Abstract
Long non-coding RNAs (lncRNAs) have emerged as key players in gene regulation. However, our incomplete understanding of the structure of lncRNAs has hindered molecular characterization of their function. Maternally expressed gene 3 (Meg3) lncRNA is a tumor suppressor that is downregulated in various types of cancer. Mechanistic studies have reported a role for Meg3 in epigenetic regulation by interacting with chromatin-modifying complexes such as the polycomb repressive complex 2 (PRC2), guiding them to genomic sites via DNA-RNA triplex formation. Resolving the structure of Meg3 RNA and characterizing its interactions with cellular binding partners will deepen our understanding of tumorigenesis and provide a framework for RNA-based anti-cancer therapies. Herein, we characterize the architectural landscape of Meg3 RNA and its interactions with PRC2 from a functional standpoint.
INTRODUCTION
Long non-coding RNAs are defined as transcripts longer than 200 nt with no protein-coding potential (1,2). The majority are transcribed by RNA pol II machinery and often spliced, 5′-capped, polyadenylated, and retained in the nucleus (3). LncRNAs constitute the major product of the human genome, greatly outnumbering protein-coding mRNAs (4). Approximately 100 000 lncRNAs have thus far been reported, although varying tissue, developmental stage, and disease-specific expression profiles make precise quantification difficult (5–7). Targeted functional studies over the past decade have identified essential roles for lncRNAs in development, differentiation and various cancers (8–18). Moreover, although overall sequence similarity is minimal (19,20), xenotypic lncRNAs have been found to exhibit micro-homology (21) and are functionally and structurally conserved (22–27). Despite these extensive research efforts, identification of new lncRNAs has far outstripped their functional characterization; for instance, out of ∼20 000 potentially functional lncRNAs reported, <2% have been experimentally investigated (28). Hence, mechanistic and functional characterization would greatly enhance the relatively new field of lncRNA structural biology.
The maternally expressed 3 (Meg3) lncRNA, a nuclear RNA (29), is functionally well-characterized. It is transcribed from the maternally expressed Meg3 gene located in the imprinted DLK1–MEG3 locus on human chromosome 14 (14q32.3) (30). The Meg3 RNA gene comprises 10 exons (30) and generates at least 12 different isoforms by alternative splicing (31,32). The predominantly expressed isoform is a ∼1600 nt transcript designated Meg3 (isoform variant 1) containing exons 1–4 and 8–10, and constituting 79–86% of all expressed Meg3 isoforms (33). Meg3 is expressed in various normal human tissues including brain, pancreas, spleen, testis, liver, ovary, placenta, mammary gland and adrenal gland, with the highest level reported in various regions of the brain (31,33). However, its expression is low or undetectable in multiple types of primary cancer and cancer cell lines (16,31,34–40). Ectopic expression of this RNA ex-vivo in these tissues or in vivo in nude mice inhibits cell/tumor growth and proliferation, establishing Meg3 as a pan-cancer tumor suppressor (41–44). Overexpression of Meg3 leads to increased p53 levels, which in turn upregulate p53-dependent gene expression, leading to G2/M cell cycle arrest and apoptosis (32,45). Meg3 antitumor activity is also mediated via p53-independent pathways, including Wnt/β-catenin (46), VEGF (44), and TGF-β (29,47). Early mechanistic studies have reported a role for Meg3 in epigenetic regulation by interacting with chromatin-modifying complexes such as the PRC2 complex, guiding them to genomic sites via DNA-RNA triplex formation (29,47,48).
Unlike proteins, the secondary structure of RNA dictates it tertiary structure through networks of triple helices, tetraloop–receptor interactions, coaxial stacking of adjacent helices at junctions, pseudoknots, kissing loops, etc. (49). As such, determining RNA secondary structure lies at the core of understanding its function. This is particularly true for lncRNAs with a less conserved primary sequence. In the current study, we report the first experimentally derived secondary structure of the 1595 nt Meg3 isoform variant 1 lncRNA using selective 2′ hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) (50–55). In SHAPE-MaP, RNAs are (i) probed with chemical reagents that selectively acylate unpaired ribonucleotides at their 2′-hydroxyl positions, and (ii) reverse transcribed under conditions that promote mutagenesis in cDNA opposite sites of modification (53,55). The sites and frequencies of these mutations are used to create SHAPE reactivity profiles indicating which RNA nucleotides are likely to be single- or double-stranded and guide the RNAstructure software (56) to generate secondary structure models ranked by Gibbs free energy. In an approach resembling those of previous studies (50,51,54), we used SHAPE-MaP to determine the secondary structures of natively folded, in vitro transcribed Meg3 RNA as well as of ectopically and transiently expressed Meg3 RNA in the U87-MG glioblastoma cell line. Our results indicate that Meg3 RNA is intricately branched and highly structured, and comparing the two structures revealed five conserved structural motifs, the functional importance of which is strongly supported by previous genetic studies (29,31). We also performed footprinting experiments to identify and characterize sites of PRC2 complex binding to co-transcriptionally folded, in vitro transcribed Meg3 RNA. Collectively, we provide a detailed structural analysis of Meg3 RNA and its interaction with PRC2 complex, laying a framework for understanding how this important lncRNA performs its biological functions.
MATERIALS AND METHODS
In vitro transcription of full length sense and antisense Meg3 RNA and its non-denaturing purification
The plasmid (pCI-flMeg3) used to generate the DNA template for in vitro transcription was created by cloning full length Meg3 DNA between XhoI and NotI sites of pCI mammalian expression vector (Addgene). The plasmid was linearized using NotI and then gel purified. Full-length Meg3 RNA was transcribed from the linearized plasmid by T7 RNA polymerase using the Megascript kit (Life Technologies) per manufacturer's guidelines. RNA was then treated with Turbo DNase I (Life Technologies) for 1 h at 37°C and immediately purified under non-denaturing conditions. Once transcribed, RNA was never frozen to maintain its native structure. Glycerol was added to the freshly transcribed RNA at a final concentration of 5%. The RNA was then fractionated on a 1.5% agarose gel in 1X TBE running buffer at a constant voltage of 5 V/cm at 4°C for ∼5 h, located by UV shadowing, gel extracted by electro-elution at 100 V constant voltage at 4°C for 12 h, and then concentrated using 3K Amicon ultra-4 centrifugal filter devices (Millipore) to a concentration of ∼1 μM (∼500 ng/μl). Purified RNA was stored at 4°C in aliquots and used no longer than week after purification.
To generate antisense Meg3 RNA (1600 nt), pCI-flMeg3 plasmid was digested by NotI and XhoI and the NotI–XhoI fragment containing Meg3 DNA was gel purified. The purified Meg3 DNA fragment was used to generate Meg3 DNA with T7 promoter sequence at the 3′ end of the sense strand by PCR. The amplified DNA was purified using PCR purification kit (Invitrogen) and then used as the template to in vitro transcribe antisense Meg3 RNA. The antisense RNA was purified and handled using the protocol developed for sense Meg3 RNA.
Determining the composition of RNA folding buffer for in vitro Meg3 RNA
The composition of the RNA folding buffer was determined experimentally by incubating 0.5 pmoles of Meg3 RNA in a total volume of 9 μl at 37°C for 30 mins with buffers varying in composition and salt concentration. The effect of the folding buffer was tested by migration of the folded RNA on a 1.5% agarose gel in 1× TBE running buffer at a constant voltage of 5 V/cm at 4°C for ∼5 h. The folding buffer (1×: 40 mM Tris, pH 8; 25 mM MgCl2; 1 mM MgCl2) that contained physiological concentration of salts and that improved/maintained the intactness of the RNA band was selected for subsequent experiments.
Chemical probing of in vitro transcribed RNA
SHAPE-MaP was performed using the amplicon workflow (55). Since mutational profiling was done using paired end sequencing using 500 cycle MiSeq reagent kit, the RNA was divided into four different overlapping zones of ∼500 nt to facilitate coverage across the entire RNA. For each zone, 0.5 pmol of Meg3 RNA was incubated with the folding buffer (1×: 40 mM Tris, pH 8; 25 mM NaCl; 1 mM MgCl2) in a final volume of 9 μl at 37°C for 30 min. The RNA was then modified with 1 μl of 100 mM 1M7. 1M7 negative control reactions were generated as above with the exception that 1 μl of DMSO was added instead of 1M7. All reactions were placed on ice and the denaturing control reaction was generated as previously described (55). Briefly, 0.5 pmol of RNA was incubated with denaturation buffer (1×: 50% formamide, 50 mM HEPES and 4 mM EDTA) in a volume of 9 μl at 95°C for 1 min. 1 μl of 100 mM 1M7 was added and the mixture incubated at 95°C for another 1 min. Thus, one each of 1M7 plus, 1M7 minus control and denaturing control were generated for each of the four zones.
Extraction and modification of in cellulo and ex vivo Meg3 RNA
Meg3 RNA transcribed inside U87-MG human glioblastoma cells was probed. This cell-line, gifted by Dr Stommel Jayne (NIH), was maintained in DMEM medium containing 10% FBS (GIBCO) and cultured at 37°C with 5% CO2. One day prior to transfection, ∼4 × 106 cells were seeded on a 15 cm Petri dish and transfected with 45 μg of pCI-Meg3 plasmid using lipofectamine 3000, following manufacturer's protocol. Twenty four hours post-transfection, the medium was replaced with fresh medium and the cells incubated for an additional day.
