Abstract
As the key enzyme of bacterial nitrogen assimilation, glutamine synthetase (GS) is tightly regulated. In cyanobacteria, GS activity is controlled by the interaction with inactivating protein factors IF7 and IF17 encoded by the genes gifA and gifB, respectively. We show that a glutamine-binding aptamer within the gifB 5′ UTR of Synechocystis sp. PCC 6803 is critical for the expression of IF17. Binding of glutamine induced structural re-arrangements in this RNA element leading to enhanced protein synthesis in vivo and characterizing it as a riboswitch. Mutagenesis showed the riboswitch mechanism to contribute at least as much to the control of gene expression as the promoter-mediated transcriptional regulation. We suggest this and a structurally related but distinct element, to be designated type 1 and type 2 glutamine riboswitches. Extended biocomputational searches revealed that glutamine riboswitches are exclusively but frequently found in cyanobacterial genomes, where they are primarily associated with gifB homologs. Hence, this RNA-based sensing mechanism is common in cyanobacteria and establishes a regulatory feedback loop that couples the IF17-mediated GS inactivation to the intracellular glutamine levels. Together with the previously described sRNA NsiR4, these results show that non-coding RNA is an indispensable component in the control of nitrogen assimilation in cyanobacteria.
INTRODUCTION
As part of amino acids, nucleobases and many other organic molecules, nitrogen (N) is a key element of life. Whereas some bacteria have evolved the ability to fix N2 gas, most organisms rely on the uptake and assimilation of organic or inorganic nitrogen sources. For microorganisms, the availability and concentrations of different N sources is highly variable requiring consecutive and tight adjustments of the assimilation machinery. Bacteria have evolved complex regulatory networks to control N uptake and the activity of assimilatory enzymes (for reviews see 1–4). The key enzyme of bacterial N assimilation is the glutamine synthetase (GS), which catalyzes the condensation of glutamate and ammonium (NH4+) to produce glutamine. To complete NH4+ assimilation, glutamine subsequently reacts with 2-oxoglutarate (2OG) producing two molecules of glutamate, a step which is catalyzed by the glutamine oxoglutarate aminotransferase (GOGAT). Expression and activity of GS is tightly regulated due to the general role of glutamine as major N source for fundamental biosynthetic pathways (5), but also due to its role as signaling molecule in some bacteria (6,7). Moreover, maintenance of glutamate homeostasis is required for optimal cell growth (8).
In enterobacteria and most other Gram-negative bacteria, GS activity is regulated by covalent modification. In response to increasing N availability, GS is inactivated via adenylylation by a specific adenylyltransferase (ATase) (for a review of GS regulation see 9). Moreover, glnA, the gene encoding GS, is transcriptionally regulated by the NtrB-NtrC two-component system (3). Under N limitation the sensory kinase NtrB phosphorylates and activates NtrC, which acts as transcriptional activator for several N assimilatory genes including glnA. The activity of both, ATase and NtrB is controlled by the regulatory protein PII (for an overview of the PII signaling system see Refs. 10–12). However, the mechanisms of N control and GS regulation are very different in other groups of bacteria such as cyanobacteria.
Cyanobacteria evolved unique mechanisms for GS regulation
Cyanobacteria are morphologically and genetically very diverse. They are the only prokaryotes performing oxygenic photosynthesis, play major roles in the global biogeochemical carbon (C) and N cycles (13–15) and are of substantial interest in green biotechnology (16–18). Compared to enterobacteria and most other Gram-negative bacteria, different mechanisms controlling N assimilation have evolved in cyanobacteria. The main transcriptional regulator involved in the coordinated up-regulation of N assimilatory genes under N-limitation is NtcA, a member of the CRP transcriptional regulator family (19). Moreover, GS activity is not modulated by covalent modification but by small inhibitory proteins, called GS inactivating factors (IFs), (20). Most cyanobacteria possess two homologous IFs, IF7 and IF17, which are encoded by the gifA and gifB genes, respectively. The expression of both genes is also regulated by NtcA, though inverse to glnA (21).
Signal perception of the N status in cyanobacteria
Although glutamine is a well-established signaling molecule for N availability in many Gram-negative bacteria, it seemed to lack any signaling function in cyanobacteria (6). Similar to other major bacterial groups, cyanobacteria sense the levels of 2OG, which connects C and N metabolism via the reactions performed by GS and GOGAT (7). In Synechocystis sp. PCC 6803 (Synechocystis), intracellular glutamine and 2OG concentrations vary with external N fluctuations, but only 2OG correlates well with NtcA-dependent gene expression (22). At the molecular level, 2OG modulates the affinity of NtcA to its recognition sequence (23,24) as well as the activity of PII which is also a key regulatory protein in cyanobacterial N control (12,25). NtcA is further activated by a 2OG dependent interaction with the transcriptional co-factor PipX (26–28). Alternatively, PipX can bind to PII under low 2OG conditions, an interaction that antagonizes PipX-dependent activation of NtcA (26).
NtcA binds as a homodimer to the conserved palindromic sequence GTA-N8-TAC with the A and T at positions 3 and 12 being somewhat variable (23,29,30). Depending on the location of the binding motif relative to the transcriptional start site, NtcA can act as activator (e.g. glnA) but also as repressor (e.g. gifA & gifB) (19). Under N excess (e.g. in presence of NH4+) the 2OG level is low and NtcA is present in an inactive form, which has poor affinity to its target sites. Under N depletion, the 2OG level increases and stimulates complex formation between PipX and NtcA which increases its binding affinity to target promoters (26,31).
Non-coding RNAs are involved in regulating N assimilation
In addition to transcriptional regulators, bacteria possess numerous and diverse small regulatory RNAs (sRNAs). These sRNAs can activate or repress gene expression at the post-transcriptional level by complementary base pairing with their mRNA targets (32,33). In cyanobacteria, the expression of several sRNAs is N-regulated (29,30,34,35). One of them, the NtcA-controlled nitrogen stress induced RNA 4 (NsiR4) impacts GS activity by interfering with the translation of the gifA mRNA (36). Another class of non-coding RNAs are riboswitches. These elements are part of untranslated regions of bacterial mRNAs and can control gene expression by ligand-induced structural modulation (37). Riboswitches are composed of an aptamer which specifically binds a ligand, and an expression platform which determines the read out of genetic information by interfering with the transcriptional or translational machinery in response to the structural modulation (38). The riboswitches identified so far respond to various ligands, including the amino acids glycine and lysine (39,40).
Previously, two RNA aptamers were reported that bind and change conformation specifically in response to glutamine (41,42). These were named the glnA and downstream-peptide (DP) aptamers (not to be confused with the glnA gene encoding glutamine synthetase), and the corresponding RNA sequence motifs were annotated as RF01739 and RF01704 in the Rfam database. Both were found to be restricted to cyanobacterial and marine metagenomic sequences and were to some extent positioned 5′ of genes involved in N metabolism (41). Although structural modulation upon glutamine binding was observed (43), a riboswitch function and its possible physiological relevance have remained undetermined.
Here, we demonstrate that both the glnA and DP aptamers indeed function as glutamine riboswitches. In the cyanobacterial model organism Synechocystis, this riboswitch regulates the expression of the major GS inactivating factor, IF17, and is necessary for the appropriate regulation of GS activity. Homologous riboswitches in other cyanobacteria are mainly associated with gifB gene homologs. Hence, glutamine riboswitches play a major role in the control of GS activity throughout the cyanobacterial phylum. Our data establish glutamine as an additional signaling molecule in cyanobacterial N control that functions through a mechanism currently not known from any other bacterial phylum.
MATERIALS AND METHODS
Strains and cultivation
A glucose-tolerant strain of Synechocystis sp. PCC 6803, originally obtained from N. Murata (Japan), was used as wild type. Cultivation was performed at 30°C under continuous white light illumination of 50–80 μmol quanta m−2 s−1 and gentle agitation in Cu2+-depleted BG11 medium (44) buffered with 20 mM TES to pH 8.0. All recombinant strains were selected on agar-solidified BG11 medium and maintained in presence of the corresponding antibiotics. However, for the in vivo experiments all mutants were grown without antibiotics, exactly as the WT.
