Abstract
Transcription initiation by archaeal RNA polymerase (RNAP) and eukaryotic RNAP II requires the general transcription factor (TF) B/ IIB. Structural analyses of eukaryotic transcription initiation complexes locate the B-reader domain of TFIIB in close proximity to the active site of RNAP II. Here, we present the first crosslinking mapping data that describe the dynamic transitions of an archaeal TFB to provide evidence for structural rearrangements within the transcription complex during transition from initiation to early elongation phase of transcription. Using a highly specific UV-inducible crosslinking system based on the unnatural amino acid para-benzoyl-phenylalanine allowed us to analyze contacts of the Pyrococcus furiosus TFB B-reader domain with site-specific radiolabeled DNA templates in preinitiation and initially transcribing complexes. Crosslink reactions at different initiation steps demonstrate interactions of TFB with DNA at registers +6 to +14, and reduced contacts at +15, with structural transitions of the B-reader domain detected at register +10. Our data suggest that the B-reader domain of TFB interacts with nascent RNA at register +6 and +8 and it is displaced from the transcribed-strand during the transition from +9 to +10, followed by the collapse of the transcription bubble and release of TFB from register +15 onwards.
INTRODUCTION
Transcription is a cyclic process in which information derived from DNA is transcribed into RNA molecules by RNA polymerases (RNAPs). Cellular RNAPs need basal transcription factors to carry out promoter-dependent RNA synthesis. The minimal preinitiation complex (PIC) of the eukaryotic RNAP II system consists of six general transcription factors (TFIID, TFIIA, TFIIB, TFIIE, TFIIF and TFIIH) representing a complex and highly regulated transcription machinery (1–4). The archaeal transcription system constitutes a simplified version of that of eukaryotes (5–8). Its preinitiation complex consists of RNAP, the TATA-box binding protein (TBP), transcription factor B (TFB) and E (TFE), which are structurally and functionally related to eukaryotic RNAP II, TBP, TFIIB and TFIIEα (9–16).
Transcription initiation starts with the recognition of the TATA-element by TBP ∼25 bp upstream of the transcription start site in the promoter region (17,18). TFB/TFIIB associates with TBP and DNA to form the TBP/TF(II)B/DNA-complex (19,20), where DNA is found in a bent conformation (21). TFB/TFIIB consists of four domains, the Zn-ribbon, B-reader, B-linker and a C-terminal core domain (Figure 1A) (22). The B-core domain at the C-terminus of TFB/TFIIB interacts with TBP (20,23,24) as well as with the BRE sequence in the promoter (25,26), to stabilize the complex and to determine the direction of transcription (27). The N-terminal Zn-ribbon domain is essential for RNAP (II) recruitment to form a PIC, in which the DNA template is still base paired and thus this complex is also referred to as closed complex (CC) (28,29). After PIC assembly the complex undergoes a structural transition to yield the open complex (OC) characterized by a region of melted DNA around the transcription start site (TSS) (30,31). A recent study showed that promoter-bound TBP and TFB are located closer to the surface of RNAP in the archaeal OC compared to the eukaryotic system. Due to the increase in torsional strain in this structural arrangement of TBP and TFB relative to the RNAP and DNA, DNA melting is most likely supported in the archael transcription system and occur in an ATP independent manner, without the need for a dedicated translocase/helicase containing factor like TFIIH, which is required for DNA melting in the RNAP II system (32–34). OC formation is further supported by the rudder of RNAP (II) and the TFB/TFIIB B-linker domain together with the RNAP (II) clamp-coiled coil domain (22,35). In addition, TFE/TFIIE can stimulate OC formation (29,36–39) and support complex stabilization by direct interaction with the non-transcribed DNA strand (NT-strand) and the RNAP (II) clamp domain, which represents a mobile element of RNAP (II). It exists in two alternative states (closed or open) and through binding of TFE the equilibrium is shifted towards the open state (39). However, this factor is not essential for transcription initiation in archaeal in vitro transcription systems (40). The resulting single-stranded part of the DNA represents the initial transcription bubble, which was estimated to be 14 nucleotides in size in the archaeal initiation complex (41) and ranges from 6 to 18 nucleotides in eukaryotic systems (36,42,43). Reannealing of the two DNA strands is prevented by interactions of the fork loop 1 and the rudder element of RNAP (II), and the TFB/TFIIB B-linker with the NT-strand (22,32,35).
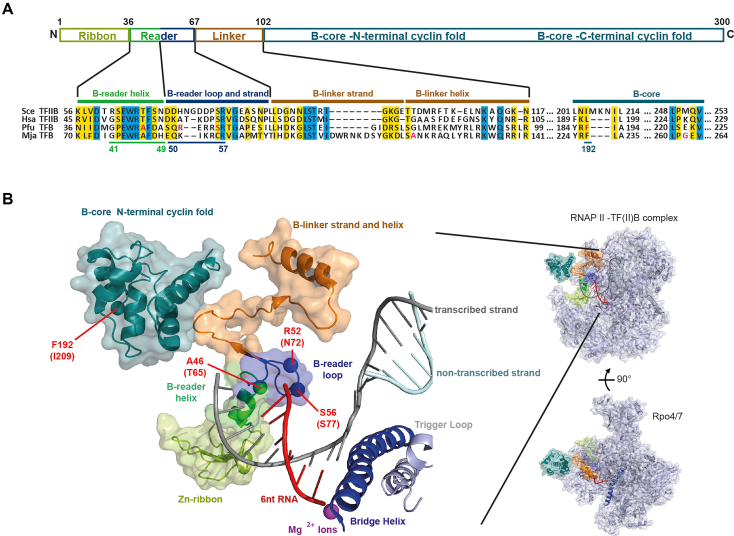
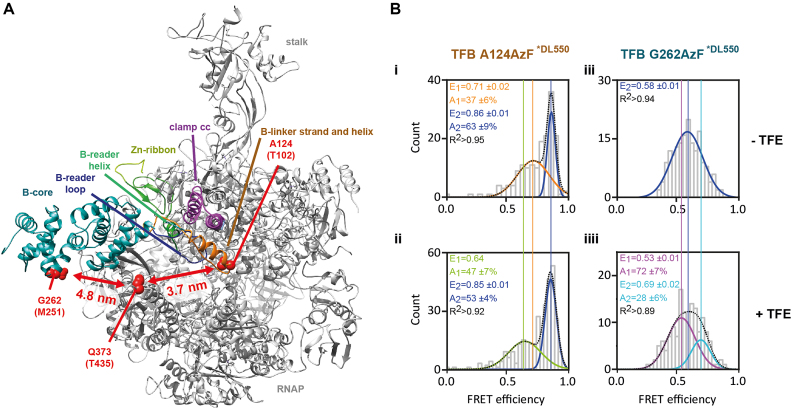
Figure 1.
Structure of TFB/TFIIB in the initially transcribing complex and schematic organization of TFB/TFIIB. (A) Schematic representation of the domain organization of TFB. The position of TFB domains like B-reader, B-linker and B-core domain is shown. Color code is as described in panel (B). Multiple sequence alignment of yeast (Saccharomyces cerevisiae; Sce) and human (Homo sapiens; Hsa) TFIIB and euryarchaeal (Pyrococcus furiosus; Pfu) TFB domains. Residues highlighted in yellow and blue indicate conserved and invariant residues respectively. The alignment was performed with Muscle (79). Bpa substituted amino acids of PfuTFB in the B-reader helix, B-reader loop and B-core domain are underlined. The corresponding PfuTFB variants used for crosslinking experiments are colured in red and the two labeled amino acids in the MjaTFB sequence used for single-molecule FRET experiments are colored in magenta. (B) Crystal structure of the initially transcribing complex including TFIIB/RNAP II and a 6nt RNA (PDB: 4BBS) (42). TFIIB domain organization is depicted in different colors: N-terminal cyclin fold of the B-core domain (cyan), B-linker (orange) and Zn-ribbon (lime green). The B-reader consists of the helix (green) and loop (blue) and is located in proximity to the template strand (gray). Bpa substituted amino acids are highlighted, the position for PfuTFB is given in bold letters, the corresponding SceTFIIB positions are in brackets, respectively. The active site of RNAP II is indicated by Mg2+-ions (magenta), RNA (red), and the bridge helix (dark blue).
Cryo-electron microscopy analysis of TFIIB in context of the eukaryotic initiation complexes showed that the linker and the reader domain of TFIIB are disordered in the CC (3,4). Further insights into structural arrangements of TFIIB as well as crosslinking studies between RNA and TFIIB revealed that the B-reader domain is located close to the active center of the polymerase in proximity to the transcribed DNA strand (T-strand) after OC formation (42,44–46). It is suggested that the B-reader loop domain stabilizes the T-strand whereas the TSS selection is supported by the B-reader helix domain (4,22). RNAP (II) starts to synthesize and release short RNA fragments during a process termed abortive initiation (47,48) until a stable DNA-RNA hybrid is formed, which further stabilizes the complex (49,50). During this event RNAP (II) pulls downstream DNA into itself, whereas the active site remains stationary, resulting in DNA scrunching (51). It is suggested that the nascent RNA is separated from DNA by charge-dependent interactions with the B-reader loop (45). The RNA further clashes with the B-reader helix because this domain is predicted from structures to be in the path of the advancing 5′end of the RNA (22,44,45). It has been hypothesized that the displacement of the B-reader by the growing RNA chain may trigger transcription bubble collapse, which takes place at position +10/+11 (41,52) and marks the end of promoter clearance in eukaryotic transcription systems (53). During bubble collapse the upstream edge of the melted DNA region closes suddenly and from that stage on the bubble has a defined size and moves along with RNAP (41,53). TFIIB and RNAP II interactions are attenuated and in the very last step, during the transition from initiation to elongation phase of transcription, the RNA is predicted to clash with the TFIIB Zn-ribbon domain at position +12/+13 (45), causing TFIIB release (54,55). Finally, RNAP (II) enters the elongation phase of the transcription cycle and moves together with the transcription bubble along the DNA until transcription is terminated (41,56).
Several mechanistic aspects of the role of TFB/TFIIB during transcription initiation were deduced from eukaryotic crystal structures and cryo-EM structures. However, so far crystal and cryo-EM structures are not available for all intermediate states that occur during the transition from the initially transcribing complex (ITC, containing a 6-base-pair (bp) DNA–RNA hybrid) to promoter release, and consequently, only limited information on the dynamic behavior of TFB domains during these steps is available. Thus, in this study we investigated interactions of the TFB B-reader and C-terminal core domains with DNA in the archaeal transcription system of Pyrococcus furiosus. We identified TFB–DNA contacts via UV-inducible crosslinking experiments based on the unnatural amino acid para-benzoyl-phenylalanine (Bpa) and analyzed the positioning and topological transitions of TFB domains during transcription initiation in the PIC and stalled transcription complexes. The crosslinking mapping data also provide evidence for the interaction of TFB with RNA, and demonstrate the displacement of the TFB B-reader domain from the T-strand, which coincides with bubble collapse.
