a Molecular Dynamics STORM Imager. Bands were quantified using the ImageQuant software. For kinetics experiments, raw data were fitted in Grace-5.1.23 using the mono-exponential function At= A∞+ A0 · exp(–k · t), where At is the radioactive RNA signal at the time point t. The A∞ values determined from the fits were used for data normalization in each experimental set. Normalized data were used to calculate the mean and standard error (SE) values shown in figures. The half-time values, t1/2, were calculated as: t1/2 = ln(2)/k.
Fluorescent assay to determine the dissociation kinetics
Assays were performed in 60 μl of TB; 50 nM of RNAP was mixed with 5 nM of the sigAP promoter fragment labeled with Cy3 at position +2 of the non-template DNA strand and incubated at 37°C for 10 min. To initiate dissociation of the MtbRNAP-promoter complexes, heparin was added to 10 ng/μl. Data were acquired using a PTI QuantaMaster spectrophotometer at room temperature. Data were fitted using the following bi-exponential equation: At = A0 + A1⋅ exp(–kfast · t)+ A2⋅ exp(–kslow · t), where At is the promoter DNA fraction bound to RNAP, calculated from the fluorescence fold change value (At = (F – F0)/F0), and kfast and kslow are the rate constants for the fast and slow phase, respectively; F is the fluorescence signal of RNAP-bound DNA, and F0 is the fluorescence signal of free DNA. The slow phase constant kslow was considered to be the rate constant kd for RPo dissociation.
Native gel electrophoresis assay to study RbpA binding
RbpA was conjugated with the sulfhydryl- reactive dye, DyLight 633 Maleimide (Thermo Scientific), as described (24). Labeled RbpA (1.6 μM) was incubated with different concentrations of the σB subunit (0.8, 1.6 and 3.2 μM) in 10 μl of TB at 37°C for 10 min. Samples were analyzed on 5–10% native PAGE in Tris–glycine buffer. Gels were scanned with a Typhoon 9400 Imager (GE Healthcare).
RESULTS
RbpA is dispensable for transcription from promoters containing the extended −10 motif
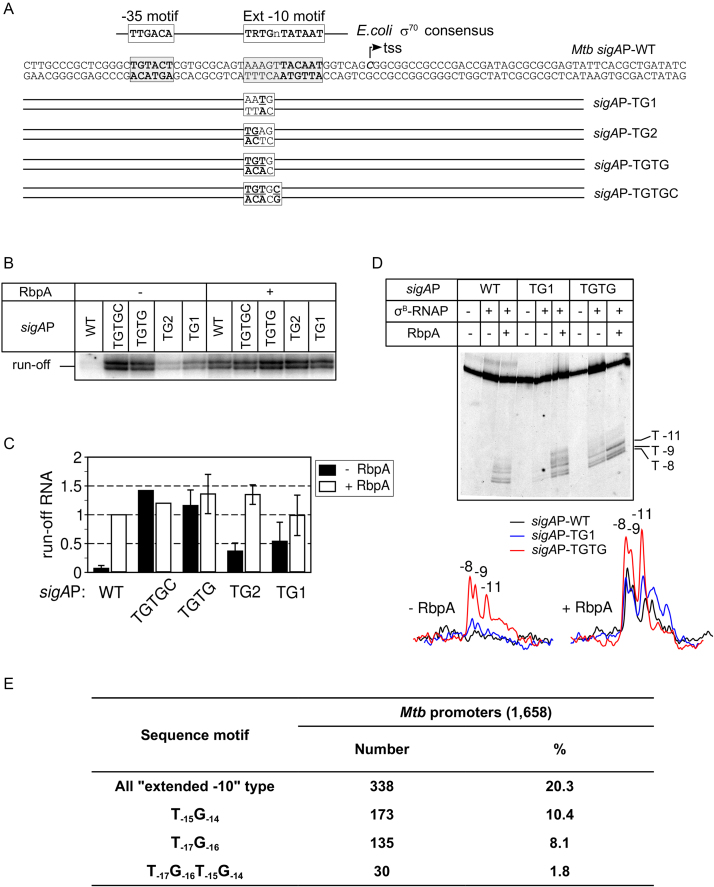
To explore the impact of the extended −10 motif on transcription initiation by the stress-response σB-MtbRNAP holoenzyme, we used four templates derived from the housekeeping, RbpA-dependent M. tuberculosis sigAP promoter (sigAP-WT) (Figure 1A). The four sigAP variants carried the TG motif at different positions: −15 to −14 (T-15G-14) in sigAP-TG1; −16, −17 (T-17G-16) in sigAP-TG2; and −14 to −17 (T-17G-16T-15G-14) in sigAP-TGTG and in sigAP-TGTGC. In addition, the sigAP-TGTGC template carried a C nucleotide at position −13. We then tested whether σB-MtbRNAP could initiate transcription from these different sigAP promoter variants using multiple-round run-off transcription assays (Figure 1B and C; Supplementary Figure S1A). In the absence of RbpA, σB-MtbRNAP was almost inactive at the sigAP-WT promoter, in agreement with our previously published results (24). Introduction of any of the TG motifs stimulated transcriptional activity. Thus, the efficiency of transcription from the sigAP-TGTG promoter in the absence of RbpA was similar to that observed at the sigAP-WT promoter in the presence of RbpA. Introduction of a C nucleotide at position −13, which is known to stimulate promoter binding by the orthologous stress-response σS subunit from E. coli (29), had no effect on σB-MtbRNAP activity. We conclude that the T-17G-16T-15G-14 motif can fully abolish RbpA requirement for transcription initiation. The fact that neither the TG1 nor the TG2 motif alone was sufficient to reach the level of transcription observed with the TGTG motif suggests that DNA bases at positions −17 to −14 interact cooperatively with σB. The RbpA-σB-MtbRNAP complex showed similar levels of transcription at all tested templates (Figure 1C), suggesting that RbpA makes MtbRNAP tolerant to sequence variations in the extended −10 motif.
Figure 1.
RbpA is dispensable for transcription from promoters containing the extended −10 motif. (A) Schematic representation of the sigAP promoter and its derivatives. Mutated bases are underlined. (B) Representative gel of the run-off [32P]-RNA products synthesized in the multiple-round transcription assay using sigAP-WT and the indicated derivatives in the absence and presence of RbpA. (C) Quantification of the run-off [32P]-RNA products obtained in the transcription assay shown in panel B (mean values ± SE of three experiments except of TGTGC values which are from one experiment). All shown products were used for quantification. Values were normalized to the value obtained with the sigAP-WT promoter in the presence of RbpA. (D) KMnO4 probing of the open complexes formed at the sigAP-WT promoter and the indicated derivatives. Promoter DNA was fluorescein-labeled on the template strand. Traces of the gel lanes are shown at the bottom (E) Number and percentage of promoters harboring the indicated extended −10 motif variants in a subset of M. tuberculosis promoters active during the exponential phase (promoters from Cortez et al. (30)). The bioinformatic analysis was performed using the UniproUGENE software (49).