Generation of recombinant strains
The 5′ UTR of gifB and 54 nt of its coding region were amplified from Synechocystis gDNA using the oligonucleotides gifB6803_NsiI_fw and gifB6803_NheI_rev. The information about the transcriptional start site (TSS) has been taken from (21). The PCR product was cloned into the plasmid pXG10-SF (45) via the NsiI and NheI sites resulting in a translational fusion of a truncated gifB ORF with the sfgfp gene. The primers used for cloning and the resulting plasmids are given in Supplemental Tables S3 and S4. The 5′ UTR of a DUF4278-encoding gene from Prochlorococcus sp. MED4 was also cloned into pXG10-SF using the primers gifBMED4_NsiI_fw/gifBMED4_NheI_rev and following the same cloning strategy (TSS extracted from (46)). Please note that this gene is covered by the Yfr6 RNA which was initially regarded as non-coding (47) but harbors an ORF. To establish point mutations within the glnA aptamer (RF01739) the pXG10-SF-gifB6803(WT) plasmid was re-amplified using the primer combination gifB_M1_fw/rev. The PCR products were introduced into Escherichia coli via transformation and the resulting plasmid pXG10-SF-gifB6803(M1) was isolated and again used as template for a subsequent PCR to establish complementary point mutation M3.
To introduce and express the gifB::sfgfp constructs in Synechocystis they were amplified from the five pXG10-SF derivatives using the primers gifB-J23101_fw and pJV863SeqRVshift. Thereby the constructs were fused to the BioBrick BBa_J23101 promoter (iGEM Registry, (48)) which was found to be constitutively active at a high level in Synechocystis (49) and expected to function in a wide range of cyanobacteria (50). The same strategy was used for the construct harboring the DP aptamer (RF01704) of Prochlorococcus sp. MED4 by using the primers gifMED4-J23101_fw/pJV863SeqRVshift. The PCR products were cloned into the replicative broad-host vector pVZ322 via the XhoI/XbaI sites thereby omitting its KmR gene. The final pVZ322-based plasmids were introduced into Synechocystis WT via conjugal transfer from E. coli. The recombinant strains were selected on BG11 agar containing 2 μg ml−1 gentamycin.
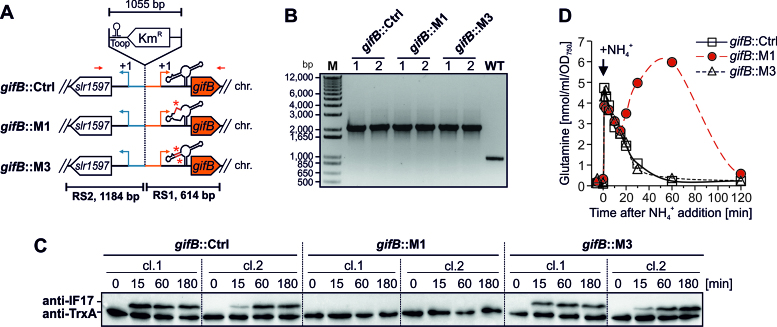
For the mutagenesis of the glutamine riboswitch in the chromosome of Synechocystis we generated constructs to exchange the WT locus of gifB via homologous recombination. First, recombination sites (RS) were amplified from gDNA using the primer combinations gifB_fwd/gifB_KmR_rev and slr1597_fwd/slr1597_rev. These carried overlapping ends with a kanamycin resistance cartridge (KmR) amplified from a customized plasmid using the primers KmR_fwd/KmR_rev. A plasmid backbone with overlapping ends to both RS was amplified from a pJET1.2 plasmid using primers pJET_fwd/pJET_rev. The RS, the KmR and the plasmid backbone were assembled using the AQUA cloning technique (51). The final plasmid pJET-gifB-RS-Ctrl was purified from a transformed E. coli DH5α strain. To establish point mutation M1 the plasmid was reamplified using primers gifB_M1_fw and gifB_M1_rev(6803). After transformation of E. coli and purification of the plasmid pJET-gifB-RS-M1, the compensatory mutation M3 was established in the same way using gifB_M3_fw/ gifB_M3_rev and resulted in the plasmid pJET-gifB-RS-M3. Because first attempts failed to establish Synechocystis mutants using the described plasmids, we extended the recombination flank partially covering slr1597 by the entire CDS. This increased the length of homologous sequence from 202 to 1184 bp. For this we amplified an extended fragment from gDNA using primer combination slr1597-NdeI_fw/slr1597-XbaI_rev. The resulting PCR product was purified and ligated into plasmids pJET-gifB-RS-Ctrl/-M1/-M3 via NdeI/XbaI restriction sites. This replaced the initial recombination site by the extended one. The resulting plasmids were used to transform the WT and established recombinant Synechocystis strains harboring either the WT gifB-5′ UTR (gifB::Ctrl) or point-mutated versions (gifB::M1, [riboswitch structure abolished] and gifB::M3 [riboswitch structure restored]). A schematic illustration of the constructs integrated into the genome is shown in Figure 4A. The recombinant strains were verified by sequencing PCR products amplified from gDNA.
Figure 4.
Mutagenesis of the glutamine riboswitch reveals its importance for gifB expression and GS regulation. (A) Schematic overview of the mutated genomic locus in Synechocystis strains carrying chromosomal point mutations in the glutamine riboswitch upstream of gifB. The kanamycin resistance (KmR) gene was introduced between the promoters of gifB and slr1597. The gifB::Ctrl strain harbors the WT sequences whereas in gifB::M1 and gifB::M3 the glutamine-binding aptamer carries point mutations M1 or M3 as shown in Figure 2A. Red arrows indicate the relative binding sites of primers used for PCR (panel B). RS - recombination site, chr. - chromosome. (B) PCR verification of the mutant strains using two biological replicates. Using WT gDNA a 977 bp fragment was amplified whereas a fragment of 2032 bp was amplified in the mutants. The point mutations M1 and M3 were verified by sequencing the PCR product. No WT allele was amplified in the mutants indicating that they are fully segregated. (C) Immunoblots showing the NH4+ induced accumulation of IF17 (upper band). At time point 0, 10 mM NH4Cl (f.c.) were added to cells grown in BG11 containing nitrate as sole N source. The TrxA protein level is shown as loading control (lower band). (D) Kinetics of intracellular glutamine levels in response to an upshift in NH4+ availability (same conditions and treatment as used for immunoblots). Data are the mean of the two biological replicates.
GFP fluorescence measurements
GFP fluorescence was measured in Synechocystis in vivo using a VICTOR3 multiplate reader (PerkinElmer). The cells were grown as described above to an OD750 of ∼0.5. For the NH4+ upshift 10 mM NH4Cl (f. c.) or the same volume of water were added to the cultures. For the measurement, 200 μl culture aliquots were filled into a 96-well microtiter plate (VWR® Tissue Culture Plates, 96 wells-F). The fluorescence was measured with the following setup: excitation filter F485, emission filter F535, measurement time 1.0 s, CW-lamp energy 21673. The cell absorbance at 750 nm (OD750) was measured in parallel (CW-lamp filter 750/8 nm, measurement time 1.0 s). A WT strain served as a negative control. From each fluorescence value the background fluorescence of the WT was subtracted and then divided by the measured OD750.
Expression analyses and glutamine determination
The Synechocystis strains gifB::Ctrl, gifB::M1 and gifB::M3 were grown in Erlenmeyer flasks under the conditions described above to an OD750 of 0.8. Samples for protein and amino acid extraction were taken prior to and in short intervals after adding NH4Cl in a final concentration of 10 mM. Proteins were extracted from cells harvested by centrifugation, separated by SDS-PAGE and transferred to nitrocellulose membranes as described previously (52). Membranes were blocked with TBS-T containing 5% milk powder and probed with sera containing anti-IF17 (1:2000) or anti-TrxA (1:5000). The antisera were obtained from M.I. Muro-Pastor and F.J. Florencio (University of Seville, Spain). As secondary antibody, anti-rabbit IgG-HRP conjugates (Thermo Fisher; catalog #65–6120) were used according to the manufacturer's protocol. Glutamine was extracted with 80% EtOH from frozen cell pellets and quantified as described previously (36,53).