MATERIALS AND METHODS
Purification of recombinant proteins
RNAP from P. furiosus cells and recombinant TBP, TFB and TFE were purified as described previously (22,40,57,58).
Mutagenesis, expression and purification of TFB
For TFB mutagenesis, the DNA sequence of tfb (PF1377) with an additional N-terminal histidine tag was cloned into the expression vector pET14b (22). Exchange of amino acid codons at the point of interest with a TAG stop codon was achieved by Phusion® High-Fidelity DNA Polymerase mutagenesis PCR using phosphorylated primers (New England Biolabs, Ipswich, USA). The linearized fragments were ligated using T4 ligase enzyme and transformed into Escherichia coli DH5α cells (New England Biolabs, Ipswich, USA). The correct nucleotide substitutions of the obtained plasmids were verified by sequencing. The plasmids were transformed into E. coli BL21 (DE3) Star (Thermo Fisher Scientific, Waltham, USA) cells harboring the pEVOL plasmid (59). This strain contains a plasmid which encodes a modified aminoacyl-tRNA synthetase (aaRS)/suppressor tRNA system under an arabinose-induced promoter, which allows incorporation of the unnatural amino acid into target protein by recognition of the TAG stop codon by the modified tRNA. Protein expression was induced with 0.5 mM IPTG and tRNA expression with 1.3 mM l-arabinose in presence of 0.2 mM p-benzoyl-l-phenylalanine (Bpa; purchased from Bachem, Bubendorf, Switzerland) (60) at an OD600 = 0.6–0.8 for simultaneous protein expression and incorporation of Bpa. The proteins were purified via Dynabeads® His-Tag Isolation (Thermo Fisher Scientific, Waltham, USA). The pellet of a 100 ml culture volume was dissolved in 3 ml binding buffer (50 mM phosphate buffer pH 8.0, 300 mM NaCl, 0.01% v/v, Tween 20) and a small pile of lysozyme was added and mixed. The suspension was incubated on ice for 45 min, and then the cells were treated with ultra sound for 6 × 1 min (50% intensity, 50% pulse length; Branson Sonifier 250) on ice and subsequently centrifuged at 4°C and 21 000g for 60 min (Hitachi himac CT15RE, VWR). The supernatant was transferred into a new tube and the samples were incubated at 70°C for 10 min to denature E. coli proteins. The centrifugation step at 4°C and 21 000g for 60 min was repeated. The supernatant was added to 200 μl beads and incubated for 30 min at 4°C under constant rotation. The magnetic particles were subsequently immobilized by a magnet particle separator and supernatant was removed. The beads were washed four times with 400 μl binding buffer by gentle pipetting for 45 s. Finally, the protein was eluted by adding 100 μl elution buffer (50 mM phosphate buffer pH 8.0, 300 mM imidazole, 300 mM NaCl, 0.01% Tween 20) to the beads and incubated at 4°C for 30 min under constant rotation. The beads were immobilized and the eluted protein in the supernatant was stored at -80°C in small portions. The concentrations of purified TFB-Bpa variants were determined by loading distinct volumes of the protein and bovine serum albumin (BSA, purchased from Roche, Mannheim, Germany) as reference protein on a denaturing 12% SDS-gel using Tris-Glycine as buffer system. The resulting bands were quantified with ImageJ software (61) and the intensities of the bands corresponding to TFB-Bpa were compared to signal intensities of BSA with known concentrations. Before usage, protein concentrations were adjusted according to a second SDS-PAGE analysis using the TFB WT protein as reference point.
Preparation of the gdh-C DNA-templates
The gdh-C-templates gdh-C6, gdh-C8, gdh-C9, gdh-C10 and gdh-C20 (5′-3′ sequence of the gdh-C20 non-transcribed strand: GCCAGGGTTTTCCCAGTCACGACGTTGT AAAACGACGGCCAGTGAA TTCGAGCTCGGTACCCGGGGATCCGAATTTTAGATTCTTTGAGCCTAATCAAATAAACAAAAGGATTTCCACTCTTGT TTACCGAAAGCTTTATATAGGCTATTGCCCAAAAATGTATCGTTAATGAGGTAATTTGGAGCAT ATGGGGGATCCTCTAGAGTCGACCTGCAGGCA TGCAAGCTTGGCGTAATCATGGTCATAGCTGT TTCCTGTGTGAAATTGTTATCCGCTC; sequences of other cassettes differ in the position of the first cytosine after the TSS at the transcribed strand (Figure 3A) used in this study consist of the strong glutamate dehydrogenase (gdh) promoter of P. furiosus and were described previously (41). Cassettes gdh-C11 to -C15 derived from the standard transcription template gdh-C20 (on pUC19 vector). The gdh-C20 template was mutated by introduction of a guanidine at the respective site on the transcribed strand via PCR mutagenesis using phosphorylated primers. After ligation with T4 ligase, transformation in E. coli DH5α, plasmid preparation and sequence validation, the templates for in vitro reactions were amplified via PCR using M13 forward and reverse primer and purified with Promega PCR-purification kit (Promega, Madison, USA).
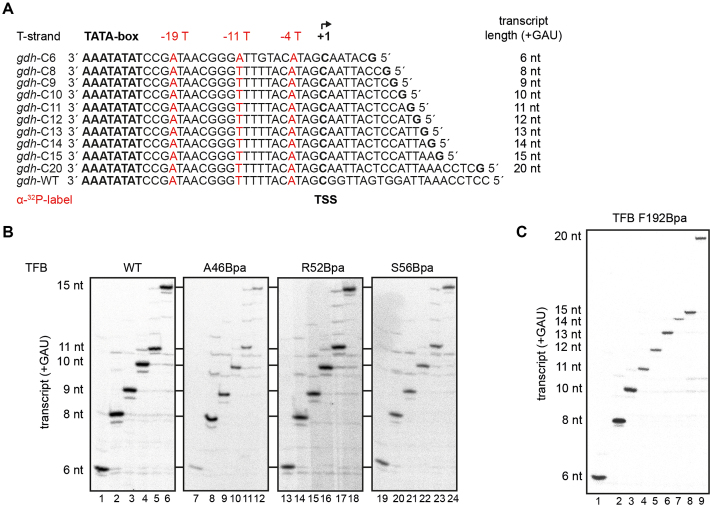
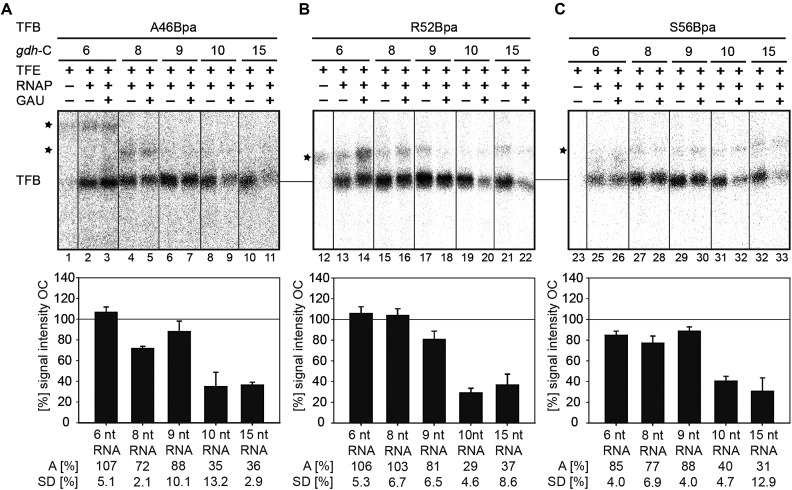
Figure 3.
Correct stalling of transcription complexes using TFB Bpa-variants. (A) Overview of the transcribed strands of the used gdh-C templates used in this study. The radiolabeled positions are highlighted in red. Additionally, the expected transcript length for each gdh-C cassette is given in case CTP is omitted from the transcription assay. (B) Transcription assays without CTP were performed at 80°C using different gdh-C cassettes and TFB WT (lanes 1–6), A46Bpa (lanes 7–12), R52Bpa (lanes 13 to 18) and S56Bpa (lanes 19 to 24). The gdh-C cassette used corresponds to the transcript length on the left. Radiolabeled RNA transcripts of different lengths between 6 and 15 nucleotides were analyzed on a 7 M urea/ 28% polyacrylamide gel. All TFB Bpa-variants showed the expected pattern in comparison to the TFB WT, indicating a specific stall position for the transcription complexes. (C) Transcription assays without CTP were performed at 80°C using different gdh-C and TFB F192Bpa. The gdh-C cassette used corresponds to the transcript length on the left. Radiolabeled RNA transcripts of the different lengths between 6 and 20 nucleotides were analyzed on a 7 M urea/ 28% polyacrylamide gel.
Electrophoretic mobility shift assays (EMSA)
Assays were carried out using 26 nM RNAP, 56 nM 5′-6-carboxyfluorescein (FAM) -labeled gdh-C20 template DNA, 475 nM TBP and 270 nM TFB in transcription buffer (40 mM HEPES–KOH pH 7.3, 250 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 0.1 mM ZnSO4) supplemented with 1 mM DTT and 0.1 mg/ml BSA and incubated at 70°C for 10 min. Subsequently, 0.2 μg poly-2′deoxyinosinic-2′-deoxycytidylic acid (poly- dIC) competitor was added to the sample and incubated for additional 10 min at 70°C. Samples were separated by electrophoresis using a Tris-Glycine buffer system and a native 4–10% gradient polyacrylamide gel.
In vitro transcription assays
Transcription initiation assays were performed in transcription buffer supplemented with 25 mM DTT and 0.25 mg/ml BSA using 10 nM gdh-C20 template DNA, 10 nM RNAP, 238 nM TBP and 135 nM TFB. Samples were incubated with 40 μM guanylyl-5′-phosphatidyl-Uracil (GpU) dinucleotide primer and 0.074 MBq [α-32P]-UTP (111 TBq/mmol) for 10 min at 70°C. 3 nt radiolabeled RNAs were extracted with phenol/chloroform/Isoamyl alcohol (PCI) and separated on a 7 M urea/28% polyacrylamide gel.
