Abstract
Alfalfa leaf curl virus (ALCV), which causes severe disease symptoms in alfalfa (Medicago sativa L.) and is transmitted by the widespread aphid species, Aphis craccivora Koch, has been found throughout the Mediterranean basin as well as in Iran and Argentina. Here we reconstruct the evolutionary history of ALCV and attempt to determine whether the recent discovery and widespread detection of ALCV is attributable either to past diagnostic biases or to the emergence and global spread of the virus over the past few years. One hundred and twenty ALCV complete genome sequences recovered from ten countries were analyzed and four ALCV genotypes (ALCV-A, ALCV-B, ALCV-C, and ALCV-D) were clearly distinguished. We further confirm that ALCV isolates are highly recombinogenic and that recombination has been a major determinant in the origins of the various genotypes. Collectively, the sequence data support the hypothesis that, of all the analyzed locations, ALCV likely emerged and diversified in the Middle East before spreading to the western Mediterranean basin and Argentina.
Keywords: Alfalfa leaf curl virus, geminivirus, alfalfa, evolutionary history
1. Introduction
Although geminiviruses have been intensively studied since the 1970s [1,2], it is only since the recent development and application of viral metagenomics-based approaches that the true diversity of this group of viruses is starting to become apparent [3,4]. The genus Capulavirus is just one of several new genera in the family Geminiviridae that have been established to accommodate some of the novel and diverse geminiviruses that have been discovered over the past ten years [5]. The genus Capulavirus currently contains four species (Alfalfa leaf curl virus, Euphorbia caput-medusae latent virus, French bean severe leaf curl virus, and Plantago lanceolata latent virus) that infect both cultivated and non-cultivated plants in southern and northern Europe, the Indian subcontinent, and South Africa [6,7,8].
Alfalfa leaf curl virus (ALCV), which causes severe disease symptoms in alfalfa (Medicago sativa L.), is transmitted by the widespread aphid species, Aphis craccivora Koch [7,9]. A study of ALCV isolates collected between 2010 and 2014 in France and Spain [10,11] revealed that both intra- and inter-species recombination has played a significant role in the evolution of ALCV. In addition, this study suggested that ALCV was probably widely distributed across the Mediterranean basin [11]. Consistent with this hypothesis, ALCV was subsequently reported in 2018 from Jordan, Syria, Lebanon, and Tunisia [12] as well as from the non-Mediterranean countries Iran [13] and Argentina [14].
The discovery of ALCV in Argentina and the Mediterranean basin so soon after its initial characterization raises questions regarding the potential global emergence of this virus: does its discovery in these far-flung regions imply that a “true” global and potentially damaging geminivirus emergence event has recently occurred, or, more prosaically, does it merely reflect the fact that broader and more intensive sampling of plant material coupled with more sensitive virus detection techniques have only recently revealed an epidemic that has been ongoing for tens (or even hundreds) of years? Interestingly, alfalfa symptoms resembling those caused by ALCV, such as plant stunting and leaf curling, crumpling, and shriveling, have been reported since the 1950s in Europe (including France, Bulgaria, Romania, and Spain) and the Middle East (Saudi Arabia) [15,16,17,18,19]. The etiological agent of this alfalfa disease, which was shown to be transmissible by both grafting and A. craccivora [18,20], was identified as a rhabdovirus, referred to as lucerne enation virus (LEV) [15]. However, further studies revealed that different types of symptoms were observed depending on the mode of transmission [19], suggesting that in several instances where alfalfa diseases have been attributed to LEV, one or more additional viruses may have been co-infecting plants together with LEV. If this “Trojan horse” hypothesis was true, the apparently sudden occurrence of ALCV could simply reflect diagnostic biases rather than the recent emergence and global spread of the virus. Addressing this question is of great importance since alfalfa is the most-cultivated perennial forage legume in temperate regions of the world [21].
Here we examine 120 ALCV full genome sequences recovered from ten countries to reveal four distinct ALCV genotypes (ALCV-A, ALCV-B, ALCV-C, and ALCV-D) with the isolates of each genotype sharing <93% genome-wide pairwise identity with those of the other three genotypes. In addition, we confirm that ALCV isolates are highly recombinogenic and that recombination has been a major determinant in their origins. Phylogenetic analyses also suggest that, of all the countries analyzed, ALCV most likely originated in Iran and diversified in the Middle East before spreading to the Mediterranean basin and then onwards to Argentina.
2. Material and Methods
2.1. Plant Sampling
Leaves from 564 alfalfa plants that were either asymptomatic or presented with conspicuous disease symptoms (including plant stunting or varying degrees of leaf curling, crumpling, and shriveling), were collected from 2010 to 2017 in Argentina (41 plant samples from 17 regions), France (73 samples, 5 regions), Greece (36 samples, 8 regions), Iran (141 samples, 10 regions), Italy (50 samples, 6 regions), Jordan (11 samples, 2 regions), Lebanon (5 samples), Namibia (25 samples, 1 region), South Africa (91 samples, 3 regions), Spain (62 samples, 3 regions), Syria (12 samples, 1 region), and Tunisia (17 samples, 2 regions).
2.2. DNA Extraction and PCR-Mediated Alfalfa Leaf Curl Virus Detection
Total DNA from the French, Greek, Italian, Jordanian, Lebanese, Namibian, South African, Spanish, Syrian, and Tunisian alfalfa samples was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Total DNA from alfalfa samples from Argentina and Iran was extracted using the cetyltrimethylammonium bromide (CTAB) method [22]. PCR-based detection of ALCV from alfalfa plants collected in all countries other than Argentina and Iran was performed using the primer pair ALCV-187F (5′-TGG AAT ATT GTG CTG CTT GG-3′) and ALCV-971R (5′-ATT TTG GGA CTT GTG CTC CA-3′), as previously described in Bernardo et al. (2016) [11]. The presence of ALCV from the Iranian alfalfa samples was tested using PCR with the primer pair Gemini F1 (5′-ATG ATG GAT AAT TCA AAC CC-3′) and Gemini R2 (5′-CAC CTC CAC TGT CTT GTC CA-3′), as described in Davoodi et al. (2018) [13]. The presence of ALCV from the Argentinean alfalfa samples was tested using PCR with the KAPA HiFi HotStart PCR Kit (Kapa Biosystems, Wilmington, MA, USA) following the manufacturer’s protocol with the primer pair ALCV CPF (5′- GAG AAC GTA TGG ATT GGT C-3′) and ALCV CPR (5′-AGT GTA TGC GTT CTT CTG G-3′). Amplification conditions consisted of: 95 °C for 3 min, 35 cycles at 98 °C for 20 s, 58 °C for 15 s, 72 °C for 45 s, and a final extension at 72 °C for 1 min.