For in cellulo probing, cells were quickly washed with 1X PBS and treated with 2.7 ml medium (without serum and antibiotic), For 1M7 plus reaction, 300 μl of 100 mM 1M7 was added directly to the medium and immediately swirled. For 1M7 minus control, cells were treated with 300 μl of DMSO instead of 1M7. Cells were incubated at 37°C for 5 min and washed twice with pre-chilled 1× PBS on ice. Fractionation into cytoplasmic and nuclear fractions used the Nuclear Extract kit (ActiveMotif) per manufacturer's guidelines. Modified RNA from both fractions was extracted using TRIzol, followed by DNase treatment and ethanol precipitation. The integrity of these fractions was confirmed by quantitating the levels of MALAT1 and U6 RNAs by real-time PCR.
For ex vivo RNA probing, cells were washed twice with pre-chilled 1× PBS on ice and fractioned into cytoplasmic and nuclear fractions using Nuclear Extract kit (ActiveMotif) per manufacturer's guidelines. RNA from the nuclear fraction was extracted gently under non-denaturing conditions to preserve the its native structure (54). Briefly, cells were resuspended in 5 ml of lysis buffer [40 mM Tris (pH 7.9), 25 mM NaCl, 6 mM MgCl2, 1 mM CaCl2, 256 mM sucrose, 0.5% Triton X-100, 1000 units/ml RNasin (Promega), and 450 units/ml DNase I (Roche)], and rotated at 4°C for 15 min. The suspension was centrifuged at 2500 g at 4°C for 2 min. to recover the cell pellet, which was then deproteinized by resuspending in 40 mM Tris (pH 7.9), 200 mM NaCl, 1.5% SDS and 500 μg/ml Proteinase K, and rotating at 20°C for 45 min. RNA was then extracted twice with an equal volume of phenol/chloroform/isoamyl alcohol (24:24:1) preequilibrated with 1× folding buffer [100 mM HEPES (pH 8.0), 100 mM NaCl and 10 mM MgCl2], followed by one extraction with chloroform. RNA was exchanged into 1.1× folding buffer using a desalting column (PD-10, GE Life Sciences) and incubated at 37°C for 30 min. Approximately 10 μg of RNA was then added to a 1/10th volume of 100 mM 1M7 in DMSO (final concentration of 10 mM) and incubated at 37°C for 5 min. Modified RNA was purified by ethanol precipitation. 1M7 negative control was prepared in the same way using equal volume of DMSO instead of 1M7.
To generate the denaturing control for ex-vivo and in-cellulo chemical probing, total RNA was extracted from transiently transfected cells using TRIzol (Ambion), followed by DNase I treatment and ethanol precipitation. Approximately 10 μg of RNA was then resuspended in 1.1× denaturing control buffer [55 mM HEPES (pH 8.0), 4.4 mM EDTA, and 55% (v/v) formamide] and incubated at 95°C for 1 min. The RNA was modified by adding 1/10th volume of 100 mM 1M7 in DMSO, and the mixture incubated at 95°C for 1 min. Modified RNA was purified by ethanol precipitation.
Mutational profiling of modified RNA
For each 1M7 plus, 1M7 minus control, and denaturing control, modified RNA was first reverse transcribed using four different reverse transcription oligos (Z1RT, Z2RT, Z3RT and Z4RT) to generate cDNA library for all four zones. Reverse transcription was performed by first annealing 2 μM of zone specific oligo (obtained by modification of 5 pmol of in vitro transcribed RNA and 1 μg of nuclear RNA) in a reaction volume of 11 μl by incubation at 65°C for 5 min followed by cooling on ice. cDNA synthesis was initiated by incubating the annealing mixture with 8 μl of 2.5× RT reaction mixture (2.5X: 125 mM Tris (pH 8.0), 187.5 mM KCl, 15 mM MnCl2, 25 mM DTT and 1.25 mM dNTPs) and 1 μl of Superscript II reverse transcriptase (Thermo Fisher, 200 U/μl) for 42°C for 3 h. Subsequently, the RNA template was hydrolyzed by adding 1 μl 2N NaOH to each reaction, then neutralized by adding 1 μl 2N HCl. The cDNA library was purified through G50 spin columns (GE healthcare).
For mutational profiling, cDNA was converted to dsDNA with Illumina adapters for high throughput sequencing on an Illumina platform. This was achieved in two consecutive PCR reactions, namely PCR1 and PCR2. The first reaction (PCR1) appends partial Illumina adapters to the ends of the amplicons. The entire cDNA library was used as template in a 100 μl PCR1 reaction (1.1 μl each of 50 pmol of forward oligo and reverse oligo's, 2 μl of 10 mM dNTPs, 20 μl of 5× Q5 reaction buffer, 1 μl of hot Start High-Fidelity DNA polymerase) using cycling conditions: 98°C for 30 s, 15 cycles of [98°C for 10 s, 50°C for 30 s, 72°C for 30 s], 72°C for 2 min. The resulting PCR product was gel purified and 1/10th used as template DNA in PCR2 reaction. The PCR2 completes the Illumina adapters while adding appropriate barcoding indices. The PCR2 reaction and cycling conditions were same as those for PCR1. The resulting amplicon library was fractionated through a 2% agarose gel and the amplicons purified from the gel slices by electro-elution at room temperature for 2 h followed by ethanol precipitation. Each amplicon library was quantitated by real time PCR using KAPA Universal Library Quantitation kit (Illumina) per manufacturer's protocol. The sequencing libraries were pooled and mixed with 20% PhiX and sequenced on a MiSeq to generate 2 × 250 paired-end reads. The sequences corresponding to the primer binding regions and the first five nt synthesized during reverse transcription were excluded from analysis for all the four zones using custom Python scripts (available upon request). The trimmed sequences from all four zones were then merged for each 1M7 plus, 1M7 minus control, and denaturing control reaction and the output sequence files fed into ShapeMapper (v1.2) software (55). SHAPE reactivity profiles were created by aligning the reads to Meg3 RNA sequence using the software with default settings. The median read depth of sequencing libraries used for generating in vitro, ex vivo and in cellulo was greater than 105 185 106 358, 108 525 respectively.
2D structure modeling
The reactivity values obtained from ShapeMapper were inputted into RNAstructure software (56) to generate the minimum free-energy secondary structure model of the RNA. The structure models were minimally adjusted locally at three places to make them comply better with the corresponding reactivity values to generate the final models presented in the study. To determine the likelihood of the RNA to form pseudoknot, the reactivity data was fed into Shapeknots (55), where the RNA was folded in 600 nt sliding windows offset by 100-nt increments. No potential pseudoknots were identified. To determine the well-structured regions within the RNA, local median SHAPE reactivity and Shannon entropy were calculated over a centered 51 nt sliding window. (50,51). Regions in which both the SHAPE reactivity and the Shannon entropy local medians were less than the global medians for at least 40 nt were designated as well-structured regions (R1-R4). Regions separated by fewer than 10 nt were combined to include all secondary structure interactions. Regions in which both the SHAPE reactivity and the Shannon entropy local medians were less than the global medians for at least 40 nt were designated as well-structured regions (R1-R4). Regions separated by fewer than 10 nt were combined to include all secondary structure interactions.
3D structure modeling
RNAcomposer is a homology-based, automated RNA modeling web server that converts RNA sequence and secondary structures into 3D structural models. Because the maximum length of input RNA for this software is 500 nt, the in vitro and ex vivo Meg3 RNA sequence and secondary structures were divided into five overlapping zones, each of which was modeled independently in RNA composer. PDB coordinates of the zones were aligned at the overlap regions in PyMol (The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA, USA) and re-saved on a global coordinate system. Using PDB_merger.py, a custom Python script that can be made available upon request, PDB files of the aligned zones were merged into in vitro and ex vivo master files containing coordinates for all atoms in the respective Meg3 RNA 3D structures. Homology region incompatibilities in RNA composer and zone alignment resulted in steric clashes in some regions of the master 3D models. These were resolved manually, and great care was taken to move structured, reliably modeled regions as blocks to logical positions consistent with the flow of the structure. The blocks were then rejoined to the whole by remodeling flexible, single stranded linking regions of RNA to accommodate the new positioning.
Identification of regions experiencing large absolute reactivity changes
The reactivity values obtained for in cellulo and ex vivo nuclear Meg3 RNA were averaged over 5 nt sliding window and the absolute differences between the averages calculated. Regions with the absolute differences greater than the global median for at least 20 consecutive nucleotides were identified as regions of differences. These regions were graphically represented as the ratio of average ex vivo to in cellulo reactivity plotted against nucleotide number.
Identification of ΔSHAPE sites
ΔSHAPE sites, defined as regions with statistically stringent reactivity difference between ex vivo and in cellulo SHAPE reactivity, were calculated as described previously (54). Briefly, the difference between ex vivo and in cellulo SHAPE reactivity were averaged over a 3 nt sliding window. The Z factor for each average was calculated. Standard errors in SHAPE reactivity measurements were also calculated. Regions undergoing significant reactivity changes (ΔSHAPE sites) were identified as regions where at least 3 nt in a 5 nt window had Z-factor >0 and absolute standard scores ≥1. ΔSHAPE sites identification in the presence of PRC2 and DNA duplexes was conducted in similar fashion with two exceptions. First, the reactivity differences were calculated by subtracting reactivity measurements determined in the presence of PRC2/DNA from those determined in their absence. Second, for duplex DNA, ΔSHAPE sites were defined as nucleotide(s) with Z-factor >0 and absolute standard scores ≥1.