In-line probing
A 104-nucleotide RNA corresponding to the Synechocystis gifB 5′ UTR as well as versions containing the point mutations M1 and M3 were generated by in vitro transcription from PCR products using T7 RNA polymerase. The respective transcription templates were PCR amplified from the pXG10-SF plasmids described above, which already harbored the gifB-UTR or its point mutations M1 and M3, using primer gifB-UTR_rev in combination with either gifB-UTR-T7_fw (WT, M3) or gifB-M1-T7_fw (M1). In vitro transcription was performed from ∼6 pmol of dsDNA in a 50 μl reaction containing 50 mM Tris-HCl (pH 7.5 at 23°C), 15 mM MgCl2, 2 mM spermidine, 5 mM DTT, 2.5 mM each of the four ribonucleoside 5′ triphosphates (NTPs) and 10 units μl−1 bacteriophage T7 RNA polymerase. The reaction was performed for 2 h at 37°C. The RNA was purified via denaturing polyacrylamide gel electrophoresis and 5′-radiolabeled with 32P as described previously (42). The radiolabeled RNA was subjected to in-line probing analyses as described previously (54). Accordingly, the RNA was incubated for 2 d at 23°C in a solution containing 50 mM Tris-HCl (pH 8.3 at 23°C), 20 mM MgCl2, 100 mM KCl and increasing concentrations of a ligand ranging from 3.2 μM to 10 mM in half-log concentration increments. As possible ligands for the 5′ UTR of gifB, we tested l-glutamine, l-glutamate and NH4Cl. After incubation the products of the in-line probing reactions were separated by denaturing gel electrophoresis (10% polyacrylamide, 8 M urea). The gels were dried and subsequently imaged using a Storm 820 PhosphorImager (GE Healthcare). The relative intensities of the various degradation products were quantified using ImageQuaNT software. Values for apparent KD were determined using a sigmoidal-dose response equation and GraphPad Prism 6.
Bioinformatics analyses
The phylogenetic tree was generated using the Neighbor-Joining method (55) and the 16S rDNA sequences of 60 representative cyanobacteria. The 16S rDNA sequences were retrieved from the whole genome RefSeq records. Alignments and evolutionary analyses were conducted in MEGA7 (56) using standard parameters. All positions containing gaps and missing data were eliminated.
In the same set of organisms, the occurrence and frequency of the RNA motifs RF01739 (glnA aptamer) and RF01704 (DP aptamer) was analyzed by performing an Infernal (v.1.1.2) (57) cmsearch for both motifs against the entire set of replicons for all species. The E-value threshold was set to 0.0001. The covariance models for the two RNA families were retrieved from the Rfam database (v.12.3, June 2017) (58). To investigate the co-occurrence of gifA and gifB genes a clustering of homologous proteins for the same 60 cyanobacteria was performed at MBGD (http://mbgd.genome.ad.jp/, release 2016/05/19) (59) with the parameters set to -o1 -HO -S -c60 -p0.5 -V0.6 -C80 -ao0.8 -ai0.95 -ne1 -EVAL = 0.00001 -SCORE = 60.
Predicted peptides downstream of retrieved RNA motifs were analyzed with PfamScan (v.1.6) against Pfam (v31.0, March 2017) (60). For this, 600 nucleotides downstream of the detected RNA motifs were extracted and translated to peptide sequences with transeq (EMBOSS v.6.5.7.0) (61). Because strand information is returned by cmsearch, the 600 nucleotides were translated only using the three reading frames corresponding to the strand where the RNA motifs are located.
The source code for the bioinformatics analyses of cyanobacterial genomes is available on github: https://github.com/PatrickRWright/gifA_gifB_RF01739_RF01704.
In a comprehensive follow-up search, all bacterial and archaeal sequences from RefSeq version 80 (62) were searched for representatives of RF01739 and RF01704 with Infernal (v.1.1.2) using the Rfam filtering parameter. Predicted downstream proteins were annotated by searching Conserved Domain Database version 3.16 (63). A collection of metagenomic sequences elsewhere described (64) was also searched for the aptamer motifs and annotated in a similar fashion.
RESULTS
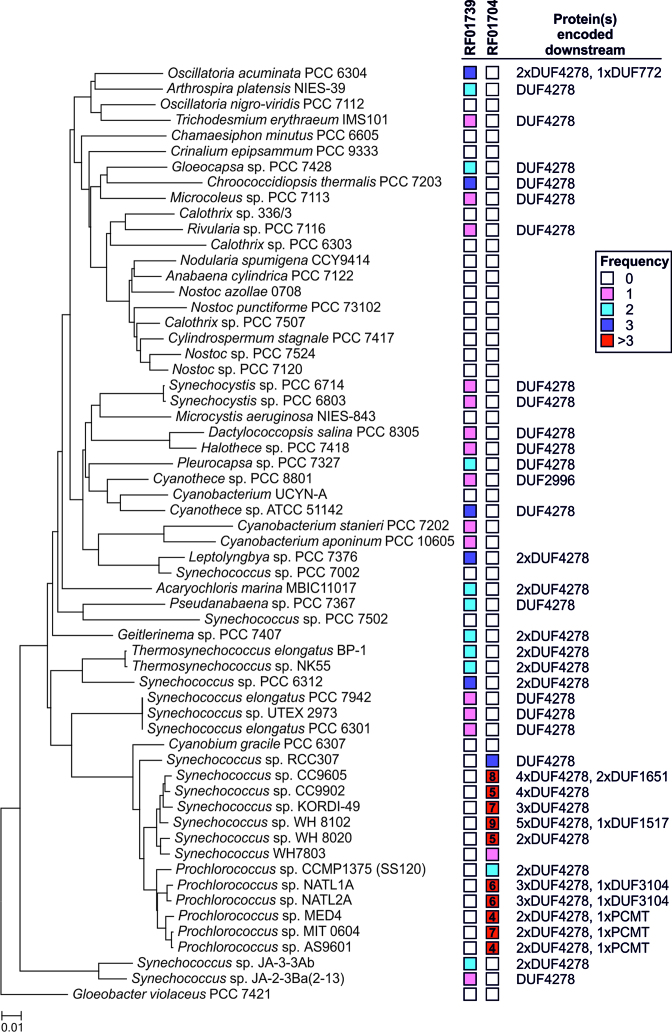
Genome sequences of 60 cyanobacterial strains representative for the major taxonomic and morphological clades were scanned for the presence of the glnA aptamer and the related DP aptamer. The glnA aptamer was detected in 28 of the 60 analyzed genomes (Figure 1, Supplementary Table S1) and hence is widely distributed throughout the cyanobacterial phylum. Whereas most of these genomes contain a single instance of the glnA aptamer, a few strains possess two or three distinct copies. Conspicuously, no homologs were found in the clade of filamentous, heterocyst-forming and N2-fixing cyanobacteria including Anabaena (Nostoc) sp. PCC 7120, Nostoc punctiforme PCC 73102 and Nodularia spumigena CCY9414. The DP aptamer, in contrast, is restricted to marine picocyanobacteria of the genera Synechococcus and Prochlorococcus, where it occurs frequently in higher numbers, e.g. nine copies in Synechococcus sp. WH8102 (Figure 1). Both aptamers are mutually exclusive, as we did not observe a single case of co-occurrence. Potentially, the DP aptamer is a derivative of the glnA aptamer, which has evolved in the monophyletic group of marine picocyanobacteria. Altogether, we found the RNA aptamers in 41/60 studied genomes. We conclude that the glnA and DP aptamers are present in the majority of cyanobacteria.
Figure 1.
Occurrence of the RF01739 (glnA aptamer) and the RF01704 (DP aptamer) motifs among cyanobacterial genomes. The phylogenetic tree was generated using the Neighbor-Joining method (55) and the 16S rDNA sequences of 60 representative cyanobacteria. Evolutionary analyses were conducted in MEGA7 (56). The RNA motif frequency within a particular genome is color coded. If the frequency was >3 the exact number of motifs was specified. Furthermore, the proteins encoded downstream of both RNA motifs were identified using the Pfam database. Note that DUF4278 is part of the GS inactivating factor IF17. Thus, the Pfam hits referring to DUF4278 most likely represent IF17 homologs or IF17-like proteins. If the number of Pfam hits was lower than the number of reported RNA motif hits, then not every downstream query returned a hit in the Pfam database. A detailed description is provided in the Material and Methods section. The complete dataset, i.e. the exact loci of the individual RNA motifs are given in Supplementary Table S1.
The RNA aptamers are frequently linked to a gene encoding a GS inhibitory protein
To elucidate the genes that are associated with the RNA aptamers, we compared the immediate downstream sequences against the Pfam database. In 25/28 genomes the glnA aptamer was located upstream of an ORF encoding a protein with a domain of unknown function (DUF4278) (Figure 1). In strains harboring more than one glnA aptamer, such as Acaryochloris marina MBIC11017 and Thermosynechococcus elongatus BP-1, it was frequently associated with a second DUF4278-encoding gene. With only a few exceptions, the DP aptamer was also associated with DUF4278-encoding genes in marine picocyanobacteria (Figure 1). However, in some cases, the aptamers were linked to other genes encoding proteins without matches in the Pfam database.