Run-off transcription assays were carried out in transcription buffer supplied with 0.25 mg/ml BSA, 25 mM DTT, 440 μM ATP, 440 μM GTP, 440 μM CTP, 2.7 μM UTP, 0.049 MBq [α-32P]-UTP (111 TBq/mmol) with 10 nM gdh-C20 template DNA, 10 nM RNAP, 238 nM TBP and 135 nM TFB and incubated at 70°C for 10 min. The 113 nt radiolabeled run off products were extracted with PCI and separated on a 7 M urea/8% acrylamide gel. The gel was transferred and fixed to a Whatman chromatography paper. Both assays were repeated in three individually performed experiments, signals were compared to wild type TFB signals and the averages as well as the standard deviation were calculated.
Transcription complexes were stalled at distinct positions in reactions that contain transcription buffer supplemented with 25 mM DTT and 0.25 mg/ml BSA and 10 nM of the respective gdh-C-cassette, 10 nM RNAP, 238 nM TBP and 135 nM TFB and were preincubated for 5 min at 80°C. A nucleotide mix containing 40 μM ATP, 40 μM GTP, 2.5 μM UTP and 0.049 MBq [α-32P]-UTP (111 TBq/mmol) together with 2 μg heparin were added and incubated for additional 5 min. The 6 nt to 20 nt radiolabeled RNA transcripts were extracted with PCI and separated on a 7 M/urea 28% acrylamide gel.
Potassium permanganate footprinting
Footprinting reactions were performed in 0.5× transcription buffer supplied with 0.1 μg/μl BSA, 1 mM DTT and 3 μl of radiolabeled and immobilized DNA, 10 nM RNAP, 238 nM TBP and 135 nM TFB and 217 nM TFE in a total volume of 25 μl. Samples were incubated at 70°C for 10 min followed by addition of 2.5 μl of a 250 mM KMnO4 solution and further incubated for 5 min. Reactions were stopped using 1.5 μl β-mercaptoethanol and 20 μl stop solution (100 mM EDTA, 1% w/v SDS) and tubes were subsequently placed on ice. Samples were placed on the magnet particle separator and the supernatant was discarded. Beads were resuspended in 18 μl H2O and mixed with 2 μl piperidine (99%). After incubation at 90°C for 30 min the supernatant was transferred into 40 μl PCI solution and mixed for 40 s. After centrifugation at 21 000g for 5 min 17 μl of the upper phase was transferred into a fresh tube and 30 μl H2O, 5μl 3 M NaAc pH 5.3, 1 μl glycogen (20 mg/ml) and 125 μl ethanol (>99.98%) were added. The samples were mixed and incubated at -80°C for 45 minutes, and centrifuged at 21 000g for 30 min. The supernatant was removed and the pellet was washed with 800 μl ethanol (70%), and centrifuged for 10 min at 21 000g. The pellet was dehumidified using a vacuum concentrator (Concentrator 5301, Eppendorf, Hamburg, Germany). DNA was dissolved in 10 μl TE-buffer and mixed with 5 μl 3× loading dye and incubated at 95°C for 3 min. Samples were separated on a 7 M urea/6% acrylamide gel.
UV-induced photochemical crosslink reactions
Radioactively labeled DNA templates for crosslinking studies were created with the respective gdh-C-cassettes. Templates for T-strand labeling were amplified with MAT2 reverse primer (5′-CCAAGCTTGCATGCCTGCAGGTCG-3′) and biotinylated M13 forward primer, for labeling the NT-strand gdhShortFOR primer (5′-TAGATTCTTTGAGCCTAATCAAAT-3′) and biotinylated M13 reverse primer were used. The 210–231 bp PCR products were purified and 1 pmol DNA was attached to 10 μg streptavidin-coated magnetic particles (Thermo Fisher Scientific, Waltham, USA) followed by generating immobilized, single-stranded DNA using alkaline treatment (62). In order to radiolabel the DNA templates in a position specific manner (63) 10 pmol of immobilized, single-stranded DNA was hybridized with 5 pmol of a complementary single stranded oligonucleotide, in which the 3′end of the oligonucleotide ends one nucleotide next to the position for labeling. The hybridized DNA was incubated with 2.5 U Klenow Fragment exo- and 2.98 MBq [α-32P] dNTP (222 TBq/mmol) for 10 min at 40°C in Klenow buffer containing 7.5 μg BSA, 30 mM Tris–Cl pH 8.0, 7 mM MgCl2, 50 mM KCl, 2 mM β-mercaptoethanol and 50% (v/v) glycerol to specifically incorporate the radioactive nucleotide. 2 mM of each dNTP and 4 U Klenow Fragment exo- was added and incubated for 10 min at 40°C to complete the strand. Immobilized templates were washed with TE buffer + 0.1% BSA and the DNA was released from the beads either with 20 U EcoRI for T-strand labeling or by heat incubation at 70°C for 5 min for NT-strand labeling (64). The supernatant containing the labeled DNA was separated from beads using magnetic particle separator, and the DNA in the supernatant was isolated with PCI, followed by ethanol precipitation. The resulting pellet was dissolved in 10 mM Tris–Cl pH 8.0. The radioactive concentration was measured with TRI-CARB 2900TR Scintillation Analyzer with QuantaSmart™ software (PerkinElmer, Rodgau-Jügesheim, Germany) by counting activity of defined sample volumes.
UV-induced photochemical crosslink reactions were performed in 25 μl sample volume with 2 nM of radiolabeled DNA, 13 nM RNAP, 238 nM TBP and 135 nM TFB in transcription buffer with 2 μg BSA and 20 mM DTT and, depending on the reaction, with additional 217 nM TFE and/or NTP Mix (40 μM ATP, 40 μM GTP and 2.7 μM UTP). The samples were covered with mineral oil and incubated 5 minutes at 80°C. 2 μg heparin was added as competitor and the samples were exposed to UV-light (Philips TUV 15W/G15T8 UV-C) for 20 min at 80°C. 15 μl of the samples were recovered and digested with 1.65 μl DNase I-mix containing 1 U DNase I, 40 mM Tris–Cl pH 8.0, 7 mM MgCl2, 100 mM NaCl, 5 mM CaCl2, 1 mM DTT, 0.1 mg/ml BSA, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin and 50% (v/v) glycerol for 10 min at 37°C. 1 μl of a 10% (w/v) SDS solution was added and reactions were incubated at 90°C for 3 min. Samples were mixed with 1.92 μl 12× Zn/HOAc-mix (0.1 mM ZnSO4, 12% (v/v) glacial acetic acid) and 1.4 μl S1 nuclease-mix containing 20 U S1 nuclease, 20 mM Tris–Cl pH 7.5, 50 mM NaCl, 0.1 mM ZnSO4, 0.1 mg/ml BSA, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml leupeptin and 50% (v/v) glycerol and incubated for 10 min at 37°C. Reactions were denatured with 6× SDS-Loading Dye (0.35 M Tris–Cl pH 6.8, 30% (v/v) glycerol, 10% (w/v) SDS, 0.6 M DTT, 0.03% (w/v) bromophenol blue) and separated on a 12% SDS-polyacrylamide gel using Tris-Glycine buffer system. The gel was transferred into fixation solution (30% (v/v) ethanol, 10% (v/v) glacial acetic acid) and incubated over night. The gel was fixed and transferred to a Whatman chromatography paper.
Analysis of radiolabeled nucleic acids
Gels with radioactive samples were exposed to an Imaging Plate for autoradiography. Signals derived from radiolabeled RNA transcripts or labeled DNA were detected with FUJIFILM FLA 5000 PhosphoImager (Fuji, Japan) and analysed with Aida Image software v4.27 (Raytest, Straubenhardt, Germany).
Single-molecule FRET measurements
Unlabeled transcription factors TBP and TFE from Methanocaldococcus jannaschii were produced as described previously (29,65,66). Subunit Rpo2″ of the archaeal RNAP from M. jannaschii were expressed, purified and labelled as described (32,39,66).
Amber codons were engineered into genes encoding C-terminally hexahistidine-tagged M. jannaschii TFB (positions A124 and G262) using the QuikChange II site-directed mutagenesis kit (Agilent). The recombinant protein was produced in Rosetta(DE3)pLysS (Novagen) cells that additionally carried the arabinose-inducible pEvol-pAzF plasmid an amber-suppressor tRNA (tRNACUA) and an engineered tyrosyl-tRNA synthetase (67). 10 ml cultures were grown in LB medium containing 100 μg/ml ampicillin and 25 μg/ml tetracycline. Sixteen hours after inoculation, cells were pelleted and the pellet was resuspended in 50 ml of M9 minimal medium supplemented with 0.4% glucose, 2 mM MgCl2, 0.1 M CaCl2, 3 nM (NH4)6Mo7O24, 400 nM H3BO3, 30 nM CoCl2, 10 nM CuS04, 80 nM MnCl2, 20 nM ZnSO4, 2 mg/ml thiamine, 0.4 mg/ml choline chloride, 0.5 mg/ml folic acid, 0.5 mg/ml nicotinamide, 1 mg/ml myo-inositol, 1 mg/ml pyridoxal, 0.05 mg/ml riboflavin, 1 mg/ml biotin and antibiotics (ampicillin and tetracycline). Cells were grown for another 24 h and pelleted (4000 g, 15 min, 4°C). The cell pellet was resuspended in 5 ml of minimal medium and expanded in 500 ml M9 minimal medium (with the supplements as described above). At an optical density of 0.6–0.8 at 600 nm, 1 mM IPTG was added to induce TFB protein expression. The cultures were grown for 4–6 h after induction in the presence of 1 mM of the unnatural amino acid p-azido-l-phenylalanine (Chem-Impex international). The cells were harvested by centrifugation, resuspended in buffer N500 (20 mM Tris-acetate pH 7.5, 10 mM Mg-acetate, 100 μM zinc sulfate, 500 mM NaCl, 10% glycerol and 0.5% Triton X-100) and subjected to sonification for cell lysis. Cell debris was removed by centrifugation (30 min, 4°C, 15 000g). TFB protein was purified by nickel-affinity chromatography on a 1 ml Ni-NTA column (GE Healthcare) following the manufacturer's protocols. After application of the cell extract supernatant, the column was washed with N500 buffer supplemented with 20 mM Imidazol to remove unspecifically bound proteins. TFB was eluted using a linear gradient from 20 to 300 mM Imidazol in N500 buffer.
MjTFB and MjRNAP subunit Rpo2’ derivatives containing site-specifically incorporated p-azido-l-phenylalanine were labelled by Staudinger ligation using phosphine derivatives of fluorescent probes (68). TFB derivatives were labeled using DyLight550-phosphine (Thermo Fisher) and MjRNAP-subunit Rpo2’ was labeled with DL650-phosphine (Thermo Fisher) as described (32,39,66).