SHAPE-MaP based footprinting to map PRC2 contact sites
Baculovirus expressed human recombinant PRC2 complex which includes full length EZH2, SUZ12, EED and RbAp46/48 (accession numbers NP_001190176.1, NP_056170, NP_003788.2, NP_002884.1 and NP_005601.1, respectively) and contains N-terminal FLAG-Tag at the N-terminus of EZH2 was purchased from ActiveMotif. For each SHAPE-MaP zone, 0.5 pmol of freshly prepared Meg3 RNA was incubated with the folding buffer (1×: 40 mM Tris, pH 8; 25 mM MgCl2; 1 mM MgCl2) in a final volume of 8 μl at 37°C for 30 min. The folded RNA was then incubated with 1 μl of PRC2 protein (2 μM) in PRC2 dilution buffer (1×: 25 mM HEPES pH 7.5, 300 mM NaCl, 5% glycerol, 0.04% Triton X-100) at 37°C for 30 min. The RNA was then modified with 1 μl of 100 mM 1M7. The PRC2 negative control reactions were prepared in similar manner except that 1 μl of PRC2 dilution buffer was added instead of PRC2. Similarly, 1M7 negative control reactions were generated as above with the exception that 1 μl of DMSO was added instead of 1M7.
Electrophoretic mobility assay
0.5 pmol of freshly prepared Meg3 RNA (∼1600 nt) and antisense Meg3 RNA (∼1600 nt) were each incubated with the folding buffer (1×: 40 mM Tris, pH 8; 25 mM NaCl; 1 mM MgCl2) in a final volume of 8 μl at 37°C for 30 min. The folded RNA was then incubated with different concentrations of 1 μl of baculovirus-expressed human recombinant PRC2 complex (ActiveMotif) in PRC2 dilution buffer (1×: 25 mM HEPES pH 7.5, 300 mM NaCl, 5% glycerol, 0.04% Triton X-100) at 37°C for 30 min. To detect, the PRC2–RNA complex formation, the mixture was allowed to migrate on a 1.5% native agarose gel in 1× TBE running buffer at a constant voltage of 5 V/cm at 4°C for ∼5 h. The fraction of the RNA bound by PRC2 was determined using ImageQuant software. The specificity of Meg3 RNA-PRC2 interaction was assessed by comparing the shift in Meg3 RNA and antisense Meg3 RNA bands.
Luciferase reporter assay
10 000 U87-MG cells/well were seeded on a 96-well plate. Twenty four hours later, cells were co-transfected with 100 ng of the reporter plasmid (pGL-luc-p53 from Addgene), 100 ng of pCI-flMeg3 plasmid/pCI-empty vector, and 25 ng of pCMV-GFP(Addgene) using Lipofectamine 3000. The medium was replaced 24 h post-transfection and the cells further incubated at 37°C for 1 day. Luciferase activity was determined using the Luciferase assay system (Promega) per manufacturer's instructions. GFP reading, used as a measure for transfection efficiency, was taken before lysing the cells for determining luciferase activity.
Comparative sequence analysis
Meg3 RNA sequences from human (Homo sapiens; NR_002766.2), orangutan (Pongo abeli; NR_037685.1), mouse (Mus musculus; NR_027651.2), rat (Rattus norvegicus; NR_131064.1) and porcine (Sus scrofa; NR_021488.1) were retrieved from NCBI browser and aligned using Clustal Omega. The sequences of Meg3 motifs were extracted from the alignment and analyzed for primary sequence and secondary structure conservation by Clustal Omega and Turbofold respectively.
RESULTS
Meg3 RNA is highly structured and contains stable functional motifs.
To structurally characterize the most physiologically relevant Meg3 RNA conformer of the predominant Meg3 RNA isoform (Supplementary Figure S13), we probed nuclear RNA extracted from U87-MG cells transiently transfected with plasmid expressing isoform variant 1 of the Meg3 gene (GenBank accession number, NR_002766.2). Cellular proteins were removed, but RNA secondary structure was preserved by gentle organic extraction (ex vivo). Meg3 RNA was expressed ectopically in this manner both to increase Meg3 expression levels and mask endogenously expressed, alternatively spliced isoforms, reverse transcription of which could obfuscate interpretation of SHAPE-MaP results. The functionality (Supplementary Figure S1) of Meg3 RNA, the absence of endogenous Meg3 isoforms (Supplementary Figure S2), and the efficient segregation of cytoplasmic and nuclear fractions (Supplementary Figure S3) were verified in a series of control experiments.
We also probed in vitro transcribed Meg3 RNA purified under native conditions, both for comparison to ex vivo RNA and as a more manageable model system for assessing PRC2 binding. To preserve conformers produced by co-transcriptional folding, this RNA was freshly prepared for all experiments and never frozen or subjected to heat denaturation. RNA thus treated was structurally homogenous, evidenced by its migration as a discrete band on native agarose gels (Supplementary Figure S4). The SHAPE reactivity profiles of the ex vivo and in vitro Meg3 RNAs are shown in Supplementary Figures S6 and S8 respectively.
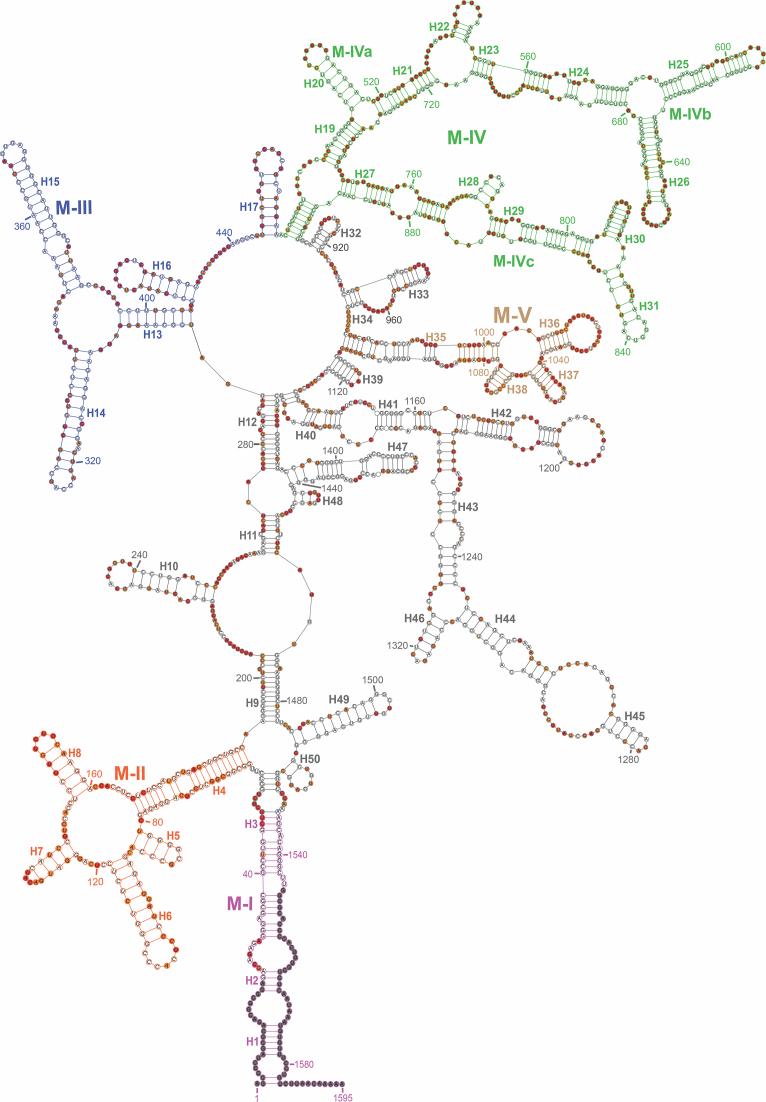
Ex vivo Meg3 RNA adopts a highly branched secondary structure organized around a flexible central junction and supporting a long-range interaction between its 5′ and 3′ termini (Figure 1). Following the helix nomenclature conventions for ribosomal RNA, helices were numbered 5′ to 3′ and separated by a multi-way junction, an internal loop comprising a total of 13 or more nt, or a bulge of more than 6 nt. Meg3 RNA is highly structured, forming 50 double-stranded helices (H1-H50) and 61.1% of nucleotides involved in Watson–Crick or wobble base-pairs. In common with other long RNAs (25,51,57), Meg3 is replete with internal loops, junctions and bulges.
Figure 1.
Ex vivo Meg3 RNA is highly structured. The figure represents the secondary structure model of ex vivo Meg3 RNA determined by SHAPE-MaP. Ectopically expressed Meg3 RNA was extracted from nuclear fraction of U87-MG cells under gentle non-denaturing conditions and probed with 1M7. 1M7 reactivity was detected by mutational profiling. Highly reactive, moderately reactive, low/unreactive nucleotides are color coded as red, orange, and white filled circles respectively. Nucleotides representing the primer binding sites for reverse transcription (RT) and PCR1 reactions, the first 5 nucleotides synthesized during reverse transcription, and nucleotides with reactivity higher than 10 (potential RT stop sites) were excluded from analysis and are denoted by gray filled circles. Helices (H1-H50) are identified and named following helix nomenclature conventions for ribosomal RNAs. Structural elements common to both the ex vivo and the in vitro transcribed Meg3 are designated as motifs (M-I to M-V). Motifs M-I, M-II, M-III, M-IV, M-V are color coded in maroon, orange, blue, green, and brown respectively. Motif M-IV is further divided into M-IVa, M-IVb, and M-IVc. Two tailed non-parametric Spearman correlation of 1M7 reactivity between two independent experiments (N) at a P value <0.0001 was 0.6544.