Weinberg et al. (41) reported a few instances in which the glnA aptamer was associated with N transporter or glutamine synthetase (glnA) genes. Indeed, we could confirm those associations in environmental DNA sequence databases, but found not a single case within the available cyanobacterial genome sequences (Supplementary Table S2). We conclude that the aptamers are mainly associated with genes encoding DUF4278-containing proteins and the link to N related transporters and the glnA gene is an exception rather than the rule.
In Synechocystis, DUF4278 characterizes the GS inactivating factor IF17 encoded by gifB. This protein has been verified as major regulatory factor of cyanobacterial N assimilation because it inhibits GS (20). Consistently, there is a good correlation between the occurrence of gifB homologs and the glnA aptamer. In 32 out of 60 analyzed genomes we found gifB homologs and in 28 of these 32 genomes the glnA aptamer was upstream. However, it should be noted that several cyanobacterial strains only harbor a gifA and no gifB homolog such as Anabaena (Nostoc) sp. PCC 7120 (65) and closely related strains (Supplementary Table S1). As mentioned above, none of the strains clustering with Anabaena sp. PCC 7120 possess the glnA aptamer, indicating that it is indeed functionally associated with gifB (Figure 1).
However, previous reports indicated that marine picocyanobacteria of the genus Prochlorococcus which exclusively harbor the DP aptamer lack full length homologs of GS inactivating factors (66). Consistently, we found no homologs to the Synechocystis IF17 in Prochlorococcus strains by using the BLASTP algorithm. Nevertheless, our analyses revealed that the ORFs downstream of the DP aptamer also frequently encode DUF4278-containing proteins. Although shorter, these proteins share partial sequence similarity with IF17 proteins from other cyanobacteria (Supplementary Figure S1). This points towards a similar function and hence they should be regarded as IF17-like proteins. Altogether, we found a tight association of the glnA and DP aptamers with genes encoding IF17 or IF17-like proteins. Hence, we hypothesized whether IF17 expression in cyanobacteria could be commonly regulated by glutamine riboswitches.
The 5′ UTR of the gifB gene from Synechocystis binds glutamine
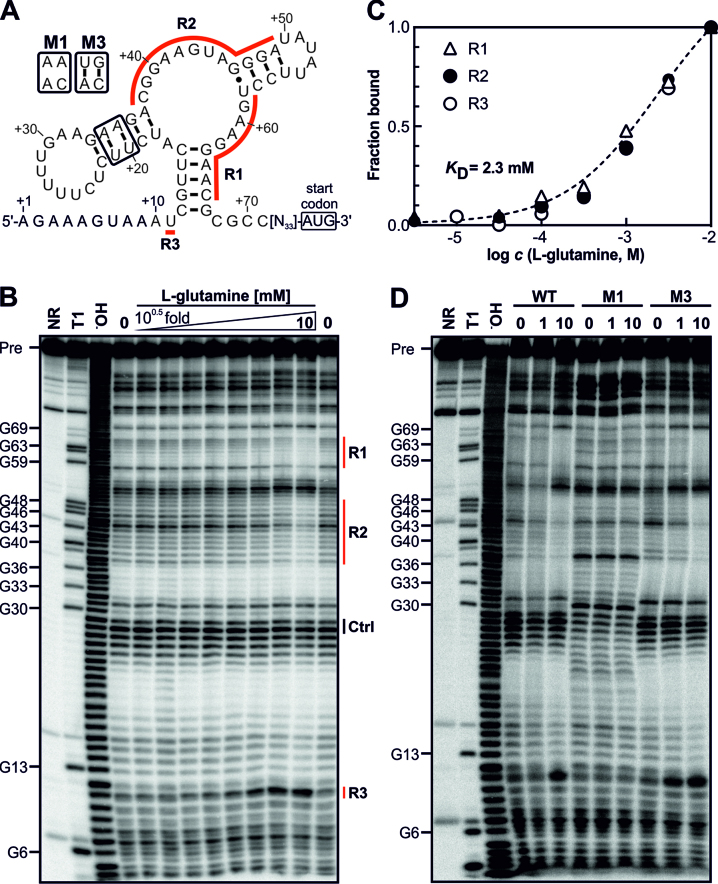
For the functional analysis of a glutamine-binding aptamer we focused on the model Synechocystis, in which the mechanism of GS inactivation via inhibitory proteins is well investigated (20). In this organism, the gifB (sll1515) gene is transcribed together with a 104 nt long 5′ UTR, which contains the predicted glnA aptamer (Figure 2A). To verify glutamine binding, in-line probing assays (54) were performed. Indeed, we observed that certain parts of the RNA construct undergo changes in the extent of spontaneous chemical degradation in presence of glutamine, which indicates that glutamine induces structural modulation (Figure 2B). The RNA exhibited an apparent dissociation constant (KD) for glutamine of ∼2.3 mM (Figure 2C). To ensure that glutamine specifically binds to the aptamer, we introduced mutations disrupting the predicted secondary structure (Figure 2A). Indeed, glutamine-dependent RNA structural changes were prevented by a mutation that disrupts conserved base-pairing (M1) and were restored by a compensatory mutation (M3, Figure 2D). The data clearly show that the 5′ UTR of the gifB gene harbors an RNA aptamer that binds and changes conformation in response to glutamine.
Figure 2.
The gifB 5′ UTR of Synechocystis harbors a glutamine-binding aptamer. (A) Predicted secondary structure of the glnA aptamer upstream of the gifB gene. The nucleotide positions refer to the transcriptional start site of the gifB gene (+1, ref. (21)). R1–3 label the regions for which structural modulation was observed (see panel B). The secondary structure is based on the secondary structure model of glnA motif RNAs published previously (42). (B) In-line probing analysis using a 5′ 32P-labeled gifB 5′ UTR. Precursor RNA (Pre) was incubated for 2 days at 23°C without a ligand (0) or in presence of increasing concentrations of L-glutamine (3.2 μM to 10 mM, half-log dilutions). The same RNA was also partially treated with Na2CO3, which mediates alkaline degradation (−OH) or with RNase T1, which cleaves after G residues (T1). A non-treated RNA served as control (NR, no reaction). (C) Densitometric evaluation of the bands corresponding to regions R1–3 whose intensities changed due to the presence of glutamine (marked by red lines in panel B). The intensities were normalized to bands which did not show glutamine-dependent intensity changes (Ctrl). (D) In-line probing using mutated RNAs. The NR, T1 and −OH samples are shown for the WT RNA.
The glutamine-binding aptamers are part of functional riboswitches that regulate gene expression
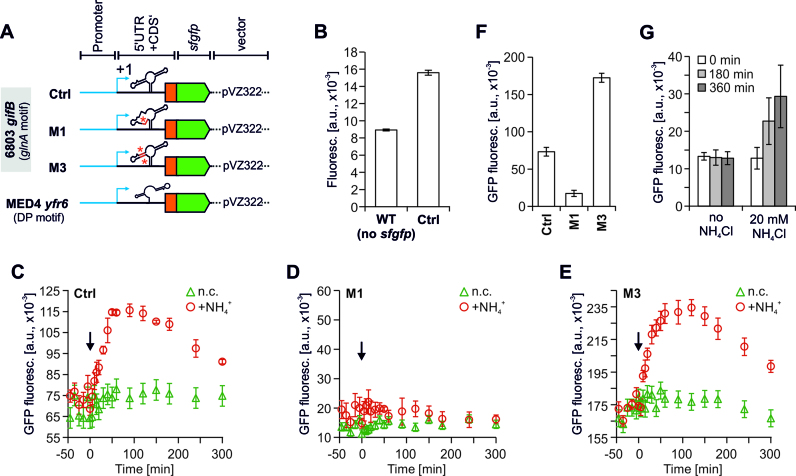
To analyze its regulatory impact, the glutamine-binding aptamer of Synechocystis was characterized in vivo. First, we performed reporter gene assays in Synechocystis using the superfolder gfp (sfgfp) gene, which was translationally fused to a truncated gifB gene and its entire 5′ UTR (Figure 3A). Transcription was driven by the synthetic BBa_J23101 promoter (iGEM Registry) which acts as a strong, constitutive promoter in Synechocystis (49). Compared to a WT strain, which did not harbor a sfgfp gene, substantial GFP fluorescence was detectable in a reporter strain carrying the J23101::gifB::sfgfp construct on a replicative plasmid (Figure 3B). These observations confirmed the activity of the synthetic promoter and sufficient translation initiation from the gifB 5′ UTR.