Pre-initiation complexes were assembled by incubating 0.1 μM promoter DNA (based on SSVT6 promoter), 10 μM TBP, 2 μM TFB, 0.67 μM RNAP, 5.9 mM DTT, 0.07 mg/ml BSA in 1x TMNE buffer (40mM Tris–HCl, pH 7.3, 250 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA and 5% glycerol) at 65°C for 15 min (39,66). To measure the PIC in presence of the third basal transcription factor TFE, 8 μM TFE was added to the PIC and further incubated at 65°C for 10 min (39). In order to prevent unspecific binding of the RNAP to DNA, heparin (final concentration of 6 μg/ml) was added to the PIC solution followed by an additional incubation step for 15 min at 65°C. The PIC complexes were immobilized via the biotin-moiety of the template DNA strand in a measurements chamber coated with Neutravidin-PEG suitable for single-molecule measurements using a total internal reflection (TIRF) microscope as described in Schulz et al. (39).
Promoter DNA strands used for this study were purchased from IBA (Göttingen) (unpaired region of the transcription bubble in the DNA heteroduplex is highlighted in red; non-template strand heteroduplex promoter DNA 5′-3′ sequence: GATTGATAGAGTAAAGTTTAAATACTTATAGAGATA GAGTATAGATAGAGGGTTGAGATGATGGTTAGGGTTGAGATGATGGTTATGGTTCATCTCGCAGCCACTCCGCA; template strand heteroduplex promoter DNA 5′-3′ sequence: TGCGGAGTGGCTG CGAGATGAACC ATAACCATCATCTCAACCCTAACCATCATCTCAACCCTCCGCTTATACTCTAT CTCTATAAGTATTTAAACTTTACTCTATCAATC – Biotin)
Single-molecule FRET measurements of immobilized pre-initiation complexes were performed as described in Schulz et al. (39) using a TIRF widefield setup. Analysis of the single-molecule data followed the protocol outlined in Schulz et al (2016) (39).
RESULTS AND DISCUSSION
TFB of P. furiosus shows a highly conserved domain organization and sequence similarity to eukaryotic TFIIB (Figure 1A) (22). It plays a role in RNAP recruitment and contributes to promoter opening, but the mechanistic details of how this occurs remained unclear, in part because the precise positioning of its conserved protein domains (e.g. the B-reader region) in the different steps of initiation is not known. Our aim was to identify TFB B-reader-DNA contacts in context of initially transcribing complexes using UV-inducible crosslinking. Therefore, a set of TFB variants was created by amino acid (AA) substitutions with the unnatural AA Bpa (59,60). This phenylalanine derivate reacts preferentially with unreactive C–H bonds when exposed to UV-light at a wavelength of 350–360 nm with a reactive spherical radius of 3.1 Å (69) and has been already successfully applied in both protein-protein and protein–DNA interaction studies (70,71). Site-specifically radiolabeled DNA templates were used to build transcription complexes containing general TFs and RNAP as indicated. After UV-exposure samples were treated with nucleases and proteins were separated by SDS-PAGE. If the Bpa-substituted AA of TFB is in close proximity to the radiolabeled position in the DNA template, a short radioactive DNA fragment covalently bound to TFB will remain after nuclease treatment, which allows detection of the crosslinking signal via SDS-PAGE and autoradiography at the molecular weight of TFB at 37 kDa. In contrast, if the Bpa-substituted AA of TFB is not in close proximity to the DNA, no crosslink occurs and no radioactive signal will be detected. Additionally, if the radiolabeled position in the DNA is too far from the crosslink position, it will be removed from TFB by the nuclease treatment and again no crosslink signal will be detectable (Supplementary Figure S1).
Selection of TFB-Bpa variants for UV-inducible crosslinking experiments
In the first step AA positions G41 to A49 of the TFB B-reader helix (E62 to N68 in Saccharomyces cerevisiae TFIIB (ScTFIIB)) and S50 to R57 of the TFB B-reader loop domain (D69 to R78 in ScTFIIB), which are located closely to the T-strand in the eukaryotic ITC (45) (Figure 1B), were substituted by Bpa. Additionally, position F192 of the TFB B-core domain was included in our study as this TFB Bpa-variant was used to to validate the reliability of the crosslinking system (Micorescu et al., to be submitted). In order to verify that the mutations do not influence the stability and function of TFB, all Bpa containing TFB variants were screened for activity in a set of in vitro assays. All selected TFB Bpa-variants could be successfully overexpressed and purified except for TFB P42Bpa (Supplementary Figure S2). In the first step, formation of the PIC was tested in gel shifts assays using fluoresecently-labeled DNA, TBP, RNAP and TFB WT or Bpa-substituted TFB variants (Figure 2A). Addition of TFB WT only or TBP and RNAP only to the promoter DNA did not result in complex formation (Figure 2A, lanes 2 and 3). In contrast, addition of TBP and TFB WT led to the formation of a stable ternary complex with DNA (Figure 2A, lanes 4 and 8) and addition of RNAP resulted in a new signal representing the PIC (CC and/or OC) (Figure 2A, lane 5 and 9). PIC formation could also be observed using TFB Bpa variants instead of TFB WT (Figure 2A, lanes 6 and 10 to 25). Thus, we also tested the ability of these PICs containing the TFB-Bpa variants to form the first phosphodiester bond as a measure of the capability of RNAP for initial RNA synthesis (Figure 2B). The B-core variant F192Bpa showed transcription activity of two-third in comparison to the TFB WT (Figure 2B, lanes 1 and 2). This AA position was chosen because of its close location to DNA (20), and the incorporated Bpa may influence the binding of TFB F192Bpa to DNA/TBP, which leads to reduced signal intensities in abortive transcription assays. Best results (>50% of WT level) were obtained for Bpa-substitutions in the B-reader helix (Figure 2B, lanes 3–11) at positions A46 and A49 and in the B-reader loop (Figure 2B, lanes 12–20) at positions R52, E53 and S56. Bpa substitutions at other positions led to strongly reduced or no abortive transcription signals. This finding suggests that the substitution of the natural AAs at these positions with Bpa influences transcription in a way that OC formation, transcription start site selection or other important interactions within the complex are affected and prevent the successful formation of the first phosphodiester bond (72–74).
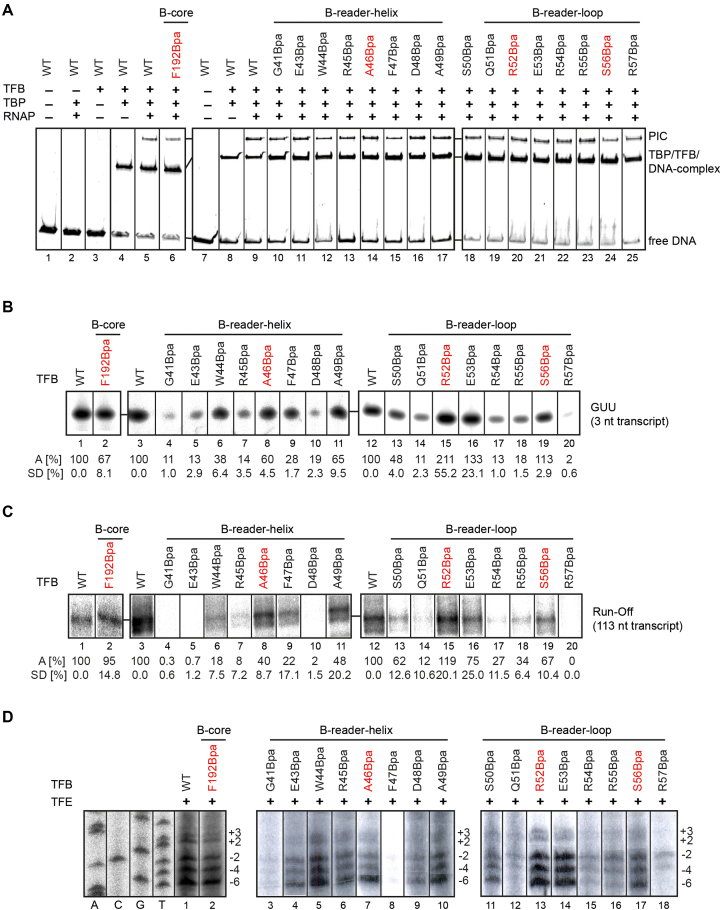
Figure 2.
Functional activity of TFB Bpa-variants. Variants used for crosslinking experiments are colored in red. (A) Electro mobility shift assays of selected TFB-Bpa variants to monitor preinitiation complex formation. 5′-FAM-labeled gdh-C20 template DNA was incubated with TBP, TFB WT or Bpa-substituted TFB variants, and RNAP as indicated and complexes were separated by electrophoresis on a native 4–10% gradient acrylamide gel. (B) Effects of Bpa substitution on transcription initiation in a first phosphodiester bond formation assay. Addition of one radiolabeled UTP to a 2 nt RNA primer (GpU) by the archaeal RNAP was analyzed on a 7 M urea/ 28% polyacrylamide (PA) gel. Intensities of resulting radiolabeled 3 nt RNA products were quantified and both average value (A) and the standard deviation (SD) were calculated from three independent experiments with the TFB WT transcript intensities set to 100%. (C) Run-off transcription assays with TFB-Bpa mutations in comparison to TFB WT. The observed specific 113 nt run-off product is only formed if TBP, TFB and RNAP are present. The resulting radiolabeled transcripts were analyzed on a 7 M urea/ 8% polyacrylamide gel. Experiments were performed three times. The intensity of full-length transcription products were quantified and compared to TFB WT to calculate the average value (A) and standard deviation (SD). (D) KMnO4 footprint analysis of the TFB-Bpa variants in the presence of TFE. The gdh-C20 template was used to assemble a complex consisting of TBP, TFB WT or the Bpa variants, TFE and RNAP. After KMnO4 treatment, samples were separated on a 6% sequencing gel. The sequencing ladder is presented on the left ranging from positions –8 to +7 relatively to the TSS and specific patterns of the cleaved radiolabeled non-template-strands are shown for KMnO4 footprint reactions using TFB WT or the Bpa variants. The positions of the T-residues within the initially melted region relatively to the TSS are depicted on the right as described previously (22,41,56).