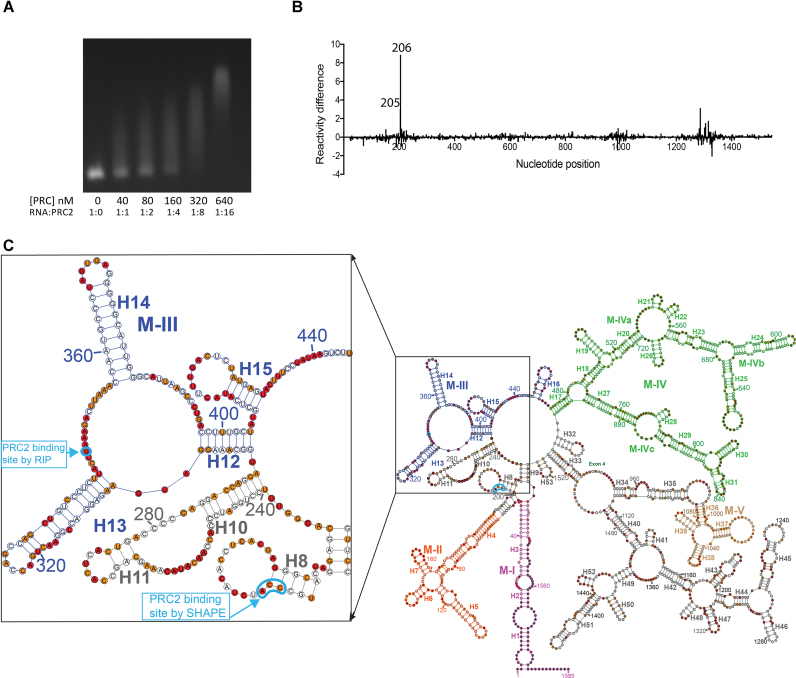
Since functionally important elements are usually housed in structurally conserved RNA elements, we visually inspected the ex vivo and in vitro (vide infra) Meg3 RNA model structures and identified common motifs (M-I to M-V) corresponding to the regions designated in Figures 1 and 3A. M-I comprises terminal helices H1–H3 formed by basepairing between nt 1–48 and 1534–1595, while M-II to M-V span nt 59–189, 294–470, 471–902 and 995–1085, respectively. M-IV can be further subdivided into M-IVa (nt 471–560, 700–745 and 894–902), M-IVb (nt 561–699) and M-IVc (nt 746–895) at logical sub-structure breakpoints. RNA segments within M-I to M-IV have previously been shown to contribute to Meg3 RNA function. Specifically, a portion of M-I comprising nt 20–38 was found to form an RNA-DNA triple helix with the TGF-β genes TGFB2, TGFBR1 and SMAD2 (29), while M-III includes a PRC2 contact point (nt 345) previously identified by RNA immunoprecipitation (RIP) in BT-549, a human breast cancer cell line (29). It has been proposed that concomitant association of Meg3 RNA with PRC2 and TGF-β gene DNA promotes downregulation of TGF-β gene expression by targeted histone methylation of promoter regions. Similarly, although the mechanistic basis for this remains unclear, regions of M-II have been demonstrated as necessary for suppression of DNA synthesis (31). Moreover, employing a compensatory mutational study, Zhang et al. reported that the secondary structure of Motif M-IVb is required for p53 driven transcriptional activation (31).
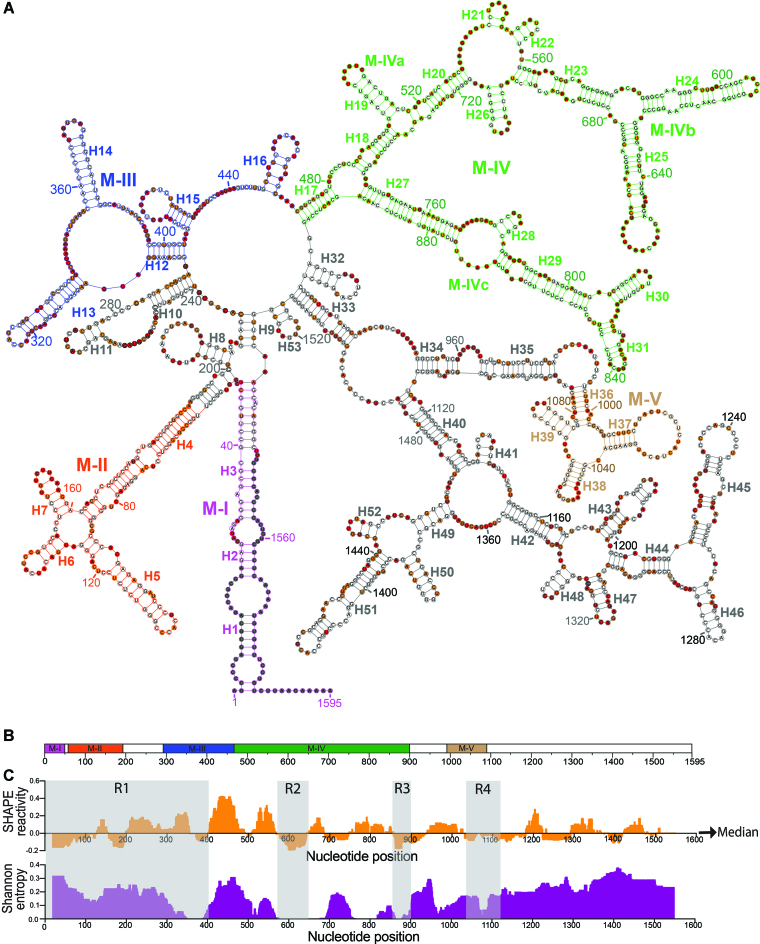
Figure 3.
In vitro transcribed native Meg3 RNA is highly structured. (A) Secondary structure model of in vitro transcribed Meg3 RNA by SHAPE-MaP. Meg3 RNA was transcribed in vitro by T7 RNA polymerase, purified under non-denaturing conditions, incubated with folding buffer containing physiological concentration of Mg2+ (1 mM), and then probed with 1M7. 1M7 reactivity was detected by mutational profiling. Highly reactive, moderately reactive, low/unreactive nucleotides are color coded as red, orange, and white filled circles respectively. Nucleotides representing the primer binding sites for reverse transcription (RT) and PCR1 reactions, the first 5 nucleotides synthesized during reverse transcription, and nucleotides with reactivity higher than 10 (potential RT stop sites) were excluded from analysis and are denoted by gray filled circles. Helices (H1–H53) are identified and named following helix nomenclature conventions for ribosomal RNAs. Structural elements common to both the ex vivo and the in vitro transcribed Meg3 are designated as motifs (M-I to M-V). Motifs M-I, M-II, M-III, M-IV, M-V are color coded in maroon, orange, blue, green, and brown respectively. Motif M-IV is further divided into M-IVa, M-IVb, and M-IVc. (B) Schematic of Meg3 RNA featuring motifs M-I to M-V. (C) Structurally stable regions of Meg3 RNA defined as a function of reactivity and Shannon entropy. Upper panel:1M7 reactivity for in vitro Meg3 RNA shown as the median reactivity over centered 55-nt sliding window, relative to the global median; regions above and below the line are more flexible or constrained than the global median respectively. Lower panel: Shannon entropy values of the RNA, smoothed over centered 55-nt sliding windows. Regions with low SHAPE reactivity and low Shannon entropy are shaded in gray and named regions R1–R4. Two tailed non-parametric Spearman correlation of 1M7 reactivity between two independent experiments (N) at a P value <0.0001 was 0.8356.
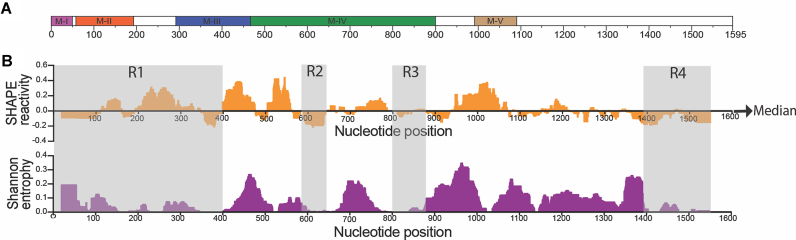
We also identified segments of RNA likely to be structurally conserved among ex vivo RNA conformers by comparing Shannon entropy and reactivity values across a 51 nt sliding window. For a given nucleotide, the Shannon entropy is inversely proportional to the frequency with which it is base paired in conformations likely to be assumed by an RNA (53,54). Thus, highly stable, well-defined RNA sub-structures are characterized by lower Shannon entropies, while regions with high Shannon entropy are likely to form alternative conformations. We identified four different regions (R1 – R4) with low Shannon entropy and reactivity values within ex vivo Meg3 RNA (Figure 2). There is significant overlap between these regions and the previously characterized motifs. Specifically, R1 encapsulates M-I through most of M-III, while R2 and R3 are embedded within M-IVb and M-IVc, respectively. The 5′ and 3′ segments of Meg3 RNA are housed in R1 and R4, indicating that the long-range interaction between the 5′ and 3′- RNA termini is stable and well conserved among alternative conformers. The importance of this interaction is also supported by mutational analysis showing that both terminal regions of Meg3 RNA are required for function (48).