2.3. Cloning and Sequencing of ALCV Full Length Genomes
Fifty-one out of the 170 alfalfa samples that tested positive for ALCV included plants sampled in France (n = 3 out of 27 samples that tested positive for ALCV), Greece (n = 3/7), Iran (n = 14/20), Italy (n = 7/9), Jordan (n = 3/11), Lebanon (n = 2/5), Spain (n = 11/27), Syria (n = 3/12), and Tunisia (n = 5/17). DNA extracted from these 51 samples was used as a template for PCR amplification of complete ALCV genomes using the HotStarTaq Plus Master Mix Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol and the pair of abutting primers, Cap-ncoIF (5′-CCA TGG CCT TCA AAG GTA GCC CAA TTC AAY ATG G-3′) and Cap-ncoIR (5′-CCA TGG GGC CTT ATY CCT CKG YGA TCG-3′), which contain an overlapping NcoI site as described in Bernardo et al. (2016) [11]. Amplification conditions were 95 °C for 5 min, 35 cycles of 94 °C for 20 s, 60 °C for 30 s, 68 °C for 165 s, and finally 72 °C for 180 s. The amplicons were gel purified using the PCR Clean-Up System (Promega, Madison, WI, USA), cloned into pGEM-T Easy (Promega, Madison, WI, USA) and Sanger sequenced using primer walking at Genewiz (South Plainfield, NJ, USA). DNA of 34 samples that tested positive for ALCV from Argentina were used as a template for PCR amplification of the complete genome using the KAPA HiFi HotStart PCR Kit (Kapa Biosystems, Wilmington, MA, USA) following the manufacturer’s protocol and the pair of abutting primers CG-F (5´-CTC AAT GAA TCC ACA TCC AAG-3´) and CG-R (5´-CGA GGA ACT CGG ACT TGG A-3´). Amplification conditions were 95 °C for 3 min, 35 cycles of 98 °C for 20 s, 57 °C for 15 s, 72 °C for 165 s, and finally 72 °C for 180 s. The amplicons were gel purified using the PCR Clean-Up System, cloned in pGEM-T Easy and Sanger sequenced using primer walking at Macrogen Inc. (Seoul, South Korea).
2.4. Pairwise-Distance, Phylogenetic, and Recombination Analyses
Genome-wide pairwise comparisons of 85 newly determined ALCV genome sequences together with 35 previously determined ALCV genome sequences were done using the Sequence Demarcation Tool SDT v1.2 [23]. The 120 ALCV sequences were aligned together with the capulavirus Euphorbia caput-medusae latent virus (EcmLV, GenBank HF921459 [6]; chosen as outgroup) using MUSCLE [24] with default settings. Evidence of potential recombination events was detected within the 120 ALCV full-genome alignment using the RDP, GENECONV, BOOTSCAN, MAXIMUM CHI SQUARE, CHIMAERA, SISCAN and 3SEQ recombination detection methods that are implemented in RDP4.94 (using default settings [25]). Only recombination events that were detected with two or more detection methods, and had significant phylogenetic support, were considered credible evidence of recombination.
A maximum likelihood (ML) phylogenetic tree of the 120 aligned full genome sequences, with recombinant regions removed, was constructed using PhyML3 [26] implemented in MEGA 7.0.26 [27] with JC + G selected as the best fit nucleotide substitution model and 1000 non-parametric bootstrap replicates. The tree was rooted with EcmLV. Branches with less than 30% bootstrap support were collapsed using TreeGraph2 [28]. In addition, a maximum likelihood (ML) phylogenetic tree of the 56 aligned ALCV-A full genome sequences, with recombinant regions removed, was constructed using PhyML3 [26] implemented in MEGA 7.0.26 [27] with T92 + I + G selected as the best fit nucleotide substitution model and 1000 non-parametric bootstrap replicates. The tree was rooted with one Iranian isolate from ALCV-C (GenBank accession number: MH085199). Branches with less than 50% bootstrap support were collapsed using TreeGraph2 [28].
The evolutionary relationships of ALCV isolates were reconstructed using replication-associated protein (Rep) and coat protein (CP) amino acid sequences. Datasets consisting of 120 predicted ALCV Rep and CP amino acid sequences together with the corresponding homologous sequence from EcmLV, chosen as a capulavirus outgroup, was used to root the Rep and CP phylogenies. Predicted Rep and CP amino acid sequences were aligned using MUSCLE [24] with default settings. Maximum likelihood phylogenetic trees of the Rep and CP were inferred using PhyML3 [26] implemented in MEGA with the Jones–Taylor–Thornton (JTT) + G amino acid substitution model chosen as the best-fit using ProtTest [29]. Five hundred bootstrap replicates were used to test the support of branches. Branches with less than 50% bootstrap support were collapsed using TreeGraph2.
2.5. Statistical Analyses
Correlation of geographic and genetic distance were assessed using a Mantel test implemented in GenAlEx [30] (with 999 random permutations used to test the significance of the correlation). Whereas the genetic distance matrix was obtained for the 56 aligned ALCV-A full genome sequences, with recombinant regions removed using MEGA 7.0.26 (CLUSTALW alignment followed by uncorrected pairwise distance estimation with pairwise deletion of gaps), the geographic distance matrix was obtained using the program Geographic Distance Matrix Simulator 1.2.3 (http://biodiversityinformatics.amnh.org/open_source/gdmg).