Figure 3.
GFP fluorescence measurements of Synechocystis strains carrying translational fusions of DUF4278-encoding genes from Synechocystis or Prochlorococcus sp. MED4 with the superfolder gfp gene. (A) Schematic illustration of the reporter constructs on a pVZ322 plasmid. The construct representing the glnA aptamer contained the entire 5′ UTR of gifB and 54 nt of its coding region in frame with the sfgfp gene. Transcription was driven by the BioBrick BBa_J23101 promoter (iGEM Registry, (48)) which was found to be constitutively active at a high level in Synechocystis (49). (B) Raw fluorescence of a WT (no sfgfp gene present) and a reporter strain carrying the Ctrl plasmid shown in panel A. Both strains had the same OD750. For each following measurement the signals measured from a simultaneously cultivated WT was subtracted from the measured fluorescence values (background correction). (C–E) Background corrected GFP fluorescence of strains carrying the control construct (Ctrl, C) or point mutated versions of the glnA aptamer (D, E). Please note that M3 is the complementary mutation to M1 (see Figure 2A). At time point 0, 10 mM NH4Cl (f.c.) were added to the cultures (indicated by arrow). In parallel, the same number of independent cultures remained untreated (negative control, n.c.). (F) Background corrected GFP fluorescence of reporter strains under standard (non-induced) growth conditions. (G) Background corrected GFP fluorescence of a Synechocystis strain carrying a DP aptamer within the 5′ UTR of the gene WP_071812974 of Prochlorococcus sp. MED4 in fusion with sfgfp (as shown in panel A). The measurement was performed under standard growth conditions and after adding 20 mM NH4Cl. Data are the mean ± SD of three biological replicates.
Sudden upshifts in NH4+ availability lead to rapid increases in intracellular glutamine levels in Synechocystis (22,36). Because a rapid increase of glutamine is accompanied by IF17 accumulation (67) in turn inactivating GS, it is reasonable to assume that the glutamine-binding aptamer upstream of gifB has a positive effect on IF17 production. To test this hypothesis we added 10 mM NH4Cl to cultures of the reporter strains. As expected, the GFP fluorescence rapidly increased after NH4+ upshift (Figure 3C). Because we used a constitutive, NtcA-independent promoter, the rise in GFP fluorescence most likely resulted from increased translation of the gifB::sfgfp mRNA mediated by the glutamine riboswitch.
To exclude the possibility that NH4+ is an additional ligand of the glnA aptamer and directly responsible for the observed changes in GFP fluorescence, we performed further in-line probing analyses using increasing concentrations of NH4Cl (Supplementary Figure S2). However, the RNA degradation patterns did not change, excluding the possibility that NH4+ is a ligand of the glnA aptamer. Hence, NH4+ cannot be directly responsible for the changes in GFP fluorescence. Moreover, because internal glutamate levels exhibit a transient negative response to NH4+ upshifts (22,36), we also tested L-glutamate as possible ligand. Again, no binding was evident from in-line probing assays (Supplementary Figure S2).
To additionally exclude the possibility of transcriptional effects that might cause the increased GFP fluorescence (e.g. due to changes in promoter activity), we introduced the same mutations used to evaluate glutamine binding by in-line probing (Figure 2). For example, the mutation M1 abolished the observed rise in GFP fluorescence after NH4+ addition (Figure 3D). Nevertheless, despite being unaffected by NH4+ upshift measurable GFP fluorescence was observed compared to the negative control (Figure 3F). Consistent with the RNA structure prediction and the in-line probing assays, the mutation M3, which restored base-pairing disrupted by M1 also restored the NH4+-induced rise in GFP fluorescence (Figure 3E). The data indicate that the structural change of the glnA aptamer strongly enhances gifB expression and adds an additional layer of control, which is glutamine dependent.
Strains harboring the DP aptamer are currently not amenable to genetic manipulation. Nevertheless, to examine its function we also fused a DP aptamer of Prochlorococcus MED4 to the sfgfp gene and the BBa_J23101 promoter. After introducing the construct into Synechocystis we performed measurements as described above. Initially, the here chosen sequence harbouring the DP aptamer was regarded as a non-coding RNA (Yfr6) (47). Later it was found that Yfr6 contains an ORF of 33 codons (68) as well as the DP aptamer in the putative 5′ UTR (41). Consistently, substantial GFP fluorescence was detected in a Synechocystis strain that transcribes this 5′ UTR in fusion with sfgfp (Figure 3G). This indicates that it indeed contains elements for translation initiation and that the corresponding IF17-like protein exists in Prochlorococcus (Figure S1). Although not as fast and sensitive as the glnA aptamer, the DP aptamer also mediates N-dependent expression regulation because an increased GFP fluorescence was observed upon NH4+ addition (Figure 3G). Altogether, the data verify that the glnA and DP aptamers are indeed part of functional riboswitches that control gene expression in a glutamine-dependent manner.
Glutamine riboswitches are important for IF17 production and GS inactivation
Our findings provide evidence that the gifB gene and IF17 production is regulated by a glutamine riboswitch. However, the expression of gifB is also controlled by the transcription factor NtcA (21). To directly address the impact of the riboswitch regulation on IF17 production in vivo, we introduced point mutations into the genomic locus upstream of gifB. For this, we inserted a kanamycin resistance cassette (KmR) in the intergenic space between gifB and the upstream gene slr1597 by homologous recombination. In addition, we generated constructs carrying the mutations M1 and M3 in the gifB 5′ UTR which were also used in the other experiments. By this strategy we obtained the strains gifB::Ctrl, gifB::M1 and gifB::M3 (Figure 4A, B).
Using an IF17-specific antibody, two clones of each strain were analyzed for gifB expression and IF17 accumulation after adding NH4Cl. Rapid IF17 accumulation was observed in the gifB::Ctrl strain (Figure 4C), matching the WT situation. However, no IF17 accumulation was observed in gifB::M1, which carries a mutation in the gifB 5′ UTR that disrupts the glutamine-binding aptamer. To verify that this did not result from disrupting a sequence that is required for expression in general, we also examined the complementary point mutation M3 in which mutation M1 is still present but the structure of the RNA aptamer was restored. Indeed, IF17 accumulation was similar between the gifB::M3 and the gifB::Ctrl strains (Figure 4C), verifying that the RNA structure itself is required for efficient IF17 production. Because we used the same mutations as in the in-line probing experiments, in which glutamine binding was abolished by M1 but restored by M3, it is very likely that the rapid IF17 accumulation in response to NH4+ upshifts is a result of glutamine-dependent RNA structural modulation.
IF17 accumulation was lacking in gifB::M1, even though the NtcA-dependent promoter was intact. Therefore, the glutamine riboswitch appears to exert the major control mechanism for IF17 production. Moreover, IF17 was reported as the functionally dominant of the two GS inactivating factors (20). Hence, the gifB-associated glutamine riboswitch might also be the most critical part for the regulation of GS activity in cyanobacteria. To test this hypothesis, we determined the glutamine levels in response to NH4+ upshifts in the strains gifB::Ctrl, gifB::M1 and gifB::M3. As expected, cells of the gifB::Ctrl strain showed rapid and high accumulation of glutamine because this amino acid is the primary product of NH4+ assimilation by GS (Figure 4D). Consistent with fast IF17 accumulation the glutamine levels started to decline immediately and reached the initial levels after about 60 min. Similar kinetics were observed in the gifB::M3 strain. In gifB::M1, however, the levels declined only transiently and increased again after 20 min pointing towards an insufficient GS regulation in that time span (Figure 4D). After 2 h also in the gifB::M1 strain similar glutamine levels were detected as observed prior to NH4+ addition. Albeit other regulatory mechanisms involved, GS activity and glutamine synthesis are also significantly determined by riboswitch-controlled IF17 synthesis.
DISCUSSION
Our findings demonstrate that the previously reported glnA and DP aptamers (41,42) function as critical elements of glutamine riboswitches in vivo. Given their secondary structure differences, we suggest renaming these elements as type 1 and type 2 glutamine riboswitches.