In addition, run-off transcription assays were performed to analyze the effect of Bpa susbstitutions in TFB on all steps of transcription from initiation to elongation (Figure 2C). Here, similar results were observed as for the initiation assays. Bpa-substitutions in the B-reader helix (Figure 2C, lanes 3–11) at positions A46 and A49 and in the B-reader loop (Figure 2C, lanes 12–20) at positions R52, E53 and S56 only modestly affected TFB function. Interestingly, the B-core position F192Bpa resulted in run-off transcripts with signal intensities of 95% as compared to TFB WT (Figure 2C, lanes 1 and 2), which is in contrast to 67% in the abortive transcription assay. This indicates that the defects during abortive transcription can be compensated so that PICs can proceed to complete transcription initiation and enter the elongation phase. Finally, to analyze if the AA substitutions affect the promoter opening event, KMnO4 footprint experiments were carried out (Figure 2D). Reactions were performed in the presence of TFE as this reflects the cellular conditions and it was demonstrated that TFB-dependent deficiencies in promoter opening can be rescued by the addition of TFE (22,29). The results showed that TFB variants G41Bpa, D48Bpa and R57Bpa are not able to open the initially melted region around the transcription start site providing a rationale for inefficient transcription in the presence of these TFB variants (Figure 2D, lanes 3, 9 and 18). TFB variants F47Bpa, Q51Bpa, R54Bpa and R55Bpa showed strongly reduced opening of the DNA (Figure 2D, lanes 8, 12, 15 and 16), whereas F192Bpa, E43Bpa, R45Bpa W44Bpa, A46Bpa, A49Bpa, S50Bpa and S56Bpa showed signal patterns comparable to TFB WT (Figure 2D, lanes 2, 4, 5, 6, 7, 10, 11 and 16). R52Bpa and E53Bpa showed increased ability to open the DNA (Figure 2D, lanes 13 and 14). These results are in good agreement with published mutational studies using TFBs from P. furiosus and other organisms (22,45,75).
Based on this analysis, TFB variants were selected for crosslinking experiments considering on the one hand the transcription activity, and on the other hand the location of the corresponding AAs in the eukaryotic ITC (Figure 1B). The catalytically fully active TFB R52Bpa variant was chosen as it is part of the B-reader loop with the closest distance to the DNA in the crystal structure (Figure 1B). Moreover, this position was already successfully used for adaption of the crosslinking approach to the P. furiosus transcription system (Micorescu et al., to be submitted) and allowed us in combination with TFB F192Bpa to create and adjust the experimental setup for crosslinking in stalled transcription complexes. Additionally, position A46Bpa was selected, which is located in the B-reader helix, and S56Bpa was chosen as an additional position in the B-reader loop, since this AA is located at the tip of the B-reader loop in the ITC model (Figure 1B).
Correct stalling of transcription complexes at registers +6 to +20 using TFB Bpa-variants
Information derived from eukaryotic structures from the PIC and ITC suggest that the B-reader domain of TFIIB is displaced during transcription initiation, when the transcript reaches a length of 10–11 bases as this domain is in path of the advancing 5′ end of the RNA (44,45). Studying TFB-DNA interactions on the WT gdh promoter is restricted to the PIC and ITC. Thus, variants of this promoter were used, which contain the first cytidine on the NT-strand at increasing distances downstream of the TSS (gdh-C cassettes) to allow stalling of transcribing complexes from position +6 to +20 (Figure 3A) (41,40,76). Transcription assays without CTP were performed to validate correct positioning of the transcription complexes on each template. Predicted RNA sizes were observed for TFB WT using 80°C as incubation temperature, indicating correct stalling (Figure 3B, lanes 1–6 and Supplementary Figure S3A). Using TFB-Bpa variants A46Bpa (B-reader helix), R52Bpa and S56Bpa (B-reader loop) and F192Bpa (B-core) yielded correctly stalled complexes as transcript lengths correspond to the pattern of reactions that included TFB1 WT (Figure 3B, lanes 7–24 and Figure 3C). Corresponding to Figure 2C reduced transcript signals were observed for TFB variants A46Bpa and S56Bpa compared to TFB WT, however the ratios between the transcripts of different length corresponds to the pattern using TFB WT. Thus, these variants were used for crosslinking experiments in stalled complexes. Additionally, we could further demonstrate that the stalled transcription complexes are still competent in transcription after 20 min, since 85% of the complexes can be chased to complete transcription indicated by the run-off signals after addition of non-labeled nucleotides to the reaction (Supplementary Figure S3B). Taken together, these results show that our in vitro transcription system using gdh-C cassettes is well suited to analyze topological transitions of TFB domains during transcription initiation.
Crosslinking experiments in the PIC
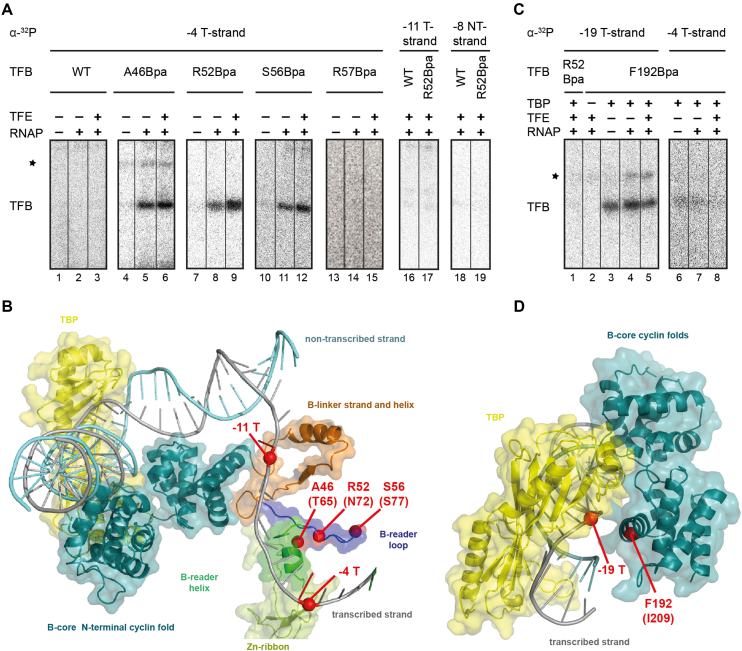
First, the crosslinking behavior of the selected TFB B-reader-Bpa variants was investigated using the gdh-C6 cassette with site-specifical radiolabels at position -4 on the T-strand. This position is in close proximity to the B-reader domain in eukaryotic crystal as well as cryo-EM structures (4,45). (Figure 4B). For TFB WT no signals were observed at the expected size of 37 kDa on the SDS gel in different crosslink reactions. Therefore we conclude that TFB without Bpa does not crosslink to DNA non-specifically (Figure 4A, lanes 1 to 3 and Supplementary Figure S4). In contrast, TFB-Bpa B-reader variants A46Bpa, R52Bpa, and S56Bpa showed no crosslink signal in the ternary complex (Figure 4A, lanes 4, 7 and 10), but a strong crosslink to position -4 on the T-strand in samples containing RNAP (Figure 4A, lanes 5, 8 and 11). This shows that the B-reader domain of archael TFB is positioned similarly as observed for eukaryotic TFIIB in the OC. Moreover, using TFE in the reactions, which is assumed to be stably associated with initiating complexes in vivo and which is able to stabilize open complex formation (14,29,40,66), increased the signal intensity of these crosslinks (Figure 4A, lanes 6, 9 and 12). Furthermore, making use of the TFB1 R57Bpa variant, which is impeded in OC formation (Figure 2D), showed the specificity of crosslinking as for this position, no crosslink signals could be observed (Figure 4A, lanes 13–15). Finally, the TFB B-reader variant R52Bpa did not crosslink to DNA radiolabeled at position -11 on the T-strand and -8 on the NT-strand (Figure 4A, lanes 17 and 19), indicating sufficient spatial resolution of the crosslinking system.
Figure 4.
Specific crosslinks of TFB B-reader and TFB core Bpa-variants in the PIC. (A) TFB WT, A46Bpa, R52Bpa, S56Bpa and R57Bpa were used in crosslinking reactions together with a gdh-C6 DNA template radiolabeled at position -4 T (lanes 1–15). The TBP/TFB/DNA-complexes for each variant are represented in the first lanes and the open complexes in the second lanes, respectively. TFE was additionally present in the samples as indicated. A radioactive signal was observed on the 12% SDS-PAGE at a size of 37 kDa, which is the result of a covalent bond between TFB and the radiolabeled DNA. Other signals appear and are marked with an asterisk, but they are not factor specific, indicating an unspecific signal. TFB WT and R52Bpa show no crosslinking signal using a ghd-C6 cassette radiolabeled at position -11 on the T-strand (lanes 16 and 17) or at position –8 on the NT-strand (lanes 18 and 19). (B) Postulated eukaryotic yeast open complex model (PDB:3K1F; (22)). TBP is coloured in yellow and TFB domains are depicted in different colors (B-core in cyan; B-reader helix in green, B-reader loop in blue, B-linker in brown) whereas RNAP is excluded. TFB-Bpa positions and radiolabeled sites are shown as red dots. (C) The TFB B-core variant F192Bpa was used in crosslinking reactions together with gdh-C6 labeled at -19 (lanes 2 to 5) or –4 (lanes 6–8) on the T-strand. Specific crosslinks to position -19 T were observed in the ternary complex and the PIC without and with TFE. TFB R52Bpa did not crosslink to position -19 T (lane 1). Unspecific signals are marked with an asterisk. (D) Crystal structure of the TFB–TBP–DNA complex from P. woesei (PDB: 1AIS; (20)). The TFB B-core is shown in cyan), TBP in yellow and the DNA containing a TATA-bix in grey (20). Amino acid F192 and DNA position –19 are indicated by red dots.
Beyond the three TFB B-reader-Bpa variants, we also used a TFB variant substituted with Bpa in the N-terminal cyclin fold of the B-core element at position F192. In the crystal structure of the TBP/TFB-DNA-complex from P. woeseii contact of TFB-F192 with DNA at the transcribed strand 19 nucleotides upstream of the transcription start site can be found (20,24) (Figure 4D), consistent with the location of the corresponding amino acid I209 of ScTFIIB in the eukaryotic crystal structure (22). A gdh-C6 cassette radiolabeled at position –19 T was designed and crosslinking experiments were performed with the B-reader domain variant R52Bpa as negative control and F192Bpa without TBP, without RNAP, with RNAP and with additional TFE (Figure 4B, lanes 2–5). We found that position TFB-F192Bpa crosslinks to DNA at the expected site in both the TBP/TFB/DNA-complex as well as in the OC with and without TFE but not in reactions performed in absence of TBP. TFB R52Bpa did not show a crosslink signal at the respective site (Figure 4B, lane 1), indicating a specific crosslink for F192Bpa. Additionally, TFB F192Bpa did not crosslink using the gdh-C cassette radiolabeled at position -4 T (Figure 4B, lanes 6–8). Therefore, TFB F192Bpa is a suitable variant to demonstrate the absence or presence of TFB at different positions in stalled complexes. In contrast to TFB-A46Bpa, R52Bpa and S56Bpa, the presence of TFE did not increase the signal intensity in experiments using F192Bpa, which indicates that the number of transcription complexes is not increased in presence of TFE. This finding leads to the assumption that the observed increase in the crosslink signal in presence of TFE is a direct effect of TFE possibly due to the increased NT-strand stabilization, which results in a closer location of the T-strand to the B-reader domain of TFB.