RbpA activates transcription by stimulating MtbRNAP capacity to form RPo (24). To assess whether the TG motif makes RbpA dispensable for transcription initiation through stimulation of RPo formation, we performed KMnO4 probing of RNAP-promoter complexes formed at equilibrium. The template DNA strand thymines at positions −11, −9, −8 of the sigAP promoter were KMnO4-reactive in RPo formed by the RbpA-σB-MtbRNAP complex (Figure 1D). In agreement with the result of the transcription assay, σB-MtbRNAP was able to open the sigAP-TGTG promoter even in the absence of RbpA. However, the amount of RPo was ∼30% of that formed in the presence of RbpA, suggesting that RbpA stimulates RPo formation even at extended −10 promoters. In the absence of RbpA, sigAP-TG1 promoter opening was barely detectable (Figure 1D), in striking contrast with the significant transcription activity of σB-MtbRNAP observed in the transcription assay (Figure 1B). This discrepancy likely indicates that RPo complexes were active but unstable (see below). We conclude that sequence-specific interaction of σB-MtbRNAP with the bases between −17 to −16 of the extended −10 motif is an essential determinant of promoter opening in the absence of RbpA.
Based on the observation that RbpA was not required for transcription from the perfect extended −10 consensus sinP3 promoter (24) and from the sigAP promoter variants harboring TG-motifs, we predict that any promoter harboring the extended −10 motif should be active in the absence of RbpA. To estimate the number of such presumably RbpA-independent or weakly RbpA-dependent gene promoters in M. tuberculosis, we performed a bioinformatic analysis using the set of 1658 promoters containing the σ70-type −10 motifs (nAnnnT) (30). These promoters were reported to be active during exponential growth (30). The analysis demonstrated that 338 of 1668 promoters contained a partial or full-length extended −10 motif (Figure 1E). Specifically, 173 (10.4%) promoters contained the T-15G-14 and 135 (8.1%) the T-17G-16 motif and therefore, they expected to be loosely RbpA-dependent. Only 30 (1.8%) promoters contained the T-17G-16T-15G-14 motif and therefore, they are expected to be constitutive. Accordingly, the majority of σB-dependent genes in M. tuberculosis are expected to be under the control of RbpA.
The extended −10 motif stabilizes MtbRNAP-promoter complexes in the absence of RbpA
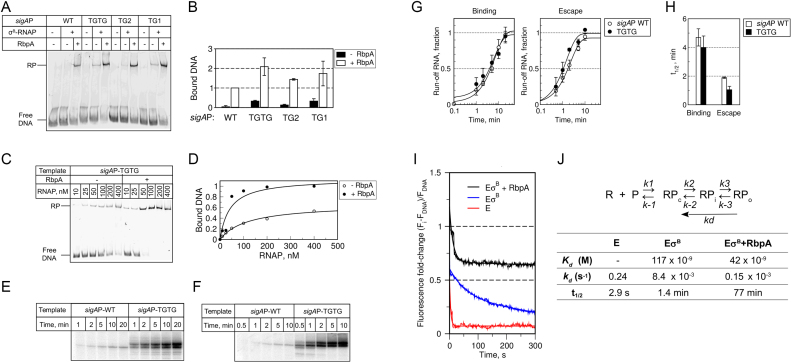
Next, we used electrophoretic mobility shift assays (EMSA) to test whether σB-MtbRNAP can form stable RPo at sigAP promoter variants in non-equilibrium conditions, in the presence of competitor poly(dI-dC) (Figure 2A and B). EMSA showed that in the absence of RbpA, the full-length extended −10 motif (T-17G-16T-15G-14) and the downstream part (T-15G-14) stabilized the σB-MtbRNAP-promoter complexes to a greater extent than the upstream T-17G-16 motif. As the A-15 → T-15 substitution was the only difference between sigAP-WT and sigAP-TG1, we conclude that the identity of the base at position −15 is critical for recognition of the extended −10 motif by the σB subunit and for stabilization of the σB-MtbRNAP-promoter complex. Titration of the sigAP-TGTG template with increasing concentrations of σB-MtbRNAP in the presence or absence of RbpA demonstrated that RbpA increased σB-MtbRNAP affinity for the promoter by ∼3-fold (calculated apparent Kd was 117 nM without RbpA and 42 nM with RbpA, Figure 2C, D, J). Thus, differently from run-off RNA synthesis, RPo formation at promoters that contain the extended −10 motif was still responsive to RbpA addition. This discrepancy is likely to arise from the different effects of RbpA and the TG motif on initiation and promoter escape in multiple-round transcription assays (see below). Similar uncoupling between RPo stability and transcriptional activity was previously reported for σA-MtbRNAP at the sinP3 and sigAP promoters where the lack of stable promoter complexes contrasted with the relatively high transcription levels (18,24).
Figure 2.
RbpA stabilizes open promoter complexes at the extended −10 promoter. (A) EMSA analysis of the promoter complex formation using σB-MtbRNAP and fluorescein-labeled sigAP promoter variants. Complexes were resolved using native 5% PAGE. (B) Quantification of the EMSA results (mean values ± SE of three experiments). (C) Effect of RbpA on sigAP-TGTG promoter binding measured by MtbRNAP titration in EMSA assays. (D) Quantification of the results shown in panel C. (E) Time-course of RPo formation monitored in a single-round run-off transcription assay. σB-MtbRNAP was incubated with the sigAP-WT and sigAP-TGTG promoters for the indicated times and then supplemented with NTPs and competitor poly(dI-dC). Representative gel showing the run-off [32P]-RNA products produced during 3 min of transcription. (F) Time-course of promoter escape monitored in a single-round transcription assay. NTPs and competitor poly(dI-dC) were added to pre-formed RPo complexes and transcription was performed for the indicated times. Representative gel showing the run-off [32P]-RNA products used for quantification. (G) Quantification of the experiments shown in E and F (mean values ± SE of three (sigAP -WT) and two (sigAP-TGTG) experiments). For each experiment, the amount of transcripts at each time-point was normalized to the plateau value. (H) The half-times of RPo formation and of promoter escape were determined from the plots shown in panel G. (I) Fluorescence fold-change during dissociation of the complexes formed by MtbRNAP at the sigAP-TGTG promoter with and without RbpA. E, MtbRNAP core enzyme (red); EσB, σB-MtbRNAP (blue); EσB + RbpA, RbpA-σB-MtbRNAP (black). The graph represents the average of three independent experiments. (J) Apparent kinetic and thermodynamic constants calculated from the data presented in panels D and I. t1/2 was calculated as: t1/2 = ln(2)/kd.