Figure 2.
Ex vivo Meg3 RNA forms structurally stable and well-defined regions defined by low SHAPE reactivity and low Shannon entropy. (A) Schematic of Meg3 RNA featuring motifs M-I to M-V. (B) Upper panel:1M7 reactivity for ex-vivo Meg3 RNA shown as the median reactivity over centered 51 nt sliding window, relative to the global median; regions above and below the line are more flexible or constrained than the global median respectively. Lower panel: Shannon entropy values of the RNA, smoothed over centered 51 nt sliding windows. Regions with low SHAPE reactivity and low Shannon entropy are shaded in gray and named regions R1–R4.
As with the ex vivo Meg3 RNA, the in vitro transcript is highly structured, and contains a unifying central junction region as well as a long-range interaction between the 5′ and 3′ termini (Figure 3A). The five shared structural motifs (M-I to M-V) are also evident in the in vitro model (Figure 3A–C) and closely resemble their ex vivo counterparts. Structural differences outside of the conserved areas likely alter three-dimensional (3D) positioning of these motifs relative to each other, perhaps indicating that their presence is sufficient for Meg3 function, while their relative positioning is, in some cases, not as important. Overall, the ex vivo and in vitro SHAPE reactivity values exhibit a moderate positive correlation (Spearman correlation, ρ = 0.59) (Supplementary Figure S5), indicating that the nuclear environment (ionic strength, RNA helicases and chaperone proteins, etc.) influences RNA folding.
Similar to the ex vivo model, four regions were particularly conserved among in vitro RNA conformers, indicated by low SHAPE reactivity and Shannon entropy values (Figure 3C). For R1–R3, there is good correlation between corresponding regions in the two models, while in vitro RNA R4 significantly overlaps motif M-V, but not the RNA 3′ terminus. Although structural conservation is predicted for the 5′ half of the long range interaction in in vitro RNA (R1), such is not the case for the 3′ half. This is likely because the predicted long range interactions for in vitro RNA is less extensive than for ex vivo RNA, and the algorithm defining conserved regions of RNA sequence uses reactivity and Shannon entropy values averaged over a 51 nt sliding window. Moreover, structural variants obtained under in vitro transcription conditions probably do not entirely match those produced in the nuclei of mammalian cells—although regions M-I to M-V are clearly conserved.
Comparative sequence analysis Meg3 transcripts from multiple species reveals evolutionarily conserved structural motifs
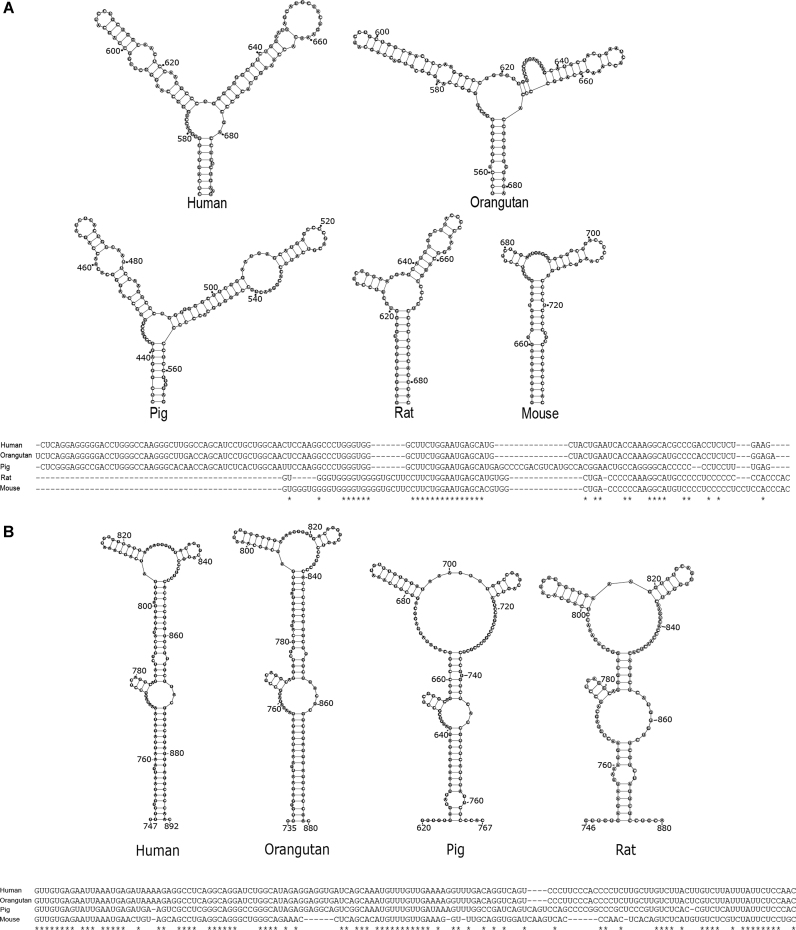
Evolutionary conservation of sequence and structural motifs among homologous lncRNAs provides robust evidence of purifying selection and crucial molecular function. We examined Meg3 RNA sequence conservation by aligning full-length human Meg3 RNA (isoform 1) sequence to the entire Meg3 RNA sequences from orangutan, rat, mouse and pig (data not shown). Significant sequence conservation was observed only in the first ∼900 nt among these species, which is in good agreement with ex vivo and in vitro RNA stable region designations R1–R3, as well as M-I through M-IV. We therefore focused on the ∼900 nt from the 5′ terminus of each of the Meg3 RNAs for downstream sequence and structure conservation analysis. Sequence alignment of this specific region revealed that the human variant shared 73%, 72%, 75% and 98% sequence identity with mouse, rat, pig and orangutan variants, respectively (Supplementary Figure S14). Strong sequence conservation of this region of RNA that houses structurally stable motifs predicted by our SHAPE-MaP experiments is a strong indication of structure conservation within this region among the mammalian species. Therefore, RNA sequences were also analyzed with Turbofold software (58), which uses an iterative probabilistic method for identifying conserved secondary structure elements among multiple homologous sequences (Supplementary Figures S10–S12). In this analysis, M-II and M-III, to which we have already ascribed important Meg3 functions, were conserved among human, orangutan and pig (Supplementary Figures S10 and S11). M-IVb was highly conserved among all species (Figure 4A), consistent with the previous observation that this motif is essential for stimulation of p53-dependent transcription (31), and M-IVc was conserved among human, orangutan, rat and pig (Figure 4B). Taken together, our SHAPE-MaP and inter-species comparative analyses show strong structural conservation of functionally important motifs both among human Meg3 RNA conformers and throughout recent evolution. Furthermore, both human Meg3 and mouse Meg3 (Gtl2) genes, the most well-studied Meg3 genes, contain 10 exons and the overall gene structure and pattern of alternative splicing between the two are well conserved (31,59). Such conservation further supports the conservation of Meg3 RNA structure proposed in our study where different parts of the processed RNA bind to each other to form conserved structures.
Figure 4.
Secondary structure of motifs M-IVb and M-IVc of Meg3 RNA are evolutionarily conserved. Secondary structure model and multiple sequence alignment of (A) Motif M-IVb and (B) Motif M-IVc among different eutherian species determined by Turbofold and Clustal Omega respectively. Asterisks represent nucleotides that were conserved across the species.
Meg3 RNA provides numerous occupancy sites for cellular factors
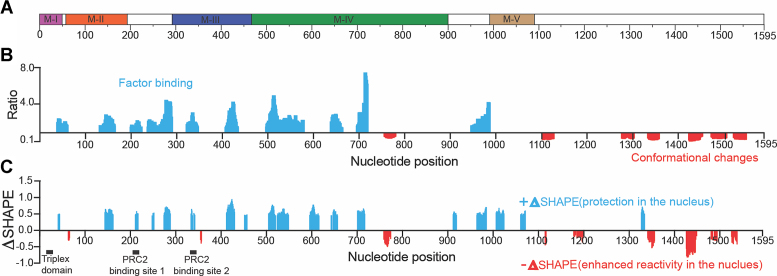
To identify contact sites of cellular factors such as proteins and nucleic acid with Meg3 RNA, we probed the RNA inside living cells (in cellulo). 1M7 was chosen as the acylating reagent in these experiments because it is cell-permeable, highly reactive and has a short half-life in aqueous solution, features that together serve to provide a ‘snapshot’ of intracellular RNA accessibility to chemical modification in the presence of cellular factors. The essence of the SHAPE-based footprinting method described here and elsewhere (50,51,54) is that reactivity values obtained from RNA in cellulo (Supplementary Figure S7) or ex vivo (Supplementary Figure S6) are compared, with large ratios/differences between corresponding sets of values indicating sites at which cellular components bind or alter the structure of the target RNA.
Meg3 RNA sequences spanned by previously characterized structural motifs are depicted together with calculated reactivity ratios and ΔSHAPE values in Figure 5A–C, respectively. The distinct mathematical transformations and significance criteria used to calculate these two metrics are described in Materials and Methods. Only values that met the significance criteria for the respective formulae are plotted, creating islands of values on the bar graphs representing potential binding sites of cellular components. Binding sites shown in Figure 5B are typically broader than those depicted in Figure 5C, in part because more consecutive nucleotides were included to calculate aggregate values for the reactivity ratio metric. The two metrics can thus be considered to represent broad and tight windows of potential cellular component binding, respectively.
Figure 5.