As TFE appears to influence the relative position of the TFB linker domain and is generally known to support open complex formation, we aimed to study the effect of TFE in more detail. To this end, we employed the closely related transcription system of Methanocaldococcus jannaschii, for which a single-molecule fluorescence resonance energy transfer (smFRET) system is available and has been shown to yield information about conformational transitions of for example the RNAP clamp throughout the transcription cycle (Figure 5) (39,77). Here, we used single-molecule FRET to monitor the distance between positions in TFB and RNA polymerase subunit Rpo2’ in the absence and presence of TFE. First, we investigated open pre-initiation complexes using a TFB variant that carried the fluorescent label at position A124. This residue is located at the end of the TFB linker domain (Figure 5Bi). Fluorophores coupled to positions in the B-reader helix or loop resulted in defective PIC formation and transcription and could not be used for smFRET measurements (data not shown). In the absence of TFE, two FRET populations could be detected. The high FRET population (FRET efficiency of 0.86) represents the dominant conformational state (Figure 5Bii). The FRET efficiency correlates to a distances of 3.8 nm, which is in good agreement with the theoretical distance of 3.7 nm between TFB A124 and Rpo2’ Q373 in the model of the archaeal open initiation complex (32). However, conformational heterogeneity can be detected as a population with lower FRET efficiency (E = 0.71, distance: 4.5 nm) is present. Addition of TFE to the open PIC results in a slight shift in FRET efficiency for the lower FRET population (E = 0.64, distance: 4.7 nm) indicating that the distance between TFB and Rpo2’ increases (Figure 5B). Also, the relative amount of molecules in the two populations shifts towards the low FRET population. Previously, we found two conformational states for the RNAP clamp in the context of the open PIC. Interestingly, TFE shifted the equilibrium towards the open clamp conformation (39). Based on these data and the fact, that the TFB linker, ribbon and reader are lining the inner surface of the clamp and protrude into the active site, it seems feasible to assume that TFB moves along with the clamp. As the clamp adopts a more open state in the open PIC in the presence of TFE, the distance between TFB and Rpo2’ also increases if TFB moves along with the clamp. This movement apparently also influences the relative distance of TFB with respect to the template DNA as evident from the crosslinking data. Additionally, we measured the distance between position G262 in the TFB core domain and Rpo2’ Q373. We assumed that the TFB core domain remains relatively static when TFE is added to the PIC. In the absence of TFE, we found a single population with a FRET efficiency of 0.58 (Figure 5Biii), which correlates to a distance of approximately 5 nm. This is in good agreement with a greater distance of 4.8 nm between these positions as deduced from the structural model of the PIC. Addition of TFE does not significantly change the FRET distribution even though the FRET efficiency histogram can be fitted better with a double Gaussian function (Figure 5Biiii). In previous studies, we found two closely spaced populations for this FRET pair (32). However, the TFB core domain does not undergo a significant structural rearrangement upon addition of TFE to the open PIC.
Figure 5.
(A) Arrangement of archaeal TFB relative to the RNA polymerase in a model of the archaeal open pre-initiation complex (see (32)). Amino acids positions in TFB (A124, G262) and in Rpo2’ (Q373) used for the incorporation of fluorescent dyes are highlighted as red spheres. Distances between TFB A124 and Rpo2’ Q373 as well as for TFB G262 and Rpo2’ Q373 are shown. The RNAP clamp coiled coil (clamp cc) is highlighted in magenta. Color code for TFB domains is the same as used in Figure 1. (B) Conformational state of TFB in the open pre-initiation complex upon association of TFE. Pre-initiation complexes were assembled using transcription initiation factors TBP, a fluorescently labelled TFB variant that carries a donor dye (panel i/ii: TFBA124AzF*DL550, panel iii/iii: TFBG262AzF*DL550), a labelled RNAP variant that carries an acceptor dye in subunit Rpo2’ (RNAP-Rpo2’Q373AzF*DL650) and a SSV T6 promoter DNA with a heteroduplex region of 4 bp surrounding the transcription start site (–3 to +1). PICs were immobilized via the biotinylated template strand on a quartz glass slide (grey) for single-molecule TIRF measurements. The histograms (i-iiii) show the smFRET efficiencies determined for the pre-initiation complex in the absence (i,iii) and presence (ii/iiii) of TFE. Histograms were fitted with a single or double Gaussian and mean FRET efficiencies (E) and the coefficient of determination (R2) are shown with standard errors.
In conclusion, crosslinking experiments under optimized conditions for stalling transcription complexes agreed well with predictions derived from crystal and cryo-EM structures of eukaryotic and archaeal transcription complexes (4,45) and thus provides a reliable system to probe the relative position of the TFB B-reader domain at different stages of transcription initiation and elongation.
TFB B-reader domain is displaced in stalled complexes at position +10
To analyze structural rearrangements of the TFB B-reader domain during transition from transcription initiation to early elongation, crosslinking experiments were performed with the TFB B-reader mutations A46Bpa, R52Bpa and S56Bpa on stalled transcription complexes. Signal intensities of the resulting TFB-DNA contacts in stalled complexes were compared to those derived from preinitiation complexes. Altered signal intensities of the crosslinks between OCs and stalled complexes can be interpreted as a change in the distance between the Bpa in the protein and the labeled DNA.
The results of the crosslink reactions with the selected gdh-templates are summarized in Figure 6. At register +6 no change in the signal intensity of the stalled complexes in comparison to the OC was detected for A46Bpa and R52Bpa (Figure 6A and B), whereas S56Bpa showed a slightly reduced signal of 85% (Figure 6C). At register +8 a reduction of the crosslinking signal between the helix position A46Bpa and DNA (72% signal intensity compared to OC) could be monitored, whereas R52Bpa and S56Bpa showed unaltered signal intensities in comparison to register +6. At register +9 signal intensities for A46Bpa and S56Bpa are only slightly decreased when compared to signals of the OC. However, R52Bpa signal intensitiy started to decrease at this position. The signal intensities of all selected TFB positions are markedly reduced at register +10 and remained low at register +15. This finding suggests a translocation of the TFB B-reader domain, which is in agreement with the postulated TFIIB B-reader displacement in eukaryotic transcription systems (45,53). We assume that the residual signal derived from stalled complexes at register +10 is background, possibly caused by complexes which could not be stalled correctly and are still positioned at the promoter start site. It has been previously described, that during stalling using gdh-C cassettes a minor fraction of the complexes is paused at position +5 and not able to reach register +6/7, which represents a major transition state in transcription and is suggested to be rate-limiting (41). Resolving transcripts smaller than 6 nt according to the experiments shown in Figure 3B for TFB R52Bpa revealed the presence of an additional transcript in the range of 5 nt for all used gdh-C cassettes (Supplementary Figure S5A), which was also previously demonstrated by Spitalny and Thomm (2003) (41). However, it could be shown that these complexes paused at register +5 can be chased by adding a full set of NTPs (41). Thus, crosslinking experiments were performed under run-off conditions using R52Bpa and the selected gdh-C templates (Supplementary Figure S5B). This showed that the crosslink signals are completely lost in the run-off crosslinks, which indicates that the residual crosslink signals at register +10 and +15 presumably does not derive from a weak TFB B-reader DNA contact at these positions.
Figure 6.
Crosslinking experiments with the TFB B-reader variants A46Bpa, R52Bpa and S56Bpa and -4 T radiolabeled DNA in complexes stalled at +6, +8, +9, +10 and +15. (A) Crosslinking reactions using the B-reader helix variant A46Bpa. (B) Crosslinking reactions using the B-reader loop variant R52Bpa. (C) Crosslinking reactions using the B-reader loop variant S56Bpa. For each figure the TBP/TFB/DNA-complex is shown using in the left lane a gdh-C6 template in the absence of RNAP, which serves as control reaction. The second lane represents the open complex, the third lane the stalled complex. For the results with gdh-cassettes –C8 to –C15 the signal originating from open initiation complexes (OC; left lanes) and the signals originating from the respective stalled complexes are shown. Signals marked with an asterisk are due to factor unspecific interactions and are not present in every lane. Results are shown for three individually experiments and the signal intensity for the stalled complexes were quantified and compared to the respective signal intensity derived from the open complex. The standard deviation (SD) and the average value (A) were calculated and are summarized in a bar diagram.
Sainsbury et al (45) postulated a RNA-DNA separation model, which is based on the charge-specific interaction of the B-reader loop and the nascent RNA. In contrast, P. furiosus does not comprise a negative charged B-reader loop. However, in accordance to the model and depending on the structure and homology between TFB and TFIIB, we assume a first interaction of RNA with S56 at a length of 6nt. If RNA is extended to 8nt, a clash of the RNA with the TFIIB B-reader helix was predicted (22). We observed a reduced signal intensity of 72% compared to the corresponding OC for the helix mutation A46Bpa at this RNA length, indicating a structural change of the B-reader helix position likely caused by a clash with the nascent RNA. The fact that the distance of R52Bpa to DNA seems not to change until position +9 suggests that this region does not interact with RNA directly and showed a steady distance to DNA. This hypothesis is further supported by the fact that R52Bpa always showed the strongest crosslink signals in our experiments compared to other selected TFB variants. This indicates that this position is located closest to DNA.