To understand the nature of these discrepancies, we tested TGTG effect on the kinetics of RPo formation and on promoter escape in single-round run-off transcription assays (Figure 2E and F). To follow RPo formation, we incubated σB-MtbRNAP and promoter DNA at 37°C for different lengths of time, before addition of nucleotides and poly(dI-dC). Transcription was performed for 3min for each time point (Figure 2E). To follow promoter escape, we supplemented pre-formed RPo complexes with nucleotides and poly(dI-dC) before transcription for various lengths of time (Figure 2F). To quantitatively characterize the process, we calculated the half-time values (t1/2) required to reach half of the maximum run-off RNA amount (Figure 2H; Supplementary Table S2). The kinetics of RPo formation on sigAP and sigAP-TGTG promoters were similar and unexpectedly slow (t1/2 ∼ 4 min) (Figure 2G and H). The kinetic of promoter escape was ∼2-fold faster for the sigAP-TGTG promoter (t1/2 ∼ 1 min) compared with the sigAP-WT promoter (t1/2 ∼ 2 min). We conclude that, in our experimental conditions, the TGTG motif does not affect the rate of RPo formation, but stimulates promoter escape. Thus, the effect of the extended −10 motif on transcription differs from that of RbpA, which accelerates RPo formation (26, 32 and see below). Based on our results, we propose that RbpA and the extended −10 motif synergistically stimulate RPo formation, probably by stabilizing the transcription bubble within the RPo.
RbpA stabilizes open promoter complexes formed at the sigAP-TGTG promoter
To quantitatively characterize the impact of RbpA on RPo formation at extended −10 promoters, we studied the dissociation kinetics of MtbRNAP-sigAP-TGTG promoter complexes using a fluorescence-based assay (31,32). We incubated the sigAP-TGTG promoter, labeled with Cy3 at position +2 of the non-template DNA strand, with MtbRNAP (E; control), σB-MtbRNAP (EσB) or the RbpA-σB-MtbRNAP complex (EσB + RbpA). Then, we monitored the fluorescence intensity change before and after addition of the competitor heparin that neutralizes free RNAP (Figure 2I). Binding of σB-MtbRNAP or RbpA-σB-MtbRNAP to the promoter DNA induced a 2-fold and 2.5-fold change in fluorescence intensity, respectively. The MtbRNAP core enzyme also induced a ∼1.8-fold increase in fluorescence, possibly due to non-specific binding and the high sensitivity of Cy3 fluorescence to the environment (33). Indeed, the core-specific signal decayed according to the single exponential decay model in <4 s after heparin addition (Figure 2I and J). As the MtbRNAP core enzyme, which cannot bind specifically to the promoter, also produced a significant change in fluorescence, we conclude that this assay can detect not only RPo, but also RPc and RPi. This result also suggests that the fluorescence signal increase observed in the presence of the holoenzyme could arise in part from non-specific binding. Indeed, after heparin addition to the complexes formed by the σB-MtbRNAP holoenzyme or RbpA-σB-MtbRNAP, we observed a first rapid fluorescence decrease (few seconds), followed by a slow decay (Figure 2I and J). Based on the results obtained with the MtbRNAP core enzyme, we attributed the first signal decrease to the dissociation of non-specific complexes (e.g., formed upon MtbRNAP binding to the DNA fragment ends), and the subsequent slow decay phase (fitted by a single exponential) to dissociation of the specific promoter-RNAP complexes. Considering the three-step model of open complex formation (Figure 2J), RNAP dissociation rate from the promoter (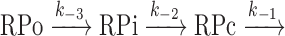 undetectable species) is determined mainly by the slow isomerization of RPo to RPc, and is characterized by the dissociation rate constant kd (kd = k-2/(1+K3)) which is determined from the exponential fit of the slow decay phase (see Methods section) (34,35). Because the RPo complex formed without RbpA dissociated ∼50-fold faster than the complex formed in its presence (Figure 2J), we conclude that RbpA acts on the isomerization step and may stabilize the final ‘open’ state of DNA in RPo (24,25).
undetectable species) is determined mainly by the slow isomerization of RPo to RPc, and is characterized by the dissociation rate constant kd (kd = k-2/(1+K3)) which is determined from the exponential fit of the slow decay phase (see Methods section) (34,35). Because the RPo complex formed without RbpA dissociated ∼50-fold faster than the complex formed in its presence (Figure 2J), we conclude that RbpA acts on the isomerization step and may stabilize the final ‘open’ state of DNA in RPo (24,25).
The synergy between RbpA and the extended −10 motif allows promoter opening at 0°C
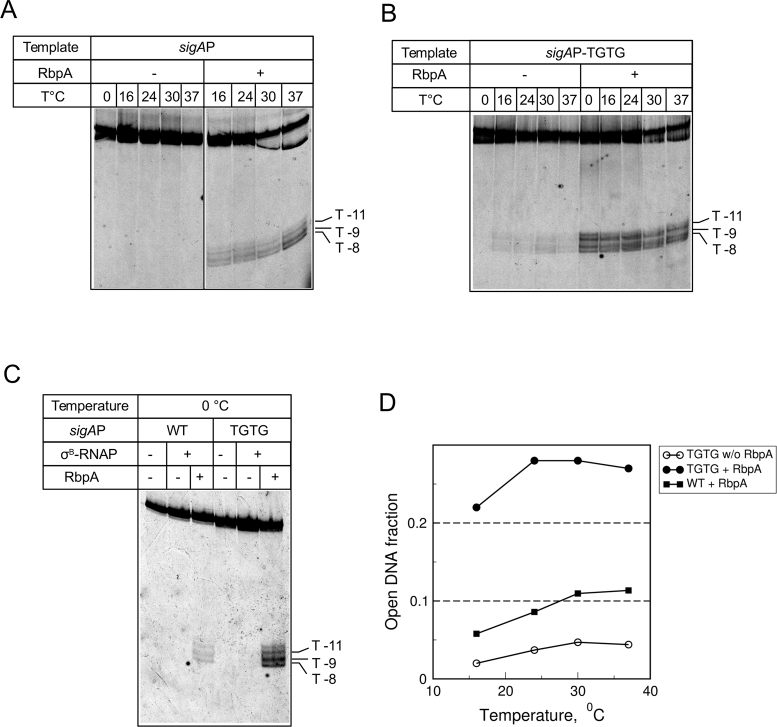
Promoter melting by E. coli RNAP containing the σ70 subunit is strictly temperature-dependent (11,36). This feature reflects RNAP capacity to undergo temperature-dependent isomerization, leading to the formation of a stable transcription bubble. To assess the effect of RbpA and the extended −10 motif on bubble formation, we performed KMnO4 probing of MtbRNAP complexes at the sigAP-WT and sigAP-TGTG promoters at increasing temperatures, from 0 to 37°C (Figure 3). RPo formation by RbpA-MtbRNAP at the sigAP-WT promoter displayed weak temperature dependence. Furthermore, RbpA-MtbRNAP could open the sigAP-WT promoter even at 0°C (Figure 3C). At the sigAP-TGTG template, DNA melting was detected at temperatures as low as 16°C even without RbpA. RbpA boosted MtbRNAP capacity to open the promoter, thus rendering it temperature-independent (Figure 3D). Indeed, the amounts of RPo formed at 0 and at 37°C were quite similar. This result suggests that MtbRNAP interaction with either RbpA or TGTG decreases the thermal energy requirement for RPo formation, while interaction with both leads to a strong cooperative effect.