A type 1 glutamine riboswitch regulates the major GS inactivating factor in the majority of cyanobacteria
Here, we show that a type 1 glutamine riboswitch is part of the N-regulatory network in Synechocystis (Figure 5). Thereby, the riboswitch is a crucial regulatory element for the major GS inactivating factor IF17. In Synechocystis, GS activity is exclusively determined by the concentration of its inactivating factors because both IF7 and IF17 can bind and inactivate GS without further modification (20). Although both factors seem to work cumulatively in vivo, IF17 appears to be more important for GS inactivation due to two reasons. First, IF17 has a higher affinity to GS compared to IF7. Second, upon NH4+ upshift GS was hardly inactivated in a mutant strain lacking IF17 (ΔgifB) whereas GS was still significantly inactivated in a mutant strain lacking IF7 (ΔgifA) (20). A mutant strain carrying the dysfunctional riboswitch showed no IF17 accumulation, albeit the NtcA-regulated promoter mediating N-dependent gifB transcription was still intact. Consistently, the same mutant was strongly deficient in restoring low glutamine levels after sudden increases in NH4+ availability pointing towards a partial loss of GS regulation. Nevertheless, after reaching the maximum of glutamine accumulation the levels started to decline in the riboswitch mutant as well (Figure 4D). This could be explained by the accumulation of IF7, the second GS inactivating factor which was still intact in gifB::M1. However, GS inhibition by IF7 alone is obviously not sufficient to reach the initial glutamine levels in the same time span as in the WT which is also consistent with literature data (20).
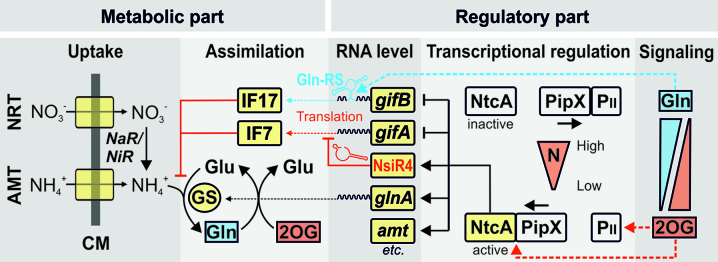
Figure 5.
Simplified overview of N control and GS regulation in the cyanobacterial model strain Synechocystis involving two different types of non-coding RNAs. AMT - ammonium transport system, NRT - nitrate transport system, CM – cell membrane, NaR - nitrate reductase, NiR - nitrite reductase, GS – glutamine synthetase, Gln - glutamine, Glu – glutamate, 2OG - 2-oxoglutarate, Gln-RS – glutamine riboswitch.
The major impact of the type 1 glutamine riboswitch was somewhat surprising because transcriptional control by NtcA was regarded as the major layer of control in the expression of both gifA and gifB, as well as the regulation of GS in cyanobacteria (21). However, our data clearly show that in addition to the NtcA-mediated transcriptional control, the regulation by a type 1 glutamine riboswitch constitutes another major control mechanism for IF17 production and GS activity, particularly after sudden changes in N availability. Similar type 1 glutamine riboswitches are frequently located upstream the ORFs of gifB homologs in other strains. Hence, this crucial regulatory mechanism appears to be common among cyanobacteria.
Nevertheless, its mode of action remains unclear to date. Structural analysis of a representative from S. elongatus PCC 7942 in a ligand-free as well as in the glutamine-bound state indicates that it might regulate translation. Ren et al. reported that high selectivity but low affinity binding of L-glutamine induces a conformational transition which includes a large change in the orientation of the P3 stem relative to P2 and P1 (43). This promotes long-range interactions, including a critical Watson-Crick base-pair interaction between G23-C60 (see Figure 4A of (43)). These conserved positions correspond to G40 and C68 in the Synechocystis riboswitch studied here (see Figure 2 and Supplementary Figure S3). Ren et al. also reported that the conformational transition and the long-range interaction melt a short helix attached to P1 which might modulate downstream events in gene expression (43). In Synechocystis, C68 and the downstream adjacent nucleotides are part of a short sequence that allows base-pairing with a putative ribosomal binding site (RBS) and might block the access of ribosomes (Supplementary Figure S3). Based on the structural data (43), we postulate that glutamine binding melts this inhibitory double strand and hence allows translation initiation (Supplementary Figure S3).
Type 2 glutamine riboswitches are associated with IF17-like proteins in marine picocyanobacteria
The type 2 glutamine riboswitch was found in the monophyletic and specialized group of marine picocyanobacteria and regulates genes encoding proteins that share the DUF4278 with IF17. Therefore, these proteins can be regarded as IF17-like, which might also function as GS inactivating factors. However, it should be noted that the IF17-like proteins in Prochlorococcus strains lack the C-terminal amino acid sequences shown to be crucial for the interaction of IF17 with GS in Synechocystis (66). Nevertheless, it is possible that the Prochlorococcus GS co-evolved structural differences allowing interaction with these truncated proteins. From an evolutionary perspective, there must be an important reason for the presence and the high copy numbers of these IF17-like proteins and their pronounced regulation by a type 2 glutamine riboswitch.
Prochlorococcus is highly abundant in ultraoligotrophic marine waters and hence plays a major role in the nutrient cycles of those habitats. It is thought that it's adaptation to these nutrient-poor conditions involved strong genome streamlining (69,70) and the replacement of protein regulators by non-coding RNAs (68). The presence and high frequency of glutamine riboswitches and of shortened versions of IF17 proteins is highly consistent with these ideas. Further evidence that these IF17-like proteins might have a function in N assimilation in marine picocyanobacteria is provided by the fact that they are most likely regulated by NtcA. A perfect NtcA-binding site overlapping the transcriptional start site was found upstream of the gene WP_071812974, which encodes an IF17-like protein in Prochlorococcus sp. MED4 (Supplementary Figure S4). NtcA binding sites usually overlap the -35 element in promoters that are activated by it (19). In contrast, NtcA binding at positions close to or overlapping with the –10 element represses transcription (21,65). Hence, it can be assumed that IF17-like proteins are also under negative NtcA control in Prochlorococcus, similar to gifB in Synechocystis and other cyanobacteria.
Altogether, we conclude that the regulation of the major GS inactivating factor by glutamine riboswitches as described in this study is crucial for the regulation of GS activity in the majority of cyanobacteria.
GS regulation in cyanobacteria involves different classes of non-coding RNAs
Controlling GS activity via inhibitory proteins is unique to cyanobacteria because other major bacterial groups use covalent modification to regulate this enzyme (9,20). Consistently, the glutamine riboswitches only occur in cyanobacteria and/or marine metagenome sequences. It should also be noted that, in metagenomic sequences of several marine samples, the type 1 glutamine riboswitch was also found upstream of glnA genes. Hence, in these organisms GS expression appears to be directly coupled to glutamine levels. Although negative regulation is reasonable, it is not known so far which effect glutamine-binding has on glnA expression in these organisms, nor what their taxonomic affiliation was.
The regulation of IF17 expression by a glutamine riboswitch adds another class of non-coding RNAs to the complex network of GS regulation in cyanobacteria. As reported previously, the regulatory sRNA NsiR4 interacts with the gifA mRNA encoding IF7, the second GS inactivating factor (36). This interaction interferes with gifA translation and thus fine-tunes IF7 accumulation kinetics. Nevertheless, transcriptional regulation remains the major level of control in this special case because the transcription of both gifA and NsiR4 is controlled by NtcA (Figure 5). Whereas NsiR4 expression is activated by NtcA under N limitation, gifA is repressed under N limitation and de-repressed under N excess (21,36). Hence, NtcA and NsiR4 jointly suppress IF7 expression when the synthesis of a GS-inactivating factor is becoming disadvantageous under N-limitation. Altogether, RNA-mediated regulation can be regarded as crucially important for GS regulation in cyanobacteria because the expression control of both IF7 and IF17 involves non-coding RNAs which are widely distributed among this bacterial group.
Glutamine is a signaling molecule in cyanobacteria
Several representatives of glutamine riboswitch aptamers have now been tested in vitro. Except for the aptamer of Synechococcus elongatus sp. PCC 7942, which also binds the non-natural D-glutamine with very poor affinity, all other representatives were verified to bind L-glutamine and none of the other tested molecules (42). The molecules tested include structural analogs of glutamine, other amino acids and also 2OG. Here, we confirmed glutamine binding by the aptamer present in the 5′ UTR of the Synechocystis gifB gene. In addition to the previously examined ligand candidates, we demonstrate that the corresponding aptamer also rejects L-glutamate as well as NH4Cl. Hence, we conclude that the regulatory effects observed in vivo after adding NH4+ result from glutamine induced structural modulation. Altogether, the data provide strong evidence that glutamine is an additional signaling molecule in cyanobacteria, which is specifically sensed by an RNA aptamer that processes the signal directly to regulate gene expression.