3. Results and Discussion
3.1. Genetic Diversity of ALCV Isolates
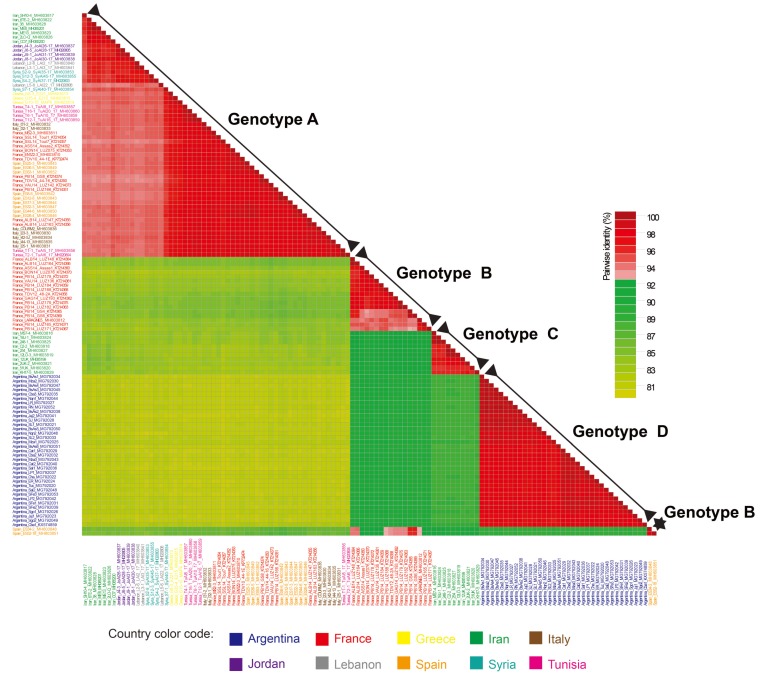
One hundred and seventy alfalfa samples tested positive for ALCV, including plants sampled from Argentina (35/41), France (27/73), Greece (7/36), Iran (20/141), Italy (9/50), Jordan (11/11), Lebanon (5/5), Spain (27/62), Syria (12/12), and Tunisia (17/17). ALCV was only ever detected from symptomatic plants but never from asymptomatic plants. All samples from Namibia and South Africa tested negative for ALCV. Overall, 85 full length genome sequences were reported in this study (GenBank accession numbers: MG792020–MG792053; MH603810–MH603860), which were analyzed together with the 35 complete genome sequences recovered in previous studies [7,11,12,13,14] (Supplementary Materials Table S1). The sizes of these 120 complete ALCV genome sequences ranged from 2712 nt to 2769 nt, and they all shared >80.2% genome-wide pairwise identity. This degree of sequence identity is above the species demarcation threshold (78%) recommended for members of the Capulavirus genus [5], and all 120 isolates were therefore considered to belong to the species Alfalfa leaf curl virus. In addition, pairwise identity distribution analysis of the 120 ALCV complete genome sequences identified two clear troughs at 87% and 93% (Supplementary Figure S1) that might indicate credible genotype demarcation thresholds. Since implementing a cutoff for ALCV genotype demarcation at 93% yielded the lowest number of genotype-level classification conflicts, we chose 93% as a tentative genotype demarcation threshold for ALCV.
Using this threshold level, four ALCV genotypes (ALCV-A, ALCV-B, ALCV-C, and ALCV-D) were clearly distinguished (Figure 1). ALCV-A isolates (n = 56) were present in almost all of the countries where ALCV has so far been reported, including France, Greece, Iran, Italy, Jordan, Lebanon, Spain, Syria, and Tunisia. By contrast, ALCV-B (n = 19) was only recovered in France and Spain and ALCV-C (n = 10) and ALCV-D (n = 35) have to date, only been found in Iran and Argentina, respectively (Figure 1). Averages of genome-wide pairwise identities between isolates of each genotype indicate that ALCV-A and ALCV-B were the most genetically diverse groups (95.7% and 96.4%, respectively) with ALCV-C isolates sharing, on average, 97.1% identity and ALCV-D isolates sharing 98.1% (Table 1). Specifically, two outlier isolates from Spain (ES34-2 and ES52-18), with strong evidence of recombination, were assigned to genotype ALCV-B because they share >93% genome-wide pairwise identity with 10 out of the 17 other isolates of genotype ALCV-B (Figure 1). While ALCV-A and ALCV-D isolates all contained at least seven recognizable open reading frames (ORFs) with more than 30 aa, including four virion-sense ORFs (V1, V2, V3, and V4) and three complementary-sense ORFs (C1, C2, and C3), ALCV-B and ALCV-C isolates lacked the V2 ORF. Both of these genomic organizations have been previously described [11].
Figure 1.
A “two-color” genome-wide pairwise identity matrix inferred using SDTv1.2 [23] showing that a tentative ALCV genotype demarcation threshold of 93% allows discrimination of the four ALCV genotypes (ALCV-A, ALCV-B, ALCV-C, and ALCV-D). Percent sequence identity is indicated by the color-coded boxes.
Table 1.
Averages of alfalfa leaf curl virus genome-wide pairwise identities.
| Group of ALCV Isolates | Averages of Genome-Wide Pairwise Identities (%) |
|---|---|
| Genotype A isolates (n = 56) | 95.7 |
| Genotype B isolates (n = 19) | 96.4 |
| Genotype C isolates (n = 10) | 97.1 |
| Genotype D isolates (n = 35) | 98.1 |
| Genotype A isolates (Middle East, n = 18) a | 96.4 |
| Genotype A isolates (Western Mediterranean basin, n = 38) b | 98.0 |
a Genotype A isolates from the Middle East include isolates from Iran, Jordan, Lebanon, and Syria. b Genotype A isolates from the western Mediterranean basin include isolates from France, Italy, Greece, Spain, and Tunisia.
3.2. Phylogenetic and Recombination Analyses
Twenty-one unique recombination events were detected (Table 2). Notably, all of the examined ALCV isolates displayed traces of recombination events with, on average, two events being evident within each analyzed isolate. Ten out of the 21 ALCV recombination events apparently involved intra-species transfers of sequences between ALCV variants whereas the other 11 detected events apparently involved inter-species sequence transfers (Table 2).
Table 2.
Recombination events detected in alfalfa leaf curl virus isolates.
| Event | Recombinant(s) | Major Parent | Minor Parent | Methods a | Breakpoints Positions b |
|---|---|---|---|---|---|
| 1 | France_VAU14_LUZ142_KT214373 | France_PB14_GS4_KT214365 | Unknown | GBMST | 1420 (nad) c–2736 (nad) |
| 2 | Jordan_J9-1_MH603839 | EcmLV_A14_HF921459 | Argentina_Cba1_KX574859 | GBMCST | 1407 (nad)–2704 (nad) |
| 3 | France_PB14_LUZ171_KT214367 | Unknown | France_LARAGNE5_MH603812 | RGBMCST | 1332 (nad)–2193 |
| 4 | Spain_ES34-2_MH603848 | France_PB14_GS4_KT214365 | Iran_SH10-4_MH603817 | RGMCST | 2097 (nad)–2196 |
| 5 | France_PB14_GS4_KT214365 | France_ALB14_LUZ148_KT214364 | Spain_ES52-18_MH603851 | RBMCT | 1750 (nad)–2053 |
| 6 | France_BON14_LUZ076_KT214370 | France_GAG14_LUZ193_KT214362 | Unknown | RGBMCST | 434–1251 |
| 7 | France_PB14_LUZ184_KT214359 | France_TV12_48-2A_KT214358 | France_LARAGNE5_MH603812 | RGBMCST | 845–1027 |
| 8 | Iran_MS7-4_MH603816 | Iran_12LO-3_MH603819 | Iran_SH10-4_MH603817 | RMCST | 277–1295 (nad) |
| 9 | France_GAG14_LUZ193_KT214362 | France_TDV12_48-2A_KT214358 | Tunisia_T6-1_MH603858 | MST | 291–1261 |
| 10 | France_SSL14_Toul7_KT214357 | France_TDV10_44-1_KP732474 | Unknown | RGMCST | 2124–2355 |
| 11 | Argentina_Sal2_MG792048 | Unknown | Spain_ES52-18_MH603851 | RBMCT | 1259–2070 (nad) |
| 12 | Spain_ES52-18_MH603851 | Unknown | France_PB14_LUZ179_KT214375 | RGMCT | 289 (nad)–1333 |
| 13 | France_PB14_LUZ179_KT214375 | France_LARAGNE5_MH603812 | Tunisia_T2-1_MH020804 | RGMCST | 868–1333 (nad) |
| 14 | France_ASS14_Assas1_KT214360 | France_PB14_LUZ171_KT214367 | Italy_COURM2-4_MH603836 | RGMCST | 901 (nad)–1029 |
| 15 | France_TDV10_44-1_KP732474 | France_LARAGNE5_MH603812 | EcmLV_A14_HF921459 | RM | 872–1053 (nad) |
| 16 | France_PB14_LUZ171_KT214367 | France_PB14_LUZ179_KT214375 | Spain_ES34-2_MH603848 | RGS | 2434–2537 |
| 17 | France_LARAGNE5_MH603812 | France_VAU14_LUZ142_KT214373 | France_BON14_LUZ076_KT214370 | RMC | 844 - 1026 |
| 18 | Argentina_Mza3_ MG792043 | Unknown | France_LARAGNE5_MH603812 | MC | 211–1261 (nad) |
| 19 | Syria_S2-9_MH603853 | Iran_38_MH603828 | Spain_ES8-6_MH603842 | MC | 1797 (nad)–2093 |
| 20 | Iran_2UK-2_MH603821 | France_PB14_LUZ184_KT214359 | EcmLV_A14_HF921459 | RG | 1976 (nad)–2016 |
| 21 | Argentina_Nqn2_MG792046 | Argentina_BsAs1_ MG792034 | Unknown | RBT | 2493 (nad)–2565 |
a RDP (R), GENECONV (G), BOOTSCAN (B), MAXIMUM CHI SQUARE (M), CHIMAERA (C), SISCAN (S) and 3SEQ (T) recombination detection methods. b Begin and end breakpoints positions in the recombinant sequence. c nad: not accurately determined.