TFB release occurs upon transcription of 15 nt long RNAs
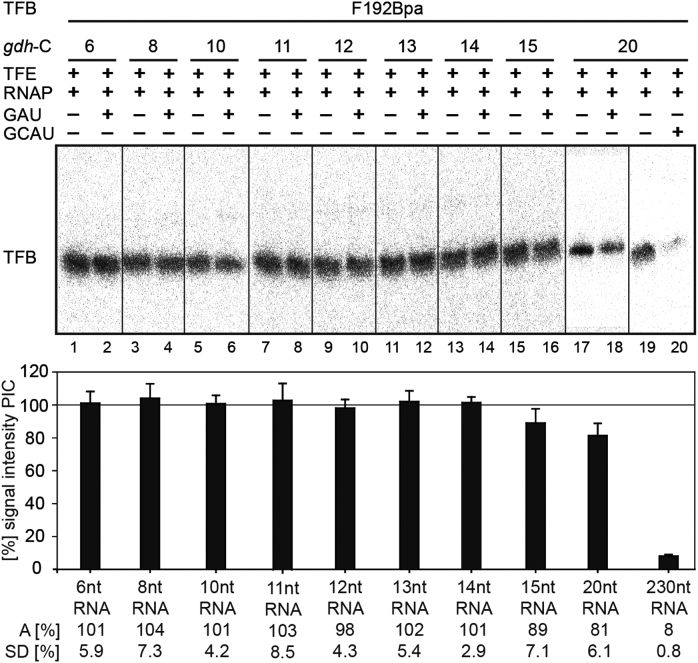
To verify that the signal reductions observed for the TFB B-reader domain Bpa variants at register +10 were not caused by an early TFB release event, the TFB Bpa-variant F192 was used in crosslink experiments. This position within TFB is located in the cyclin fold domain far away from the active site, and therefore it should not be affected by structural rearrangements of the B-reader elements (22). The results of the crosslink experiments with TFB F192 in stalled complexes are summarized in Figure 7. Poly-dAdT was additionally used in the reactions in order to trap TFB to prevent re-association of TFB to the promoter site after a possible TFB release event. The experiments showed that the signal intensities of TFB-DNA interactions in stalled complexes at registers +6 to +14 do not change in comparison to the signals derived from the respective PICs, indicating that TFB is not released at these positions. At position +15 and +20 a slight decrease of the signal intensity was observed, whereas crosslinking experiments under run-off conditions showed only background signals.
Figure 7.
Crosslinking experiments with the TFB variant F192Bpa and radiolabeled DNAs at position -19 T in complexes stalled at +6 to +20 as indicated. TFB is present at register +6 to +14 and crosslinking signals decrease at register +15 to +20. No crosslinking signal is detectable under run-off conditions. TFB-F192Bpa was crosslinked in initiation complexes (PIC; left lanes for each cassette) and in stalled complexes at the respective site (right lane for each cassette) with gdh-C cassettes containing a radiolabeled nucleotide at position -19 on the T-strand. Crosslinking under run-off conditions was performed on the gdh-C20 cassette. All reactions were performed in triplicates and the signals derived from crosslink experiments in halted complexes were compared to the signals of the respective PIC. The standard deviation (SD) as well as the average (A) was calculated and summarized in the bar diagram.
These data shows that TFB is present in complexes transcribed to register +10 and demonstrate further that the loss of interactions of the B-reader domain and DNA mentioned previously are not due to a TFB release event, but is a result of TFB B-reader translocation. We therefore conclude that the TFB B-reader domain is displaced at register +10, which causes a bubble collapse at this position consistent with predictions derived from eukaryotes (53). Our observations are further in accordance with previous KMnO4 footprint experiments which revealed a reduced size of the open DNA region at registers +10/+11 in the P. furiosus transcription system (41).
For the human system, TFIIB release was described to take place at an RNA length of 12/13 nt in vitro (55). It was also postulated that a 12 nt RNA clashes with the Zn-ribbon domain of TFIIB in a crystal structure of an initially transcribing S. cerevisiae TFIIB/RNAP II complex, since the Zn-ribbon blocks the exit pore of RNAP II (45). Additionally, Xie et al. (78) showed that TFB is released in in vitro reactions lacking TFE in the archaeal M. thermoautotrophicus system when complexes where chased to position +24. Moreover, in a human in vitro transcription system TFIIB release was shown to take place between +6 and +16 (54). Therefore, we expected a TFB release event between registers +12 to +14 but in our in vitro system we could not monitor this event at the respective sites. In contrast, we observed that the amount of TFB started to decrease at register +15 whereas the signal is further reduced at register +20, and TFB appears to be completely absent in run-off experiments. These findings lead us to the assumption that TFB is destabilized between position +14 and +15 and the contacts between TFB and RNAP, TBP and DNA are further destabilized from these positions onwards. Due to the fact that the signal intensity does not drop from one nucleotide position to the next we further assume that the exact point of release of TFB cannot be pinpointed to a distinct nucleotide position in our in vitro transcription system. This may rely on the presence of a too high background signal deriving from complexes, which did not build the PIC, or from complexes, which are still positioned at the promoter start site in the open complex state or abortive transcription state. In addition, it remains unclear if the nascent RNA in stalled complexes has enough kinetic energy to induce the release of TFB from the complex, and if TFB release is an instantaneous event, or if this process happens slowly due to the persisting interactions between TFB and DNA, TBP and RNAP. In addition, the stabilizing effect of TFE and its impact on TFB release is still unclear.
CONCLUSION
In this study, we made use of a highly specific crosslinking method to unravel the conformational changes of the general transcription factor TFB in archaeal transcription during the initiation to elongation phase transition. We demonstrated that the TFB B-reader domain is located in close proximity to the active site of the transcription machinery as TFB residues of A46Bpa, R52Bpa and S56Bpa can be specifically crosslinked to the template DNA at position -4 T in the OC. Additionally, TFB-F192Bpa of the B-core element specifically crosslinks to DNA labeled at -19 T in the ternary, as well as in the open initiation complex. These observations are in agreement with published structural models of eukaryotic transcription complexes (4,3,45), further supporting the conservation of initiation complex architecture between the archaeal and eukaryotic domain of life. For that reason we conclude that the topology of the archaeal TFB is highly similar to that of eukaryotic TFIIB. Our crosslinking system further allowed us to investigate dynamic structural transitions of TFB by analyzing the respective TFB-DNA contacts in stalled complexes that mimic the stepwise transition from the initiation to elongation complex. We could show that transcription complexes with the selected TFB variants can be stalled correctly at the distinct sites on the gdh-C templates. Our experiments provide evidence that nascent RNA interacts with the B-reader loop at a RNA length of six nucleotides, and clashes with the B-reader helix at a length of 8 nucleotides. We further could demonstrate that the B-reader domain is distant from its DNA site at register +10, resulting in the collapse of the transcription bubble. Using the B-core variant TFB F192Bpa we showed that TFB is present at registers +6 to +14, whereas the amount of TFB in the complexes were reduced from register +15 onward and is completely absent in crosslinked run-off samples. We assume that after B-reader displacement and transcription bubble collapse at register +10, TFB is destabilized within the complex, which in turn induces TFB release and promoter escape (summarized in Supplementary Figure S6).
Single-molecule FRET experiments furthermore suggest that TFB linker-reader moves along with the RNAP clamp and that the TFE-induced conformational change towards an open RNAP clamp in the OC also influences the position of TFB linker-reader module.
Taken together, our data presented here provide more detailed insights into structural rearrangements of the transcription factor B during the transition from initiation to early elongation of transcription on a biochemical level. It further complements the structural information derived from crystal structures and cryo-EM analysis of eukaryotic transcription complexes and provides a better understanding of the transcription initiation mechanism. The crosslinking method established in this work is highly specific and can be applied to address open questions regarding molecular mechanisms of transcription, e.g. to monitor DNA scrunching, which has not been observed yet in archaeal transcription.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Winfried Hausner for reading the manuscript carefully and for his support and Patrick Cramer for initial ideas to this work. In addition, we thank Mirijam Zeller for performing initial experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB960 project A3 to S.D., R.R. and M.T. and SFB960 project A7 to D.G]; National Institutes of Health Academic Research Enhancement Award [R15 GM083306 to M.B.]. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1. He Y., Fang J., Taatjes D.J., Nogales E.. Structural visualization of key steps in human transcription initiation. Nature. 2013; 495:481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sainsbury S., Bernecky C., Cramer P.. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015; 16:129–143. [DOI] [PubMed] [Google Scholar]
- 3. Plaschka C., Hantsche M., Dienemann C., Burzinski C., Plitzko J., Cramer P.. Transcription initiation complex structures elucidate DNA opening. Nature. 2016; 533:353–358. [DOI] [PubMed] [Google Scholar]
- 4. He Y., Yan C., Fang J., Inouye C., Tjian R., Ivanov I., Nogales E.. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016; 533:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bell S.D., Jackson S.P.. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 2001; 4:208–213. [DOI] [PubMed] [Google Scholar]
- 6. Carlo S. de, Lin S.-C., Taatjes D.J., Hoenger A.. Molecular basis of transcription initiation in Archaea. Transcription. 2010; 1:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grohmann D., Werner F.. Recent advances in the understanding of archaeal transcription. Curr. Opin. Microbiol. 2011; 14:328–334. [DOI] [PubMed] [Google Scholar]
- 8. Decker K.B., Hinton D.M.. Transcription regulation at the core: similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu. Rev. Microbiol. 2013; 67:113–139. [DOI] [PubMed] [Google Scholar]
- 9. Bell S.D., Jackson S.P.. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 1998; 6:222–228. [DOI] [PubMed] [Google Scholar]
- 10. Werner F., Grohmann D.. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev.. Microbiol. 2011; 9:85–98. [DOI] [PubMed] [Google Scholar]
- 11. Jun S.-H., Reichlen M.J., Tajiri M., Murakami K.S.. Archaeal RNA polymerase and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2011; 46:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brindefalk B., Dessailly B.H., Yeats C., Orengo C., Werner F., Poole A.M.. Evolutionary history of the TBP-domain superfamily. Nucleic Acids Res. 2013; 41:2832–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jun S.-H., Hirata A., Kanai T., Santangelo T.J., Imanaka T., Murakami K.S.. The X-ray crystal structure of the euryarchaeal RNA polymerase in an open-clamp configuration. Nat. Commun. 2014; 5:5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blombach F., Smollett K.L., Grohmann D., Werner F.. Molecular mechanisms of transcription initiation-structure, function, and evolution of TFE/TFIIE-Like factors and open complex formation. J. Mol. Biol. 2016; 428:2592–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burton S.P., Burton Z.F.. The sigma enigma: bacterial sigma factors, archaeal TFB and eukaryotic TFIIB are homologs. Transcription. 2014; 5:e967599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adachi N., Senda T., Horikoshi M.. Uncovering ancient transcription systems with a novel evolutionary indicator. Sci. Rep. 2016; 6:27922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hausner W., Frey G., Thomm M.. Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNA(Val) gene of Methanococcus vannielii. J. Mol. Biol. 1991; 222:495–508. [DOI] [PubMed] [Google Scholar]