Figure 3.
MtbRNAP forms open promoter complexes at 0°C. (A–C) Temperature-dependence of promoter melting by MtbRNAP probed with KMnO4. DNA was labeled with fluorescein on the template strand. MtbRNAP-promoter complexes were formed at the indicated temperatures in the absence (–) or presence (+) of RbpA. (D) Quantification of the results shown in panels A and B. The open DNA fractions were calculated as the cleaved DNA to total DNA ratio.
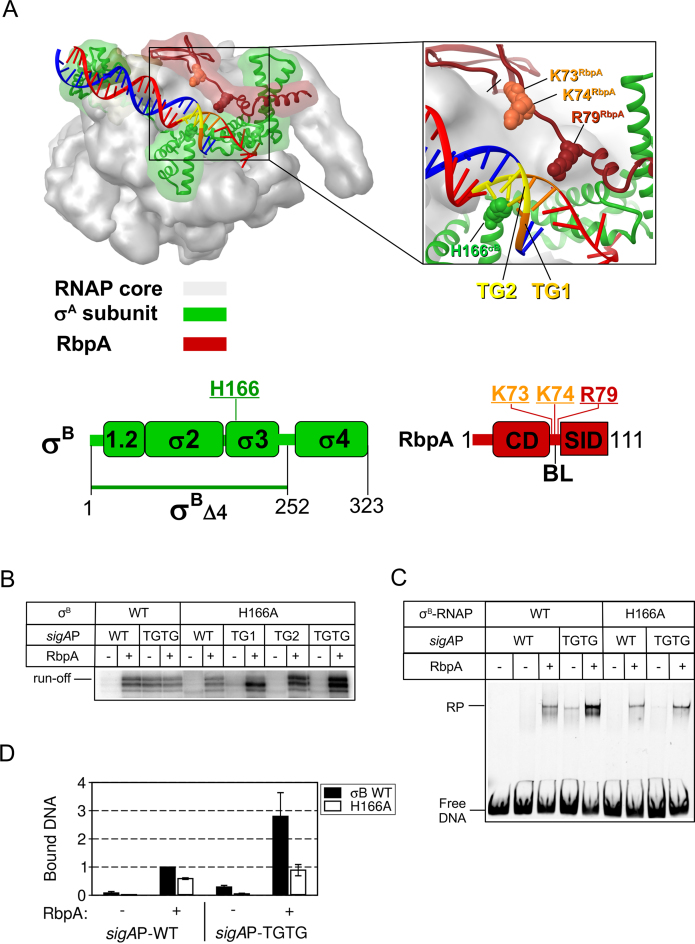
The σH166A mutant abolishes MtbRNAP-promoter interaction at the extended −10 motif
The H455 residue in the σ70 subunit domain 3 (σ3) interacts with the −17GC base pair (C on the template strand) of the extended −10 motif (8,37). On the basis of the structure of Mycobacterium smegmatis RNAP, the homologous residue H166 in σB should interact with the upstream (T-17G-16; TG2) and downstream (T-15G-14; TG1) parts of the TGTG motif in the major DNA groove (see model Figure 4A). RbpA interacts with the TGTG motif on the opposite DNA face, and could affect its interaction with σ3. To determine whether RbpA affected the σ3-TG motif interaction, we generated the σB subunit containing the His → Ala substitution in position 166. In the absence of RbpA, the H166A substitution abolished run-off transcription from the sigAP promoter variants that contain the extended −10 motif (Figure 4B and D; Supplementary Figure S1B). Addition of RbpA restored the activity of σH166A-MtbRNAP to a level even higher than that of the wild type holoenzyme, possibly due to increased RNAP recycling. In agreement with the result of the transcription assay, EMSA showed that the H166A substitution abolished RPo formation at the sigAP-TGTG promoter (Figure 4C). Compared with the wild type holoenzyme, σH166A-RNAP formed less RPo at the sigAP-TGTG promoter also in the presence of RbpA (Figure 4C and D). Thus, we conclude that the sequence-specific interaction between the H166 residue of the σB subunit and the TGTG motif is pivotal for efficient transcription initiation by σB -MtbRNAP at promoters of the extended −10 class. This interaction enhances RbpA capacity to stabilize RPo, but is not essential for RbpA-mediated transcription activation.
Figure 4.
The substitution H166A in region 3 of σB abolishes MtbRNAP interaction at the extended −10 motif. (A) Structural model of Mycobacterium smegmatis RNAP in complex with RbpA and promoter DNA (PDB code: 5TW1). Red ribbon, RbpA; green ribbon, σA subunit; gray semitransparent molecular surface, RNAP core; blue, DNA template strand; red, DNA non-template strand; orange, TG1-motif (T-15G-14); yellow, TG2-motif (T-17G-16). Residues in σB (H166) and RbpA (K73, K74, R79) that were mutated are shown in CPK rendering. Schematic representations of the RbpA and σB domains are shown at the bottom. The positions of the mutated residues are indicated. (B) Run-off [32P]-RNA products synthesized in run-off transcription assays using sigAP promoter derivatives in the presence or not of RbpA. (C) EMSA analysis of promoter complex formation by σB-MtbRNAP and fluorescein-labeled sigAP promoter variants. Complexes were resolved in native 5% PAGE. (D) Quantification of the experiment shown in panel C (mean values ± SE of two experiments).
Domain σ4 is essential for transcription initiation from extended −10 promoters
Interaction of domain 4 of σ70 (σ4) with the −35 element in the promoter is dispensable for transcription from promoters of the extended −10 class (9,38). To test whether the σ4/-35 element interaction contributes to transcription initiation in the presence of RbpA, we introduced mutations in the −35 motif of the sigAP-WT and sigAP-TGTG promoters (Figure 5A). EMSA and multiple-round run-off transcription assays demonstrated that substitutions in the −35 element did not significantly affect promoter binding and transcription (Figure 5B–E; Supplementary Figure S2A), suggesting that sequence-specific recognition of the −35 element by σ4 is dispensable for transcription initiation in the presence of RbpA or of the TGTG-motif. However, we cannot exclude that, in the absence of the perfect −35-motif, a non-specific interaction of σ4 with promoter through contacts with the DNA phosphate backbone (39) contributes to transcription initiation.
Figure 5.