Effects of nuclear environment on Meg3 RNA structure. Intracellular Meg3 RNA (in cellulo) and cell free Meg3 RNA (ex vivo) expressed in U87-MG cells were probed with IM7 and their reactivity difference compared to define factor binding regions and conformationally flexible regions of the RNA. (A) Schematic of Meg3 RNA showing motifs location. (B) Ratio between positive (cyan) and negative (red) reactivity difference within regions of substantial reactivity change. (C) Positive and negative Δ SHAPE sites. Cyan and red peaks represent regions within the RNA where protection or enhanced reactivity were most significant, respectively.
Reactivity ratio calculations were used to identify 17 such windows located throughout the Meg3 RNA sequence but concentrated between nt 50–780 and 1270–1550 (Figure 5B). This distribution suggest that cellular factors bind primarily to conserved structural regions 1 through 4 (R1–R4) and motifs M-I to M-IV, including the stem structure created by long range 5′-to-3′-interactions. Of the 17 windows, 10 represent sites in which in cellulo reactivity values were less than ex vivo values. This is consistent with classic nucleic acid footprinting, where direct protein or ligand binding shields RNA from modifying or cleavage agents. Although the false positive detection rate of protein binding by positive ΔSHAPE sites is low (54), these positive ΔSHAPE sites can also result from allosteric changes in the secondary structure of the RNA itself. The remaining seven binding windows exhibited in cellulo reactivity greater than that of ex vivo-probed RNA, suggesting that cellular factor binding directly or indirectly results in greater exposure of the RNA in these windows to acylation. These RNA segments are primarily located between nt 1100–1550, a region that in mutational studies was determined to be indispensable (48), yet is predicted to largely be structurally variable (Figure 2B). The region also encompasses most of the 3′ half of the 5′-3′ long range interaction predicted in the ex vivo structural model (Figure 1). It is therefore tempting to speculate that windows near the 3′ terminus do not reflect direct binding of cellular factors but are instead indicative of structural rearrangements that help accommodate binding of such factors at sites closer to the 5′ terminus. More specifically, the 5′ half of the long range interaction possibly contributes to formation of the triple helices that tether Meg3 to DNA at select sites, and in doing so releases the 3′ half from base pairing interactions that protect it from acylation. Indeed, previously published work suggests that the Meg3 RNA component of functional DNA-RNA triple helices involves nt 20–38 of M-1 (29), and these nucleotides cannot engage DNA unless the 5′-3′ long range interactions predicted in our structural models are disrupted.
With a few exceptions, there is good correlation between windows of interaction predicted using the reactivity ratio (Figure 5B) and ΔSHAPE calculations (Figure 5C). Twenty-six ΔSHAPE sites were identified using the latter approach. Seventeen of these were positive, nine were negative and, as with the reactivity ratio calculations, the sites were distributed toward the 5′ and 3′ termini of Meg3 RNA, respectively. Although ΔSHAPE calculations can theoretically be used to map binding sites of specific proteins or complexes to an RNA in question, the results here serve primarily to support our reactivity ratio calculations and conclusions regarding Meg3 RNA structure and function.
PRC2 binding mapped by SHAPE-based footprinting
A unifying theme in lncRNA biology is that these elements play an important role in epigenetic regulation. Like other lncRNAs, Meg3 RNA provides a scaffold for assembly of chromatin-modifying complexes such as PRC2, tethering them to target genes to affect repressive histone modification. Human PRC2 is comprised of multiple subunits, including EHZ2 (the catalytic subunit), SUZ21, EED and RbAp46/48 (60), of which EZH2 and SUZ12 have been implicated in RNA binding (61,62)
Here, we characterize the binding of purified human PRC2 to in vitro transcribed, natively folded Meg3 RNA by electrophoretic mobility shift analysis (EMSA) and SHAPE-MaP footprinting (Figure 6). Using the former method, we were able to establish that the two components formed stable ribonucleoprotein complexes under in vitro solution conditions, and the complexes formed were not non-specific aggregates (Figure 6A, Supplementary Figure S9). For SHAPE-based footprinting, we mixed MEG3 RNA with purified PRC2 at a ratio of 1:4, which our EMSA results indicate shifts 36–83% of the total RNA and minimizes non-specific PRC2 binding. By employing the high-stringency ΔSHAPE method and calculations, we observed significant protection at nt C204, C205, and C206 in the presence of PRC2 (Figure 6B). These three nucleotides are located in a loop adjacent to the extended stems formed by long range 5′-3′ interactions in both the ex vivo and in vitro Meg3 RNA structures (Figures 1 and 6C). In contrast, a previous photoactivatable ribonucleoside-enhanced crosslinking and RNA immunoprecipitation study utilizing antibody against EZH2 mapped a single Meg3 RNA-PRC2 contact point at U345 (29), located in the single-stranded valley of the M-III Y-like motif. Although considerably separated in RNA primary sequence, the different contact points are not necessarily mutually exclusive, but instead probably reflect limitations in the two methods, as well as the relative sizes of the PRC2 complex and Meg3 RNA. For instance, site-specific photocrosslinking is relatively inefficient, only occurs in proximity to select targets, and in this instance, can only occur at sites of 4-thiouridine incorporation in Meg3 RNA, while the sensitivity of SHAPE-based footprinting is generally greatest in regions where the unliganded RNA is single stranded. Moreover, the relative sizes of the PRC2 complex and Meg3 RNA predicts that a stable interaction would almost certainly involve protein contact with nucleotides across multiple motifs or sub-structures. Hence, although neither result comprehensively defines the PRC2 binding site on Meg3 RNA, the two together suggest that (i) PRC2 most readily binds to single stranded RNA segments within Meg3 RNA motifs, and (ii) the motifs that collectively house the contact sites may be in proximity in 3D and contribute to a larger binding domain.
Figure 6.
PRC2 specifically binds the 5′ terminus of native in vitro Meg3 RNA. (A) EMSA showing Meg3 RNA shift in the presence of increasing concentration of human PRC2 protein on 1.5% native agarose gel. N = 3. (B) Reactivity difference plot generated by subtracting reactivity measurements in the presence of PRC2 from those in absence of PRC2. Nucleotides corresponding to Δ SHAPE sites in the presence PRC2 complex are shown on top of the reactivity peaks. (C) Mapping of PRC2 induced Δ SHAPE sites (enclosed in cyan) on SHAPE derived Meg3 RNA secondary structure model.
Modeling Meg3 RNA structure in 3D
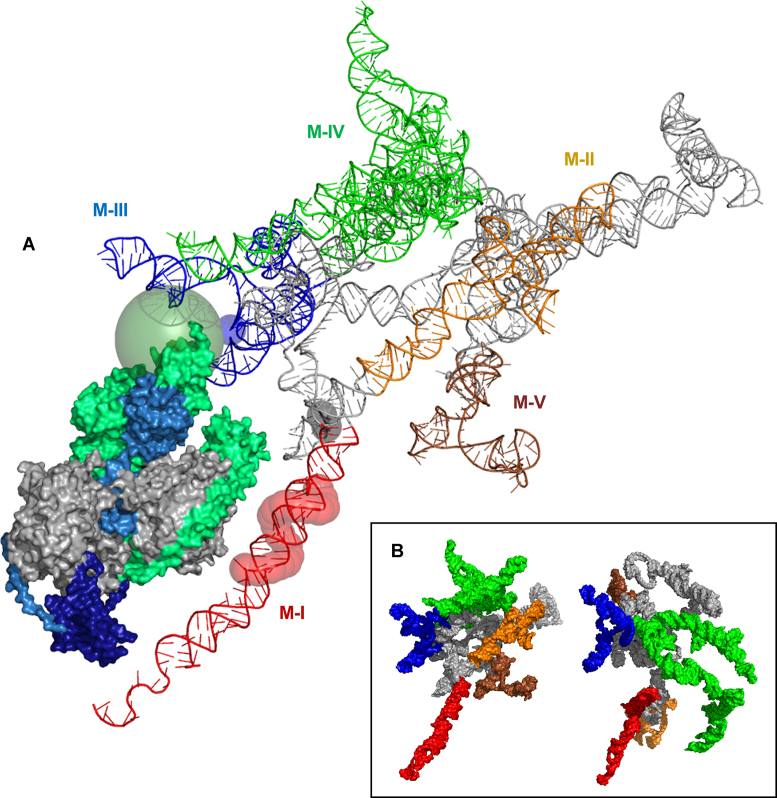
3D models of ex vivo and in vitro Meg3 RNA structure (Figure 7) were generated using RNAcomposer (63), an automated RNA 3D structure modeling server, the respective secondary structure models derived from SHAPE-MaP (Figures 1 and 3A), PyMol molecular viewing and manipulation software (The PyMOL Molecular Graphics System (2002) DeLano Scientific, San Carlos, CA, USA), and custom python scripts to assemble sets of atomic coordinates into complete PDB files. Conserved structural motifs M-I to M-V, PRC2 contact sites predicted by CLIP or ΔSHAPE, and the nucleotide implicated in triple helix formation (29) are all indicated (Figure 7A). The overall structure is highly furcated and lacks an easily definable center of density. As in the secondary structure model, the extended 5′-3′ interaction housing M-I is the most extended, isolated substructure. Although not entirely visible in this view, the Y-shaped motifs comprising M-III, M-IVb and parts of M-IVc and the unstructured regions of the RNA are also notable. These motifs are predicted to emerge from the poorly defined center of the structure into the surrounding space, perhaps serving as docking points for proteins or protein complexes with loose, structure-based binding specificities. Indeed, nt 345, a potential PRC2 contact site (29), is located in the single strand region linking the two branches of the M-III Y-shaped motif. Nucleotides 204–206, the PRC2 contact sites identified by ΔSHAPE, are located at the junction of M-I and M-II, while the predicted nidus of triple helix formation is within M-I. The shapes and relative positioning of M-I and M-III would be compatible with PRC2 binding, and it is certainly feasible that nt 204–206 and 345 can simultaneously contact the large protein complex. It also seems reasonable to suggest that histones proximal to the DNA-RNA triple helix might be ideally positioned for PRC2-mediated methylation in the proposed structural context.