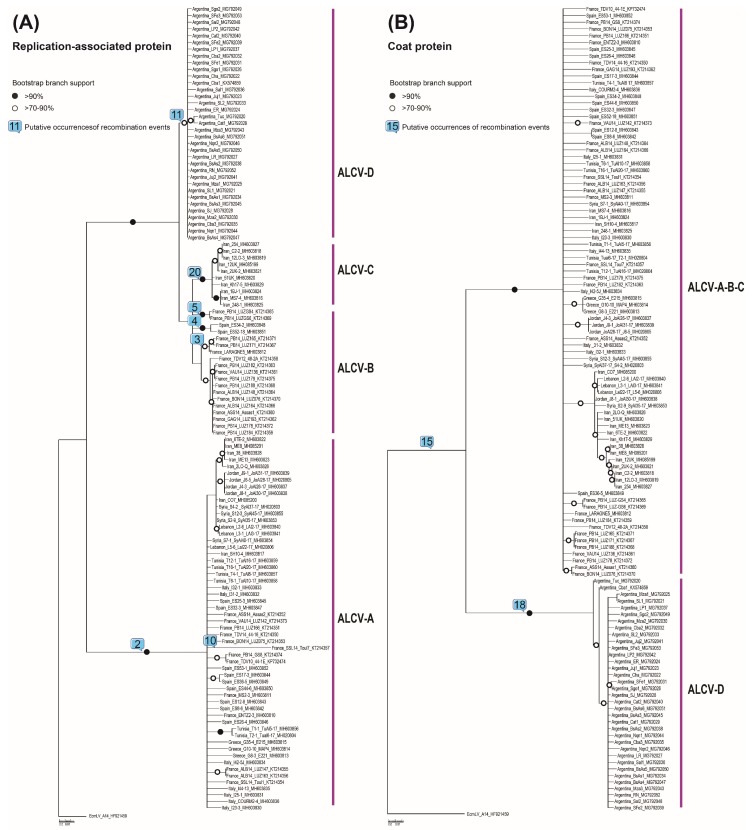
Interestingly, event 2 (corresponding with event 1 in Bernardo et al. (2016) [11]), which involved an inter-species sequence transfer of the entire replication-associated protein (rep) gene, accounted for the clear divergence of the ALCV-A isolates from all of the other ALCV genotypes (Figure 2A). At least five recombination events involving the partial replacement of the rep gene (events 3, 4, 5, 11, and 20) also appear to be the primary causes of the genetic differences found between ALCV-B, -C, and -D isolates (Figure 2A).
Figure 2.
Maximum-likelihood phylogenetic trees of predicted replication-associated protein (Rep) (A) and coat protein (CP) (B) amino acid sequences of 120 ALCV isolates, both rooted with Euphorbia caput-medusae latent virus CP and Rep amino acid sequences. Branches with less than 50% bootstrap support were collapsed. Branches associated with a black dot have bootstrap supports above 90% whereas those with white dots have bootstrap supports above 70%. Putative occurrences of recombination events (numbers within squares correspond to the number of the event as listed in Table 2) are depicted on branches of both phylogenetic trees.
The coat protein (cp) genes of the ALCV isolates have apparently been less affected by inter-species sequence exchanges than the rep genes of these isolates (Table 2); a factor that was likely responsible for the ALCV CP proteins displaying less variability than the ALCV REP proteins. The ALCV CP protein is likely to play a central role in insect transmission and virus genome packaging [31] and, as appears to be the case for begomoviruses [32], the cp gene is either less prone to recombination than the rep gene, or is more prone to recombination-induced functional impairments than the rep gene such that viruses with recombinant cp genes are selected against. However, event 15 (corresponding with event 9 in Bernardo et al. (2016) [11]), which is detectable in all analyzed ALCV isolates other than those in the ALCV-D genotype (all from Argentina) and eleven of the ALCV-B isolates from France and Spain, involved the transfer of a fragment of the cp. If this recombination event predated the most recent common ancestor of the analyzed ALCV isolates, then, in the ALCV-D isolates and the eleven ALCV-B isolates, evidence of this event may have been obscured by subsequent recombination events 6, 7, 13, 14, and 17 (Table 2).
The ALCV-D genotype has also probably arisen following two major recombination events collectively involving transfers of approximately 2/3 of the genome: one involving acquisition of a rep gene (event 11, Figure 2A) and the other involving acquisition of the cp gene (event 18, Figure 2B).
These results indicate that, as is the case with other geminiviruses, ALCV (and probably the other capulaviruses) are likely highly recombinogenic and, as a consequence, have the potential for rapid adaptive evolution [33,34,35,36]. Given that ALCV is transmitted by A. craccivora, which feeds on a diverse range of host plant species [37], the host range encounters that ALCV might naturally have with other geminiviruses is probably high. This alone might account for 38% of the recombination events that were identified here as involving unknown geminiviruses. In addition, while A. craccivora has been reported on all continents except the Antarctic [38], and displays some morphological and genetic evidence of differentiation into host races [39], one can expect that geographical populations of this aphid could have driven the microevolution of the viruses that they carry and, by extension, promoted the rapid adaptive evolution of these viruses by recombination: as is suggested by the diversity of the Argentinean ALCV isolates.