Impact of σ4 and the –35 element on MtbRNAP activity. (A) Scheme showing the −35 motif of the sigAP promoter with the introduced mutations underlined. (B) Run-off [32P]-RNA products synthesized by wild type σB-MtbRNAP from sigAP-WT and sigAP-TGTG and the respective variants lacking the −35 element (Δ-35). (C) Quantification of the results of the experiment shown in panel B (mean values ± SE of two experiments). (D) EMSA analysis of promoter complex formation by σB-MtbRNAP using the sigAP-WT and sigAP-TGTG promoters and the respective variants lacking the −35 element (Δ-35). (E) Quantification of the results shown in panel D (mean values ± SE of two experiments). (F) Analysis of the RbpA-σB subunit interaction by native gel electrophoresis. RbpA, labeled with DyLight 633, was incubated with increasing concentrations (0.8, 1.6, 3.2 μM) of wild type σB (WT) or the mutant in which domain 4 residues 252–323 were deleted (Δ4). (G) Abortive transcription activity of wild type σB-MtbRNAP (WT) and mutant σBΔ4-MtbRNAP (Δ4) on the lacUV5-bubble template harboring a heteroduplex region. (H) Run-off [32P]-RNA products synthesized in the presence of wild type σB-MtbRNAP (WT) or mutant σBΔ4-MtbRNAP (Δ4) and the sigAP-WT, sigAP-TGTG or B. subtilis sinP3 promoter that lacks the −35 element.
If the σ4/-35 element interaction is dispensable for transcription initiation in the presence of RbpA, σ4 deletion should not affect the activity of the MtbRNAP-RbpA complex. To test this hypothesis, we generated a σB subunit mutant in which the C-terminal residues 252–323 were deleted (σBΔ4) (Figure 4A). Using a native PAGE-based protein-protein interaction assay (24), we demonstrated that this deletion did not affect the σB subunit capacity to form a stable complex with RbpA (Figure 5F). To test whether mutant σBΔ4-MtbRNAP was catalytically active, we performed abortive transcription assay using the synthetic lacUV5 promoter harboring a heteroduplex region between position −11 and −5 (28) (Figure 5G). Initiation of transcription on lacUV5 promoter by addition of dinucleotide RNA primer, ApA, and two nucleotides (GTP and [α32P]-UTP) resulted in formation of short RNA products, up to 7nt in length (14). In these experimental conditions, wild type σB-MtbRNAP initiated transcription (with similar efficiency) both in the presence and absence of RbpA, which is in line with the fact that RbpA acts at the promoter melting step. The mutant σBΔ4-MtbRNAP displayed reduced (∼18%), but detectable catalytic activity, compared with the wild type enzyme. This could be caused by defects in promoter binding, holoenzyme σBΔ4-MtbRNAP assembly, and structural disturbance in the σB region 3.2, which is implicated in transcription initiation and promoter escape (40,41). As expected, the mutant σBΔ4-MtbRNAP holoenzyme was inactive in run-off transcription assays performed with the sigAP-WT promoter in the absence and presence of RbpA (Figure 5H). Surprisingly, the mutant σBΔ4-MtbRNAP holoenzyme was also inactive at the sigAP-TGTG promoter and at the sinP3 promoter (24) that harbors the perfect extended −10 consensus motif (5′-T-17GTGcTATAAT-7–3′) and lacks the −35 element. Addition of RbpA partially restored σBΔ4-MtbRNAP activity at the sinP3, but not at the sigAP-TGTG promoter, possibly due to differences in promoter architectures. Because mutant MtbRNAP showed low but detectable activity on the lacUV5 bubble template, irrespective of RbpA, the lack of transcription at the sigAP promoter in the presence of RbpA and at the sigAP-TGTG in the absence of RbpA cannot be explained only by defects in initiation of RNA synthesis. Altogether, these results suggest that RbpA cannot compensate for the lack of σ4 interaction with the core enzyme or/and the promoter, and that σ4 per se is an essential component of the RbpA-mediated activation mechanism at the −10/-35 class promoters. Based on these results we hypothesize that even for promoters of the extended −10 class, interaction of the σ4 domain with the MtbRNAP core enzyme, and/or non-specific interaction with DNA contribute to transcription initiation.
RbpA-BL modulates σB-MtbRNAP selectivity for initiating transcription substrates
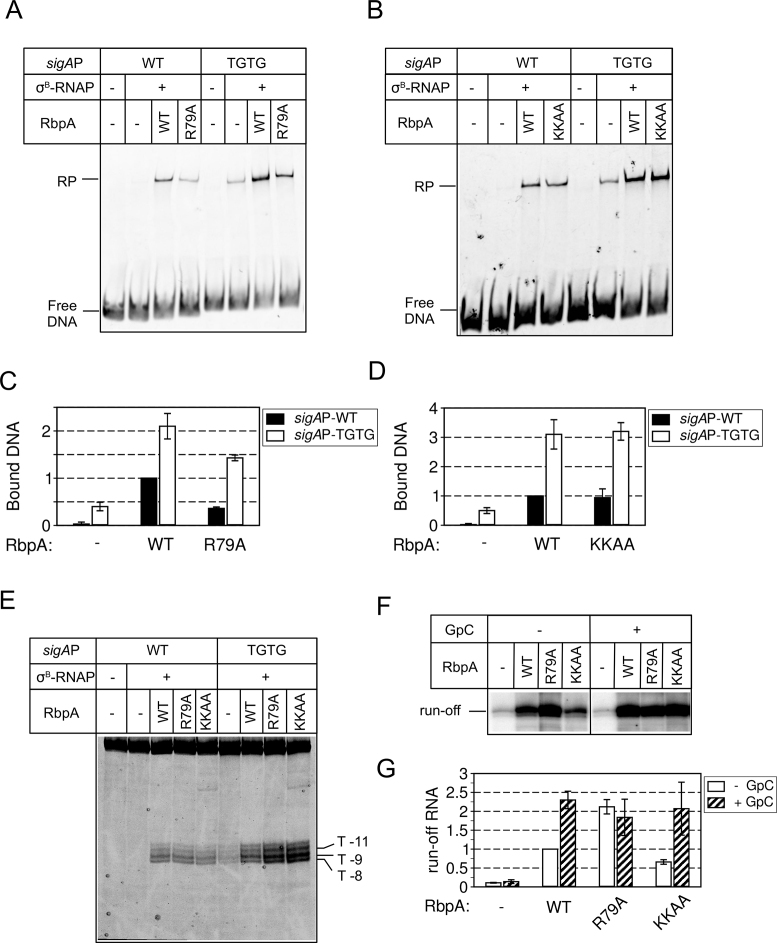
Based on the structure of RPo and RbpA-SID fragment, it was proposed that the residues R79 (in contact with nucleotides −13 and −14 of the non-template DNA strand), K73 and K74 in RbpA-BL interact with DNA at the TGTG element in the DNA minor groove (25,26). We assessed whether mutations in these residues (R79A and the double substitution K73A, K74A (KKAA)) affected RPo formation at the sigAP-WT and sigAP-TGTG promoters. The EMSA results showed that in agreement with previous findings (25), the R79A substitution in RbpA decreased RPo stability by ∼2-fold at the sigAP-WT and by ∼1.5-fold at the sigAP-TGTG promoter (Figure 6A and C). Conversely, the KKAA substitutions had no effect on RPo stability (Figure 6B and D). Furthermore, KMnO4 probing demonstrated that neither the R79A nor the KKAA substitution hinders sigAP-WT promoter opening (Figure 6E). Opening of the sigAP-TGTG promoter was even enhanced in the presence of the RbpA mutants. This discrepancy between KMnO4 probing and EMSA results can be explained by the formation of an unstable RPo that dissociates in the non-equilibrium conditions of EMSA, but can be detected in the equilibrium conditions of KMnO4 probing. Thus, we conclude that R79 contributes to RPo stabilization, while K73 and K74 are dispensable.