Figure 7.
MEG3 RNA 3D structure models. (A) MEG3 RNA 3D structure (helix-ladder) predicted from natively folded, in vitro transcribed RNA secondary structure. Conserved motifs I–V are shown in red, orange, blue, green, and brown, respectively. Phosphate atoms of nucleotides predicted to interact with the PRC2 complex by CLIP (blue) or SHAPE-MaP (gray) are depicted as semi-transparent spheres scaled to 4-times the expected van der Waals radii to increase visibility. The phosphates in M-I nucleotides previously found to interact with DNA duplex are also shown (red spheres). The PRC2 surface representation was generated from cryo-EM model structure PDB coordinates (PMID: 29348366; PDB accession number 6c24). The EZH2 and SUZ12 subunits are highlighted (lime green, sky blue) together with elements of those subunits implicated in RNA binding. Specifically, RNA-binding EZH2 residues 342–368 are housed within a larger segment (residues 307–422) that is unresolved in the model structure and approximated here by a 25 Å radius, semitransparent forest green sphere. The ‘foot’ region of SUZ12, depicted here in dark blue, is also implicated in RNA binding, perhaps to the extreme 3′ terminus of M-I. (B) Surface representation of native, in vitro transcribed (left) and nuclear ex-vivo (right) MEG3 RNAs. Color coding for motifs I–V matches that in (A).
The 3D structure models of in vitro and ex vivo Meg3 RNA are compared in Figure 7B. The relative positioning of M-I and M-III is remarkably similar in the two structures, suggesting that the means by which intracellular Meg3 RNA accommodates PRC2 and the DNA-RNA triple helix in the cell are likely to closely resemble those predicted by our in vitro structure model. In contrast, positioning of the other conserved motifs in the ex vivo structure varies significantly from that predicted in the in vitro 3D model, indicating that the presence of conserved structural elements, such as the Y-like motifs, may be more critical to Meg3 function than their relative positioning.
DISCUSSION
In this study, we performed extensive structural analysis of Meg3 lncRNA produced by in vitro transcription or derived from living cells. The latter approach is particularly challenging, given that the molecular environments, and thus RNA structures and binding partners, are likely to vary. We overcame these challenges in two ways: First, expression of Meg3 RNA synchronizes intracellular milieus by inducing G1 to M cell cycle arrest (45,64). Second, we used 1M7, a highly reactive SHAPE electrophile with a short half-life, for intracellular probing, thus limiting the extent of RNA conformational sampling or protein binding/dissociation during the modification reaction. Another challenge associated with in vivo structure determination is the low abundance of lncRNA relative to ribosomal RNAs and mRNAs. Fortunately, the cDNA libraries produced by reverse transcription of lncRNAs are amplified by PCR in the SHAPE-MaP protocol, rendering the sensitivity and specificity of this method sufficient for lncRNA structural analysis.
Overall, our data revealed that the 5′ portion of Meg3 RNA, spanning nt 1–912, was highly conserved in sequence and structure. This is in good agreement with observations that this region is rich in cellular factor binding sites and contains elements important for Meg3 RNA function (29,31). With some exceptions, much of the remaining Meg3 sequence was found to be structurally variable and not evolutionary conserved. The 3′ region in particular harbored few factor-binding sites, and its structure was highly influenced by cellular factors. Taken together, our results indicate that (i) Meg3 RNA contains highly stable, functionally relevant substructures interspersed among more structurally fluid sequence elements, and (ii) the conserved elements are maintained regardless of the method of RNA preparation and purification. While it is intuitive to consider the Meg3 structural organization as merely a collection of conserved protein or DNA binding sites amid a fluid background, the functional importance of the more structurally variable regions remains an important question. More specifically, for a pleiotropic lncRNA such as Meg3, with roles in cellular differentiation, angiogenesis, senescence and various cancers, the capacity for conformational sampling conferred by regions of variable RNA structure might provide diverse scaffolding interfaces for assembly of distinct RNPs. RNA with multiple isoforms are also good candidates for hypermethylation (65), the patterns of which could influence Meg3 RNA structure, stability and function.
Although many proteins have been shown by to interact with Meg3 RNA, we chose to focus on the interaction of full-length Meg3 with the PRC2 complex. PRC2 primarily di/trimethylates histone H3K27, represses transcription of genes involved primarily in development and cellular differentiation (66,67), and also interacts with other well studied lncRNAs including HOTAIR and Xist (57,68). Meg3 and other lncRNAs may regulate this function by binding PRC2 and tethering it to DNA regulatory elements by contributing to site-specific DNA-RNA triple helices. Our ΔSHAPE analysis identified a PRC2 contact point with Meg3 RNA at nt 204–206 in M-I which, in 3D, is predicted to be near nt 345, the M-III contact point with EZH2 (29). The two observations are compatible with a large binding interface between PRC2 and Meg3 RNA involving M-I and M-III proximal to the reported site of triplex formation (29) (Figure 6). Indeed, although Meg3-DNA triplex formation is independent of PRC2 binding (29), the close relative positioning of PRC2 and DNA during association with Meg3 RNA might facilitate local histone methylation. Given that the spatial separation between the two predicted contact sites is significant, the nt 345 site was identified using anti-EZH2 monoclonal antibody, and PRC2 likely contacts Meg3 RNA at several points, the site identified by ΔSHAPE likely reflects contact with the SUZ12 subunit (61,69,70) of PRC2, which has also been implicated in RNA binding.
Collectively, our study answers basic questions regarding Meg3 structure and activity, providing a template for functional and mechanistic studies of other lncRNAs. These findings also lay a framework for efficiently harnessing the tumor suppressive property of the RNA for potential anti-cancer therapies. Meg3 RNA is by far the best-characterized tumor suppressor lncRNA. In various cancer cell lines and clinical samples, Meg3 is either not expressed or is expressed at low levels, and exogenous expression of Meg3 RNA in such cells can slow growth and induce apoptosis. Targeted restoration or overexpression of Meg3 RNA in affected cells, therefore, offers a promising avenue for cancer treatment. Toward this end, delivery of Meg3 RNA using MS2 virus-like particles (VLPs) has recently proven safe, fast, and effective in significantly attenuating cell growth in vitro and in vivo (71), although the 1.6 kb length of Meg3 RNA limits delivery options for this approach. Using data presented in this study, one can easily envisage engineering Meg3 RNA constructs that are much shorter than the native version yet, because they contain critical, conserved, well-defined structural motifs, retain full functionality in tumor suppression.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Dr Jayne Stommel (NIH) for providing U87-MG cell-line, Dr Vineet Kewalramani (NIH) and Dr KyeongEun Lee (NIH) for allowing C.S. to use the tissue culture facility, and Dr Mary Kearney (NIH) and Dr Valerie Boltz (NIH) for allowing C.S. to operate the Illumina MiSeq instrument for this study.
Author Contributions: Conceptualization and design, C.S.; methodology, C.S and J.W.R.; investigation, C.S.; writing – original draft, C.S.; writing – review & editing, C.S., J.W.R. and S.F.J.L.G.; funding acquisition, S.F.J.L.G.; resources, S.F.J.L.G.; supervision, S.F.J.L.G.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research Training Award Fellowship (to C.S.); NIH; Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services [ZIA BC 010493 to S.F.J.L.G.]. Funding for open access charge: Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services [ZIA BC 010493].
Conflict of interest statement. None declared.