3.3. Geographic Distribution of ALCV
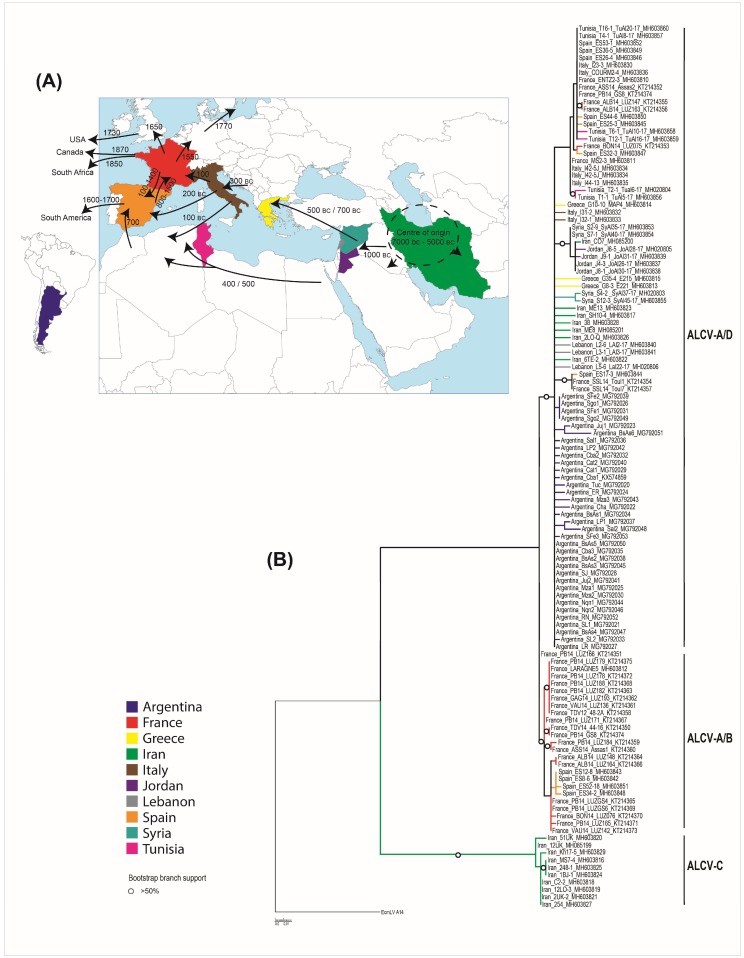
While ALCV was initially reported from southern France and northern Spain, we show here that ALCV is found in diverse geographical and climatic zones of both the Old and New Worlds (Figure 3A), including temperate oceanic (Rodez/France or Buenos Aires/Argentina), continental (Entzeim/France), mountainous (Courmayeur/Italy), cold semi-desert (Isfahan/Iran), hot semi-desert (Kerman/Iran), or subtropical (Jujuy/Argentina) climatic zones. Given the worldwide distribution of A. craccivora and the “climatic flexibility” of ALCV, it is plausible that few natural barriers exist that would effectively limit the global spread of this virus [38]. Even the presently known distribution of ALCV suggests that it is capable of infecting a range of alfalfa varieties within the M. sativa species complex, including M. sativa subsp. sativa and probably M. sativa subsp. falcata.
Figure 3.
(A) Map of Europe, the Middle East, and South America indicating the different routes and approximate dates of spread of cultivated alfalfa from its center of origin (this map is adapted from Prosperi et al. (2014) [44]). Countries from which ALCV has been reported are highlighted in several colors corresponding to those used in the phylogenetic tree. (B) A maximum likelihood phylogenetic tree of the 120 aligned ALCV recombination-free genome sequences, with JC + G selected as the best fit nucleotide substitution model and 1000 non-parametric bootstrap replicates. The tree was rooted with Euphorbia caput-medusae latent virus. Branches with less than 30% bootstrap support were collapsed. Branches associated with a dotted circle have bootstrap support values above 50%.
Although the natural host range of ALCV is unknown, it is plausible that even without alternative hosts, ALCV could persist in “wild” alfalfa populations that are commonly found within a variety of unmanaged habitats. If ALCV has a broad host range, as is commonly the case with geminiviruses, then it will be very difficult to control the local spread of the virus wherever it is introduced to in the world. Identification of alfalfa cultivars that are tolerant or resistant to A. craccivora could constitute a sustainable strategy to moderate the impacts of ALCV wherever it occurs.
It must be emphasized that the currently known geographical range of ALCV excludes some of the regions that were investigated in this study. While the virus was detected in every country within the Mediterranean basin and the Middle East from which samples were obtained, it was not detected in samples from South Africa and Namibia, where alfalfa has been grown since the mid 1800 s (Figure 3A).
By contrast, we found that ALCV was widespread in Argentina, occurring in all 17 alfalfa-growing regions of the country from where samples were obtained. The 35 genome sequences from Argentinean samples collected between 2010 and 2017 were genetically highly homogeneous, sharing an average of 98.1% genome-wide pairwise identity. While this degree of diversity is consistent with the hypothesis that the Argentinean ALCV population was founded by a single introduced ALCV variant, our recombination analysis indicates that the descendants of this founder virus have undergone two significant inter-species recombination events that replaced 2/3 of the original genome, including the entire rep and cp genes with sequences from either a distantly related ALCV genotype or from a different Capulavirus species. One of the Argentinean isolates harbors traces of an additional minor recombination event (event 21, Table 2) that occurred in the large intergenic region. Collectively, the widespread distribution of ALCV in Argentina, the low diversity of the Argentinean ALCV population, and the fact that all of the isolates appear to have descended from the same recombinant ancestor, suggest that the virus was probably introduced only once and has subsequently spread throughout the country. This may have involved the efficient, large scale, and long-range transmission of the virus by A. craccivora. This is plausible since studies focusing on another persistently A. craccivora transmitted circular ssDNA virus (subterranean clover stunt virus, SCSV, Nanoviridae) have revealed that A. craccivora in Australia can migrate over several hundred kilometers from the coastal areas to cause SCSV re-infestation of pastures in the arid regions of southeast Australia [40]. Another possibility is that the same ALCV variant may have been introduced throughout Argentina in infected planting material such as seeds. Although seed-transmission has never been demonstrated for capulaviruses, this scenario cannot be completely ruled out as several recent studies have confirmed that geminiviruses belonging to at least three genera (Begomovirus, Becurtovirus, and Curtovirus) can be seed-transmitted [41,42,43].