Our findings contribute to the general understanding of sensing the N status in bacteria. Albeit the specific regulatory mechanisms are diverse, it is widely accepted that bacteria perceive the cellular N status via the intracellular concentration of 2OG (7). Nevertheless, glutamine is also central for N-signaling because several bacterial groups possess a second mode of signal perception via the glutamine sensing and PII-modifying enzyme GlnD (for a review of glutamine signaling see ref. (6)). Moreover, in those bacteria that lack homologs of GlnD, independent glutamine signaling cascades evolved. For instance, in Bacillus subtilis the GS itself is the major glutamine sensor that transduces the signal to a transcriptional regulator (71). Accordingly, the Bacillus GS shows feedback inhibition by glutamine which is in contrast to the GS of proteobacteria that is inhibited by various metabolites but not by glutamine (72). In cyanobacteria, GS is neither inhibited by glutamine (73) nor regulated via a glutamine-dependent PII signaling pathway due to the lack of GlnD homologs (6). Accordingly, for a long time it was believed that glutamine does not contribute to the perception of the N status in cyanobacteria. However, our data confute that assumption as we show that cyanobacteria evolved an alternative mechanism of sensing and processing the glutamine signal by using a riboswitch. This sensing mechanism establishes a feedback loop by inducing the accumulation of IF17, which in turn inactivates GS and glutamine synthesis (Figure 5).
The glutamine riboswitch aptamer examined in this study exhibited poor ligand affinity compared to the aptamers of other riboswitch classes. This is consistent with KD values between 0.5 and 5 mM previously reported for both types of glutamine aptamers (42). Much lower KD values (i.e. higher affinities) have been reported, for instance, for glycine (KD ∼10 μM) or lysine (KD ∼1 μM) riboswitch aptamers (39,40). However, it should be noted that the intracellular glutamine level is rather high compared to other amino acids and highly dynamic in response to changes in N availability. In our experiments, the glutamine levels increased ∼40 fold when cells grown on nitrate as the sole N source were suddenly exposed to NH4+. Similar dynamics have been reported for other bacteria such as E. coli (74) or Salmonella typhimurium (75). Moreover, the absolute glutamine concentration in glucose-fed, exponentially growing E. coli cells was measured to be 3.8 mM (76). Assuming a similar concentration range in cyanobacteria and taking the dramatic increases up to 40-fold into account, the poor ligand affinity of the glutamine riboswitch seems to be perfectly tuned to the physiologically relevant concentration of its ligand, perhaps to avoid continuous glutamine binding and switching into the ON state conformation.
Interestingly, similar affinities have been reported for other glutamine-sensing systems. Biochemical analyses of the bifunctional enzyme GlnD from E. coli which reversibly uridylylates/deuridylylates the PII protein revealed that the reaction responded strongly to changes in the glutamine levels in the range between 0.1 and 4 mM (77), which is also within the physiological range of the reported cellular glutamine concentrations (76). Moreover, in green algae, mosses and higher plants, glutamine is sensed by a C-terminal extension of the PII protein, the Q loop (78). For several representatives, KD values for glutamine were estimated ranging from 2.4 to 9.2 mM (78) which are again within the range of glutamine concentrations reported in several plant species (79–81). This emphasizes how the independent and diverse glutamine-sensing elements have evolved to perfectly match the cellular levels of glutamine.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Klaudia Michl for technical assistance during amino acid measurements and M.I. Muro-Pastor and F.J. Florencio (University of Seville, Spain) for providing the antisera used in this study. Moreover, we thank Karl Forchhammer (University of Tübingen, Germany) for critical reading.
Author Contributions: S.K. designed the study. S.K., P.B. and R.M.A. performed experiments. P.R.W. and K.I.B. performed bioinformatics. M.H. contributed the amino acid analysis. S.K., P.R.W., R.M.A., K.I.B., R.R.B. and W.R.H. analyzed data. S.K. wrote the manuscript with contributions from all authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wissenschaftliche Gesellschaft Freiburg im Breisgau and the Grünewald-Zuberbier-Stiftung [to S.K.]; National Science Foundation Graduate Research Fellowship Program [DGE1122492 to R.M.A]; Howard Hughes Medical Institute; National Institutes of Health [GM022778 to R.R.B.]. Funding for open access charge: Helmholtz-Centre for Environmental Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. van Heeswijk W.C., Westerhoff H.V., Boogerd F.C.. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol. Mol. Biol. Rev. 2013; 77:628–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Magasanik B. Genetic control of nitrogen assimilation in bacteria. Annu. Rev. Genet. 1982; 16:135–168. [DOI] [PubMed] [Google Scholar]
- 3. Merrick M.J., Edwards R.A.. Nitrogen control in bacteria. Microbiol. Rev. 1995; 59:604–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003; 57:155–176. [DOI] [PubMed] [Google Scholar]
- 5. Zalkin H., Smith J.L.. Enzymes utilizing glutamine as an amide donor. Adv. Enzymol. Relat. Areas Mol. Biol. 1998; 72:87–144. [DOI] [PubMed] [Google Scholar]
- 6. Forchhammer K. Glutamine signalling in bacteria. Front. Biosci. 2007; 12:358–370. [DOI] [PubMed] [Google Scholar]
- 7. Leigh J.A., Dodsworth J.A.. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 2007; 61:349–377. [DOI] [PubMed] [Google Scholar]
- 8. Yan D., Ikeda T.P., Shauger A.E., Kustu S.. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:6527–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stadtman E.R. The story of glutamine synthetase regulation. J. Biol. Chem. 2001; 276:44357–44364. [DOI] [PubMed] [Google Scholar]
- 10. Arcondéguy T., Jack R., Merrick M.. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 2001; 65:80–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ninfa A.J., Atkinson M.R.. PII signal transduction proteins. Trends Microbiol. 2000; 8:172–179. [DOI] [PubMed] [Google Scholar]
- 12. Forchhammer K. P(II) signal transducers: novel functional and structural insights. Trends Microbiol. 2008; 16:65–72. [DOI] [PubMed] [Google Scholar]
- 13. Capone D.G., Zehr J.P., Paerl H.W., Bergman B., Carpenter E.J.. Trichodesmium, a globally significant marine cyanobacterium. Science. 1997; 276:1221–1229. [Google Scholar]
- 14. Montoya J.P., Holl C.M., Zehr J.P., Hansen A., Villareal T.A., Capone D.G.. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature. 2004; 430:1027–1032. [DOI] [PubMed] [Google Scholar]
- 15. Zehr J.P., Waterbury J.B., Turner P.J., Montoya J.P., Omoregie E., Steward G.F., Hansen A., Karl D.M.. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature. 2001; 412:635–638. [DOI] [PubMed] [Google Scholar]
- 16. Ducat D.C., Way J.C., Silver P.A.. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011; 29:95–103. [DOI] [PubMed] [Google Scholar]
- 17. Hagemann M., Hess W.R.. Systems and synthetic biology for the biotechnological application of cyanobacteria. Curr. Opin. Biotechnol. 2017; 49:94–99. [DOI] [PubMed] [Google Scholar]
- 18. Savakis P., Hellingwerf K.J.. Engineering cyanobacteria for direct biofuel production from CO2. Curr. Opin. Biotechnol. 2015; 33:8–14. [DOI] [PubMed] [Google Scholar]
- 19. Herrero A., Muro-Pastor A.M., Flores E.. Nitrogen control in cyanobacteria. J. Bacteriol. 2001; 183:411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. García-Domínguez M., Reyes J.C., Florencio F.J.. Glutamine synthetase inactivation by protein–protein interaction. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:7161–7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. García-Domínguez M., Reyes J.C., Florencio F.J.. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 2000; 35:1192–1201. [DOI] [PubMed] [Google Scholar]
- 22. Muro-Pastor M.I., Reyes J.C., Florencio F.J.. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 2001; 276:38320–38328. [DOI] [PubMed] [Google Scholar]
- 23. Luque I., Flores E., Herrero A.. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994; 13:2862–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanigawa R., Shirokane M., Maeda Si S., Omata T., Tanaka K., Takahashi H.. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate. in vitro. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:4251–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fokina O., Chellamuthu V.-R., Forchhammer K., Zeth K.. Mechanism of 2-oxoglutarate signaling by the Synechococcus elongatus PII signal transduction protein. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:19760–19765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Espinosa J., Forchhammer K., Burillo S., Contreras A.. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 2006; 61:457–469. [DOI] [PubMed] [Google Scholar]
- 27. Espinosa J., Rodríguez-Mateos F., Salinas P., Lanza V.F., Dixon R., de la Cruz F., Contreras A.. PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2423–E2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forcada-Nadal A., Forchhammer K., Rubio V.. SPR analysis of promoter binding of Synechocystis PCC6803 transcription factors NtcA and CRP suggests cross-talk and sheds light on regulation by effector molecules. FEBS Lett. 2014; 588:2270–2276. [DOI] [PubMed] [Google Scholar]