REFERENCES
- 1. Banfai B., Jia H., Khatun J., Wood E., Risk B., Gundling W.E. Jr, Kundaje A., Gunawardena H.P., Yu Y., Xie L. et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012; 22:1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P.. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015; 43:D174–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L.. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011; 25:1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012; 338:1435–1439. [DOI] [PubMed] [Google Scholar]
- 9. Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., Sanchez-Gomez D.B., Hacisuleyman E., Li E., Spence M. et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013; 2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pauli A., Valen E., Lin M.F., Garber M., Vastenhouw N.L., Levin J.Z., Fan L., Sandelin A., Rinn J.L., Regev A. et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012; 22:577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pauli A., Rinn J.L., Schier A.F.. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011; 12:136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013; 152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dorji T., Monti V., Fellegara G., Gabba S., Grazioli V., Repetti E., Marcialis C., Peluso S., Di Ruzza D., Neri F. et al. Gain of hTERC: a genetic marker of malignancy in oral potentially malignant lesions. Hum. Pathol. 2015; 46:1275–1281. [DOI] [PubMed] [Google Scholar]
- 14. Tao R., Hu S., Wang S., Zhou X., Zhang Q., Wang C., Zhao X., Zhou W., Zhang S., Li C. et al. Association between indel polymorphism in the promoter region of lncRNA GAS5 and the risk of hepatocellular carcinoma. Carcinogenesis. 2015; 36:1136–1143. [DOI] [PubMed] [Google Scholar]
- 15. Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X., Gejman R., Mahta A., Zhong Y., Rice K.A., Zhou Y., Cheunsuchon P., Louis D.N., Klibanski A.. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010; 70:2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown C.J., Hendrich B.D., Rupert J.L., Lafreniere R.G., Xing Y., Lawrence J., Willard H.F.. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992; 71:527–542. [DOI] [PubMed] [Google Scholar]
- 18. Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S.. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992; 71:515–526. [DOI] [PubMed] [Google Scholar]
- 19. Pang K.C., Frith M.C., Mattick J.S.. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006; 22:1–5. [DOI] [PubMed] [Google Scholar]
- 20. Wang J., Zhang J., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K.. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004; 431:758. [PubMed] [Google Scholar]
- 21. Hezroni H., Koppstein D., Schwartz M.G., Avrutin A., Bartel D.P., Ulitsky I.. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015; 11:1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P.. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011; 147:1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quinn J.J., Zhang Q.C., Georgiev P., Ilik I.A., Akhtar A., Chang H.Y.. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016; 30:191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P.. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010; 3:ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Novikova I.V., Hennelly S.P., Sanbonmatsu K.Y.. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012; 40:5034–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith M.A., Gesell T., Stadler P.F., Mattick J.S.. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 2013; 41:8220–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Theimer C.A., Blois C.A., Feigon J.. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005; 17:671–682. [DOI] [PubMed] [Google Scholar]
- 28. Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017; 543:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E. et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015; 6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyoshi N., Wagatsuma H., Wakana S., Shiroishi T., Nomura M., Aisaka K., Kohda T., Surani M.A., Kaneko-Ishino T., Ishino F.. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000; 5:211–220. [DOI] [PubMed] [Google Scholar]
- 31. Zhang X., Rice K., Wang Y., Chen W., Zhong Y., Nakayama Y., Zhou Y., Klibanski A.. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010; 151:939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R., Ansell P.J., Zhao J., Weng C., Klibanski A.. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007; 282:24731–24742. [DOI] [PubMed] [Google Scholar]
- 33. Zhang X., Zhou Y., Mehta K.R., Danila D.C., Scolavino S., Johnson S.R., Klibanski A.. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 2003; 88:5119–5126. [DOI] [PubMed] [Google Scholar]
- 34. Wang C., Yan G., Zhang Y., Jia X., Bu P.. Long non-coding RNA MEG3 suppresses migration and invasion of thyroid carcinoma by targeting of Rac1. Neoplasma. 2015; 62:541–549. [DOI] [PubMed] [Google Scholar]
- 35. Greife A., Knievel J., Ribarska T., Niegisch G., Schulz W.A.. Concomitant downregulation of the imprinted genes DLK1 and MEG3 at 14q32.2 by epigenetic mechanisms in urothelial carcinoma. Clin. Epigenet. 2014; 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao J., Dahle D., Zhou Y., Zhang X., Klibanski A.. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J. Clin. Endocrinol. Metab. 2005; 90:2179–2186. [DOI] [PubMed] [Google Scholar]
- 37. Gejman R., Batista D.L., Zhong Y., Zhou Y., Zhang X., Swearingen B., Stratakis C.A., Hedley-Whyte E.T., Klibanski A.. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J. Clin. Endocrinol. Metab. 2008; 93:4119–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Braconi C., Kogure T., Valeri N., Huang N., Nuovo G., Costinean S., Negrini M., Miotto E., Croce C.M., Patel T.. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011; 30:4750–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yao H., Sun P., Duan M., Lin L., Pan Y., Wu C., Fu X., Wang H., Guo L., Jin T. et al. microRNA-22 can regulate expression of the long non-coding RNA MEG3 in acute myeloid leukemia. Oncotarget. 2017; 8:65211–65217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang W., Dong K., Li K., Dong R., Zheng S.. MEG3, HCN3 and linc01105 influence the proliferation and apoptosis of neuroblastoma cells via the HIF-1alpha and p53 pathways. Sci. Rep. 2016; 6:36268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chunharojrith P., Nakayama Y., Jiang X., Kery R.E., Ma J., De La Hoz Ulloa C.S., Zhang X., Zhou Y., Klibanski A.. Tumor suppression by MEG3 lncRNA in a human pituitary tumor derived cell line. Mol. Cell. Endocrinol. 2015; 416:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Modali S.D., Parekh V.I., Kebebew E., Agarwal S.K.. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Mol. Endocrinol. 2015; 29:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kawakami T., Chano T., Minami K., Okabe H., Okada Y., Okamoto K.. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Hum. Mol. Genet. 2006; 15:821–830. [DOI] [PubMed] [Google Scholar]
- 44. Gordon F.E., Nutt C.L., Cheunsuchon P., Nakayama Y., Provencher K.A., Rice K.A., Zhou Y., Zhang X., Klibanski A.. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010; 151:2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu K.H., Li W., Liu X.H., Sun M., Zhang M.L., Wu W.Q., Xie W.P., Hou Y.Y.. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013; 13:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao Y., Lu X.. Decreased expression of MEG3 contributes to retinoblastoma progression and affects retinoblastoma cell growth by regulating the activity of Wnt/beta-catenin pathway. Tumour Biol. 2016; 37:1461–1469. [DOI] [PubMed] [Google Scholar]
- 47. Terashima M., Tange S., Ishimura A., Suzuki T.. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J. Biol. Chem. 2017; 292:82–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iyer S., Modali S.D., Agarwal S.K.. Long noncoding RNA MEG3 is an epigenetic determinant of oncogenic signaling in functional pancreatic neuroendocrine tumor cells. Mol. Cell. Biol. 2017; 37:e00278-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blythe A.J., Fox A.H., Bond C.S.. The ins and outs of lncRNA structure: How, why and what comes next. Biochim. Biophys. Acta. 2016; 1859:46–58. [DOI] [PubMed] [Google Scholar]
- 50. Smola M.J., Christy T.W., Inoue K., Nicholson C.O., Friedersdorf M., Keene J.D., Lee D.M., Calabrese J.M., Weeks K.M.. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sztuba-Solinska J., Rausch J.W., Smith R., Miller J.T., Whitby D., Le Grice S.F.J.. Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res. 2017; 45:6805–6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lu Y.F., Mauger D.M., Goldstein D.B., Urban T.J., Weeks K.M., Bradrick S.S.. IFNL3 mRNA structure is remodeled by a functional non-coding polymorphism associated with hepatitis C virus clearance. Sci. Rep. 2015; 5:16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Siegfried N.A., Busan S., Rice G.M., Nelson J.A., Weeks K.M.. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods. 2014; 11:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smola M.J., Calabrese J.M., Weeks K.M.. Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry. 2015; 54:6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smola M.J., Rice G.M., Busan S., Siegfried N.A., Weeks K.M.. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 2015; 10:1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H.. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013; 41:W471–W474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Somarowthu S., Legiewicz M., Chillon I., Marcia M., Liu F., Pyle A.M.. HOTAIR forms an intricate and modular secondary structure. Mol. Cell. 2015; 58:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harmanci A.O., Sharma G., Mathews D.H.. TurboFold: iterative probabilistic estimation of secondary structures for multiple RNA sequences. BMC Bioinformatics. 2011; 12:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schuster-Gossler K., Bilinski P., Sado T., Ferguson-Smith A., Gossler A.. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev. Dyn. 1998; 212:214–228. [DOI] [PubMed] [Google Scholar]
- 60. Margueron R., Reinberg D.. The Polycomb complex PRC2 and its mark in life. Nature. 2011; 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kasinath V., Faini M., Poepsel S., Reif D., Feng X.A., Stjepanovic G., Aebersold R., Nogales E.. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018; 359:940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Long Y., Bolanos B., Gong L., Liu W., Goodrich K.J., Yang X., Chen S., Gooding A.R., Maegley K.A., Gajiwala K.S. et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife. 2017; 6:e31558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W.. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012; 40:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Qin R., Chen Z., Ding Y., Hao J., Hu J., Guo F.. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013; 60:486–492. [DOI] [PubMed] [Google Scholar]
- 65. Ke S., Alemu E.A., Mertens C., Gantman E.C., Fak J.J., Mele A., Haripal B., Zucker-Scharff I., Moore M.J., Park C.Y. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015; 29:2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y.. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002; 298:1039–1043. [DOI] [PubMed] [Google Scholar]
- 67. Talbert P.B., Henikoff S.. Spreading of silent chromatin: inaction at a distance. Nat. Rev. Genet. 2006; 7:793–803. [DOI] [PubMed] [Google Scholar]
- 68. Sarma K., Cifuentes-Rojas C., Ergun A., Del Rosario A., Jeon Y., White F., Sadreyev R., Lee J.T.. ATRX directs binding of PRC2 to xist RNA and polycomb targets. Cell. 2014; 159:1228. [DOI] [PubMed] [Google Scholar]
- 69. Betancur J.G., Tomari Y.. Cryptic RNA-binding by PRC2 components EZH2 and SUZ12. RNA Biol. 2015; 12:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cifuentes-Rojas C., Hernandez A.J., Sarma K., Lee J.T.. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell. 2014; 55:171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu S., Zhu J., Jiang T., Zhong Y., Tie Y., Wu Y., Zheng X., Jin Y., Fu H.. Identification of lncRNA MEG3 binding protein using MS2-Tagged RNA affinity purification and mass spectrometry. Appl. Biochem. Biotechnol. 2015; 176:1834–1845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.