3.4. Geographical Origin of ALCV
The ML phylogenetic tree of all 120 aligned ALCV complete genome sequences (with recombinationally-derived genome fragments removed and rooted with EcmLV) indicated that, among the locations from which samples were analyzed, the most recent common ancestor (MRCA) was probably located in Iran (Figure 3B). The three main ALCV lineages diverged from this MRCA, to subsequently form the ALCV-C, ALCV-B, ALCV-A, and ALCV-D genotypes (Figure 3B). The genotype A/D lineage probably experienced three major recombination events: Event 2 that involved the acquisition of a rep gene sequence from an unknown capulavirus and yielded genotype ALCV-A, and Events 11 and 18 that respectively involved the transfers of rep and cp genes from a divergent currently undiscovered ALCV lineage or a currently undiscovered Capulavirus species, which together yielded genotype ALCV-D. Two recombination events between ancestral ALCV-A and ALCV-B viruses (Events 1 and 3 in our analysis) yielded a sub-clade within the genotype A/B lineage that contains rep gene sequences that are today found in viruses that would otherwise be classified as belonging to the ALCV-A genotype (represented by eight isolates in our analysis).
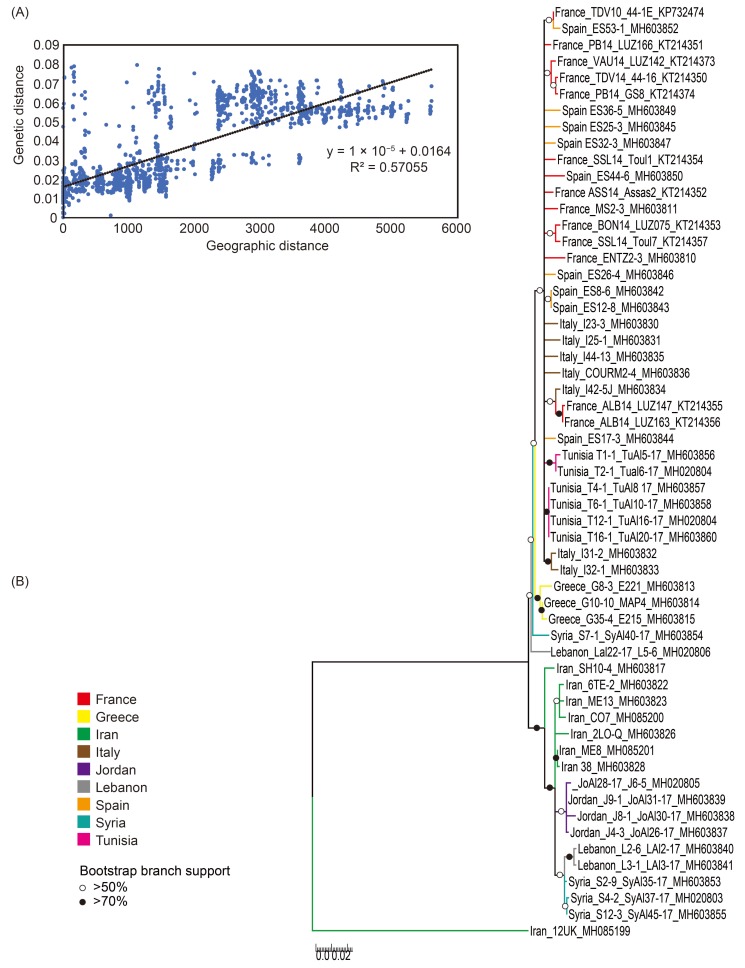
Interestingly, the averages of genome-wide pairwise identities of ALCV-A isolates from the Middle East (96.4%; isolates from Iran, Jordan, Lebanon, and Syria, Table 1) was lower than that of isolates from the western/central Mediterranean basin (98.0%; isolates from France, Italy, Greece Spain, and Tunisia, Table 1); this supports the hypothesis that genotype ALCV-A originated in the Middle East before spreading further westward. A Mantel test of association between total genetic distance and geographic distance revealed a strong correlation for the ALCV-A isolates (Rxy: correlation coefficient of Mantel test = 0.755; P (rxy-rand ≥ rxy-data: probability of positive autocorrelation (one tailed) = 0.001). This result implies a high degree of genetic isolation by distance within the Mediterranean and Middle East countries (Figure 4A).
Figure 4.
(A) Relationship of pairwise genetic distance of the 56 aligned ALCV-A full genome sequences with pairwise geographical distance (km). SSx (sum of products of x matrix elements) = 3.143 × 109; SSy (sum of products of y matrix elements) = 0.657; SPxy (sum of cross products of corresponding elements of the x and y matrices) 3.433 × 104; Rxy (Mantel correlation coefficient) = 0.755 and P (rxy-rand ≥ rxy-data); probability of Rxy based on 999 standard permutations across the full dataset = 0.001. (B) A maximum likelihood phylogenetic tree of the 56 aligned ALCV-A recombination-free genome sequences, with T92 + I + G selected as the best fit nucleotide substitution model and 1000 non-parametric bootstrap replicates. The tree was rooted with one Iranian isolate from ALCV-C (GenBank accession number: MH085199). Branches with less than 50% bootstrap support were collapsed. Branches associated with white and black dots have bootstrap support values above 50% and above 70%, respectively.
In addition, the ML phylogenetic tree of all 56 aligned ALCV-A complete genome sequences (with recombinationally-derived genome fragments removed and rooted with an ALCV-C isolate), indicated that the ALCV-A MRCA was probably located in the Middle East (Figure 4B) and that the Greek isolates followed by the western Mediterranean isolates (from France, Italy, Spain, and Tunisia) became successively more divergent from the Middle Eastern isolates (Figure 4B). Collectively these results suggested that ALCV-A isolates originated in the Middle East and spread further westward.
Finally, the average genome-wide pairwise identity of the ALCV-B isolates, (96.4%; where all examined isolates have so far only been found in France and Spain), was lower than that of ALCV-C (97.1%), ALCV-D (98.1%), and ALCV-A from the western/central Mediterranean basin (98.0%); this supports the hypothesis that ALCV-B has been circulating for longer in Western Europe than the other ALCV genotypes. Collectively, the sequence data support the hypothesis that ALCV emerged and diversified in the Middle East (with Iran being the most probable origin of all the analyzed locations) before spreading possibly in at least two waves, to the Mediterranean basin and onwards from there to Argentina. This “two-waves” hypothesis fits well with the known domestication and spread history of alfalfa. While the first evidence of alfalfa cultivation can be traced to 7000 BC in Iran and/or central Asia [44], the plant was probably only domesticated in central Asia in approximately 5000 BC [45]. Alfalfa cultivation then spread to the Middle East around 1000 BC and from there to Greece by the Medes armies between 500 and 700 BC, and finally to Italy by 300 BC and the rest of the Roman Empire by 100 AD (Figure 3A). Alfalfa cultivation in Europe then declined during the Middle Ages but was later reintroduced there via Spain by the Arabs in approximately 700 AD (Figure 3A). Thereafter, alfalfa was introduced to South America by the Spanish in the sixteenth century (Figure 3A).