- 18. Ouhammouch M., Hausner W., Geiduschek E.P.. TBP domain symmetry in basal and activated archaeal transcription. Mol. Microbiol. 2009; 71:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nikolov D.B., Chen H., Halay E.D., Usheva A.A., Hisatake K., Lee D.K., Roeder R.G., Burley S.K.. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995; 377:119–128. [DOI] [PubMed] [Google Scholar]
- 20. Kosa P.F., Ghosh G., DeDecker B.S., Sigler P.B.. The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. PNAS. 1997; 94:6042–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gietl A., Holzmeister P., Blombach F., Schulz S., von Voithenberg L.V., Lamb D.C., Werner F., Tinnefeld P., Grohmann D.. Eukaryotic and archaeal TBP and TFB/TF(II)B follow different promoter DNA bending pathways. Nucleic Acids Res. 2014; 42:6219–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kostrewa D., Zeller M.E., Armache K.-J., Seizl M., Leike K., Thomm M., Cramer P.. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009; 462:323–330. [DOI] [PubMed] [Google Scholar]
- 23. Bagby S., Kim S., Maldonado E., Tong K.I., Reinberg D., Ikura M.. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995; 82:857–867. [DOI] [PubMed] [Google Scholar]
- 24. Littlefield O., Korkhin Y., Sigler P.B.. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. PNAS. 1999; 96:13668–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lagrange T., Kapanidis A.N., Tang H., Reinberg D., Ebright R.H.. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes & Development. 1998; 12:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qureshi S.A., Jackson S.P.. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell. 1998; 1:389–400. [DOI] [PubMed] [Google Scholar]
- 27. Bell S.D., Kosa P.L., Sigler P.B., Jackson S.P.. Orientation of the transcription preinitiation complex in archaea. PNAS. 1999; 96:13662–13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen H.-T., Hahn S.. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell. 2003; 12:437–447. [DOI] [PubMed] [Google Scholar]
- 29. Werner F., Weinzierl R.O.J.. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol. Cell. Biol. 2005; 25:8344–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pan G., Greenblatt J.. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J. Biol. Chem. 1994; 269:30101–30104. [PubMed] [Google Scholar]
- 31. Hausner W., Thomm M.. Events during initiation of archaeal transcription: open complex formation and DNA-protein interactions. J. Bacteriol. 2001; 183:3025–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagy J., Grohmann D., Cheung A.C.M., Schulz S., Smollett K., Werner F., Michaelis J.. Complete architecture of the archaeal RNA polymerase open complex from single-molecule FRET and NPS. Nat. Commun. 2015; 6:6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fishburn J., Tomko E., Galburt E., Hahn S.. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. PNAS. 2015; 112:3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grunberg S., Warfield L., Hahn S.. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat. Struct. Mol. Bol. 2012; 19:788–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naji S., Bertero M.G., Spitalny P., Cramer P., Thomm M.. Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res. 2008; 36:676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holstege F.C., van der Vliet P C, Timmers H.T.. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996; 15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 37. Naji S., Grünberg S., Thomm M.. The RPB7 orthologue E' is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J. Biol. Chem. 2007; 282:11047–11057. [DOI] [PubMed] [Google Scholar]
- 38. Blombach F., Salvadori E., Fouqueau T., Yan J., Reimann J., Sheppard C., Smollett K.L., Albers S.V., Kay C.W.M., Thalassinos K. et al. . Archaeal TFEalpha/beta is a hybrid of TFIIE and the RNA polymerase III subcomplex hRPC62/39. eLife. 2015; 4:e08378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schulz S., Gietl A., Smollett K., Tinnefeld P., Werner F., Grohmann D.. TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. PNAS. 2016; 113:E1816–E1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grünberg S., Bartlett M.S., Naji S., Thomm M.. Transcription factor E is a part of transcription elongation complexes. J. Biol. Chem. 2007; 282:35482–35490. [DOI] [PubMed] [Google Scholar]
- 41. Spitalny P., Thomm M.. Analysis of the open region and of DNA-protein contacts of archaeal RNA polymerase transcription complexes during transition from initiation to elongation. J. Biol. Chem. 2003; 278:30497–30505. [DOI] [PubMed] [Google Scholar]
- 42. Liu X., Bushnell D.A., Wang D., Calero G., Kornberg R.D.. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010; 327:206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tomko E.J., Fishburn J., Hahn S., Galburt E.A.. TFIIH generates a six-base-pair open complex during RNAP II transcription initiation and start-site scanning. Nat. Struct. Mol. Biol. 2017; 24:1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bushnell D.A., Westover K.D., Davis R.E., Kornberg R.D.. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004; 303:983–988. [DOI] [PubMed] [Google Scholar]
- 45. Sainsbury S., Niesser J., Cramer P.. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013; 493:437–440. [DOI] [PubMed] [Google Scholar]
- 46. Bick M.J., Malik S., Mustaev A., Darst S.A.. TFIIB is only approximately 9 A away from the 5′-end of a trimeric RNA primer in a functional RNA polymerase II preinitiation complex. PLoS One. 2015; 10:e0119007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carpousis A.J., Gralla J.D.. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980; 19:3245–3253. [DOI] [PubMed] [Google Scholar]
- 48. Munson L.M., Reznikoff W.S.. Abortive initiation and long ribonucleic acid synthesis. Biochemistry. 1981; 20:2081–2085. [DOI] [PubMed] [Google Scholar]
- 49. Nudler E., Mustaev A., Lukhtanov E., Goldfarb A.. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997; 89:33–41. [DOI] [PubMed] [Google Scholar]
- 50. Kireeva M.L., Komissarova N., Waugh D.S., Kashlev M.. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000; 275:6530–6536. [DOI] [PubMed] [Google Scholar]
- 51. Revyakin A., Liu C., Ebright R.H., Strick T.R.. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006; 314:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luse D.S. Promoter clearance by RNA polymerase II. Biochim. Biophys. Acta. 2013; 1829:63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pal M., Ponticelli A.S., Luse D.S.. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell. 2005; 19:101–110. [DOI] [PubMed] [Google Scholar]
- 54. Tran K., Gralla J.D.. Control of the timing of promoter escape and RNA catalysis by the transcription factor IIb fingertip. J. Biol. Chem. 2008; 283:15665–15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cabart P., Ujvari A., Pal M., Luse D.S.. Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. PNAS. 2011; 108:15786–15791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fouqueau T., Zeller M.E., Cheung A.C., Cramer P., Thomm M.. The RNA polymerase trigger loop functions in all three phases of the transcription cycle. Nucleic Acids Res. 2013; 41:7048–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hausner W., Wettach J., Hethke C., Thomm M.. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J. Biol. Chem. 1996; 271:30144–30148. [DOI] [PubMed] [Google Scholar]
- 58. Waege I., Schmid G., Thumann S., Thomm M., Hausner W.. Shuttle vector-based transformation system for Pyrococcus furiosus. Appl. Environ. Microbiol. 2010; 76:3308–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Young T.S., Ahmad I., Yin J.A., Schultz P.G.. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010; 395:361–374. [DOI] [PubMed] [Google Scholar]
- 60. Kauer J.C., Erickson-Viitanen S., Wolfe H.R. Jr, DeGrado W.F.. p-Benzoyl-L-phenylalanine, a new photoreactive amino acid. Photolabeling of calmodulin with a synthetic calmodulin-binding peptide. J. Biol. Chem. 1986; 261:10695–10700. [PubMed] [Google Scholar]
- 61. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Espelund M., Stacy R.A., Jakobsen K.S.. A simple method for generating single-stranded DNA probes labeled to high activities. Nucleic Acids Res. 1990; 18:6157–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bartholomew B., Tinker R.L., Kassavetis G.A., Geiduschek E.P.. Photochemical cross-linking assay for DNA tracking by replication proteins. Methods Enzymol. 1995; 262:476–494. [DOI] [PubMed] [Google Scholar]
- 64. Holmberg A., Blomstergren A., Nord O., Lukacs M., Lundeberg J., Uhlen M.. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005; 26:501–510. [DOI] [PubMed] [Google Scholar]
- 65. Werner F., Weinzierl R.O.J.. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002; 10:635–646. [DOI] [PubMed] [Google Scholar]
- 66. Grohmann D., Nagy J., Chakraborty A., Klose D., Fielden D., Ebright R.H., Michaelis J., Werner F.. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell. 2011; 43:263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chin J.W., Martin A.B., King D.S., Wang L., Schultz P.G.. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. PNAS. 2002; 99:11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saxon E., Bertozzi C.R.. Cell surface engineering by a modified Staudinger reaction. Science. 2000; 287:2007–2010. [DOI] [PubMed] [Google Scholar]
- 69. Dorman G., Prestwich G.D.. Benzophenone photophores in biochemistry. Biochemistry. 1994; 33:5661–5673. [DOI] [PubMed] [Google Scholar]
- 70. Lee H.S., Dimla R.D., Schultz P.G.. Protein-DNA photo-crosslinking with a genetically encoded benzophenone-containing amino acid. Bioorg. Med. Chem. Lett. 2009; 19:5222–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xie J., Schultz P.G.. A chemical toolkit for proteins–an expanded genetic code. Nat. Rev. Mol. Cell Biol. 2006; 7:775–782. [DOI] [PubMed] [Google Scholar]
- 72. Bangur C.S., Pardee T.S., Ponticelli A.S.. Mutational analysis of the D1/E1 core helices and the conserved N-terminal region of yeast transcription factor IIB (TFIIB): identification of an N-terminal mutant that stabilizes TATA-binding protein-TFIIB-DNA complexes. Mol. Cell. Biol. 1997; 17:6784–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pinto I., Wu W.H., Na J.G., Hampsey M.. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 1994; 269:30569–30573. [PubMed] [Google Scholar]
- 74. Pardee T.S., Bangur C.S., Ponticelli A.S.. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J. Biol. Chem. 1998; 273:17859–17864. [DOI] [PubMed] [Google Scholar]
- 75. Wiesler S.C., Weinzierl R.O.J.. The linker domain of basal transcription factor TFIIB controls distinct recruitment and transcription stimulation functions. Nucleic Acids Res. 2011; 39:464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Grünberg S., Reich C., Zeller M.E., Bartlett M.S., Thomm M.. Rearrangement of the RNA polymerase subunit H and the lower jaw in archaeal elongation complexes. Nucleic Acids Res. 2010; 38:1950–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schulz S., Kramm K., Werner F., Grohmann D.. Fluorescently labeled recombinant RNAP system to probe archaeal transcription initiation. Methods. 2015; 86:10–18. [DOI] [PubMed] [Google Scholar]
- 78. Xie Y., Reeve J.N.. Transcription by Methanothermobacter thermautotrophicus RNA polymerase in vitro releases archaeal transcription factor B but not TATA-box binding protein from the template DNA. J. Bacteriol. 2004; 186:6306–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.