Figure 6.
Effect of mutations in RbpA-BL on RPo formation and transcription initiation. (A) EMSA analysis of promoter complex formation by σB-MtbRNAP in the presence of the RbpAR79A mutant. (B) EMSA analysis of promoter complex formation by σB-MtbRNAP in the presence of the RbpAKKAA mutant. (C, D) Quantification of the results (mean value ± SE of two experiments) shown in panel A and B, respectively. (E) KMnO4 probing of MtbRNAP-promoter complexes formed in the presence of the indicated RbpA mutants. (F) Run-off [32P]-RNA products synthesized during run-off transcription assay from the sigAP promoter by σB-MtbRNAP in the absence or presence of the indicated RbpA variants. Transcription was performed with or without RNA primer (GpC). (G) Quantification of the results shown in panel F (mean values ± SE of two experiments).
Next, we tested whether the RbpA mutants could stimulate de novo transcription. In multiple-round transcription assay, RbpAR79A stimulated transcription from the sigAP promoter more than wild type RbpA (Figure 6F and G; Supplementary Figure S2B). This effect could arise from the weaker promoter binding that in turn could facilitate promoter escape and consequently MtbRNAP recycling. In the same conditions, the RbpAKKAA mutant stimulated MtbRNAP less efficiently than wild type RbpA (the amount of RNA produced by MtbRNAP was reduced by 2-fold). Short RNA primers can bypass defects in de novo transcription initiation (39,40). To determine whether short RNAs could rescue the σB-MtbRNAP activity in the presence of RbpA mutants, we repeated the run-off transcription experiments in the presence of 100 μM GpC primer, which is complementary to the positions −1/+1 of the sigAP promoter. Addition of GpC fully rescued the transcription defect induced by RbpAKKAA. Furthermore, the run-off RNA levels in all RbpA-containing reactions were equal.
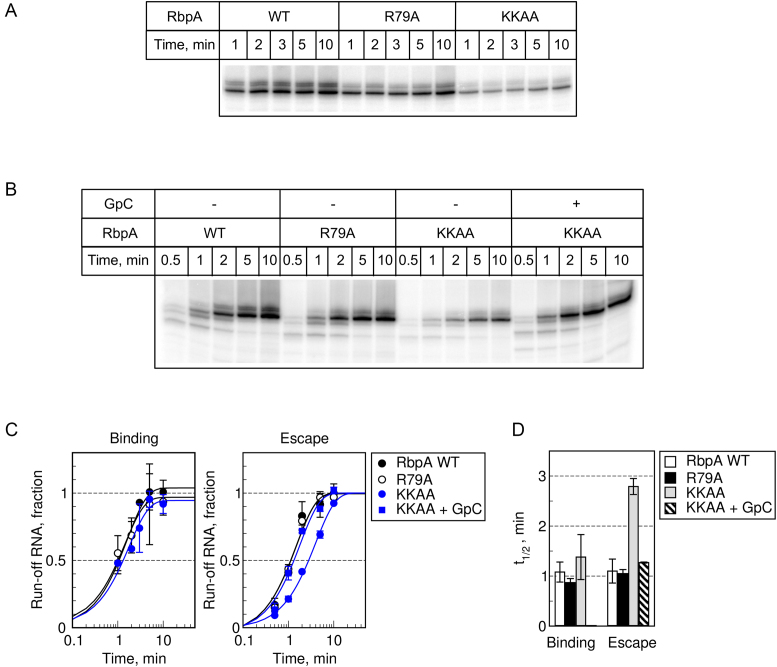
To better understand the effect of substitutions in RbpA on RPo formation and promoter escape, we performed single-round transcription assays as described above (Figure 7). The half-time of RPo formation in the presence of RbpA was ∼4-fold higher than without RbpA (compare Figure 2G, H and Figure 7C, D). The half-time of promoter escape was also 2-fold greater in the presence of RbpA (Figure 7, Table S2), suggesting that RbpA regulates not only promoter melting but also the initial transcription. We detected no effect of the R79A substitution on RPo activation and escape rates, at least in the time resolution range of our assay. Therefore, we cannot presently explain why this substitution affects the yield of run-off RNA in multiple-round transcription assays. In agreement with the result of the multiple-round transcription assays, the KKAA substitutions led to a ∼3-fold decrease in escape rate (t1/2 = 2.8 min), while addition of GpC stimulated promoter escape. Based on our results and on the recent structure of the MtbRNAP-RbpA complex (42), we propose that RbpA, can indirectly modulate MtbRNAP selectivity for the priming substrates (NTPs and short RNAs) through contact with the σ3.2 finger (42) which controls transition from initial transcription to productive elongation (28,40,41).
Figure 7.
Effect of mutations in RbpA-BL on the kinetics of RPo formation and promoter escape. (A) RPo formation was monitored in single-round run-off transcription assays. (B) Promoter escape was monitored in single-round run-off transcription assays. Representative gels showing run-off [32P]-RNA products synthesized from the sigAP-WT promoter by σB-MtbRNAP in the presence of the indicated RbpA variants. (C) Quantification of the experiments shown in A and B (mean values ± SE of three experiments). All shown RNA products were used for quantification. (D) The half-times of RPo formation and promoter escape were determined from the plots shown in panel C.
DISCUSSION
Transcription initiation from most of the bacterial promoters requires simultaneous binding of the RNAP σ subunit to the −10 and −35 elements. The weak conservation of the −35 element in M. tuberculosis promoters (30) predicts the existence of compensatory mechanisms. Here, we found that RbpA abolishes the requirement of sequence-specific interactions between the domains 3 and 4 of the σ subunit and promoter DNA upstream of the −10 element. Therefore it converts MtbRNAP into a hyperactive enzyme with promiscuous promoter selection. Moreover, the presence of the T-17G-16T-15G-14 motif fully abolishes RbpA requirement for transcription initiation, suggesting that mycobacterial promoters bearing the extended −10 motif are constitutive. Our estimation is that at least 2% of the known M. tuberculosis promoters belong to this group. Moreover, ∼14% of M. tuberculosis promoters contain the T-15G-14 motif (43), and thus should be loosely dependent on RbpA, or hyperactive in the presence of RbpA. Based on the similarities between the effects of RbpA and of the TGTG motif on transcription, we propose that RbpA could strengthen the interaction of the σ subunit domains 3 and 4 with promoter DNA upstream of the −10 element. This hypothesis is supported by our smFRET study showing that σB in the MtbRNAP holoenzyme adopts a conformation incompatible with binding to −10/-35 promoters and that RbpA stabilizes σB in a conformation compatible with binding (27). The finding that σ4 was still required for transcription initiation in the context of the extended −10 sinP3 and sigAP-TGTG promoters even in the presence of RbpA suggests that σ4 may have additional roles in initiation, probably in organizing RNAP clamp or β subunit flap domains for RPo formation, as proposed for E. coli σ70 (44). The σ4/β-flap contact may be essential for correct positioning of σ3 (e.g., region 3.2) in RPo, which in turn affects RPo stability and RNA synthesis initiation (28,40).