It is important to stress, however, that it remains to be determined whether the timescales of ALCV dissemination throughout the Mediterranean mirror those of alfalfa dissemination. Given that all the alfalfa samples examined here have been collected over the past eight years (2010–2017) there was insufficient temporal signal in our data sets to infer accurate and precise nucleotide substitution rates that would enable the estimation of the dates when ancestral sequences likely arrived in the countries from which they were sampled. Samples of ALCV-infected alfalfa plants collected in the early to mid-1900s might yield ALCV genome sequences that could provide the temporal signal that is necessary to infer whether ALCV did indeed disseminate together with the spread of alfalfa cultivation throughout the Mediterranean.
Supplementary Materials
The following are available online at http://www.mdpi.com/1999-4915/10/10/542/s1, Table S1: Features of the 120 ALCV isolates analyzed in this study. Figure S1: Pairwise identity frequency distribution plot of 120 complete genome sequences of alfalfa leaf curl virus.
Author Contributions
P.R., N.B., and D.F. conceived and designed the experiments; Z.D., N.B., C.R., D.F., S.G.K., E.K.C, S.G., C.J., S.S., V.T., F.G., E.F.-O., J.N.-C., N.A., A.R.M., Jo.H., S.M., Ja.H., H.M., and P.R. collected the alfalfa samples, Z.D., N.B., C.R., S.G.K., S.G., C.J., S.S., V.T., and F.G. performed the experiments; Z.D., N.B., C.R., D.F., S.G.K., V.T., F.G., A.V., R.G.D., G.W.H., D.P.M., and P.R. analyzed the data; D.P.M. and P.R. wrote the paper that was edited and approved by all authors.
Funding
This study was funded by the Agropolis Fondation (E-Space flagship program) grant number 1504-004. D.P.M. has received a research grant from the National Research Foundation of South Africa. S.S. and R.G.D. were jointly supported by the Queensland Department of Agriculture and Fisheries and the University of Queensland through the Queensland Alliance for Agriculture and Food Innovation, and by AgriFutures Australia project PRJ-009751. N.B., V.T., and F.G. were partially supported by INTA through the PNPV 1135022 project. J.H. was supported by the Iran National Science Foundation (INSF) under grant number 96008997. J.N.C. and E.F.O. were supported by Grant AGL2016-75819-C2-2-R from the Ministerio de Economía, Industria y Competitividad (MINECO, Spain) co-financed by the European Regional Development Fund.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bock K.R., Guthrie E.J., Woods R.D. Purification of maize streak virus and its relationship to viruses associated with streak diseases of sugarcane and panicum maximum. Ann. Appl. Biol. 1974;77:289–296. doi: 10.1111/j.1744-7348.1974.tb01405.x. [DOI] [Google Scholar]
- 2.Duffus J.E., Gold A.H. Infectivity neutralization used in serological tests with partially purified beet curly top virus. Phytopathology. 1973;63:1107–1110. doi: 10.1094/Phyto-63-1107. [DOI] [Google Scholar]
- 3.Claverie S., Bernardo P., Kraberger S., Hartnady P., Lefeuvre P., Lett J.M., Galzi S., Filloux D., Harkins G.W., Varsani A., et al. From spatial metagenomics to molecular characterization of plant viruses: A geminivirus case study. Adv. Virus Res. 2018;101:55–83. doi: 10.1016/bs.aivir.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Roossinck M.J., Martin D.P., Roumagnac P. Plant virus metagenomics: Advances in virus discovery. Phytopathology. 2015;105:716–727. doi: 10.1094/PHYTO-12-14-0356-RVW. [DOI] [PubMed] [Google Scholar]
- 5.Varsani A., Roumagnac P., Fuchs M., Navas-Castillo J., Moriones E., Idris A., Briddon R.W., Rivera-Bustamante R., Zerbini F.M., Martin D.P. Capulavirus and grablovirus: Two new genera in the family geminiviridae. Arch. Virol. 2017;162:1819–1831. doi: 10.1007/s00705-017-3268-6. [DOI] [PubMed] [Google Scholar]
- 6.Bernardo P., Golden M., Akram M., Naimuddin, Nadarajan N., Fernandez E., Granier M., Rebelo A.G., Peterschmitt M., Martin D.P., et al. Identification and characterisation of a highly divergent geminivirus: Evolutionary and taxonomic implications. Virus Res. 2013;177:35–45. doi: 10.1016/j.virusres.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Roumagnac P., Granier M., Bernardo P., Deshoux M., Ferdinand R., Galzi S., Fernandez E., Julian C., Abt I., Filloux D., et al. Alfalfa leaf curl virus: An aphid-transmitted geminivirus. J. Virol. 2015;89:9683–9688. doi: 10.1128/JVI.00453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Susi H., Laine A.L., Filloux D., Kraberger S., Farkas K., Bernardo P., Frilander M.J., Martin D.P., Varsani A., Roumagnac P. Genome sequences of a capulavirus infecting plantago lanceolata in the aland archipelago of finland. Arch. Virol. 2017;162:2041–2045. doi: 10.1007/s00705-017-3298-0. [DOI] [PubMed] [Google Scholar]
- 9.Zerbini F.M., Briddon R.W., Idris A., Martin D.P., Moriones E., Navas-Castillo J., Rivera-Bustamante R., Roumagnac P., Varsani A., Ictv Report C. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017;98:131–133. doi: 10.1099/jgv.0.000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernardo P., Charles-Dominique T., Barakat M., Ortet P., Fernandez E., Filloux D., Hartnady P., Rebelo T.A., Cousins S.R., Mesleard F., et al. Geometagenomics illuminates the impact of agriculture on the distribution and prevalence of plant viruses at the ecosystem scale. ISME J. 2018;12:173–184. doi: 10.1038/ismej.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernardo P., Muhire B., Francois S., Deshoux M., Hartnady P., Farkas K., Kraberger S., Filloux D., Fernandez E., Galzi S., et al. Molecular characterization and prevalence of two capulaviruses: Alfalfa leaf curl virus from france and euphorbia caput-medusae latent virus from south africa. Virology. 2016;493:142–153. doi: 10.1016/j.virol.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Kumari S.G., Moukahel A.R., Richet C., Galzi S., Filloux D., Roumagnac A., Asaad N., Hijazi J., Mghandef S. First report of alfalfa leaf curl virus affecting alfalfa (medicago sativa l.) in jordan, lebanon, syria and tunisia. Plant Dis. 2018;102:2052. doi: 10.1094/PDIS-03-18-0404-PDN. [DOI] [PubMed] [Google Scholar]
- 13.Davoodi Z., Heydarnejad J., Massumi H., Richet C., Galzi S., Filloux D., Roumagnac A. First report of alfalfa leaf curl virus from alfalfa in iran. Plant Dis. 2018 doi: 10.1094/PDIS-04-18-0571-PDN. [DOI] [PubMed] [Google Scholar]