- 29. Giner-Lamia J., Robles-Rengel R., Hernández-Prieto M.A., Muro-Pastor M.I., Florencio F.J., Futschik M.E.. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017; 45:11800–11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mitschke J., Vioque A., Haas F., Hess W.R., Muro-Pastor A.M.. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20130–20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vázquez-Bermúdez M.F., Herrero A., Flores E.. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 2002; 512:71–74. [DOI] [PubMed] [Google Scholar]
- 32. Storz G., Vogel J., Wassarman K.M.. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011; 43:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wagner E.G.H., Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 34. Ionescu D., Voss B., Oren A., Hess W.R., Muro-Pastor A.M.. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J. Mol. Biol. 2010; 398:177–188. [DOI] [PubMed] [Google Scholar]
- 35. Kopf M., Klähn S., Scholz I., Matthiessen J.K.F., Hess W.R., Voss B.. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014; 21:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klähn S., Schaal C., Georg J., Baumgartner D., Knippen G., Hagemann M., Muro-Pastor A.M., Hess W.R.. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6243–E6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mandal M., Breaker R.R.. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004; 5:451–463. [DOI] [PubMed] [Google Scholar]
- 38. Garst A.D., Edwards A.L., Batey R.T.. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol. 2011; 3:a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.M., Ruzzo W.L., Breaker R.R.. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004; 306:275–279. [DOI] [PubMed] [Google Scholar]
- 40. Sudarsan N., Wickiser J.K., Nakamura S., Ebert M.S., Breaker R.R.. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003; 17:2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weinberg Z., Wang J.X., Bogue J., Yang J., Corbino K., Moy R.H., Breaker R.R.. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010; 11:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ames T.D., Breaker R.R.. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011; 8:82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ren A., Xue Y., Peselis A., Serganov A., Al-Hashimi H.M., Patel D.J.. Structural and dynamic basis for low-affinity, high-selectivity binding of L-glutamine by the glutamine riboswitch. Cell Rep. 2015; 13:1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y.. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979; 111:1–61. [Google Scholar]
- 45. Corcoran C.P., Podkaminski D., Papenfort K., Urban J.H., Hinton J.C.D., Vogel J.. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 2012; 84:428–445. [DOI] [PubMed] [Google Scholar]
- 46. Voigt K., Sharma C.M., Mitschke J., Lambrecht S.J., Voß B., Hess W.R., Steglich C.. Comparative transcriptomics in two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 2014; 8:2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Axmann I.M., Kensche P., Vogel J., Kohl S., Herzel H., Hess W.R.. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005; 6:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. parts.igem.org 2017; The iGEM registry of standard biological parts.
- 49. Camsund D., Heidorn T., Lindblad P.. Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. J. Biol. Eng. 2014; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Camsund D., Lindblad P.. Engineered transcriptional systems for cyanobacterial biotechnology. Front. Bioeng. Biotechnol. 2014; 2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beyer H.M., Gonschorek P., Samodelov S.L., Meier M., Weber W., Zurbriggen M.D.. AQUA cloning: a versatile and simple enzyme-free cloning approach. PLoS ONE. 2015; 10:e0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baumgartner D., Kopf M., Klähn S., Steglich C., Hess W.R.. Small proteins in cyanobacteria provide a paradigm for the functional analysis of the bacterial micro-proteome. BMC Microbiol. 2016; 16:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hagemann M., Vinnemeier J., Oberpichler I., Boldt R., Bauwe H.. The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Biol. (Stuttg). 2005; 7:15–22. [DOI] [PubMed] [Google Scholar]
- 54. Regulski E.E., Breaker R.R.. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008; 419:53–67. [DOI] [PubMed] [Google Scholar]
- 55. Saitou N., Nei M.. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 56. Kumar S., Stecher G., Tamura K.. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016; 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nawrocki E.P., Burge S.W., Bateman A., Daub J., Eberhardt R.Y., Eddy S.R., Floden E.W., Gardner P.P., Jones T.A., Tate J. et al. . Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015; 43:D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Uchiyama I., Mihara M., Nishide H., Chiba H.. MBGD update 2015: microbial genome database for flexible ortholog analysis utilizing a diverse set of genomic data. Nucleic Acids Res. 2015; 43:D270–D276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rice P., Longden I., Bleasby A.. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000; 16:276–277. [DOI] [PubMed] [Google Scholar]
- 62. Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J.. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016; 44:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R. et al. . CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017; 45:D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weinberg Z., Lünse C.E., Corbino K.A., Ames T.D., Nelson J.W., Roth A., Perkins K.R., Sherlock M.E., Breaker R.R.. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res. 2017; 45:10811–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Galmozzi C.V., Saelices L., Florencio F.J., Muro-Pastor M.I.. Posttranscriptional regulation of glutamine synthetase in the filamentous cyanobacterium Anabaena sp. PCC 7120: Differential expression between vegetative cells and heterocysts. J. Bacteriol. 2010; 192:4701–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Saelices L., Galmozzi C.V., Florencio F.J., Muro-Pastor M.I.. Mutational analysis of the inactivating factors, IF7 and IF17 from Synechocystis sp. PCC 6803: critical role of arginine amino acid residues for glutamine synthetase inactivation. Mol. Microbiol. 2011; 82:964–975. [DOI] [PubMed] [Google Scholar]
- 67. Galmozzi C.V., Fernández-Avila M.J., Reyes J.C., Florencio F.J., Muro-Pastor M.I.. The ammonium-inactivated cyanobacterial glutamine synthetase I is reactivated in vivo by a mechanism involving proteolytic removal of its inactivating factors. Mol. Microbiol. 2007; 65:166–179. [DOI] [PubMed] [Google Scholar]
- 68. Steglich C., Futschik M.E., Lindell D., Voss B., Chisholm S.W., Hess W.R.. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008; 4:e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dufresne A., Garczarek L., Partensky F.. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005; 6:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Giovannoni S.J., Cameron Thrash J., Temperton B.. Implications of streamlining theory for microbial ecology. ISME J. 2014; 8:1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wray L.V., Zalieckas J.M., Fisher S.H.. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell. 2001; 107:427–435. [DOI] [PubMed] [Google Scholar]
- 72. Woolfolk C.A., Stadtman E.R.. Regulation of glutamine synthetase. 3. Cumulative feedback inhibition of glutamine synthetase from. Escherichia coli. Arch. Biochem. Biophys. 1967; 118:736–755. [DOI] [PubMed] [Google Scholar]
- 73. Orr J., Haselkorn R.. Kinetic and inhibition studies of glutamine synthetase from the cyanobacterium Anabaena 7120. J. Biol. Chem. 1981; 256:13099–13104. [PubMed] [Google Scholar]
- 74. Yuan J., Doucette C.D., Fowler W.U., Feng X.-J., Piazza M., Rabitz H.A., Wingreen N.S., Rabinowitz J.D.. Metabolomics-driven quantitative analysis of ammonia assimilation in E. coli. Mol. Syst. Biol. 2009; 5:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ikeda T.P., Shauger A.E., Kustu S.. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 1996; 259:589–607. [DOI] [PubMed] [Google Scholar]
- 76. Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D.. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009; 5:593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jiang P., Peliska J.A., Ninfa A.J.. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry. 1998; 37:12782–12794. [DOI] [PubMed] [Google Scholar]
- 78. Chellamuthu V.-R., Ermilova E., Lapina T., Lüddecke J., Minaeva E., Herrmann C., Hartmann M.D., Forchhammer K.. A widespread glutamine-sensing mechanism in the plant kingdom. Cell. 2014; 159:1188–1199. [DOI] [PubMed] [Google Scholar]
- 79. Winter H., Robinson D.G., Heldt H.W.. Subcellular volumes and metabolite concentrations in barley leaves. Planta. 1993; 191:180–190. [Google Scholar]
- 80. Winter H., Robinson D.G., Heldt H.W.. Subcellular volumes and metabolite concentrations in spinach leaves. Planta. 1994; 193:530–535. [Google Scholar]
- 81. Fritz C., Mueller C., Matt P., Feil R., Stitt M.. Impact of the C-N status on the amino acid profile in tobacco source leaves. Plant Cell Environ. 2006; 29:2055–2076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.