We observed that RbpA increases MtbRNAP affinity for promoters bearing the extended −10 motif (∼3 fold decrease in apparent Kd) and stabilizes RPo (∼50-fold decrease in kd). Thus, we propose that RbpA acts on two initiation steps: promoter binding (RPc formation), and RPc isomerization to RPo. Our result differs from a previous study reporting no effect of RbpA on the kd of Mycobacterium bovis RNAP (MboRNAP) binding at the rrnAP3 and vapB10 promoters (26). We found that these promoters harbor the T-17G-16 motif and therefore, belong to the extended −10 class. Consequently, the properties of these promoter templates should be close to those of the sigAP promoter templates used in our study. However, Hubin et al. used MboRNAP assembled with the σA and not the σB subunit. The difference in the structures of these two σ subunits could explain the discrepancy in the observed RbpA effect on kd. As the σA and σB residues that interact with the extended −10 and −35 elements are almost identical (supplementary Figure S3 in Hu et al. (24)), they are unlikely to explain their different behavior. The major difference in structure between σA and σB is the long N-terminus (202 amino acids) of σA that could be involved in RPo formation, as reported for the N-terminus of E. coli σ70 (45). In addition, the N-terminus of RbpA (RbpANTT) interacts with the variable region σ3.2 (42) which controls bubble stability in RPo and transition from initial transcription to productive elongation (28,40,41).
Role of the extended −10 motif in promoter melting at suboptimal temperatures
Studies on the E. coli σ70-RNAP and B. subtilis σA-RNAP holoenzymes demonstrated that the extended −10 motif stabilizes RPo and allows promoter melting triggered by RNAP at low (6 and 10°C), suboptimal temperatures (46,47). Also it has been proposed that the T-15G-14 motif can decrease the thermal energy requirement for RPo formation by MtbRNAP (48). Our results demonstrate that the identity of the nucleotides at positions −17 to −16 is critical for melting of promoters of the extended −10 class. The combination of TGTG motif and RbpA strongly decreased the thermal energy requirement for promoter melting and allowed MtbRNAP to form open complexes even at 0°C. The effect of RbpA on Tm supports its action on isomerization of the closed-to-open complex. We speculate that interaction at the TGTG motif could promote DNA bending around RNAP. This will direct promoter DNA to the downstream channel in RNAP, thus facilitating distortion of the −10 element DNA by the region 2 of σ and then formation of the transcription bubble.
Role of RbpA-BL in transcription initiation
Previous studies on σA-MboRNAP suggested that interaction of the residue R79 in RbpA-BL with promoter DNA at positions −13, −14 is critical for RbpA function in vitro and in vivo (25,26). We observed a moderate effect of this mutation on RPo stability and no effect on promoter opening and on transcriptional activity. Furthermore, the RbpAR79A mutant was even more active than wild type RbpA in multiple-round transcription assays without a 2-mer RNA primer. This discrepancy indicates that the σA-MtbRNAP and σB-MtbRNAP holoenzymes respond differently to RbpA. Indeed, differently from σB-MtbRNAP, the σA–MtbRNAP holoenzyme cannot form stable RPo at the extended −10 sinP3 promoter without RbpA and at the sigAP promoter in the presence of RbpA (18,24). The molecular basis of these differences, lying in the above mentioned structural properties of σA and σB, is an intriguing subject for future studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank L. Makrini for technical assistance in protein purification.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French National Research Agency [MycoMaster ANR-16-CE11-0025-01]; CNRS [PRC Russie CNRS/RFBR to K.B.]; Fondation pour la recherche médicale (FRM) (to Y.H.) during his stay in the laboratory of K.B. ; ERASMUS MUNDUS Svaagata fellowship (to A.S.P.); Infectiopole Sud (to R.K.V.). Funding for open access charge: MycoMaster ANR-16-CE11-0025-01.
Conflict of interest statement. None declared.
REFERENCES
- 1. Feklístov A., Sharon B.D., Darst S.A., Gross C.A.. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014; 68:357–376. [DOI] [PubMed] [Google Scholar]
- 2. Borukhov S., Nudler E.. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008; 16:126–134. [DOI] [PubMed] [Google Scholar]
- 3. Rodrigue S., Provvedi R., Jacques P., Gaudreau L., Manganelli R.. The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2006; 30:926–941. [DOI] [PubMed] [Google Scholar]
- 4. Gruber T.M., Gross C.A.. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003; 57:441–466. [DOI] [PubMed] [Google Scholar]
- 5. Graves M.C., Rabinowitz J.C.. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for “extended” promoter elements in gram-positive organisms. J. Biol. Chem. 1986; 261:11409–11415. [PubMed] [Google Scholar]
- 6. Voskuil M.I., Chambliss G.H.. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 1998; 26:3584–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitchell J.E., Zheng D., Busby S.J.W., Minchin S.D.. Identification and analysis of ‘extended −10′ promoters in Escherichia coli. Nucleic Acids Res. 2003; 31:4689–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barne K.A., Bown J.A., Busby S.J., Minchin S.D.. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ‘extended-10′ motif at promoters. EMBO J. 1997; 16:4034–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar A., Malloch R.A., Fujita N., Smillie D.A., Ishihama A., Hayward R.S.. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J. Mol. Biol. 1993; 232:406–418. [DOI] [PubMed] [Google Scholar]
- 10. Helmann J.D. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995; 23:2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buc H., McClure W.R.. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985; 24:2712–2723. [DOI] [PubMed] [Google Scholar]
- 12. Rogozina A., Zaychikov E., Buckle M., Heumann H., Sclavi B.. DNA melting by RNA polymerase at the T7A1 promoter precedes the rate-limiting step at 37°C and results in the accumulation of an off-pathway intermediate. Nucleic Acids Res. 2009; 37:5390–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saecker R.M., Record M.T.J., Dehaseth P.L.. Mechanism of bacterial transcription initiation: RNA polymerase − promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011; 412:754–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brodolin K., Zenkin N., Severinov K.. Remodeling of the sigma70 subunit non-template DNA strand contacts during the final step of transcription initiation. J. Mol. Biol. 2005; 350:930–937. [DOI] [PubMed] [Google Scholar]