- 14.Bejerman N., Trucco V., de Breuil S., Pardina P.R., Lenardon S., Giolitti F. Genome characterization of an Argentinean isolate of alfalfa leaf curl virus. Arch. Virol. 2018;163:799–803. doi: 10.1007/s00705-017-3673-x. [DOI] [PubMed] [Google Scholar]
- 15.Alliot B., Signoret P.A., Giannott J. Presentation of bacilliform virus-particles associated with enation disease of alfalfa (medicago-sativa l) Cr. Acad. Sci. D Nat. 1972;274:1974–1976. [Google Scholar]
- 16.Blattný C. Virus papillosity of the leaves of lucerne. Folia Microbiol. 1959;4:212–215. doi: 10.1007/BF02928312. [DOI] [Google Scholar]
- 17.Cook A.A., Wilton A.C. Alfalfa enation virus in the Kingdom of Saudi Arabia. FAO Plant Prot. Bull. 1984;32:139–140. [Google Scholar]
- 18.Leclant F., Alliot B., Signoret P.A. Transmission et épidémiologie de la maladie à énations de la luzerne (lev). Premiers résultats. Ann. Phytopathol. 1973;5:441–445. [Google Scholar]
- 19.Rodriguez Sardiña J., Novales Lafarga J. Una virosis de la alfalfa con produccíon de “enations”. An. INIA/Ser. Prot. Veg. 1973;3:132–146. [Google Scholar]
- 20.Alliot B., Signoret P.A. La “maladie à énations de la luzerne”, une maladie nouvelle pour la france. Phytopath. Z. 1972;74:69–73. doi: 10.1111/j.1439-0434.1972.tb04647.x. [DOI] [Google Scholar]
- 21.Annicchiarico P. Alfalfa forage yield and leaf/stem ratio: Narrow-sense heritability, genetic correlation, and parent selection procedures. Euphytica. 2015;205:409–420. doi: 10.1007/s10681-015-1399-y. [DOI] [Google Scholar]
- 22.Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 23.Muhire B.M., Varsani A., Martin D.P. Sdt: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R.C. Muscle: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D.P., Murell B., Golden M., Khoosal A., Muhire B. Rdp4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1 doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of phyml 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S., Stecher G., Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stover B.C., Muller K.F. Treegraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abascal F., Zardoya R., Posada D. Prottest: Selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 30.Peakall R., Smouse P.E. Genalex 6.5: Genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fondong V.N. Geminivirus protein structure and function. Mol. Plant Pathol. 2013;14:635–649. doi: 10.1111/mpp.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lima A.T.M., Silva J.C.F., Silva F.N., Castillo-Urquiza G.P., Silva F.F., Seah Y.M., Mizubuti E.S.G., Duffy S., Zerbini M.F. The diversification of begomovirus populations is predominantly driven by mutational dynamics. Virus Evol. 2017;3 doi: 10.1093/ve/vex005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffy S., Holmes E.C. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus tomato yellow leaf curl virus. J. Virol. 2008;82:957–965. doi: 10.1128/JVI.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harkins G.W., Delport W., Duffy S., Wood N., Monjane A.L., Owor B.E., Donaldson L., Saumtally S., Triton G., Briddon R.W., et al. Experimental evidence indicating that mastreviruses probably did not co-diverge with their hosts. Virol. J. 2009;6:104. doi: 10.1186/1743-422X-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefeuvre P., Martin D.P., Harkins G., Lemey P., Gray A.J., Meredith S., Lakay F., Monjane A., Lett J.M., Varsani A., et al. The spread of tomato yellow leaf curl virus from the middle east to the world. PLoS Pathog. 2010;6:e1001164. doi: 10.1371/journal.ppat.1001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monjane A.L., Harkins G.W., Martin D.P., Lemey P., Lefeuvre P., Shepherd D.N., Oluwafemi S., Simuyandi M., Zinga I., Komba E.K., et al. Reconstructing the history of maize streak virus strain a dispersal to reveal diversification hot spots and its origin in Southern Africa. J. Virol. 2011;85:9623–9636. doi: 10.1128/JVI.00640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackman R.L., Eastop V.F. Aphids as Crop Pests. In: van Emden H.F., Harrington R., editors. Taxonomic Issues. CABI; Wallingford, UK: 2007. pp. 1–29. [Google Scholar]
- 38.CIE . Distribution Maps of Plant Pests. CAB International; Wallingford, UK: 1983. [Google Scholar]
- 39.Coeur d’acier A., Jousselin E., Martin J.F., Rasplus J.Y. Phylogeny of the genus aphis linnaeus, 1758 (homoptera: Aphididae) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2007;42:598–611. doi: 10.1016/j.ympev.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Gutierrez A.P., Morgan D.J., Havenstein D.E. The ecology of Aphis craccivora koch and subterranean clover stunt virus. I. The phenology of aphid populations and the epidemiology of virus in pastures in South-East Australia. J. Appl. Ecol. 1971;8:699–721. doi: 10.2307/2402678. [DOI] [Google Scholar]
- 41.Anabestani A., Behjatnia S.A.A., Izadpanah K., Tabein S., Accotto G.P. Seed transmission of beet curly top virus and beet curly top iran virus in a local cultivar of petunia in iran. Viruses. 2017;9:299. doi: 10.3390/v9100299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kil E.J., Kim S., Lee Y.J., Byun H.S., Park J., Seo H., Kim C.S., Shim J.K., Lee J.H., Kim J.K., et al. Tomato yellow leaf curl virus (TYLCV-IL): A seed-transmissible geminivirus in tomatoes. Sci. Rep. 2016;6:19013. doi: 10.1038/srep19013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J., Kil E.J., Kim S., Seo H., Byun H.S., Park J., Chung M.N., Kwak H.R., Kim M.K., Kim C.S., et al. Seed transmission of sweet potato leaf curl virus in sweet potato (ipomoea batatas) Plant Pathol. 2015;64:1284–1291. doi: 10.1111/ppa.12366. [DOI] [Google Scholar]
- 44.Prosperi J.M., Jenczewski E., Muller M.H., Fourtier S., Sampoux J.P., Ronfort J. Alfalfa domestication history, genetic diversity and genetic resources. Legume Perspect. 2014:13–14. [Google Scholar]
- 45.Muller M.H., Poncet C., Prosperi J.M., Santoni S., Ronfort J. Domestication history in the medicago sativa species complex: Inferences from nuclear sequence polymorphism. Mol. Ecol. 2006;15:1589–1602. doi: 10.1111/j.1365-294X.2006.02851.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.