- 15. Feklistov A., Darst S.A.. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011; 147:1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M.X., Arnold E., Ebright R.H.. Structural basis of transcription initiation. Science. 2012; 338:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srivastava D.B., Leon K., Osmundson J., Garner A.L., Weiss L.A., Westblade L.F., Glickman M.S., Landick R., Darst S.A., Stallings C.L. et al. Structure and function of CarD, an essential mycobacterial transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu Y., Morichaud Z., Chen S., Leonetti J., Brodolin K.. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the σA-containing RNA polymerase holoenzyme. Nucleic Acids Res. 2012; 40:6547–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bortoluzzi A., Muskett F.W., Waters L.C., Addis P.W., Rieck B., Munder T., Schleier S., Forti F., Ghisotti D., Carr M.D. et al. Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors. J. Biol. Chem. 2013; 288:14438–14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forti F., Mauri V., Dehò G., Ghisotti D.. Isolation of conditional expression mutants in Mycobacterium tuberculosis by transposon mutagenesis. Tuberculosis (Edinb.). 2011; 91:569–578. [DOI] [PubMed] [Google Scholar]
- 21. Betts J.C., Lukey P.T., Robb L.C., McAdam R.A., Duncan K.. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002; 43:717–731. [DOI] [PubMed] [Google Scholar]
- 22. Provvedi R., Boldrin F., Falciani F., Palù G., Manganelli R.. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology. 2009; 155:1093–1102. [DOI] [PubMed] [Google Scholar]
- 23. Tabib-Salazar A., Liu B., Doughty P., Lewis R.A., Ghosh S., Parsy M.-L., Simpson P.J., O’Dwyer K., Matthews S.J., Paget M.S. et al. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res. 2013; 41:5679–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu Y., Morichaud Z., Perumal A.S., Roquet-Baneres F., Brodolin K.. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res. 2014; 42:10399–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hubin E.A., Tabib-Salazar A., Humphrey L.J., Flack J.E., Olinares P.D.B., Darst S.A., Campbell E.A., Paget M.S.. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7171–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hubin E.A., Fay A., Xu C., Bean J.M., Saecker R.M., Glickman M.S., Darst S.A., Campbell E.A.. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife. 2017; 6:e22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vishwakarma R.K., Cao A.M., Morichaud Z., Perumal A.S., Margeat E., Brodolin K.. Single-molecule analysis reveals the mechanism of transcription activation in M. tuberculosis. Sci. Adv. 2018; 4:eaao5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morichaud Z., Chaloin L., Brodolin K.. Regions 1.2 and 3.2 of the RNA polymerase σ subunit promote DNA melting and attenuate action of the antibiotic lipiarmycin. J. Mol. Biol. 2016; 428:463–476. [DOI] [PubMed] [Google Scholar]
- 29. Becker G., Hengge-Aronis R.. What makes an Escherichia coli promoter sigma(S) dependent? Role of the −13/-14 nucleotide promoter positions and region 2.5 of sigma(S). Mol. Microbiol. 2001; 39:1153–1165. [DOI] [PubMed] [Google Scholar]
- 30. Cortes T., Schubert O.T., Rose G., Arnvig K.B., Comas I., Aebersold R., Young D.B.. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013; 5:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rammohan J., Ruiz Manzano A., Garner A.L., Prusa J., Stallings C.L., Galburt E.A.. Cooperative stabilization of Mycobacterium tuberculosis rrnAP3 promoter open complexes by RbpA and CarD. Nucleic Acids Res. 2016; 44:7304–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ko J., Heyduk T.. Kinetics of promoter escape by bacterial RNA polymerase: effects of promoter contacts and transcription bubble collapse. Biochem. J. 2014; 463:135–144. [DOI] [PubMed] [Google Scholar]
- 33. Stennett E.M.S., Ciuba M.A., Lin S., Levitus M.. Demystifying PIFE: The photophysics behind the protein-induced fluorescence enhancement phenomenon in Cy3. J. Phys. Chem. Lett. 2015; 6:1819–1823. [DOI] [PubMed] [Google Scholar]
- 34. Kontur W.S., Saecker R.M., Capp M.W., Record M.T. Jr. Late steps in the formation of E. coli RNA polymerase-lambda P R promoter open complexes: characterization of conformational changes by rapid [perturbant] upshift experiments. J. Mol. Biol. 2008; 376:1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruff E.F., Drennan A.C., Capp M.W., Poulos M.A., Artsimovitch I., Record M.T. Jr. E. coli RNA polymerase determinants of open complex lifetime and structure. J. Mol. Biol. 2015; 427:2435–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roe J.H., Burgess R.R., Record M.T. Jr. Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J. Mol. Biol. 1985; 184:441–453. [DOI] [PubMed] [Google Scholar]
- 37. Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., Darst S.A.. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife. 2015; 4:doi:10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar A., Grimes B., Fujita N., Makino K., Malloch R.A., Hayward R.S., Ishihama A.. Role of the sigma 70 subunit of Escherichia coli RNA polymerase in transcription activation. J. Mol. Biol. 1994; 235:405–413. [DOI] [PubMed] [Google Scholar]
- 39. Campbell E.A., Muzzin O., Chlenov M., Sun J.L., Olson C.A., Weinman O., Trester-Zedlitz M.L., Darst S.A.. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002; 9:527–539. [DOI] [PubMed] [Google Scholar]
- 40. Kulbachinskiy A., Mustaev A.. Region 3.2 of the sigma subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006; 281:18273–18276. [DOI] [PubMed] [Google Scholar]
- 41. Duchi D., Bauer D.L., Fernandez L., Evans G., Robb N., Hwang L.C., Gryte K., Tomescu A., Zawadzki P., Morichaud Z., Brodolin K. et al. RNA polymerase pausing during initial transcription. Mol. Cell. 2016; 63:939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boyaci H., Chen J., Lilic M., Palka M., Mooney R.A., Landick R., Darst S.A., Campbell E.A.. Fidaxomicin jams Mycobacterium tuberculosis RNA polymerase motions needed for initiation via RbpA contacts. Elife. 2018; 7:e34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hubin E.A., Lilic M., Darst S.A., Campbell E.A.. Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures. Nat. Commun. 2017; 8:16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Minakhin L., Severinov K.. On the role of the Escherichia coli RNA polymerase sigma 70 region 4.2 and alpha-subunit C-terminal domains in promoter complex formation on the extended −10 galP1 promoter. J. Biol. Chem. 2003; 278:29710–29718. [DOI] [PubMed] [Google Scholar]
- 45. Vuthoori S., Bowers C.W., McCracken A., Dombroski A.J., Hinton D.M.. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 2001; 309:561–572. [DOI] [PubMed] [Google Scholar]
- 46. Voskuil M.I., Chambliss G.H.. The TRTGn motif stabilizes the transcription initiation open complex. J. Mol. Biol. 2002; 322:521–532. [DOI] [PubMed] [Google Scholar]
- 47. Burns H.D., Belyaeva T.A., Busby S.J., Minchin S.D.. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended −10 sequences. Biochem. J. 1996; 317:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Agarwal N., Tyagi A.K.. Role of 5′-TGN-3′ motif in the interaction of mycobacterial RNA polymerase with a promoter of ‘extended −10′ class. FEMS Microbiol. Lett. 2003; 225:75–83. [DOI] [PubMed] [Google Scholar]
- 49. Okonechnikov K., Golosova O., Fursov M.. UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012; 28:1166–1167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.